Abstract
Lipoic acid is a covalently bound cofactor found throughout the domains of life that is required for aerobic metabolism of 2-oxoacids and for C1 metabolism. Utilization of exogenous lipoate is catalyzed by a ligation reaction that proceeds via a lipoyl-adenylate intermediate to attach the cofactor to the ϵ-amino group of a conserved lysine residue of protein lipoyl domains. The lipoyl ligases of demonstrated function have a large N-terminal catalytic domain and a small C-terminal accessory domain. Half of the members of the LplA family detected in silico have only the large catalytic domain. Two x-ray structures of the Thermoplasma acidophilum LplA structure have been reported, although the protein was reported to lack ligase activity. McManus et al. (McManus, E., Luisi, B. F., and Perham, R. N. (2006) J. Mol. Biol. 356, 625–637) hypothesized that the product of an adjacent gene was also required for ligase activity. We have shown this to be the case and have named the second protein, LplB. We found that complementation of Escherichia coli strains lacking lipoate ligase with T. acidophilum LplA was possible only when LplB was also present. LplA had no detectable ligase activity in vitro in the absence of LplB. Moreover LplA and LplB were shown to interact and were purified as a heterodimer. LplB was required for lipoyl-adenylate formation but was not required for transfer of the lipoyl moiety of lipoyl-adenylate to acceptor proteins. Surveys of sequenced genomes show that most lipoyl ligases of the kingdom Archaea are heterodimeric. We propose that the presence of an accessory domain provides a diagnostic to distinguish lipoyl ligase homologues from other members of the lipoate/biotin attachment enzyme family.
Lipoic acid is a covalently bound cofactor that conveys activated reaction intermediates between different active sites of multienzyme complexes (1). Lipoate is essential for aerobic metabolism of 2-oxoacids and for glycine cleavage. In its active form lipoate is attached to the ϵ-amino group of a small (∼80-residue) well conserved lipoyl domain (LD)2 lysine residue via an amide bond. LDs are typically found at the N termini of the E2 subunits of 2-oxoacid dehydrogenase complexes. In the 2-oxoacid complexes, lipoylated LD receives the decarboxylated acid from the E1 subunit active site in thioester linkage to a lipoate thiol. The acyl thioester is then converted to the corresponding CoA thioester by thioester exchange catalyzed by the E2 subunit active site. The dihydrolipoamide dehydrogenase subunit (E3) then oxidizes the dihydrolipoyl-LD back to the lipoyl-LD to reset the catalytic cycle. In the glycine cleavage (also called glycine decarboxylase and glycine dehydrogenase) system, the lipoyl domain exists as a free protein designated H. Lipoyl-LD receives the product of glycine decarboxylation, methylamine, from the P protein. The methylamine is then transferred to the T protein to produce methylenetetrahydrofolate that is typically used to synthesize serine from a second molecule of glycine. In Escherichia coli, lipoic acid is essential for aerobic growth because of the need for pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase. The glycine cleavage system is not required for growth of wild type E. coli strains but is required for growth of Arabidopsis where the H protein can be present at millimolar concentrations in photosynthetic cells (2).
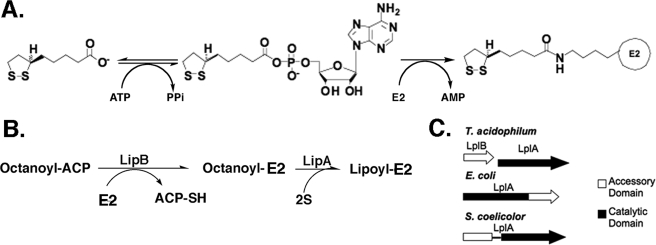
The reactions whereby lipoic acid-modified proteins are produced are best understood in E. coli. The most straightforward pathway is via lipoate-protein ligase, an activity first described by L. J. Reed et al. (3) (see Fig. 1). These workers postulated that lipoate was attached to protein by a two-step ATP-dependent reaction with lipoyl-AMP as an activated intermediate (Fig. 1). Although the lipoate-protein ligases were key reagents in demonstration of the role of lipoic acid in the 2-oxoacid dehydrogenase reactions (3, 4), the protein was not purified to homogeneity, and thus the proposed mechanism could not be considered proved. The E. coli lplA gene was the first gene encoding a lipoate-protein ligase isolated, and LplA was the first such ligase purified to homogeneity (5, 6). The isolation of null mutants in lplA showed that LplA does not play a role in de novo lipoic acid synthesis but rather acts to scavenge lipoic acid from the environment (6, 7).
FIGURE 1.
Lipoic acid metabolism in E. coli. Panel A, the lipoyl ligase (LplA) reaction that proceeds through the lipoyl-adenylate intermediate. In E. coli LplA acts to scavenge lipoic acid from the growth medium. Panel B, schematic of lipoic acid synthesis in E. coli. LipB transfers an octanoyl moiety from the fatty acid biosynthetic intermediate, octanoyl-acyl carrier protein, to the LD domain of a lipoate-accepting protein (in this case the E2 subunit of a 2-oxoacid dehydrogenase). The octanoylated LD domain is the substrate of LipA, an S-adenosylmethionine radical enzyme that replaces one hydrogen atom on each of octanoate carbons 6 and 8 with sulfur atoms. Panel C, the differing arrangements of genes and domains found in lipoate ligases in T. acidophilum, E. coli, and Streptomyces coelicolor. Only a single nucleotide lies between the T. acidophilum LplB and LplA coding sequences.
LplA is a 38-kDa monomeric protein (5). Assays with a fully defined system have demonstrated that LplA plus lipoate and Mg-ATP are sufficient to reconstitute lipoylation in vitro and that lipoyl-AMP is a reaction intermediate (5, 6, 8, 9). Thus, it is clear that LplA catalyzes both the ATP-dependent activation of lipoate to lipoyl-AMP as well as the transfer of this activated lipoyl species to apoprotein with concomitant release of AMP. The E. coli LplA enzyme has been shown to be capable of utilizing lipoate and several lipoate analogues such as octanoate as donors for the post-translational modification of E2 apoproteins in vivo (5, 6).
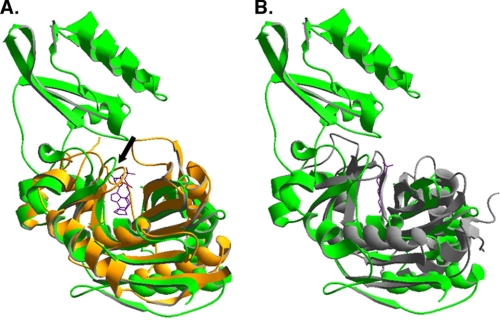
Recently crystal structures of E. coli LplA and of LplA homologues have been reported including an E. coli LplA-lipoic acid complex (10–12). The reported structures of the unliganded proteins agree well and show E. coli LplA to be a two-domain protein consisting of a large N-terminal domain and a small C-terminal domain (Figs. 1 and 2). However, the E. coli LplA-lipoic acid complex is difficult to interpret because lipoic acid molecules were heterogeneously bound to LplA molecules within the crystals and were poorly resolved. In one case the lipoic acid carboxyl was hydrogen-bonded to Ser-72, whereas in another case Arg-140 was the hydrogen bond donor (10). Because enzymes rarely show such plasticity and lipoic acid is a hydrophobic molecule, it seemed possible that the observed association of the cofactor with a hydrophobic LplA surface in the interdomain cavity was artifactual. Moreover in prior work K. E. Reed et al. (13) had isolated LplA mutants resistant to inhibition by an analogue of lipoic acid in which the sulfur atoms had been replaced with selenium. Because this is a very discrete modification of the LplA substrate, the mutant protein would be expected to have an alteration close to the pocket that binds the lipoic acid thiolane ring. However, the site of this mutation (Gly-76 to serine (7)) was distal from the lipoate binding site reported. This dilemma was resolved by two lipoic acid-containing structures of an LplA homologue from the archaeon Thermoplasma acidophilum (11, 12) that can be readily superimposed on the E. coli LplA structure except that the T. acidophilum protein lacks the E. coli LplA C-terminal domain (Fig. 2). In both T. acidophilum structures the lipoate thiolane ring was adjacent to the glycine residue that corresponds to E. coli Gly-76, the residue giving resistance to the selenium analogue, and a plausible reorganization of the molecule to prevent binding of the slightly larger analogue was proposed (12). Moreover addition of lipoic acid to a complex of the T. acidophilum LplA with ATP gave lipoyl-AMP showing that the lipoic acid was bound in a physiologically meaningful manner (11). The lipoyl-AMP was bound in a U-shaped pocket and was well shielded from solvent. Thus, it seems that the locations of the lipoate moieties in the two T. acidophilum LplA structures indicate that these represent catalytically competent lipoate binding sites (rather than the sites of E. coli LplA where lipoate bound). A caveat was that the T. acidophilum LplA was inactive in catalysis of the overall LplA reaction (12). Because T. acidophilum LplA lacks the C-terminal domain (CTD) of E. coli LplA (11, 12), this suggested that the missing domain was required for activity, and a second protein was proposed to interact with T. acidophilum LplA to allow the complete reaction (12). If this were the case, the T. acidophilum lipoyl ligase would provide an unusually facile system to investigate the role of the CTD in lipoate-protein ligases.
FIGURE 2.
Structural alignments of LplA and LipB structures. Previously published crystal structures were aligned using DeepView (37). Panel A, E. coli LplA (Protein Data Bank code 1X2H in green) aligned with T. acidophilum LplA (Protein Data Bank code 2ART in orange). The lipoyl-adenylate intermediate bound to T. acidophilum LplA is shown in purple. The adenylate binding loop is indicated with an arrow. Panel B, M. tuberculosis LipB (Protein Data Bank code 1W66 in gray) is aligned with the E. coli LplA structure of panel A. The purple line denotes the covalent decanoate adduct present in the M. tuberculosis LipB structure. The substrate binding pocket is conserved among members of the protein family. The accessory domain is not part of the binding pocket and appears to play an indirect role in catalysis.
If the lipoate-protein ligase reaction can be catalyzed by a heteromeric protein this may allow better discrimination of lipoyl ligases from acyl carrier protein:protein octanoyltransferases. In E. coli de novo lipoic acid biosynthesis is accomplished by two enzymes, the LipB octanoyltransferase and the LipA lipoyl synthase (14) (Fig. 1). LipB transfers the octanoate moiety from the octanoyl-acyl carrier protein intermediate of fatty acid biosynthesis to the ϵ-amine of the conserved LD lysine residue resulting in amide-linked octanoate (Fig. 1). LipA then catalyzes replacement of a hydrogen atom on each of octanoate carbons 6 and 8 with sulfur atoms derived from a LipA iron-sulfur center via an S-adenosylmethionine-dependent radical mechanism (14, 15). That is, lipoic acid is assembled on its cognate proteins (16).
Although the two classes of LD-modifying enzymes, LplA and LipB, show very low amino acid sequence conservation and utilize different chemistries, the proteins surprisingly show structural conservation and have related active site architectures (17, 18) (Fig. 2). The Mycobacterium tuberculosis LipB and T. acidophilum LplA can be superimposed by using all matching Cα positions with a root mean square deviation of ≈2.5 Å with good topological matching of most secondary structural elements (18). Hence in length and structure LipBs resemble LplAs that lack the C-terminal domain. Although the E. coli LipB and LplA sequences align very poorly, a large number of proteins in the data bases have similarities to both proteins, and therefore annotation of a given protein as a ligase or octanoyltransferase is not straightforward. If an LplA CTD can be a separate protein, an additional criterion to distinguish lipoate ligases and octanoyltransferases would be available. It should be noted that biotin ligases also show structural (but not sequence) conservation with LipB and LplA, and this group of proteins comprises the Pfam family PF03099 (19). However, all known biotin ligases have a C-terminal domain that greatly aids in their annotation. We report that, as predicted by McManus et al. (12), the CTD function essential for lipoate-protein ligase activity is encoded by a gene located immediately upstream of T. acidophilum lplA that we call lplB.
EXPERIMENTAL PROCEDURES
Chemicals, Bacterial Strains, and Growth Media
LB rich and M9 minimal media were prepared as described previously (20). Bacterial cultures were grown at 37 °C. T. acidophilum DSM 128 genomic DNA was purchased from the ATCC. Oligonucleotides were synthesized by Integrated DNA Technologies and are shown in Table 1. PCR amplification was performed using Pfu polymerase (Stratagene), and A overhangs were added with Taq polymerase (New England Biolabs). The TOPO TA Cloning kit was used for cloning PCR products into the pCR2.1 vector supplied (Invitrogen), and the Topoisomerase Cloning kit was used for pET101 (Invitrogen). DNA constructs were sequenced by the Roy J. Carver Biotechnology Center of the University of Illinois. Plasmids, strains, and primers used in this study are listed in Table 1. All reagents and biochemicals were obtained from Sigma-Aldrich unless otherwise noted. Radiolabeled [α-32P]ATP and l-[35S]methionine were purchased from American Radiolabeled Chemicals.
TABLE 1.
Strains, plasmids, and primers used in this work
CGSC, The Coli Genetic Stock Center.
| Strain/plasmid | Relevant characteristics | Source or Ref. |
|---|---|---|
| Strains | ||
| MG1655 | CGSC | |
| JK1 | rpsL (StrepR) | 7 |
| TM131 | rpsL lipA ::Tn1000 lplA ::Tn10 | 7 |
| Acella DE3 | ompT hsdSB(rB− mB−) gal dcm (DE3) ΔendA ΔrecA | Edge Biosystems |
| Rosetta2 DE3 | ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2 | Novagen |
| ZX221 | rpsL ΔlipB ::FRT ::aac | This study |
| QC144 | ΔlplA ::FRT ::kan | This study |
| QC145 | ΔlplA ::FRT ::kan ΔlipB ::FRT ::cat | This study |
| QC146 | ΔlplA ::FRT ΔlipB ::FRT | This study |
| QC038 | rpsL lipA ::Tn1000 lplA ::Tn10/pBAD322G | This study |
| QC090 | rpsL lipA ::Tn1000 lplA ::Tn10/pQC005 | This study |
| QC091 | rpsL lipA ::Tn1000 lplA ::Tn10/pQC021 | This study |
| QC092 | rpsL lipA ::Tn1000 lplA ::Tn10 pQC022 | This study |
| QC064 | ompT hsdSB(rB− mB−) gal dcm (DE3) ΔendA ΔrecA /pQC024 | This study |
| QC096 | ompT hsdSB(rB− mB−) gal dcm (DE3) ΔendA ΔrecA/pQC035 | This study |
| QC049 | ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2, pQC017 | This study |
| QC108 | ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2, pQC043 | This study |
| QC164 | ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2, pQC017, pQC056 | This study |
| QC165 | ompT hsdSB(rB− mB−) gal dcm (DE3)/pRARE2, pQC043, pQC055 | This study |
| QC166 | ΔlplA ::FRT ΔlipB ::FRT/pTara/pQC035 | This study |
| Plasmids | ||
| pBAD322G | GmR arabinose-inducible expression vector | 22 |
| pTARA | ChlR arabinose-inducible T7 polymerase | 39 |
| pET19b | AmpR T7 expression vector | Novagen |
| pET28b+ | KnR T7 expression vector | Novagen |
| pET30b+ | KnR T7 expression vector | Novagen |
| pET101/TOPO | AmpR T7 expression vector | Invitrogen |
| pRSF-1b | KnR T7 expression vector with RSF origin | Novagen |
| pCR2.1 | TOPO TA cloning vector | Invitrogen |
| pQC005 | pBAD322G encoding a TA0513-4 operon | This study |
| pQC017 | pET101 encoding N-terminal hexahistidine-tagged TA0514 | This study |
| pQC019 | pCR2.1 with TA0514 insert | This study |
| pQC020 | pCR2.1 with TA051 insert | This study |
| pQC021 | pBAD322G encoding TA0514 | This study |
| pQC022 | pBAD322G encoding TA0513 | This study |
| pQC028 | pCR2.1 encoding E2 LD gene | This study |
| pQC024 | pET30(a)+ encoding the E2 LD | This study |
| pQC034 | pCR2.1 encoding C-terminal hexahistidine-tagged GcvH | This study |
| pQC035 | pET28(b)+ encoding C-terminal hexahistidine-tagged GcvH | This study |
| pQC043 | pET19b encoding N-terminal hexahistidine-tagged TA0513m | This study |
| pQC055 | pRSF1b encoding TA0514 | This study |
| pQC056 | pRSF1b encoding TA0513 | This study |
| Oligonucleotides | Sequence | |
| Q007 | tagccatggttctcaattatactatgcat | |
| Q008 | tacaagcttacagggatatcgagacgtt | |
| Q017 | agcgagaaaaaagagtgacccattactacaagaaaggaaatcgttgtgtaggctggagctgcttc | |
| Q018 | aaaatccggcaaatcgaagagaaagttgcccgcatgggcgggtaacatatgaatatcctccttag | |
| Q019 | cccccacttttactcattctccacggagatgccgttttgtatcagtgtgtaggctggagctgcttcgaa | |
| Q020 | gtaatgacccagtgtaaattgggccattgatgtatggaattaagccatatgaatatcctcctt | |
| Q025 | tcagatcaccctcaaagc | |
| Q026 | caccatgcatcatcatcatcatcatatggaaggcaggcttctt | |
| Q027 | tagccatggaaggcaggcttctt | |
| Q032 | aatcatatgtacgaattcaaactgccagacataggtg | |
| Q033 | attgagctcttaaggctgctgtaccggagcc | |
| Q049 | tcatgacagaggtaccagagggttt | |
| Q050 | ttaatgatgatgatgatgatgttgtattaacttcctgtactccgatg | |
| Q068 | actcgagtcagatcaccctcaaagc | |
| Q069 | ttctagataaggaggagaccaatgcatcaccatcaccatcacatgcatatgatgtacagc | |
All E. coli strains were derivatives of E. coli K-12. Strain QC146 was constructed by the method of Datsenko and Wanner (21). The lplA gene was replaced with a kanamycin resistance cassette by transformation of the PCR product obtained using primers Q017 and Q018. The lipB gene was replaced with a chloramphenicol resistance cassette using the PCR product obtained using primers Q019 and Q020. The insertions were transduced with phage P1 into the wild type strain MG1655, the antibiotic cassettes were removed using the Flp protein encoded by the temperature-sensitive plasmid pCP20 (21), and the lipB lplA phenotype was verified on M9 0.2% glucose medium comparing growth with 5 μg/ml lipoic acid or 5 mm acetate plus 5 mm succinate. The strains are described in Table 1.
Plasmid Constructions
The TA0513 and TA0514 open reading frames were amplified as a single fragment by PCR from T. acidophilum genomic DNA using primers Q007 and Q008, which added terminal NcoI and HindIII sites. This product was directly cloned into the same sites of pBAD322G (22) under the control of an arabinose-inducible promoter to give pQC005. TA0514 was amplified with primers Q027 and Q008, which added NcoI and HindIII sites, and TA cloned into pCR2.1 to give pQC019 followed by insertion between the NdeI and HindIII site of pBAD322G to give pQC021. TA0513 was amplified with primers Q007 and Q025 and TA cloned into pCR2.1 to give pQC020 followed by insertion between the NdeI and XbaI sites of pBAD322G to give pQC022. A derivative of TA0514 having an N-terminal hexahistidine tag was amplified from genomic DNA using primers Q026 and Q008, which also added the N-terminal hexahistidine tag sequence. The product was TOPO cloned directly into pET101 to give pQC017. TA0513m was amplified using primers Q069 and Q068, which added an EcoRI site, a ribosome binding site, an N-terminal hexahistidine tag, and an XhoI site. This product was directly cloned between the EcoRI and XhoI sites of pET19b to give pQC043.
The LD of the E2 subunit of the branched chain dehydrogenase was determined by alignment to the LD derived by proteolytic digestion of Bacillus stearothermophilus pyruvate dehydrogenase E2 subunit (23). The domain contains the first 86 amino acids of the branched chain dehydrogenase E2 from open reading frame TA1436. The domain was amplified from genomic DNA with primers Q032 and Q033 creating an NdeI restriction site within the initiation codon and a stop codon following codon 86. This was TA cloned into pCR2.1 to give pQC028. The gene was then ligated between the NdeI and SacI sites of pET30(a)+ to give pQC024. A putative T. acidophilum gcvH, open reading frame TA1366m, was found by homology and was amplified with primers Q050 and Q049, which introduced a C-terminal hexahistidine tag and a BspHI site that overlapped the initiation codon. The product was TA cloned into pCR2.1 to give pQC034 and subsequently inserted into the NcoI and XbaI sites of pET28(b)+ to give pQC035. TA0514 was cut from pQC021 with NcoI and HindIII and inserted between the same sites of pRSF-1b to give pQC055. TA0513 was digested from pQC022 and ligated into the NcoI and XbaI sites of pRSF-1b to give pQC056.
Bioinformatics Analyses
Identification and bioinformatics characterization of single domain homologues of LplB were done using the Berkeley Phylogenomics Website (24). Twenty- six homologues were identified using the TA0513m protein sequence from 10 iterations of SHMM (subfamily hidden Markov model) and PSI-BLAST (position-specific iterative basic local alignment search tool). Genome comparison, homology searches, and lipoic acid subsystem analyses were performed using the National Microbial Pathogen Database Resource (25). Alignments were generated using ClustalW2 (26).
Complementation and 2-Oxoacid Dehydrogenase Assays
Strains JK1 and TM131 containing pBAD322G expressing the indicated ligase proteins were grown in LB medium supplemented with 0.2% glucose, 5 mm acetate, 5 mm succinate, and 5 mg/liter lipoic acid. The cells were harvested at an A600 of 0.6 by centrifugation and resuspended in 10 mm Tris-HCl buffer (pH 7.5) at ∼100 mg of wet weight/ml. Cells were lysed by two passages through a French pressure cell at ∼20,000 p.s.i. Lysates were cleared by centrifugation at 27,000 × g for 15 min. Assays of the pyruvate dehydrogenase and oxoglutarate dehydrogenase complex activities were a modification of previous continuous spectrophotometric assays (27, 28). The reaction mixture contained 150 mm Tris-HCl buffer (pH 8.5), 3 mm l-cysteine, 0.1 mm CoA, 0.5 mm thiamin pyrophosphate, and 2 mm acetylated NAD analogue (2-acetylpyridine adenine dinucleotide). For assays of pyruvate dehydrogenase, 5 mm MgCl2 was also included to give maximal activity. Cell extract protein contents were determined using the using the Bio-Rad Protein Assay kit. The amount of extract assayed varied from 100 to 1 μg/ml final concentration. Each assay began by addition of 50 mm substrate (pyruvate or 2-oxoglutarate) at 25 °C. The reduction of 2-acetylpyridine adenine dinucleotide was measured at 366 nm, and the extinction coefficient of the reduced form in the assay buffer was experimentally determined to be 7.0 mm−1 cm−1.
Purification of LplA and LplB
Purification of hexahistidine-tagged versions of LplA and LplB was done using Ni2+ affinity chromatography followed by anion exchange chromatography. Plasmids QC048 and QC049 were transformed into strain Rosetta2 DE3, and the transformants were grown at 37 °C in 2 liters of LB medium with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. At an A600 of 0.6 expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h before harvest and storage of the cell pellets at −80 °C. All chromatography steps were performed at 4 °C. The cell pellets were resuspended in lysis buffer (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, 1 mm dithiothreitol, and 10 mm imidazole) and lysed using a French pressure cell. The lysate was cleared at 48,000 × g for 15 min, and 5 ml of a 50% slurry of Ni2+-NTA-agarose resin (Qiagen) was added to the cleared lysate and incubated for 1 h at 4 °C with mixing. The resin was packed into a 0.75-inch column and washed with at least 10 column volumes of wash buffer (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 20 mm imidazole) by gravity flow. The protein was eluted with the same buffer (elution buffer) containing 250 mm imidazole. The eluate was dialyzed overnight in 50 mm Tris-HCl (pH 8.0) and subjected to anion exchange using a 1-ml POROS HQ 20 column with a gradient of 10–500 mm NaCl using an AKTA Purifier 10 FPLC run at 5 ml/min. Fractions containing pure protein, as judged by SDS gel electrophoresis followed by Coomassie Blue staining, were dialyzed overnight in storage buffer that contained 10 mm sodium phosphate buffer (pH 7.0), 100 mm NaCl, 1 mm dithiothreitol, and 10% glycerol. The proteins were concentrated using Vivaspin concentrators (Sartorius), flash frozen in dry ice and ethanol, and stored at −80 °C. The masses of purified proteins were determined after dialysis against 10 mm ammonium acetate followed by drying under vacuum. The samples were submitted to the University of Illinois at Urbana-Champaign Mass Spectrometry Laboratory for matrix-assisted laser desorption ionization mass spectrometry analysis.
Purification of Lipoyl Domain Substrates
The E. coli E2p hybrid domain was a gift from Dr. Xin Zhao of this laboratory and had been purified by precipitation and ion exchange chromatography as described previously (16). Plasmid pQC024 in Acella λDE3 cells (Edge Biosystems) was used to express the E2 domain in LB medium with 50 μg/ml kanamycin. The culture was induced at an A600 of 0.5 for 4 h. The collected cell pellet was washed with 10 mm sodium phosphate buffer (pH 7.0) and stored at −80 °C. Purification of T. acidophilum E2 domain of the branched chain dehydrogenase was performed as described previously for E. coli E2p domain (16). This anion exchange protocol allows resolution of the apo and holo forms of the domain as shown by 20% native PAGE. Pure apo domain was dialyzed and stored as described for LplA and LplB. The masses of apo- and holo-LDs were verified as described for LplA and LplB except that electrospray mass spectrometry was performed. All E2 domain preparations were found to lack the N-terminal methionine residue.
Plasmids pQC035 and pTara were used to express the putative glycine cleavage H protein TA1366m in the LD modification-deficient strain QC146 grown in LB medium containing 0.1% glucose, 5 mm acetate, 5 mm succinate, 50 μg/ml kanamycin, and 25 μg/ml chloramphenicol. When the cells reached an A600 of 0.6, 0.2% arabinose was added as an inducer, and cells were harvested by centrifugation 4 h later. GcvH was purified by Ni2+ affinity and anion exchange as described above for LplA and LplB.
Assay of Lipoyl Domain Modification
Protein concentrations were determined using extinction coefficients calculated from the ProtParam program on the ExPASy Tools Website. The assays contained 50 mm sodium phosphate buffer, 1 mm sodium lipoate, 5 mm disodium ATP, 5 mm dithiothreitol, 1 mm MgCl2, and 20 μm apo-LD. The 2 μm LplA and 20 μm LplB were added as indicated. Three different apo-LDs were used. Reactions (20 μl) were incubated at 55 °C for 1 h in the case of the T. acidophilum proteins and 16 h for the E. coli LD domain. Lipoyl domain modification was determined by gel shift by native polyacrylamide gel electrophoresis using 20% Tris-glycine gels.
Lipoyl-AMP was synthesized by the method of L. J. Reed et al. (3). Lipoyl-AMP was weighed and dissolved in 100 mm sodium phosphate buffer (pH 7.0) before use as a substrate for lipoyl domain modification by LplA and LplB. The reaction contained 50 mm sodium phosphate (pH 7.0), 1 mm tris(2-carboxyethyl)phosphine, 0.l mm MgCl2, 20 μm apo-LD, and 1 mm lipoyl-AMP. LplA (2 μm) and LplB (20 μm) were added as indicated. Reactions (20 μl) were incubated at 55 °C for 30 min. Lipoyl domain modification was determined as described above.
Assay of Enzymatic Lipoyl-adenylate Intermediate Formation
Formation of lipoyl-AMP was assayed using radiolabeled [α-32P]ATP. The reactions contained 50 mm sodium phosphate (pH 7.0), 1 mm tris(2-carboxyethyl)phosphine, 10 μm [α-32P]ATP, 10 μm MgCl2, and 0.1 mm sodium lipoate. 15 μm LplB and 10 μm LplA were added as indicated. The reaction was incubated for 30 min at 55 °C. 1 μl of the reaction was subjected to cellulose thin layer chromatography on plates containing a fluorescent indicator developed in isobutyric acid:NH4OH:water (66:1:33). Lipoyl-AMP had an Rf of 0.68, whereas the Rf of AMP was 0.41. The thin layer chromatograms were dried and exposed to a phosphorimaging plate for 10 min to 10 h and visualized using a Fujifilm FLA-3000. The same reaction as above was also run with 0.1 μm [α-32P] ATP and 1 μm LplA. Additionally 3 μm apo-LD was added to this and preincubated for 15 min without ATP to remove any enzyme-bound ATP or adenylate.
Isolation of enzyme-bound lipoyl-adenylate was performed after the reaction above with LplA and LplB using a 10-kDa Microcon centrifugal filter device (Millipore). The reaction was diluted with 600 μl of 50 mm sodium phosphate buffer (pH 7.0), applied to the filter, and centrifuged at 14 × g for 15 min. This was followed by four 400-μl buffer washes of the filter after which the protein was recovered. The fractions were analyzed by TLC and phosphorimaging as described above.
Determination of the Size and Stoichiometry of the LplA-LplB Complex
Strains QC049, QC108, QC164, and QC165 were used to determine the size and stoichiometry of the lipoyl ligase complex. These strains have T. acidophilum lipoyl ligase proteins expressed from a T7 promoter. All proteins were hexahistidine-tagged except for TA0513 in QC164 and TA0514 in QC165. LplA and LplB were expressed together in Rosetta2 DE3 cells using plasmids with compatible origins of replication. Strain QC164 contained pQC017 and pQC056 that expressed a hexahistidine-tagged LplA and native LplB. Strain QC165 contained pQC043 and pQC055 that expressed a hexahistidine-tagged LplB and native LplA. The strains were grown in 50 ml of LB medium with 100 μm ampicillin and 50 μg/ml kanamycin where appropriate. Cultures were grown to an A600 of 0.6, and expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. The cells were harvested by centrifugation and stored at −80 °C. Cellytic Express (Sigma) was dissolved at 50 mg/ml in 10 mm Tris-HCl (pH 8.0) and used to resuspend cells at ∼400 mg/ml wet weight. Lysis occurred after incubation at room temperature for 20 min. The extract was cleared by centrifugation at 18,000 × g for 20 min. The extract was then applied twice to a pre-equilibrated Ni2+-NTA spin column (Qiagen). The column was washed thrice with 600 μl of wash buffer. Finally the products were eluted with 100 μl of elution buffer and were analyzed by SDS-PAGE or gel filtration chromatography.
Gel filtration chromatography was performed in 50 mm sodium phosphate buffer (pH 7.0) containing 150 mm NaCl using a Superdex 200 10/300 GL column on an AKTA purifier 10 FPLC run at 0.5 ml/min. The void volume was found to be 7.7 ml using blue dextran. Low range molecular weight standards (GE Healthcare) were also run to establish a standard curve. The partition coefficient, Kav, was calculated from Kav = (Ve − V0)/(Vt − V0) where V0 is the void volume, Ve is the elution volume, and Vt is the total column volume.
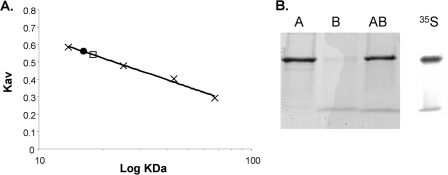
The ratio of LplA to LplB was determined by protein labeling with [35S]methionine (29). Strain QC164 was grown in M9 medium with 0.4% glucose, 50 μg/ml l-methionine, 50 μg/ml ampicillin, 25 μg/ml kanamycin, and 12 μg/ml chloramphenicol. Cells were grown to an A600 of 0.6, 1 mm isopropyl 1-thio-β-d-galactopyranoside was added, and 10 min later cells were washed thrice with M9 medium. Cells were added to M9 medium with 0.4% glucose, 1 mm isopropyl 1-thio-β-d-galactopyranoside, and 0.2 μg/ml rifampicin. After 15 min 5 μCi/ml l-[35S]methionine and 50 μg/ml l-cysteine were added to label methionine residues and prevent labeling of cysteine residues. Cultures were allowed to grow for 2 h until harvest by centrifugation. The cells were lysed using Cellytic Express (Sigma) as above. The complex was purified using a Ni2+-NTA spin column (Qiagen), and the eluate was subjected to gel filtration as described above. Fractions were subjected to SDS-PAGE followed by Coomassie Blue staining. The gel was dried and analyzed using a Fujifilm FLA-3000 and phosphorimaging plate. The ratio of LplA to LplB was determined by comparing the ratio of the intensities of each band. The ratio was corrected for the number of methionine residues in each protein (14 for hexahistidine-tagged LplA and five for native LplB).
RESULTS
Complementation of E. coli lplA Mutants by T. acidophilum lplA and lplB
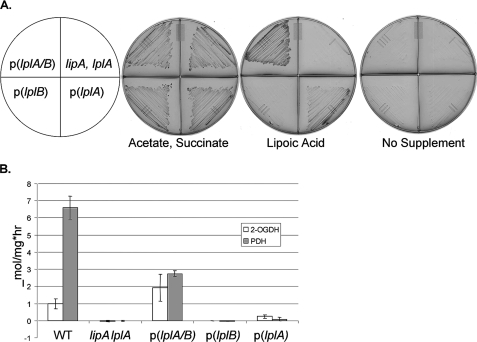
An E. coli lipA lplA mutant was used for complementation studies because the lack of lipoyl ligase has no phenotype when lipoic acid biosynthesis is intact (6, 7). The lplA null mutation of the E. coli host was complemented when lplA and lplB of T. acidophilum were coexpressed but not when the genes were expressed separately. This was shown by restoration of growth on minimal medium supplemented with lipoic acid (Fig. 3, panel A). Complementation was due to activation of 2-oxoacid complexes, an activity expected of a lipoyl ligase (Fig. 3, panel B). A slight increase in 2-oxoglutarate dehydrogenase activity was seen with LplA alone suggesting that the protein has some ability to modify lipoyl domains. This was also seen by the very weak complementation of growth by LplA alone (Fig. 3, panel A). This was unexpected because previous in vitro assays demonstrated no activity (12) and suggested that LplA has low levels of activity in the absence of LplB. The greater modification of 2-oxoglutarate dehydrogenase suggests that T. acidophilum lipoyl ligase may have a greater relative affinity for this E2 domain than does E. coli LplA.
FIGURE 3.
Complementation of an E. coli lipoyl ligase null mutant strain by T. acidophilum lplA and lplB. The lipA lplA strain, TM131, was transformed with pBAD322G-derived plasmids, p(lplA), p(lplB), and p(lplA plus B), expressing the T. acidophilum genes (as indicated) from an arabinose-inducible promoter. The wild type (WT) and lipA lplA both with an empty vector are the control strains. The complementation assays were performed on M9 minimal agar containing 0.2% arabinose and 0.1% vitamin assay casamino acids. Where indicated, 5 mm acetate plus 5 mm succinate or 5 μg/ml lipoic acid was added. To prevent carryover of lipoic acid, before testing the strains were grown on the same medium containing 5 mm acetate, 5 mm succinate, and 24 mg/liter gentamicin. Panel A, ability of expression of LplA and LplB alone or together to restore growth of E. coli strain TM131 (lplA lipA) when supplemented with lipoic acid. Representative plates of three replicate experiments are shown. Panel B, activation of the pyruvate dehydrogenase (PDH) and 2-oxoglutarate dehydrogenase (OGDH) complexes upon expression of LplA and LplB alone or together was assayed using an acetylated NAD analogue as a substrate for the overall reaction of the dehydrogenase complex. Results are reported as μmol of NAD analogue reduced/mg of cell extract h−1. The error bars denote twice the S.D. from at least four assays.
Requirement of LplB in Lipoyl Ligation
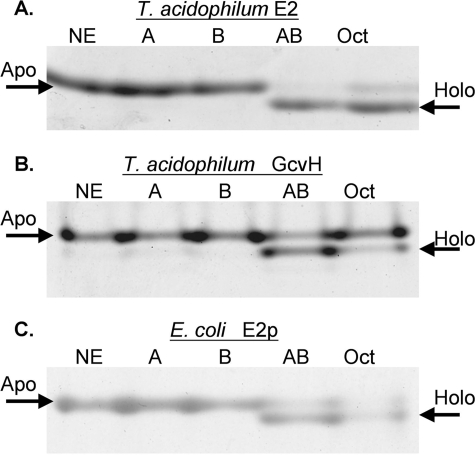
The LplA/LplB ligation reaction was reconstituted in vitro. Purified hexahistidine-tagged LplA and LplB proteins were tested for the ability to modify the T. acidophilum E2 and GcvH LDs in the presence of lipoate and ATP. Lipoylation was observed only when both LplA and LplB were present (Fig. 4). Although as mentioned above a very weak activity was observed with LplA alone in vivo, we detected no activity in vitro in agreement with a prior report (12). Octanoate was also a substrate for LplA-LplB as was demonstrated previously with E. coli lipoyl ligase (5, 6). The E. coli E2p LD was a substrate, but full modification required very long incubations.
FIGURE 4.
Ligase activity of LplA and LplB with two acceptor LDs measured by the gel shift assay. The modified lipoyl domain loses a charge upon modification of the target lysine residue resulting in more rapid migration on native gels than the unmodified domain. Representative gels of three independent experiments are shown. Lane designations given in parentheses below are the same in all three panels. Lane 1, no enzyme (NE); lane 2, LplA (A); lane 3, LplB (B); lane 4, LplA plus LplB (AB); lane 5, LplA plus LplB with octanoate in place of lipoate (Oct). Reaction components are listed under “Experimental Procedures.” Panel A, modification of the T. acidophilum branched chain dehydrogenase E2 lipoyl domain by lipoate or octanoate attachment. Panel B, modification of the putative T. acidophilum GcvH. Panel C, modification of the E. coli hybrid E2p domain. Note that efficient lipoylation of the E. coli domain required a 16-fold longer incubation time than the T. acidophilum domains.
The Role of LplB in Lipoyl-AMP Formation
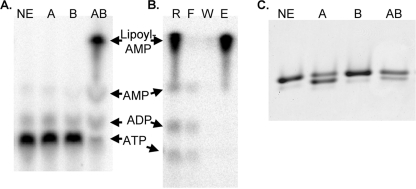
LplB provides a test for the function of the CTD of the canonical single subunit lipoyl ligases. Based on the reported synthesis of adenylate within LplA crystals (11) we hypothesized that LplB would be required for transfer of lipoate from lipoyl-AMP but not for adenylate formation. To test this hypothesis the interactions of LplA and LplB with the lipoyl-adenylate intermediate were tested by two approaches: by direct measurement of adenylate formation (Fig. 5, panel A) and by use of chemically synthesized lipoyl-adenylate as a substrate (Fig. 5, panel C). We found that lipoyl-adenylate was formed with [α-32P]ATP as substrate in the absence of LD acceptor. The adenylate intermediate was resolved from other products by thin layer chromatography using the solvent system used for chemical lipoyl-adenylate synthesis (3) followed by autoradiography and phosphorimaging (Fig. 5, panel A). About 1 eq of lipoyl-adenylate/eq of the LplA-LplB ligase was formed after 30 min of incubation, a result similar to that obtained for E. coli biotin ligase (30). The lipoyl-AMP intermediate fractionated with the ligase complex indicating that the intermediate remained protein-bound (Fig. 5, panel B). We also attempted to demonstrate association of the synthesized lipoyl-adenylate using Ni2+ affinity column fractionation. However, elution resulted in significant lipoyl-AMP hydrolysis due to nucleophilic attack of lipoyl-AMP by the imidazole eluant (31). This result suggests that little or no lipoyl-AMP remained bound to the Ni2+ affinity-purified enzyme. To our surprise, LplB was required for adenylate formation (Fig. 5, panel A). No adenylate was formed with LplA alone even in the presence of 100-fold molar excesses of ATP and lipoic acid. To eliminate the possibility that the active site contained unlabeled adenylate or ATP, the reaction was pretreated with apo domain (equimolar with enzyme) before addition of [32P]ATP (data not shown). The results obtained were similar to those shown, and no lipoyl-adenylate was detected unless both LplA and LplB were present.
FIGURE 5.
The role of LplB in synthesis of lipoyl-adenylate and transfer of the lipoyl moiety. Panel A, synthesis of 32P-labeled lipoyl-adenylate from [α-32P]ATP was analyzed by cellulose thin layer chromatography and visualized by autoradiography. Lane 1, no enzyme (NE); lane 2, LplA (A); lane 3, LplB (B); lane 4, LplA plus LplB (AB). Panel B, demonstration of enzyme-bound lipoyl-AMP with a centrifugal filter device. Lane 1, reaction (R); lane 2, flow-through (F); lane 3, final wash (W); lane 4, retained enzyme fraction (E). Panel C, transfer of lipoate from synthetic lipoyl-adenylate to LD assayed by gel shift. Lanes 1–4 are labeled as in panel A.
We also tested transfer of lipoate from chemically synthesized lipoyl-AMP to an LD. The results (Fig. 5, panel C) demonstrated that only the LplA protein was required for transfer of the lipoyl moiety and that LplB is not required for interaction with the LD. This latter finding is not surprising because the related LipB octanoyltransferases interact with LDs although they lack a CTD (Fig. 2). Lipoic acid and AMP were also seen among the reaction products because of the lability of the lipoyl-AMP mixed anhydride. Based on findings with biotinyl-AMP, lipoyl-AMP could chemically modify the domain (32). However, little such modification was seen under our reaction conditions (Fig. 5, panel C).
Properties of the LplA-LplB Complex
The LplA-LplB complex was recovered during Ni2+ affinity, anion exchange, and size exclusion chromatographic separations indicating reasonably stable interaction. Affinity-tagged LplA bound native LplB (Fig. 6). In contrast tagged LplB coexpressed with native LplA resulted in little or no protein upon elution of the Ni2+ affinity columns. This is presumably due to the poor solubility of LplB and the low efficiency of the LplB affinity tag that may be further decreased when LplB is complexed with LplA. Size exclusion chromatography indicated an LplA-LplB complex size that was considerably smaller than the molecular weight calculated for an equimolar A-B complex. However, LplA and LplB when chromatographed alone also appeared smaller than their calculated masses such that a 1:1 complex is a reasonable interpretation. We have no explanation for this aberrant behavior but note that similar atypical elution was reported for another member of this protein family, E. coli LipB (33). Specific biosynthetic labeling of the proteins with [35S]methionine allowed the ratio of LplA to LplB to be determined. An LplA to LplB ration of 1.6 ± 0.2:1 was obtained by size exclusion chromatography. However, the elution positions of LplA and the complex overlap such that excess LplA present in the chromatographed extract could have falsely increased the ratio. Excess LplA might be expected because of greater relative expression resulting from the higher copy number of pET101 relative to pRSF-1b. In a second approach we purified the [35S]methionine-labeled complex by Ni2+ affinity chromatography and separated the subunits by denaturing gel electrophoresis (Fig. 6, panel B). Phosphorimaging analysis of the gel gave a ratio of LplA to LplB of 1.36:1. This result suggests that the complex is somewhat unstable and together with the gel filtration data indicate that an A-B heterodimer was the best fit of the data.
FIGURE 6.
Characterization of the LplA-LplB complex. Panel A, size exclusion chromatography of the purified proteins. The calibration curve was prepared using (in order of ascending molecular mass) ribonuclease A, chymotrypsinogen, ovalbumin, and bovine serum albumin as standards, shown as “×” symbols. Hexahistidine-tagged LplA is shown as a circle and was estimated to have a mass of 16.6 ± 0.5 kDa, whereas mass spectrometry gave a mass of 30.8 kDa. Hexahistidine-tagged LplB eluted after the linear range of the column (<10 kDa), although it had a mass of 11.2 kDa as determined by mass spectrometry. The lipoyl ligase (LplA-LplB) complex is shown as a square and elutes soon after LplA and has an estimated mass of 18.2 ± 0.4 kDa and a mass calculated from the individual mass spectra of 41.1 kDa. The estimated sizes are the average of four runs with independent protein samples. The S.D. is also reported. Panel B, demonstration of the lipoyl ligase complex by Ni2+ affinity chromatography. The elution products were subjected to SDS-PAGE on a 4–20% gradient Tris-glycine gel and visualized with Coomassie Blue R-250. LplB copurified with hexahistidine-tagged LplA (lane AB). Hexahistidine-tagged LplA and LplB were also purified individually as references (lanes A and B, respectively). The rightmost lane is a phosphorimaging scan of l-[35S]methionine-labeled lipoyl ligase complex after SDS-PAGE on a 4–20% gradient Tris-glycine gel.
DISCUSSION
This study confirms the hypothesis of McManus et al. (12) that LplB is the missing CTD of the T. acidophilum LplA. Although LplB is the defining member of the “bacterial lipoate-protein ligase C terminus” family (Pfam PF10437), it is doubly misnamed because it is not of bacterial origin, nor is it always a C-terminal domain. Many actinomycete LplA homologues have a domain at their N termini homologous to a CTD suggesting that they are circularly permuted LplAs (this is currently under investigation). As a result of the misnaming of the PF10437 family, it contains only one single domain sequence: that of T. acidophilum LplB. We suggest this family might better be called the lipoyl ligase accessory domain family. We detected other single domain LplB candidates using a Phylobuilder global-global homology search. LplB homologues are present in many Archaea and Bacteria but are not necessarily encoded near the lplA gene. Alignments of selected sequences are shown in Fig. 7. Although the number of sequenced Archaea is few, the lplB-lplA gene arrangement appears to be the typical form of archaeal lipoyl ligases. Exceptions are the putative lipoyl ligases of Sulfolobus that encode putative LplAs that both have and lack an LplB-like CTD. There are few LplB homologues in Bacteria with the exception that all sequenced Bordetella strains contain an LplB homologue and a T. acidophilum-like LplA. Although cultured methanogens do not require lipoic acid, a putative two-subunit lipoyl ligase is found in the genome of the uncultured methanogen RC1.
FIGURE 7.
ClustalW alignment of representative LplB homologues. The first six sequences are representative single domain LplB homologues. Black shading denotes identical residues whereas gray shading denotes residues of similar properties. The putative S. coelicolor (Sc) ligase has an N-terminal LplB domain. E. coli LplA (Ec) is the canonical C-terminal domain ligase. The putative S. pneumoniae (Sp) LplA also has a C-terminal domain. The Bos taurus (Bt) bLT has a C-terminal domain of unknown function. Conserved Gly residues correspond to flexible loops in the E. coli LplA structure (10). Most conserved residues are predicted to be in the interior of the protein and probably serve structural roles. Close relatives to LplB, including single domain proteins, contain a GDFF motif. The aspartate residue of this motif (Asp-41 in LplB) is well conserved among LplB homologues. The Genbank accession numbers of the aligned sequences (all previously published) are (from top to bottom) NP_393989, NP_579363.1, NP_880065, YP_687213.1, ZP_02512737, ACB07333.1, CAA18910, NP_345629, AAC77339, and BAA24354. Ta, T. acidophilum; Pf, Pyrococcus furiosus; Bp, Bordetella pertussis; M, methanogen, uncultured; Mm, Mycoplasma mycoides; Kc, Korarchaeum cryptofilum.
The existence of two classes of LplAs, with and without accessory domains, was proposed previously (12). The results of our study show that lipoyl-AMP formation requires the accessory domain. Because LipB octanoyltransferases have only a catalytic domain but bind and modify their protein substrate, we hypothesize that lipoyl ligase homologues lacking an accessory domain in the genome perform a different reaction. This is the case for the mammalian lipoyl ligase homologues, which have a CTD that has almost no sequence similarity with LplB (Fig. 7). Like T. acidophilum LplA, these enzymes are unable to catalyze lipoyl-adenylation but transfer the lipoyl moiety when lipoyl-AMP is provided (34). This raises the question of the function of the mammalian CTD if it is not involved in adenylation. We found that both LplB and LplA are required for efficient complementation of an lplA mutant of E. coli and for efficient activation of the pyruvate and 2-oxoglutarate dehydrogenases. In vitro both we and McManus et al. (12) found that LplA failed to modify the E2p lipoyl domain. However, Kim et al. (11) reported that upon first soaking LplA crystals in ATP and then in lipoate the crystals contained lipoyl-AMP. We were unable to detect lipoyl-AMP formation by LplA despite use of a sensitive radiochemical assay. Moreover because LplA alone can catalyze lipoyl transfer from exogenously supplied lipoyl-AMP to LD (Fig. 3, panel C), we and McManus et al. (12) should have observed lipoylation activity if lipoyl-AMP had been formed. The difference between the crystal and solution studies of LplA cannot be due to lipoyl-AMP stability. If this were the case we should have observed increased AMP accumulation in the lipoyl-AMP synthesis reaction; this was not the case (Fig. 5, panel A). It therefore seems possible that the function of LplB is to modulate the structure of LplA such that lipoyl-AMP synthesis can occur and that perhaps crystal packing somehow mimicked this modulation. However, the weak modification of the 2-oxoglutarate dehydrogenase seen in the presence of only LplA (Fig. 3) suggests that some lipoyl-AMP is formed in vivo. This could be due to trace ligase activity by LplA alone or by the presence of an E. coli factor that can mimic LplB. The E. coli pyruvate dehydrogenase domains seem poor substrates for the T. acidophilum ligase. Expression of LplA alone or LplA and LplB together in E. coli results in more 2-oxoglutarate dehydrogenase activity than pyruvate dehydrogenase activity, although the latter enzyme has three LDs per E2 subunit, and a single lipoylated LD is sufficient for activity (1). Indeed the E. coli E2p domain was a poor substrate in vitro (Fig. 4).
The available structures of lipoyl ligases provide a possible mechanism for LplB action. The structure of E. coli LplA shows that the conserved CTD Asp (Asp-42 in LplB) residue faces the catalytic domain and is close to the loop formed by residues 72–82, highly conserved residues that line the lipoyl-AMP binding pocket (10). The G76S mutation conferring resistance to selenolipoic acid is also present in this loop, consistent with its role in substrate binding (13). In an unpublished Streptococcus pneumonia LplA structure (Protein Data Bank accession code 1VQZ), the conserved Asp of the accessory domain is also in close proximity to conserved residue Gln-47, and both residues are close to the conserved loop that lines the active site. A structure of a catalytically active lipoyl ligase of known activity complexed with lipoyl-adenylate is not yet available, but an analogous biotin ligase structure has been solved (35). Although some LplB residues are conserved in the biotin ligase accessory domain, it appears that the interaction of the CTD with the catalytic domain is different. The biotin ligase accessory domain is shorter and lacks the two outward facing α helices that are present in the lipoyl ligase accessory domain.
Because of their unique ATP binding motif, it has been argued that members of the biotin ligase superfamily originally evolved to bind small molecules and that adenylation activity arose independently (35). The role of the accessory domain in adenylation suggests that it may be important for binding of a small molecule substrate. Indeed the CTD of biotin ligase has also been shown to be important for binding ATP (36). Within the lipoyl ligase family the lipoyl ligase accessory domain can be found as an N-terminal domain, a CTD, or as a separate protein or is not found at all (Fig. 8). The accessory domains found as separate proteins are closely related (Fig. 7), so the variable domain architecture of these lipoate ligases could be a recent development. The advantage of a multimeric lipoyl ligase complex is not obvious. Perhaps the accessory domain was initially absent and was later added during evolution of ligase function. This is reasonable given that the LplA protein family is rooted in such sequences (Fig. 8). If so, the T. acidophilum lipoyl ligase complex may be an evolutionary relic. The activity of the T. acidophilum LplA and LplB complex with octanoate in place of lipoate was first reported for the E. coli enzyme (6). However in E. coli the octanoyl-LD can be converted to lipoyl-LD by lipoyl synthase (LipA). In contrast, T. acidophilum lacks a recognizable lipoyl synthase homologue, and thus octanoylation would render the LD inactive. Hence to obtain function of its lipoic acid-requiring enzymes T. acidophilum would need an environment rich in lipoic acid and deficient in octanoic acid.
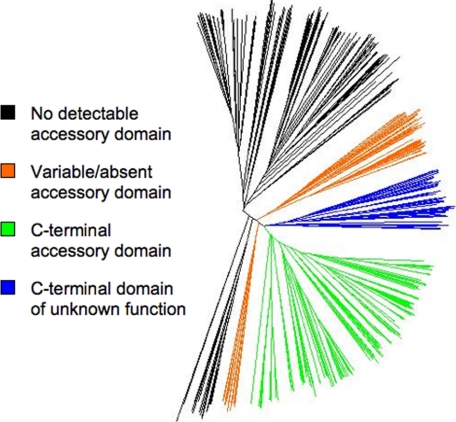
FIGURE 8.
The lipoyl ligase subtree of the BPL_LplA_LipB Pfam family (PF03099) displayed using Dendroscope (38). Domain architecture is annotated by color. Black clade proteins contain a catalytic domain but no detectable LplB homologue in the genome. The green clade proteins have a C-terminal accessory domain. Blue clade proteins are found only in mammalian genomes and have a C-terminal domain of unknown function that has only background sequence similarity with the green clade C-terminal accessory domains (Fig. 7). The orange clade proteins have a variable domain architecture including N-terminal accessory domains and independently coded accessory domains. The deeply branching and multiclade presence of LplAs with independent LplBs suggests that this architecture is an evolutionary relic. The tree root, determined with biotin ligases and octanoyltransferases as outgroups, is within the black clades. This suggests that the catalytic domain was originally independent of any other domains in the common ancestral protein.
Acknowledgments
We thank Stewart Gardner and Dr. Xin Zhao for help in strain construction.
This work was supported, in whole or in part, by National Institutes of Health Grants 5 T32 GM070421, a Ruth L. Kirschstein National Research Service Award from the NIGMS, and AI15650 from the NIAID.
- LD
- lipoyl domain
- CTD
- C-terminal domain
- NTA
- nitrilotriacetic acid
- E1
- E2, and E3, the three subunits of the 2-oxoacid complexes.
REFERENCES
- 1.Perham R. N. (2000) Annu. Rev. Biochem. 69, 961–1004 [DOI] [PubMed] [Google Scholar]
- 2.Engel N., van den Daele K., Kolukisaoglu U., Morgenthal K., Weckwerth W., Pärnik T., Keerberg O., Bauwe H. (2007) Plant Physiol. 144, 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed L. J., Leach F. R., Koike M. (1958) J. Biol. Chem. 232, 123–142 [PubMed] [Google Scholar]
- 4.Koike M., Reed L. J. (1960) J. Biol. Chem. 235, 1931–1938 [PubMed] [Google Scholar]
- 5.Green D. E., Morris T. W., Green J., Cronan J. E., Jr., Guest J. R. (1995) Biochem. J. 309, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris T. W., Reed K. E., Cronan J. E., Jr. (1994) J. Biol. Chem. 269, 16091–16100 [PubMed] [Google Scholar]
- 7.Morris T. W., Reed K. E., Cronan J. E., Jr. (1995) J. Bacteriol. 177, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reche P., Perham R. N. (1999) EMBO J. 18, 2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reche P., Li Y. L., Fuller C., Eichhorn K., Perham R. N. (1998) Biochem. J. 329, 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara K., Toma S., Okamura-Ikeda K., Motokawa Y., Nakagawa A., Taniguchi H. (2005) J. Biol. Chem. 280, 33645–33651 [DOI] [PubMed] [Google Scholar]
- 11.Kim D. J., Kim K. H., Lee H. H., Lee S. J., Ha J. Y., Yoon H. J., Suh S. W. (2005) J. Biol. Chem. 280, 38081–38089 [DOI] [PubMed] [Google Scholar]
- 12.McManus E., Luisi B. F., Perham R. N. (2006) J. Mol. Biol. 356, 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed K. E., Morris T. W., Cronan J. E., Jr. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cronan J. E., Zhao X., Jiang Y. (2005) Adv. Microb. Physiol. 50, 103–146 [DOI] [PubMed] [Google Scholar]
- 15.Cicchillo R. M., Booker S. J. (2005) J. Am. Chem. Soc. 127, 2860–2861 [DOI] [PubMed] [Google Scholar]
- 16.Zhao X., Miller J. R., Jiang Y., Marletta M. A., Cronan J. E. (2003) Chem. Biol. 10, 1293–1302 [DOI] [PubMed] [Google Scholar]
- 17.Kim do J., Lee S. J., Kim H. S., Kim K. H., Lee H. H., Yoon H. J., Suh S. W. (2008) Proteins 70, 1620–1625 [DOI] [PubMed] [Google Scholar]
- 18.Ma Q., Zhao X., Nasser Eddine A., Geerlof A., Li X., Cronan J. E., Kaufmann S. H., Wilmanns M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J. (1972) Experiments in Molecular Genetics, pp. 431–433, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cronan J. E. (2006) Plasmid 55, 152–157 [DOI] [PubMed] [Google Scholar]
- 23.Packman L. C., Perham R. N., Roberts G. C. (1984) Biochem. J. 217, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glanville J. G., Kirshner D., Krishnamurthy N., Sjölander K. (2007) Nucleic Acids Res. 35, W27–W32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeil L. K., Reich C., Aziz R. K., Bartels D., Cohoon M., Disz T., Edwards R. A., Gerdes S., Hwang K., Kubal M., Margaryan G. R., Meyer F., Mihalo W., Olsen G. J., Olson R., Osterman A., Paarmann D., Paczian T., Parrello B., Pusch G. D., Rodionov D. A., Shi X., Vassieva O., Vonstein V., Zagnitko O., Xia F., Zinner J., Overbeek R., Stevens R. (2007) Nucleic Acids Res. 35, D347–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27.Amarasingham C. R., Davis B. D. (1965) J. Biol. Chem. 240, 3664–3668 [PubMed] [Google Scholar]
- 28.Guest J. R., Creaghan I. T. (1973) J. Gen. Microbiol. 75, 197–210 [DOI] [PubMed] [Google Scholar]
- 29.Tabor S., Richardson C. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Beckett D. (1994) Biochemistry 33, 7354–7360 [DOI] [PubMed] [Google Scholar]
- 31.Jencks W. P. (1957) Biochim. Biophys. Acta 24, 227–228 [DOI] [PubMed] [Google Scholar]
- 32.Streaker E. D., Beckett D. (2006) Protein Sci. 15, 1928–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nesbitt N. M., Baleanu-Gogonea C., Cicchillo R. M., Goodson K., Iwig D. F., Broadwater J. A., Haas J. A., Fox B. G., Booker S. J. (2005) Protein Expr. Purif. 39, 269–282 [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara K., Hosaka H., Matsuda M., Okamura-Ikeda K., Motokawa Y., Suzuki M., Nakagawa A., Taniguchi H. (2007) J. Mol. Biol. 371, 222–234 [DOI] [PubMed] [Google Scholar]
- 35.Wood Z. A., Weaver L. H., Brown P. H., Beckett D., Matthews B. W. (2006) J. Mol. Biol. 357, 509–523 [DOI] [PubMed] [Google Scholar]
- 36.Chapman-Smith A., Mulhern T. D., Whelan F., Cronan J. E., Jr., Wallace J. C. (2001) Protein Sci. 10, 2608–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 38.Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wycuff D. R., Matthews K. S. (2000) Anal. Biochem. 277, 67–73 [DOI] [PubMed] [Google Scholar]