Abstract
Similar to phosphorylation, GlcNAcylation (the addition of O-GlcNAc to Ser(Thr) residues on polypeptides) is an abundant, dynamic, and inducible post-translational modification. GlcNAcylated proteins are crucial in regulating virtually all cellular processes, including signaling, cell cycle, and transcription. Here we show that calcium/calmodulin-dependent kinase IV (CaMKIV) is highly GlcNAcylated in vivo. In addition, we show that upon activation of HEK293 cells, hemagglutinin-tagged CaMKIV GlcNAcylation rapidly decreases, in a manner directly opposing its phosphorylation at Thr-200. Correspondingly, there is an increase in CaMKIV interaction with O-GlcNAcase during CaMKIV activation. Furthermore, we identify at least five sites of GlcNAcylation on CaMKIV. Using site-directed mutagenesis, we determine that the GlcNAcylation sites located in the active site of CaMKIV can modulate its phosphorylation at Thr-200 and its activity toward cAMP-response element-binding transcription factor. Our results strongly indicate that the O-GlcNAc modification participates in the regulation of CaMKIV activation and function, possibly coordinating nutritional signals with the immune and nervous systems. This is the first example of an O-GlcNAc/phosphate cycle involving O-GlcNAc transferase/kinase cross-talk.
The post-translational modification (PTM)3 of proteins with O-GlcNAc was discovered over 2 decades ago (1). Unlike “traditional glycosylation,” O-GlcNAc is not elongated into more complex structures and is localized mostly in nucleocytoplasmic compartments. Similar to phosphorylation, GlcNAcylation is abundant, dynamic, and inducible and occurs on Ser and Thr residues (for review see Ref. 2). To date, all O-GlcNAc-modified proteins are also phosphoproteins. Interestingly, many phosphorylation sites are also known GlcNAcylation sites (e.g. endothelial nitric-oxide synthase (3), c-Myc (4), estrogen receptor-β (5), and RNA polymerase II (6)); however, the relationship between the two PTMs is not simply reciprocal. Several proteins can be concomitantly phosphorylated and GlcNAcylated or the adjacent phosphorylation and GlcNAcylation sites can regulate the addition of the other moiety (e.g. p53 (2, 7)). Recently, different groups were able to show an extensive interplay between GlcNAcylation and phosphorylation using proteomic approaches (8–11).
Unlike phosphorylation, where more than 600 enzymes regulate the turnover of phosphate (12), O-GlcNAc cycling in mammals is regulated by only one known gene encoding the catalytic subunit for O-GlcNAc transferase (13, 14) and one known gene encoding the catalytic subunit for removal, O-GlcNAcase (15). However, in a process similar to that used by Ser/Thr phosphatases (16), both O-GlcNAc transferase and O-GlcNAcase transiently associate with binding partners to dynamically produce holoenzymes with unique properties and specificities (17–21).
GlcNAcylation is crucial in regulating virtually all cellular processes, including signaling, cell cycle, and transcription, affecting protein-protein interactions, activity, stability, and expression (2). O-GlcNAc transferase deletion in mice led to embryonic lethality (22), highlighting the importance of OGT and GlcNAcylation for cell viability. In addition, dysfunctional protein GlcNAcylation/phosphorylation seems to be important for the pathology of diabetes and Alzheimer disease (23). GlcNAcylation is involved in several signaling pathways, including regulating cellular responses to insulin (24), cell cycle control (25), stress protection (26), and calcium cycling (27). Recently, several studies have shown a direct link between GlcNAcylation and Ca2+ homeostasis in cardiomyocytes (28, 29).
Calcium (Ca2+) is a central secondary messenger involved in regulating cytoskeletal proteins, ion pumps, and activities of enzymes, including Ca2+/calmodulin-dependent protein kinases (CaMKs). Calcium/calmodulin-dependent kinase IV (CaMKIV) is a multifunctional Ser/Thr kinase found predominantly in the brain, T-cells, and testis. CaMKIV is activated in the presence of increased intracellular [Ca2+], acting as a potent mediator of Ca2+-induced gene expression. The regulation of CaMKIV activity is complex and involves a multistep process to achieve full activation (30, 31). First, Ca2+/CaM binds to CaMKIV, exposing its activation loop to allow calcium/calmodulin-dependent kinase kinase (CaMKK) (32) to phosphorylate Thr-200 within this activation loop, resulting in a 10–20-fold increase in total activity to generate Ca2+/CaM-independent activity (33). CaMKIV may then also undergo an autophosphorylation event on Ser-11 and Ser-12 in its N terminus (34).
CaMKIV has been shown to phosphorylate and regulate the activity of several proteins, such as c-AMP-response element-binding protein (CREB), oncogene 18, AP-1 (activator protein 1), MEF2 (myocyte enhancer factor 2), and O-GlcNAc transferase, among others. Interestingly, KCl-induced depolarization of neuroblastoma cells rapidly activates O-GlcNAc transferase to increase GlcNAcylation levels, suggesting a potential regulatory role for O-GlcNAc during Ca2+ signaling (35).
Here we show that CaMKIV is highly GlcNAcylated in vivo. We also show that upon ionomycin treatment, CaMKIV GlcNAcylation decreases, whereas its interaction with O-GlcNAcase increases in a manner directly opposing the phosphorylation of Thr-200 on CaMKIV. Furthermore, we identify at least five sites of GlcNAcylation on CaMKIV. Using site-directed mutagenesis, we have determined that the GlcNAcylation sites located in the active cleft of CaMKIV modulate the phosphorylation of CaMKIV at Thr-200 and its activity toward CREB. Our results strongly indicate that the O-GlcNAc modification participates in the regulation of CaMKIV activation and function.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293A cells were grown in Dulbecco's modified Eagle's medium (25 mm glucose; Mediatech) containing 10% (v/v) fetal bovine serum (Gemini Bio-Products) and penicillin/streptomycin (Mediatech). Jurkat human T-lymphocytes were maintained in RPMI 1640 medium, 10 mm sodium pyruvate, nonessential amino acids, penicillin/streptomycin, and 10% fetal bovine serum. Cells were stimulated with 1 μm ionomycin for the indicated times.
Rat Cerebellum Extract
Cerebella were dissected from rat brains (Pel-Freez) and homogenized in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1 μm PUGNAc with protease and phosphatase inhibitors. The homogenate was clarified by centrifugation and filtration.
DNA Transfection and Plasmids
HEK293A cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The human CaMKIV cDNA was obtained from the ATCC and subcloned into pEF-HA (36) to create HA-CaMKIV. CaMKIV point mutants T57A/S58A, S137A, S189A, S344A/S345A, S356A, T200A, and T200E were created using the QuikChange site-directed mutagenesis system (Stratagene). The mutations were confirmed by DNA sequencing by the Synthesis & Sequencing Facility at Johns Hopkins University. The GST-CREB prokaryotic expression plasmid was created by subcloning CREB from pCMV-mycCREB (37) kindly provided by Dr. Ginty (Johns Hopkins University) into pGEX-4T2 (GE Healthcare). Recombinant GST-CREB was expressed and purified from Escherichia coli BL21 cells according to the manufacturer's instructions.
Immunoprecipitation
Cells were washed with phosphate-buffered saline and collected into lysis buffer (0.5% Nonidet P-40 (Sigma) in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1 μm PUGNAc with protease and phosphatase inhibitors). Cell lysates were sonicated and centrifuged to remove debris. Immunoprecipitations were performed with the indicated antibodies and captured with GammaBind G-Sepharose (GE Healthcare). The immunoprecipitates were then washed with lysis buffer and submitted for enzymatic assays or eluted in Laemmli buffer for immunoblot analysis.
Immunoblotting
Samples were separated on Criterion precast SDS-polyacrylamide gels (Bio-Rad), and the gels were subsequently electroblotted to nitrocellulose (Bio-Rad). The membranes were blocked in Tris-buffered saline with 0.1% (v/v) Tween 20 with either 3% (w/v) bovine serum albumin or 3% (w/v) nonfat dry milk. The blocked membranes were then incubated overnight at 4 °C with primary antibodies against O-GlcNAc (CTD110.6), O-GlcNAcase, HA (HA.11; Covance), CaMKIV (Cell Signaling), or pT200 CaMKIV (Santa Cruz Biotechnology). CTD110.6 O-GlcNAc immunoblots were performed using bovine serum albumin as the blocking agent. Streptavidin-conjugated horseradish peroxidase (Pierce) was used to probe for biotin labeling. The blots were then washed, incubated with the appropriate secondary antibody, developed using ECL (GE Healthcare), and exposed to Hyperfilm ECL (GE Healthcare). NIH Image or ImageJ software were used for densitometric analysis of immunoblots, and all measurements were normalized against HA loading controls.
β-Elimination, γ-Phosphatase, and β-GlcNAcase Treatments
Immunoprecipitated HA-CaMKIV was washed with lysis buffer, washed once with water, and submitted to the following treatments. β-Elimination was performed overnight at 4 °C using the GlycoProfile β-elimination kit (Sigma) according to the manufacturer's instructions. γ-Phosphatase (New England Biolabs) treatment was performed according to the manufacturer's instructions. β-GlcNAcase (Sigma) treatment was performed in 20 mm MES, pH 5.6, 50 mm NaCl for 2 h at 30 °C, followed by 12 h at 4 °C. All reactions were stopped and eluted with Laemmli buffer for immunoblot analysis.
Enzymatic Labeling of O-GlcNAc Sites
HA-CaMKIV was immunoprecipitated, washed with lysis buffer, and washed twice with reaction buffer containing 20 mm HEPES, pH 7.9, 50 mm NaCl, 1 μm PUGNAc, and 5 mm MnCl2 with protease and phosphatase inhibitors. Next, 2 μl of Gal-T1 Y289L (Invitrogen) and 2 μl of 0.5 mm UDP-GalNAz (Invitrogen) were added to a reaction volume of 20 μl. The reaction was performed overnight at 4 °C. The beads were washed twice with reaction buffer to remove excess UDP-GalNAz. The samples were then reacted with biotin alkyne (Invitrogen) or tetramethyl-6-carboxyrhodamine (TAMRA) alkyne (Invitrogen) according to the manufacturer's instructions. Galactosyltransferase labeling with UDP-[3H]galactose was performed as described previously (38). The enzymatic reactions were eluted with Laemmli buffer for immunoblot analysis or gel-purified for site-mapping analysis. In-gel fluorescence was imaged and measured using a Typhoon Variable Mode Imager (GE Healthcare).
Site-mapping Analysis
Immunoprecipitated HA-CaMKIV was labeled with GalNAz and reacted with biotin alkyne as described above (supplemental Fig. S1). The sample was gel-purified, trypsin-digested, and enriched for O-GlcNAc-containing peptides over streptavidin-agarose (Invitrogen). Biotin-containing peptides were eluted from the streptavidin-agarose by β-elimination followed by Michael addition (BEMAD) (39), to replace the GlcNAc-GalNAz-biotin moiety with DTT (Fig. 2A), and analyzed by an LTQ mass spectrometer (Thermo Finnigan) coupled with a nano-two-dimensional liquid chromatography pump (Eksigent Technologies). The mass spectrometer was programmed to record a full precursor scan, followed by fragmentation and MS/MS scans of the top eight most intense ions. The data were analyzed using Mascot (Matrix Science) against the full human protein data base with DTT-modified Ser/Thr as variable modifications.
FIGURE 2.
GlcNAcylation of CaMKIV opposes its phosphorylation at Thr-200 . A, lysates from HEK293 cells transfected with HA-CaMKIV and treated with 1 μm ionomycin for the indicated times were immunoprecipitated (IP) for HA and immunoblotted (IB) for O-GlcNAc, Thr(P)-200 (pT200), and HA. B, relative fold change in Thr(P)-200/HA signal (diamonds) and O-GlcNAc/HA signal (circles) during ionomycin treatment (normalized to 0 h time point). C, lysates from HEK293 cells transfected with HA-CaMKIV and treated with or without 1 μm ionomycin for 2 min were immunoprecipitated for HA and immunoblotted for O-GlcNAcase, Thr(P)-200, and HA. D, relative fold change in O-GlcNAcase/HA signal with ionomycin treatment (normalized to untreated control). E, lysates from HEK293 cells transfected with HA-CaMKIV wild type, HA-CaMKIV T200A, or HA-CaMKIV T200E were immunoprecipitated for HA and immunoblotted for O-GlcNAc and HA. F, relative fold change in O-GlcNAc/HA signal for T200A and T200E mutants (normalized to wild-type control). Asterisk is used to note p < 0.05 for samples compared with control or 0-h time point.
Homology Modeling of CaMKIV Kinase Domain
The automated comparative modeling program SWISS-MODEL was used for homology modeling (40). Using the first approach automated mode, SWISS-MODEL selected CaMKIG from the RCSB Protein Data Bank as best template by sequence similarity. Using the x-ray crystal structure of CaMKIG (Protein Data Bank code 2JAM)4 as template, SWISS-MODEL created a predicted model of the kinase domain of CaMKIV. The resulting Protein Data Bank code coordinates are found in supplemental Fig. S3. Figures and other representations of the predicted CaMKIV kinase domain structure were created using MacPyMol (DeLano Scientific). Sequence alignments, percent identity, and percent similarity scores were generated using Clone Manager (Sci-Ed software) and BLAST2 sequences using the BLOSUM62 scoring matrix.
ATP Binding Analysis
Cells expressing various forms of HA-CaMKIV were lysed in buffer containing 20 mm Tris-HCl, pH 7.5, 0.5 mm DTT, 0.1% Tween 20, 5 mm MgCl2, 1 mm CaCl2, 1 μm CaM, and 1 μm PUGNAc with protease and phosphatase inhibitors. Lysates were sonicated and centrifuged to remove debris. Clarified lysates were incubated with 10 μl of Kinase-Bind γ-phosphate-linked high substitution ATP resin (Innova Biosciences) for 2 h at 4 °C. The binding reactions were washed with lysis buffer and eluted with Laemmli buffer for immunoblot analysis.
CaMKIV Activity toward CREB
HA-CaMKIV and its mutants were immunoprecipitated, washed with lysis buffer, washed once with lysis buffer containing 500 mm LiCl, washed twice with lysis buffer, and then washed twice with reaction buffer containing 20 mm Tris-HCl, pH 7.4, 0.1% Nonidet P-40, 8 mm MgCl2, 1 mm CaCl2, 1 μm CaM, and 1 μm PUGNAc with protease and phosphatase inhibitors. Recombinant GST-CREB (2 μg), 2 μCi of [32P]ATP, and 2 μm ATP were added to each reaction and incubated for 20 min at 30 °C. The reactions were stopped by addition of Laemmli buffer and subjected to autoradiography and immunoblot analysis. For experiments without ionomycin treatment, samples were incubated for 90 min at 30 °C, and the length of exposure to film was significantly longer than ionomycin-stimulated samples.
Statistical Analysis
Immunoblots shown are representative of at least three independent experiments. All experiments, excluding site-mapping analysis, were performed a minimum of three times. Error bars in all charts represent the mean ± S.E. The paired two-tailed Student's t test was used to determine the statistical significance of any differences between the experimental samples and the control or 0-h samples. p values less than 0.05 were deemed statistically significant at the 95% confidence level. p values are indicated in each figure legend.
RESULTS
CaMKIV Is GlcNAcylated
To determine whether CaMKIV was GlcNAcylated, we used several approaches and controls to detect the O-GlcNAc modification. First, we transfected HEK293 cells with an HA-tagged form of CaMKIV, immunoprecipitated using anti-HA tag antibodies, and performed galactosyltransferase reactions using UDP-[3H]galactose. In this reaction, the radioactive galactose moiety is specifically transferred to terminal GlcNAc residues. Overexpressed HA-CaMKIV was labeled with [3H]galactose indicating the presence of terminal GlcNAc (Fig. 1A).
FIGURE 1.
CaMKIV is GlcNAcylated. A–D, lysates from HEK293 cells transfected with empty HA plasmid or HA-CaMKIV were immunoprecipitated for HA. A, subjected to galactosyltransferase labeling in the presence of UDP-[3H]galactose for autoradiography. B, labeled in the presence of UDP-GalNAz and reacted with TAMRA alkyne for detection by in-gel fluorescence. C, treated with β-elimination. IP, immunoprecipitation; IB, immunoblot. D, treated with γ-phosphatase (γ-PPase) or GlcNAcase prior to immunoblotting for O-GlcNAc and HA. A, prior to autoradiography, the gel was stained for total protein using Coomassie Brilliant Blue G-250 (CBB G250). B, TAMRA fluorescence was detected in-gel prior to staining for total protein using SYPRO Ruby. E, rat cerebellum extract was immunoprecipitated for CaMKIV, subjected to galactosyltransferase labeling in the presence of UDP-GalNAz, reacted with biotin alkyne, and immunoblotted for CaMKIV or biotin (using streptavidin-horseradish peroxidase). F, lysates from Jurkat cells were immunoprecipitated for CaMKIV or using nonspecific (ns) mouse antibodies and immunoblotted for O-GlcNAc or CaMKIV.
Recently, a chemoenzymatic approach to detect O-GlcNAc was described using a mutant galactosyltransferase GalT1 Y289L and an azide derivative of UDP-GalNAc (UDP-GalNAz) as donor substrate (41). The resulting azide-labeled proteins were chemically tagged with TAMRA alkyne for detection by in-gel fluorescence. HA-CaMKIV was strongly labeled with TAMRA (Fig. 1B).
We were also able to detect GlcNAcylation of HA-CaMKIV using the O-GlcNAc-specific antibody CTD110.6 (42) (Fig. 1C). As a specificity control, we performed a reductive β-elimination reaction under mild alkaline conditions to remove O-linked but not N-linked sugars. As expected, β-elimination treatment completely abolished the O-GlcNAc signal on CaMKIV (Fig. 1C), showing the high specificity of the CTD110.6 antibody for O-GlcNAc. Under these mild β-elimination conditions, very little protein degradation was observed.
To further confirm that CaMKIV is GlcNAcylated, we treated HA-CaMKIV with β-GlcNAcase and γ-phosphatase. β-GlcNAcase treatment was sufficient to remove most of the O-GlcNAc from CaMKIV without changing its protein levels, whereas γ-phosphatase treatment completely abolished the slower migrating species, indicating that phosphorylation is responsible for altering CaMKIV mobility on an SDS-polyacrylamide gel (Fig. 1D). In this experiment, the phosphorylated species of HA-CaMKIV does not appear to include phosphorylation at Thr-200 (data not shown), consistent with the cells being in a basal unstimulated state.
Next, we investigated the presence of O-GlcNAc on endogenously expressed CaMKIV. Toward this end, we immunoprecipitated CaMKIV from rat cerebella and Jurkat human T-lymphocytes, because it had been shown previously that CaMKIV is highly expressed in these tissues (43, 44). CaMKIV was found to be GlcNAcylated in both rat cerebella (Fig. 1E) and Jurkat cells (Fig. 1F).
Interplay between the GlcNAcylation of CaMKIV and Its Phosphorylation at Thr-200
The regulation of CaMKIV activity is complex and involves binding to Ca2+/CaM, auto-phosphorylation on multiple Ser residues at its N terminus, and phosphorylation in its activation loop on Thr-200 by the upstream kinase CaMKK (32, 33). The phosphorylation at Thr-200 by CaMKK increases the CaMKIV activity about 10-fold by decreasing the Km value for its substrates (34). The phosphorylation of Thr-200 of CaMKIV has been shown to be fast and transient in response to ionomycin (45). Consistent with previous reports, we observed increased CaMKIV Thr-200 phosphorylation upon ionomycin stimulation, peaking at 2 min and decreasing after 5 min (Fig. 2, A and B). Interestingly, CaMKIV GlcNAcylation rapidly decreased at 2 min and returned to basal levels after 10 min, in a manner that directly opposed its phosphorylation at Thr-200 (Fig. 2, A and B).
Based upon this observation, we sought to determine whether CaMKIV interacts with O-GlcNAcase during activation. Indeed, CaMKIV interacted with O-GlcNAcase, and this interaction increased with ionomycin stimulation (Fig. 2, C and D), indicating that O-GlcNAcase is recruited to CaMKIV to facilitate the removal of O-GlcNAc from CaMKIV during its activation.
To determine whether phosphorylation at Thr-200 affects the GlcNAcylation of CaMKIV, we used site-directed mutagenesis to create Ala and Glu substitutions at Thr-200, to prevent and mimic constitutive phosphorylation at Thr-200, respectively. Indeed, the GlcNAcylation levels of the T200A mutant was increased 2-fold over wild-type CaMKIV, whereas the T200E mutant displayed less O-GlcNAc (Fig. 2, E and F), supporting a reciprocal relationship between the GlcNAcylation of CaMKIV and its phosphorylation at Thr-200.
Determining the GlcNAcylation Sites on CaMKIV
To explain how GlcNAcylation of CaMKIV could affect its phosphorylation at Thr-200, we identified the sites of GlcNAcylation on CaMKIV. Mapping O-GlcNAc sites has been one of the major obstacles in studying protein GlcNAcylation, mostly because O-GlcNAc is extremely labile during mass spectrometry analysis. Here we modified the newly developed chemoenzymatic labeling protocol, combining it with β-elimination followed by Michael addition with DTT (BEMAD) for O-GlcNAc site-mapping (Fig. 3A) (10, 39). Immunoprecipitated HA-CaMKIV was labeled with GalNAz and reacted with a biotin alkyne to covalently attach a biotin tag to the GlcNAc site (supplemental Fig. S1). The biotinylated O-GlcNAc peptides were then enriched over streptavidin-agarose prior to the BEMAD reaction, which replaced the labile GlcNAc-GalNAz-biotin moiety with a stable DTT tag to facilitate identification, using collision-induced fragmentation on a conventional ion trap mass spectrometer.
FIGURE 3.
Identification of GlcNAcylation sites on CaMKIV. A, work flow scheme for labeling and enrichment of GlcNAcylated CaMKIV peptides for O-GlcNAc site identification by liquid chromatography-MS/MS. B, MS/MS spectra for unambiguous assignment of Ser-137. C, Ser-189. D, Ser-356 as GlcNAcylation sites in vivo. E, diagram of CaMKIV displaying location of known phosphorylation sites (red), GlcNAcylation sites (blue), kinase domain (filled black box), autoinhibitory domain (open box), and Ca2+/CaM-binding domain (filled gray box).
Using this approach, we were able to identify at least five O-GlcNAc sites on CaMKIV (Thr-57/Ser-58, Ser-137, Ser-189, Ser-344/Ser-345, and Ser-356). The MS/MS spectra of selected O-GlcNAc sites are shown in Fig. 3B (Ser-137), Fig. 3C (Ser-189), and Fig. 3D (Ser-356). CaMKIV is also known to be phosphorylated at multiple sites (46). A diagram of CaMKIV illustrates the location of the known sites of phosphorylation and GlcNAcylation (Fig. 3E).
Removal of O-GlcNAc Sites on CaMKIV Reduces Its GlcNAcylation Levels
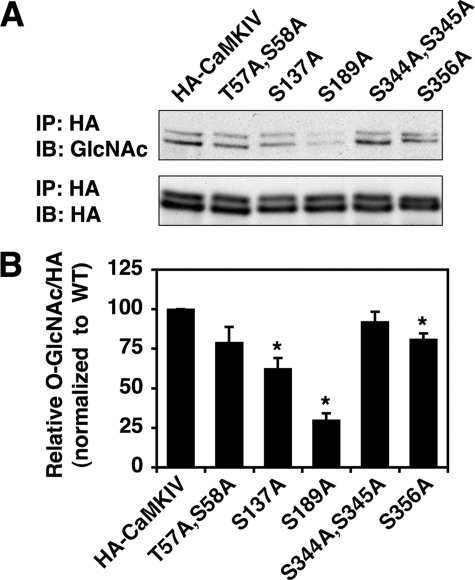
To confirm the identified O-GlcNAc sites on CaMKIV, we mutated each site to Ala and analyzed its effect on CaMKIV GlcNAcylation levels (Fig. 4). First, removal of Ser-344 and Ser-345 did not seem to significantly reduce total CaMKIV GlcNAcylation levels. Removal of Ser-356 or Thr-57 and Ser-58 gave slight reductions in CaMKIV GlcNAcylation levels, as compared with wild type. However, mutation of Ser-137 to Ala resulted in a 40% reduction in total CaMKIV GlcNAcylation levels. Interestingly, replacement of Ser-189 with Ala drastically reduced the GlcNAcylation levels by 70%, clearly indicating that Ser-189 is the major site of GlcNAcylation on CaMKIV under basal conditions. In addition, Ser-189 is conserved in all human CaMKI isoforms, suggesting the possibility that they are also GlcNAcylated under basal conditions (supplemental Fig. S2).
FIGURE 4.
Removal of O-GlcNAc sites on CaMKIV reduces its GlcNAcylation levels. A, lysates from HEK293 cells transfected with HA-CaMKIV wild type or its indicated mutants were immunoprecipitated (IP) for HA and immunoblotted (IB) for O-GlcNAc and HA. B, relative fold change in O-GlcNAc/HA signal for each CaMKIV mutant (normalized to wild-type (WT) control). Asterisk is used to note p < 0.05 for samples compared with control.
Removal of O-GlcNAc Sites on CaMKIV Alters Its Phosphorylation at Thr-200 during Activation
To determine whether any of the O-GlcNAc sites on CaMKIV can modulate its phosphorylation at Thr-200, we transfected HEK293 cells with each of the mutants and stimulated the cells with ionomycin (Fig. 5). Our results demonstrate that the S344A, S345A, and S356A mutants did not significantly change Thr-200 phosphorylation, whereas S137A slightly increased Thr-200 phosphorylation. Interestingly, the S189A mutation significantly increased CaMKIV phosphorylation at Thr-200, consistent with an opposing reciprocal relationship between GlcNAcylation and Thr-200 phosphorylation. However, the double mutant T57A/S58A showed a drastic 90% reduction in Thr-200 phosphorylation, indicating that removal of Thr-57 and Ser-58 prevents CaMKIV from being phosphorylated at Thr-200. Altogether, these results suggest that GlcNAcylation of CaMKIV may differentially affect its phosphorylation at Thr-200.
FIGURE 5.
Removal of O-GlcNAc sites on CaMKIV alters Thr-200 phosphorylation during ionomycin treatment. A, lysates from HEK293 cells transfected with HA-CaMKIV wild type or its indicated mutants and treated with 1 μm ionomycin for 2 min were immunoprecipitated (IP) for HA and immunoblotted (IB) for Thr(P)-200 (pT200) or HA. B, relative fold change in Thr(P)-200/HA signal for each CaMKIV mutant (normalized to wild-type control treated with ionomycin). As a control, a duplicate sample transfected with HA-CaMKIV wild type (WT) was left untreated to show the magnitude of Thr(P)-200 induced by ionomycin treatment. Asterisk is used to note p < 0.05 for samples compared with control.
Predicted Structure of CaMKIV Kinase Domain
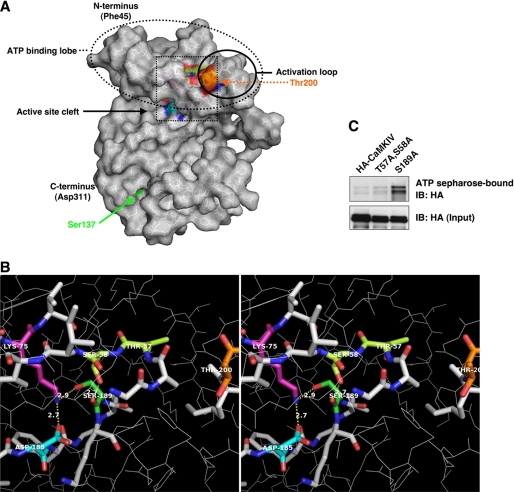
In an attempt to clarify the ways that GlcNAcylation may affect CaMKIV, we decided to determine the location of the O-GlcNAc sites in its three-dimensional structure. However, because the structure of CaMKIV has not been solved, we instead used homology modeling to find the approximate locations of the GlcNAcylation sites. Using SWISS-MODEL, a recently solved x-ray crystal structure of CaMKIG (2JAM) was chosen as the modeling template because of its high identity (50%) and similarity (68% using BLOSUM62 scoring matrix) to CaMKIV in its kinase domain (supplemental Fig. S2). SWISS-MODEL was then able to create a predicted structure for CaMKIV (Fig. 6A, supplemental Fig. S3, and supplemental movie). The predicted CaMKIV structure was evaluated using a number of tools, including ANOLEA, ProQRes, DFire, QMEAN, GROMOS, WhatCheck, and Procheck (40, 47–53). These software tools gave reasonably good scores (data not shown), consistent with a good quality predicted model with very little disallowed bond lengths and angles or unfavorable packing environments.
FIGURE 6.
Locations of the GlcNAcylation sites on a predicted CaMKIV structure and within its active site cleft. A, predicted structure of the CaMKIV kinase domain (residues 45–311) based on homology modeling using CaMKIG as template. B, area shown within dotted box is magnified. B, stereo image of active site cleft, displaying residues known to be important for ATP binding (Lys-75 in cyan and Asp-185 in magenta), the phosphorylation site on the activation loop (Thr-200 in orange), and the identified GlcNAcylation sites (Thr-57, Ser-58, and Ser-189 in green). Distances shown are in angstroms and were calculated using MacPyMol. C, lysates from HEK293 cells transfected with HA-CaMKIV wild type, T57A/S58A, or S189A were tested for the ability to bind ATP resin. ATP-resin bound proteins were immunoblotted (IB) for HA.
Using this model, we were able to predict that Ser-189, Thr-57, and Ser-58 are all localized within the catalytic cleft of CaMKIV, in close proximity to Lys-75 and Asp-185, key residues responsible for ATP binding (Fig. 6, A and B). This motivated us to determine whether the S189A or T57A/S58A double mutants are able to bind to ATP. To address this point, we incubated extracts from cells overexpressing wild-type CaMKIV, the double mutant T57A/S58A, or the S189A mutant with ATP-agarose. Interestingly, the T57A/S58A double mutant seemed to have the same binding affinity to ATP-agarose as wild-type CaMKIV, whereas the S189A mutant displayed increased binding affinity to the ATP-agarose (Fig. 6C), indicating a possibly highly active form of CaMKIV. Also, these data suggest that Ser-189 GlcNAcylation of CaMKIV may negatively affect its ability to bind ATP in the basal unstimulated state.
CaMKIV Activity toward CREB
Next, we decided to determine whether any of the O-GlcNAc sites on CaMKIV modulate its kinase activity. Toward this end, we immunoprecipitated each of the CaMKIV mutants expressed from HEK293 cells and assayed their kinase activity toward recombinant CREB. We determined the activity of the mutants in the basal state and after 2 and 5 min of ionomycin stimulation (Fig. 7).
FIGURE 7.
Removal of O-GlcNAc sites on CaMKIV alters its kinase activity. A, lysates from HEK293 cells transfected with HA-CaMKIV wild type or its indicated mutants were immunoprecipitated (IP) for HA and assayed for kinase activity toward recombinant GST-CREB. CBB, Coomassie Brilliant Blue; IB, immunoblot. B, relative fold change in CREB phosphorylation for each CaMKIV mutant (normalized to wild-type (WT) control). C and D, same as A and B but after treatment with ionomycin for 2 min. E and F, same as A and B but after treatment with ionomycin for 5 min. As a negative control, empty vector was included with this time point. Asterisk is used to note p < 0.05 for samples compared with control. Note: because A, C, and E represent different exposure times, samples between time points cannot be directly compared with one another.
Strikingly, the T57A/S58A double mutant displayed no detectable activity even after 5 min of ionomycin stimulation, indicating that O-GlcNAc at these sites may shut down the kinase activity or Thr-57 and/or Ser-58 are necessary for catalysis. We discarded the possibility of O-GlcNAc at these sites being necessary for activity, because the GlcNAcylation level of T57A/S58A is just slightly lower than wild type (Fig. 4). Compared with wild-type CaMKIV, the S356A mutant did not significantly change the kinase activity, whereas the S344A/S345A double mutant slightly reduced the activity in the basal state and after 2 min of ionomycin stimulation. In contrast, the S137A mutation increased the activity about 2-fold in the basal state and 1.5-fold after ionomycin stimulation for 2 min.
Interestingly, the S189A mutation dramatically increased the activity 5-fold in the basal state, decreasing to 3.5-fold after 2 min and displaying no significant difference after 5 min of ionomycin stimulation. In fact, it appears that the S189A mutation may be a constitutively active form of CaMKIV. These data indicate that GlcNAcylation of CaMKIV at Ser-189 may negatively regulate its activity in basal conditions and during early ionomycin stimulation. It is noteworthy that after 5 min of ionomycin stimulation, all mutants except the T57A/S58A double mutant displayed similar kinase activity toward CREB, as compared with wild type.
DISCUSSION
Despite GlcNAcylation being discovered over 24 years ago, we are just beginning to understand its extent and function as a result of the advancement of new research tools. The role of O-GlcNAc in cellular signaling is at least due in part to its complex interplay with phosphorylation by direct competition (same site occupancy) (e.g. c-Myc) or via steric hindrance by reciprocal modification at proximal sites (e.g. p53 (7)). Recently, our group showed an extensive interplay at the site level between GlcNAcylation and phosphorylation by using inhibitors of O-GlcNAcase and protein phosphatases (11). However, the interplay between GlcNAcylation and phosphorylation is likely to be more complex, and two new mechanisms have emerged as follows: (i) regulation of OGT or O-GlcNAcase by phosphorylation of their regulatory/targeting subunits, and (ii) regulation of kinases and phosphatases by GlcNAcylation.
Recently, Song et al. (35) showed that OGT is activated by CaMKIV-dependent phosphorylation under calcium flux in neuroblastoma cells. Here we show for the first time that CaMKIV is highly GlcNAcylated in vivo. Altogether, these data suggest a complex regulatory cycle and cross-talk between CaMKIV and OGT. This type of relationship may be quite common among different kinases and OGT, supporting the complex and extensive cross-talk between these two PTMs.
The regulation of CaMKIV activity upon calcium flux is a complex process that involves several steps. First, Ca2+/CaM binds to CaMKIV, which displaces PP2A from the auto-regulatory domain to generate basal activity. Then the phosphorylation of Thr-200 by a CaMKK generates a highly active form of CaMKIV with considerable Ca2+-independent activity. Subsequently, Ca2+ levels decrease, enabling dissociation of Ca2+/CaM and re-association of PP2A, which dephosphorylates CaMKIV to attenuate its activity.
The CaMKIV activation-inactivation cycle has been shown to be brief and tightly regulated (45, 54). Here the phosphorylation of Thr-200 on CaMKIV induced by ionomycin was rapid and transient, with most of the phosphate removed within 10 min of stimulation. Interestingly, the O-GlcNAc levels on CaMKIV rapidly decreased, and its interaction with O-GlcNAcase increased during early activation, in a manner directly opposing the phosphorylation of Thr-200 and its PP2A interaction (45, 55). In addition, GlcNAcylation of CaMKIV was restored to basal levels after 10 min, indicating that OGT is recruited to CaMKIV to restore the O-GlcNAc levels after Thr-200 dephosphorylation. Unfortunately, the interaction between OGT and CaMKIV during ionomycin stimulation was not detected, likely due a transient or weak interaction (data not shown).
Furthermore, to determine whether phosphorylation at Thr-200 affected the GlcNAcylation of CaMKIV, we substituted Thr-200 with Ala or Glu to prevent or constitutively mimic the charge effect of phosphorylation, respectively. Indeed, the GlcNAcylation levels of the T200A mutant was increased 2-fold over wild-type CaMKIV, whereas the T200E mutant displayed less O-GlcNAc, confirming a direct interplay between the GlcNAcylation of CaMKIV and its phosphorylation at Thr-200.
Mapping the sites of GlcNAcylation within proteins has been a major challenge, mostly because O-GlcNAc is extremely labile during mass spectrometry analysis. Recently, the advancement of new tools coupled with proteomic approaches have significantly increased the number of identified O-GlcNAc sites (8–10, 56). Here we combined the newly developed chemoenzymatic labeling protocol for tagging and enrichment of O-GlcNAc peptides (10) with BEMAD (39) for downstream mass spectrometric analysis. Using this combined approach, we were able to identify at least five O-GlcNAc sites on CaMKIV. The sites identified were Thr-57/Ser-58, Ser-137, Ser-189, Ser-344/Ser-345, and Ser-356. It is worth noting that, for the site-mapping experiments, we used cells grown under normal basal conditions. However, our data indicate that CaMKIV GlcNAcylation is dynamic and changes upon stimulation. Therefore, we do not exclude the possibility of additional O-GlcNAc sites on CaMKIV, occurring under different conditions.
CaMKIV is also highly phosphorylated (31). However the O-GlcNAc sites described here are not known phosphorylation sites, excluding the possibility of direct competition by same site occupancy. Despite the increasing number of O-GlcNAc sites recently described, just a few O-GlcNAc sites have been studied by mutational analysis (e.g. p53 (7), CRTC2 (57), and Foxo (58)). Unfortunately, unlike phosphorylation, O-GlcNAcylation cannot be mimicked by any naturally occurring amino acid. At best, Ala mutagenesis of specific glycosylation sites within proteins can be used to effect changes in its function. Here we mutated each identified site on CaMKIV to Ala and individually analyzed its effect on CaMKIV GlcNAcylation levels, Thr-200 phosphorylation levels, and kinase activity toward CREB.
The removal of Ser-344 and Ser-345 did not significantly reduce total CaMKIV GlcNAcylation levels or change Thr-200 phosphorylation, but it slightly reduced its activity, both in basal state and upon 2 min of ionomycin stimulation. In contrast, the removal of Ser-356 gave slight reductions in CaMKIV GlcNAcylation levels, as compared with wild type, but did not significantly change Thr-200 phosphorylation or its kinase activity. The fact that the removal of these residues did not significantly decrease the GlcNAcylation on CaMKIV indicates that the stoichiometry of O-GlcNAc at these sites is low and thus not sufficient to reduce global O-GlcNAc levels when mutated. Similarly, this phenomenon was also observed when an O-GlcNAc site on p53 (Ser-149) was mutated to Ala, highlighting the extreme difficulty of observing reductions in total GlcNAcylation levels on proteins that contain multiple O-GlcNAc sites (7).
In contrast, substitution of Ser-137 to Ala resulted in a 40% reduction in total CaMKIV GlcNAcylation levels, indicating that stoichiometry of this site is relatively high in basal conditions. In addition, the S137A mutation increased CaMKIV kinase activity 2-fold in the basal state and 1.5-fold upon 2 min of ionomycin stimulation, without significantly increasing Thr-200 phosphorylation.
Structural studies have been of great importance for the understanding of protein function. Here, in an attempt to clarify the ways that GlcNAcylation may affect CaMKIV, we decided to determine the location of the O-GlcNAc sites on its three-dimensional structure. Because the structure of CaMKIV has not been solved, we used homology modeling (SWISS-MODEL) using a solved x-ray structure of the CaMKIG kinase domain as the modeling template, due its high identity and homology to CaMKIV. Using this approach, we were able to predict that Ser-189, Thr-57, and Ser-58 are all localized within the catalytic cleft of CaMKIV. The removal of Thr-57 and Ser-58 did not seem to significantly reduce total CaMKIV GlcNAcylation levels, but it drastically reduced its phosphorylation at Thr-200. In addition, the T57A/S58A double mutant displayed no detectable kinase activity even after 5 min of ionomycin stimulation. In all of our experiments, 5 min of ionomycin stimulation was sufficient to achieve a high level of kinase activity for the wild-type enzyme, as well as the other O-GlcNAc site mutants. We believe this result indicates that Thr-57 and Ser-58 are residues important for catalysis, despite the fact that this mutant was still able to bind to ATP-agarose.
Remarkably, our data suggest that Ser-189 is the major site of GlcNAcylation on CaMKIV in the basal state, because mutation of this site drastically reduces its GlcNAcylation levels by 70% in basal conditions. This result indicates that O-GlcNAc at Ser-189 is extremely abundant with the stoichiometry nearly 100%. Furthermore, the S189A mutant displayed a robust increased binding to ATP-agarose and drastically increased the kinase activity 5-fold in the basal unstimulated state, suggesting that Ser-189 GlcNAcylation of CaMKIV may negatively affect its ability to bind ATP and catalyze phosphorylation. Interestingly, upon ionomycin stimulation, the higher basal activity of the S189A mutant gradually diminished to levels comparable with that of the wild-type enzyme, displaying only a 3.5-fold increase after 2 min and no significant difference in activity after 5 min. In addition, we showed that the S189A mutant displayed a slight increase in Thr-200 phosphorylation after 2 min of ionomycin stimulation, indicating that GlcNAcylation at Ser-189 is likely to participate in interplay with phosphorylation at Thr-200. We are currently developing the site-specific Ser-189 O-GlcNAc antibody to further investigate the role of GlcNAcylation at this site.
Taken together, we showed that CaMKIV is highly GlcNAcylated in vivo. In addition, we showed that upon ionomycin treatment, CaMKIV GlcNAcylation rapidly decreases, and its interaction with O-GlcNAcase increases, in a manner directly opposing its phosphorylation at Thr-200. Furthermore, we identified at least five sites of GlcNAcylation on CaMKIV. Using site-directed mutagenesis, we have determined that the GlcNAcylation levels, Thr-200 phosphorylation levels, and kinase activity can vary according the O-GlcNAc site mutated. In addition, we showed that Ser-189 is GlcNAcylated under basal conditions, and its removal increased kinase activity and Thr-200 phosphorylation. Altogether, our results suggest that GlcNAcylation of CaMKIV may differentially affect its regulation at multiple steps, leading to a new model for CaMKIV activation and additional points for controlling its activity.
Physiologically and mechanistically speaking, GlcNAcylation of CaMKIV may serve to coordinate nutritional and metabolic signals with the immunological and nervous response, because CaMKIV is found predominantly in cells of the immune and nervous systems. CaMKIV has previously been shown to be involved in T-cell activation and secretion of interleukin-2 (31, 59, 60). In fact, knockdown of either CaMKIV (61) or OGT (62) in T-cells impairs activation and subsequent secretion of interleukin-2. Furthermore, overexpression of CaMKIV (63) or elevating global GlcNAcylation in neurons (64) enhances long term potentiation in vivo.
CaMKIV is a multifunctional Ser/Thr kinase that has several substrates, including CREB, serum-response factor, CREB-binding protein, AP-1, MEF2, and OGT among others. Interestingly, O-GlcNAc has been found on CREB (65), serum-response factor (66), CREB-binding protein (67), and even OGT (68), indicating several common substrates between CaMKIV and OGT. The fact that OGT and CaMKIV can modulate each other, combined with the fact that they share several common substrates and phenotypes, indicates that the relationship between CaMKIV and OGT is most likely complex and involves different signaling pathways. Although there have been many studies indicating the extensive cross-talk between GlcNAc and phosphate, this is the first definitive example of a kinase being regulated directly by O-GlcNAc and the first example of an O-GlcNAc/phosphate cycle involving OGT/kinase cross-talk.
Supplementary Material
Acknowledgment
We thank members of the laboratory for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA42486 and DK61671 and Contract N01-HV-28180 from NHLBI. G. W. H. receives a share of royalty received by the University on sales of the CTD 110.6 antibody. Terms of this arrangement are managed by The Johns Hopkins University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and a movie.
J. E. Debreczeni, A. Bullock, T. Keates, F. H. Niesen, E. Salah, L. Shrestha, C. Smee, F. Sobott, A. C. W. Pike, G. Bunkoczi, F. von Delft, A. Turnbull, J. Weigelt, C. H. Arrowsmith, A. Edwards, M. Sundstrom, and S. Knapp, unpublished data.
- PTM
- post-translational modification
- GlcNAcylation
- addition of O-GlcNAc to Ser(Thr) residues on polypeptides
- CaMKIV
- calcium/calmodulin-dependent kinase IV
- O-GlcNAcase
- O-GlcNAc selective β-N-acetylglucosaminidase
- CREB
- cAMP-response element-binding transcription factor
- OGT
- UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyltransferase (O-GlcNAc transferase)
- CaM
- calmodulin
- CaMKK
- calcium/calmodulin-dependent kinase kinase
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenyl carbamate (an inhibitor of β-N-acetylglucosaminidase)
- HA
- YPYDVPDYA influenza hemagglutinin-HA (epitope)
- HEK293
- human embryonic kidney cells 293
- pEF-HA
- plasmid vector based upon eukaryotic translation elongation factor 1 promoter with HA tag
- GST
- glutathione S-transferase
- pCMV
- plasmid based upon cytomegaloviral promoter
- Gal-T1 Y289L
- mutant β1,4-galactosyltransferase with larger binding pocket
- UDP-GalNAz
- uridine diphosphate-N-azidoacetylgalactosamine
- TAMRA
- 5(6)-carboxytetramethylrhodamine
- GalNAz
- N-azidoacetylgalactosamine
- DTT
- dithiothreitol
- BEMAD
- β-elimination, Michael Addition, dithiothreitol
- MES
- 4-morpholineethanesulfonic acid
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1.Torres C. R., Hart G. W. (1984) J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 2.Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 3.Du X. L., Edelstein D., Dimmeler S., Ju Q., Sui C., Brownlee M. (2001) J. Clin. Invest. 108, 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamemura K., Hayes B. K., Comer F. I., Hart G. W. (2002) J. Biol. Chem. 277, 19229–19235 [DOI] [PubMed] [Google Scholar]
- 5.Cheng X., Hart G. W. (2001) J. Biol. Chem. 276, 10570–10575 [DOI] [PubMed] [Google Scholar]
- 6.Comer F. I., Hart G. W. (2001) Biochemistry 40, 7845–7852 [DOI] [PubMed] [Google Scholar]
- 7.Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 8.Khidekel N., Ficarro S. B., Peters E. C., Hsieh-Wilson L. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13132–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Pandey A., Hart G. W. (2007) Mol. Cell. Proteomics 6, 1365–1379 [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Gucek M., Hart G. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 13.Kreppel L. K., Blomberg M. A., Hart G. W. (1997) J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 14.Lubas W. A., Frank D. W., Krause M., Hanover J. A. (1997) J. Biol. Chem. 272, 9316–9324 [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. (2001) J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 16.Cohen P. T. (2002) J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 17.Yang X., Zhang F., Kudlow J. E. (2002) Cell 110, 69–80 [DOI] [PubMed] [Google Scholar]
- 18.Iyer S. P., Hart G. W. (2003) J. Biol. Chem. 278, 24608–24616 [DOI] [PubMed] [Google Scholar]
- 19.Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. (2008) J. Biol. Chem. 283, 33935–33941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke A. J., Hurtado-Guerrero R., Pathak S., Schüttelkopf A. W., Borodkin V., Shepherd S. M., Ibrahim A. F., van Aalten D. M. (2008) EMBO J. 27, 2780–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., Evans R. M. (2008) Nature 451, 964–969 [DOI] [PubMed] [Google Scholar]
- 22.Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dias W. B., Hart G. W. (2007) Mol. Biosyst. 3, 766–772 [DOI] [PubMed] [Google Scholar]
- 24.Copeland R. J., Bullen J. W., Hart G. W. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. (2005) J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 26.Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. (2004) J. Biol. Chem. 279, 30133–30142 [DOI] [PubMed] [Google Scholar]
- 27.Clark R. J., McDonough P. M., Swanson E., Trost S. U., Suzuki M., Fukuda M., Dillmann W. H. (2003) J. Biol. Chem. 278, 44230–44237 [DOI] [PubMed] [Google Scholar]
- 28.Jones S. P., Zachara N. E., Ngoh G. A., Hill B. G., Teshima Y., Bhatnagar A., Hart G. W., Marbán E. (2008) Circulation 117, 1172–1182 [DOI] [PubMed] [Google Scholar]
- 29.Ramirez-Correa G. A., Jin W., Wang Z., Zhong X., Gao W. D., Dias W. B., Vecoli C., Hart G. W., Murphy A. M. (2008) Circ. Res. 103, 1354–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Means A. R. (2000) Mol. Endocrinol. 14, 4–13 [DOI] [PubMed] [Google Scholar]
- 31.Racioppi L., Means A. R. (2008) Trends Immunol. 29, 600–607 [DOI] [PubMed] [Google Scholar]
- 32.Anderson K. A., Means R. L., Huang Q. H., Kemp B. E., Goldstein E. G., Selbert M. A., Edelman A. M., Fremeau R. T., Means A. R. (1998) J. Biol. Chem. 273, 31880–31889 [DOI] [PubMed] [Google Scholar]
- 33.Soderling T. R. (1999) Trends Biochem. Sci. 24, 232–236 [DOI] [PubMed] [Google Scholar]
- 34.Chatila T., Anderson K. A., Ho N., Means A. R. (1996) J. Biol. Chem. 271, 21542–21548 [DOI] [PubMed] [Google Scholar]
- 35.Song M., Kim H. S., Park J. M., Kim S. H., Kim I. H., Ryu S. H., Suh P. G. (2008) Cell. Signal. 20, 94–104 [DOI] [PubMed] [Google Scholar]
- 36.Cheung W. D., Hart G. W. (2008) J. Biol. Chem. 283, 13009–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riccio A., Alvania R. S., Lonze B. E., Ramanan N., Kim T., Huang Y., Dawson T. M., Snyder S. H., Ginty D. D. (2006) Mol. Cell 21, 283–294 [DOI] [PubMed] [Google Scholar]
- 38.Slawson C., Lakshmanan T., Knapp S., Hart G. W. (2008) Mol. Biol. Cell 19, 4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mol. Cell. Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 40.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 41.Khidekel N., Arndt S., Lamarre-Vincent N., Lippert A., Poulin-Kerstien K. G., Ramakrishnan B., Qasba P. K., Hsieh-Wilson L. C. (2003) J. Am. Chem. Soc. 125, 16162–16163 [DOI] [PubMed] [Google Scholar]
- 42.Comer F. I., Vosseller K., Wells L., Accavitti M. A., Hart G. W. (2001) Anal. Biochem. 293, 169–177 [DOI] [PubMed] [Google Scholar]
- 43.Ohmstede C. A., Jensen K. F., Sahyoun N. E. (1989) J. Biol. Chem. 264, 5866–5875 [PubMed] [Google Scholar]
- 44.Frangakis M. V., Chatila T., Wood E. R., Sahyoun N. (1991) J. Biol. Chem. 266, 17592–17596 [PubMed] [Google Scholar]
- 45.Anderson K. A., Noeldner P. K., Reece K., Wadzinski B. E., Means A. R. (2004) J. Biol. Chem. 279, 31708–31716 [DOI] [PubMed] [Google Scholar]
- 46.Anderson K. A., Kane C. D. (1998) Biometals 11, 331–343 [DOI] [PubMed] [Google Scholar]
- 47.Benkert P., Tosatto S. C., Schomburg D. (2008) Proteins 71, 261–277 [DOI] [PubMed] [Google Scholar]
- 48.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 49.Melo F., Feytmans E. (1998) J. Mol. Biol. 277, 1141–1152 [DOI] [PubMed] [Google Scholar]
- 50.Wallner B., Elofsson A. (2006) Protein Sci. 15, 900–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H., Zhou Y. (2002) Protein Sci. 11, 2714–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christen M., Hünenberger P. H., Bakowies D., Baron R., Bürgi R., Geerke D. P., Heinz T. N., Kastenholz M. A., Kräutler V., Oostenbrink C., Peter C., Trzesniak D., van Gunsteren W. F. (2005) J. Comput. Chem. 26, 1719–1751 [DOI] [PubMed] [Google Scholar]
- 53.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 54.Hanissian S. H., Frangakis M., Bland M. M., Jawahar S., Chatila T. A. (1993) J. Biol. Chem. 268, 20055–20063 [PubMed] [Google Scholar]
- 55.Westphal R. S., Anderson K. A., Means A. R., Wadzinski B. E. (1998) Science 280, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 56.Rexach J. E., Clark P. M., Hsieh-Wilson L. C. (2008) Nat. Chem. Biol. 4, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dentin R., Hedrick S., Xie J., Yates J., 3rd, Montminy M. (2008) Science 319, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 58.Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., Hart G. W. (2008) J. Biol. Chem. 283, 16283–16292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raman V., Blaeser F., Ho N., Engle D. L., Williams C. B., Chatila T. A. (2001) J. Immunol. 167, 6270–6278 [DOI] [PubMed] [Google Scholar]
- 60.Anderson K. A., Means A. R. (2002) Mol. Cell. Biol. 22, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan F., Means A. R., Liu J. O. (2005) EMBO J. 24, 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golks A., Tran T. T., Goetschy J. F., Guerini D. (2007) EMBO J. 26, 4368–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L. J., Zhang X. H., Fukushima H., Zhang F., Wang H., Toyoda H., Li B. M., Kida S., Zhuo M. (2008) Eur. J. Neurosci. 27, 1923–1932 [DOI] [PubMed] [Google Scholar]
- 64.Tallent M. K., Varghis N., Skorobogatko Y., Hernandez-Cuebas L., Whelan K., Vocadlo D. J., Vosseller K. (2009) J. Biol. Chem. 284, 174–181 [DOI] [PubMed] [Google Scholar]
- 65.Lamarre-Vincent N., Hsieh-Wilson L. C. (2003) J. Am. Chem. Soc. 125, 6612–6613 [DOI] [PubMed] [Google Scholar]
- 66.Reason A. J., Morris H. R., Panico M., Marais R., Treisman R. H., Haltiwanger R. S., Hart G. W., Kelly W. G., Dell A. (1992) J. Biol. Chem. 267, 16911–16921 [PubMed] [Google Scholar]
- 67.Tai H. C., Khidekel N., Ficarro S. B., Peters E. C., Hsieh-Wilson L. C. (2004) J. Am. Chem. Soc. 126, 10500–10501 [DOI] [PubMed] [Google Scholar]
- 68.Kreppel L. K., Hart G. W. (1999) J. Biol. Chem. 274, 32015–32022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.