Abstract
Protein arginine methyltransferase 6 (PRMT6) is known to catalyze the generation of asymmetric dimethylarginine in polypeptides. Although the cellular role of PRMT6 is not well understood, it has been implicated in human immunodeficiency virus pathogenesis, DNA repair, and transcriptional regulation. PRMT6 is known to methylate histone H3 Arg-2 (H3R2), and this negatively regulates the lysine methylation of H3K4 resulting in gene repression. To identify in a nonbiased manner genes regulated by PRMT6 expression, we performed a microarray analysis on U2OS osteosarcoma cells transfected with control and PRMT6 small interfering RNAs. We identified thrombospondin-1 (TSP-1), a potent natural inhibitor of angiogenesis, as a transcriptional repression target of PRMT6. Moreover, we show that PRMT6-deficient U2OS cells exhibited cell migration defects that were rescued by blocking the secreted TSP-1 with a neutralizing peptide or blocking α-TSP-1 antibody. PRMT6 associates with the TSP-1 promoter and regulates the balance of methylation of H3R2 and H3K4, such that in PRMT6-deficient cells H3R2 was hypomethylated and H3K4 was trimethylated at the TSP-1 promoter. Using a TSP-1 promoter reporter gene, we further show that PRMT6 directly regulates the TSP-1 promoter activity. These findings show that TSP-1 is a transcriptional repression target of PRMT6 and suggest that neutralizing the activity of PRMT6 could inhibit tumor progression and therefore may be of cancer therapeutic significance.
Protein arginine methyltransferases (PRMTs)3 catalyze the addition of one or two methyl groups to the guanidino nitrogen atoms of arginine resulting in asymmetric and symmetric dimethylarginines using S-adenosylmethionine as a methyl donor (1, 2). Protein arginine methylation is a common post-translational modification (3), and it has been shown to regulate many cellular processes, including nuclear export (4), protein-protein interactions (5, 6), ribosome biogenesis (7, 8), DNA repair (9, 10), genomic stability (11), pre-mRNA splicing (12–15), and gene transcription (16–19).
PRMT6 was identified by homology to other PRMTs, has unique methylation properties, and can methylate arginines that reside within glycine-arginine-rich motifs (20). However, PRMT6 tends to methylate arginines in non-glycine-arginine-rich motifs (20) as observed, for example, in HIV TAT protein (21) and DNA polymerase β (10). Kinetic analysis has revealed that PRMT6 exhibits a distributive mechanism for multiple methylation of a single arginine residue (22). PRMT6 is predominantly nuclear (20), but its function is not well understood. By virtue of its ability to methylate TAT and other HIV proteins, PRMT6 is implicated in HIV pathogenesis by acting as a restriction factor for viral replication (21, 23–25). PRMT6 has been shown to methylate the HMGA1a protein in the second AT hook and likely regulates chromatin structure organization (26, 27). The fact that PRMT6 methylates DNA polymerase β indicates a role in DNA repair (10).
It is well known that histone methylation influences gene expression (28, 29). Recently, it was shown that asymmetric dimethylation of histone H3 Arg-2 (H3R2me2a) interferes with the protein-protein interactions of plant homology domain finger domain (30, 31) and the WD40 repeat domain with methylated H3K4 (32–34). The presence of H3R2me2a in promoter sequences prevents the recruitment of the lysine methyltransferases, thereby abrogating H3K4 trimethylation. Thus the presence of H3R2me2 and H3K4me3 is mutually exclusive (30, 32–34). In mammals, it was shown that PRMT6 is responsible for the asymmetrical dimethylation of H3R2 (32–34). These findings suggest that PRMT6 is a negative regulator of gene expression; however, the knowledge of its physiological targets remains limited.
To answer these questions, a microarray approach was used to identify physiological genes regulated by PRMT6. We report that thrombospondin-1 (TSP-1) is stimulated in PRMT6-deficient cells. TSP-1 is a high molecular weight homotrimeric glycoprotein composed of three identical disulfide-linked 180-kDa subunits (35, 36). TSP-1 is synthesized by many cell types and secreted into the extracellular matrix (37–41). TSP-1 is a potent natural inhibitor of angiogenesis and endothelial cell migration (42, 43). TSP-1 inhibits cell migration by binding cell surface receptors, including CD36 with its type 1 sequence repeat domains (44, 45). The TSP-1 gene is transcriptionally regulated by serum (46) and p53 (47). Epigenetic changes have also been reported, and notably the TSP-1 gene is regulated by DNA methylation (48). What is less understood is the epigenetic histone regulation of the TSP-1 gene.
In this study, we show that a total of 51 genes are significantly up-regulated (p < 0.05 and fold change >2) and 28 genes down-regulated (p < 0.05 and fold change <2) in PRMT6-deficient U2OS cells. One of the up-regulated genes was TSP-1, which is therefore a putative repression target of PRMT6. Arginine methylation of the H3R2 mark was validated as the mechanism by which TSP-1 gene expression was regulated, indicating an epigenetic mode of TSP-1 regulation.
EXPERIMENTAL PROCEDURES
Materials
The antibodies against PRMT6 (Western blot, A300-929A; immunoprecipitation, A300-928A) were from Bethyl Laboratories (Montgomery, TX), and antibodies against H3R2me2a (catalog number 07-585) and H3K4me3 (catalog number 9751) were from Upstate Biotechnology, Inc. (Lake Placid, NY) and Cell Signaling Technology (Pickering, Ontario, Canada), respectively. The antibody against α-tubulin and other biochemical reagents were from Sigma. TSP-1 antibody (Ab-6, NeoMarkers, Union City, CA) used for Western blot was kindly provided by Dr. Béliveau from the Université du Québec à Montréal, and the neutralizing TSP-1 antibody used during the cell movement assays was from Novus Biologicals (Littleton, CO). GGWSHW and GGYSHW peptides were synthesized by W. M. Keck Biotechnology Resource Center (New Haven, CT). The pVAX1-myc vector expressing human PRMT6 wild type and catalytically inactive PRMT6 VLD:KLA, have been already described (21).
Control and PRMT6-deficient U2OS Cells
The human osteosarcoma cells (U2OS) were obtained from American Type Culture Collection (Manassas, VA). Transient siRNA-mediated PRMT6 knockdown was performed using PRMT6 siRNA that targets nucleotides 996–1114 (5′-GCA AGA CAC GCA CGU UUC A-3′) from Dharmacon Research (Lafayette, CO.). A control oligonucleotide targeting GFP was also purchased from Dharmacon, and the sequence of the GFP siRNA was 5′-AAU UGC CAC AAC AGG GUC GUG-3′. U2OS cells were then transfected with GFP-siRNA (control, siGFP) or the PRMT6-siRNA (siPRMT6) using RNAi MAX Lipofectamine (Invitrogen) according to the manufacturer's protocol. The cells were used in subsequent assays after a 72-h incubation at 37 °C with the siRNA, and experiments with siRNA-transfected cells were performed as indicated in the figure legends. The PRMT6 expression was analyzed by immunoblotting.
Western Blot Analysis
After 48 h of siRNA-mediated PRMT6 knockdown, cells were exposed to serum-free cell culture medium. After 18 h of incubation, conditioned media were removed and concentrated (20×), whereas the cells were solubilized in lysis buffer (20 mm Tris, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 mg/ml leupeptin, 1 mm phenylmethanesulfonyl fluoride, pH 7.5). Cell lysates were subjected to SDS-PAGE, and separated proteins were then transferred to nitrocellulose membranes (Bio-Rad). Following transfer, immunodetection analysis was performed.
cRNA Preparation, Illumina Microarray Hybridization, and Scanning
RNA samples were prepared with TRIzol reagent from Invitrogen using standard procedures, and the RNA quality and integrity were then checked by Agilent Bioanalyzer. cRNA amplification and labeling with biotin were performed using Illumina TotalPrep RNA amplification kit (Ambion, Inc., Austin, TX) with 250 ng of total RNA as input material. cRNA yields were quantified with Agilent Bioanalyzer, and 1.5 μg of cRNAs were hybridized to the Sentrix HumanRef-8 version 3.0 Expression BeadChips (Illumina, Inc., San Diego). Each chip contains six arrays, and each array contains >48,000 gene transcripts, of which 46,000 were derived from human genes at the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) and UniGene databases. All reagents and equipment used for hybridization were purchased from Illumina. According to the manufacturer's protocol, cRNA was hybridized to arrays for 16 h at 58 °C before being washed and stained with streptavidin-Cy3. Then the bead chips were centrifuged to dry and were scanned on the Illumina BeadArray Reader confocal scanner. The statistical analysis of microarray data was then realized using the FlexArray 1.1.3 software package (Génome Québec, Montréal, Québec, Canada). Data were also analyzed through the use of Ingenuity Pathway Analysis Ingenuity Systems (Redwood City, CA).
Cell Movement Assays
siGFP- and siPRMT6-treated U2OS cell migration assays were performed using Transwell filters (Costar; 8-μm pore size) precoated with 0.15% gelatin. Briefly, 1 × 105 cells were resuspended in 100 μl of serum-free medium with or without neutralizing peptide GGWSHW (20 μm), control peptide GGYSHW (20 μm), monoclonal antibody against TSP-1 (10 μg/ml), or a nonspecific IgG (10 μg/ml) and added into the upper chamber of each Transwell (the lower chamber of the Transwell contained 10% serum as well as peptides, monoclonal antibody against TSP-1, or a nonspecific IgG). The plates were then placed at 37 °C in 5% CO2, 95% air atmosphere during 6 h. Cells that migrated to the lower surface of the filters were fixed with 4% paraformaldehyde in PBS and stained with 0.1% crystal violet, 20% MeOH. Migrating cells were visualized at a magnification of ×100 using a digital Canon PowerShot G6 camera (Canon Canada, Mississauga, Ontario, Canada) attached to a Carl Zeiss Axiovert 40 CFL microscope (Carl Zeiss Canada Ltd., Toronto, Ontario, Canada), and the average number of migrated cells per field was assessed by measuring the cell density of at least five random fields per filter using Scion Image software Alpha version 4.0.3.2 (Scion Corp., Frederick, MD). For cell invasion assays, we used Transwell filters (Costar; 8 μm pore size) precoated with 50 μg of air-dried Matrigel matrix (BD Biosciences). For wound healing assay, siGFP- and siPRMT6-treated U2OS cells were seeded on 6-well culture plates coated overnight at 4 °C with 1 μg/ml fibronectin (F4759, Sigma), and the cells were maintained until they reached confluence. The confluent cell monolayer was wounded with pipette tips. After washing with warmed PBS, the cells were incubated in fresh culture medium, and wound healing was surveyed 24 h later, and the cells were fixed with 4% paraformaldehyde in PBS and stained with 0.1% crystal violet, 20% MeOH. Cell motility was monitored at ×25 magnification using a digital Canon PowerShot G6 camera attached to a Carl Zeiss Axiovert 40 CFL microscope.
Immunofluorescence
To visualize cell morphology, siGFP- and siPRMT6-treated U2OS cells were first plated on coverslips, and the cells were then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 1% Triton X-100 in PBS for 10 min at room temperature. The reagents used for fluorescence microscopy were α-vinculin antibodies (V9131, Sigma) and Alexa Fluor-546 phalloidin (A12380, Invitrogen). Cells were mounted in Immuno-Mount (Thermo Shandon, Pittsburgh, PA), and cellular morphology was visualized at a ×400 magnification using a Carl Zeiss Axio Imager microscope. Images were processed in Adobe Photoshop CS prior to publication.
TSP-1 Promoter Activity Assays
The pMTSP-1 vector containing −2,800 to +48 bp of the murine TSP-1 promoter upstream of firefly luciferase reporter gene has been described (49) and was kindly provided by Dr. Paul Bornstein (University of Washington, Seattle) via Addgene (Addgene plasmid 12409; Cambridge, MA). U2OS cells were first plated in 24-well plates. U2OS cells were transfected with siRNAs (siGFP and siPRMT6) and were then transfected 24 h after siRNA transfection with the TSP-1 reporter construct along with the Renilla luciferase plasmid pRL-TK (Promega, Madison, WI.) for 48 h. Also, U2OS cells were transfected with PRMT6 expression vectors (pVAX1-empty, pVAX1-PRMT6 wild type, and pVAX1-PRMT6 VLD:KLA) as well as TSP-1 reporter construct along with the Renilla luciferase plasmid pRL-TK for 48 h. The firefly and Renilla luciferase activities were measured in cell lysates using the Dual-Luciferase® reporter assay system (Promega) according to the manufacturer's recommendations. The luciferase activities in the control conditions (siGFP and pVAX1-empty) were normalized to 1, and the subsequent ratio of Firefly luciferase on Renilla luciferase activities was expressed as fold induction.
Chromatin Immunoprecipitation (ChIP) Analysis
U2OS cells at 80% confluency were transfected with either siGFP control or siRNA against PRMT6. 72 h post transfection, ChIP analysis was performed following the Upstate ChIP Assay kit protocol (catalog number 7-295). For the immunoprecipitation, 4 μg of the αPRMT6, αH3R2me2a, and αH3K4me3 was used for each condition and incubated with the cross-linked complexes overnight at 4 °C. PCR of the TSP-1 promoter on the input and isolated DNA was performed using IllustraTM TaqDNA polymerase (GE Healthcare). As described previously (50), the TSP-1 promoter primer sequences were as follows: forward, 5′-AAC GAA TGG CTC TCT TGG TGT-3′ and reverse 5′-CTT TCC AGC TAG AAA GTG AA-3′.
RESULTS
PRMT6-deficient U2OS Cells Have Altered Expression of Genes Involved in Cell Movement
To investigate the cellular role and function of PRMT6, we reduced the expression of PRMT6 using siRNA interference strategies. The osteosarcoma cell line U2OS was utilized, as these cells have the presence of wild-type p53 (51) and retinoblastoma protein (52) cell cycle regulators. U2OS cells were transiently transfected with control double-stranded RNAs (siGFP) or siRNAs that target PRMT6 (siPRMT6). A PRMT6 knockdown of ∼90% was observed in U2OS cells by immunoblotting (Fig. 1A). Gene expression profiling between control and PRMT6 siRNA-treated U2OS cells was examined by Illumina microarray technology. Total RNA was isolated from asynchronously growing control and PRMT6 siRNA-treated U2OS cells and analyzed in quadruplicate (supplemental Table 1). The statistical analysis of microarray data using the FlexArray software version 1.2 package showed a total of 51 genes that were significantly up-regulated (p < 0.05 and fold change >2) and 28 genes that were down-regulated (p < 0.05 and fold change <2) in PRMT6-deficient U2OS cells. The PRMT6 mRNA expression level was reduced to 24%, consistent with the ∼90% knockdown observed at the protein level (Fig. 1A). These findings are consistent with a role of PRMT6 as a negative regulator of gene expression, albeit of a select group of genes in U2OS cells. The data were analyzed through the use of Ingenuity Pathway Analysis software, and we entered the 79 entries of which 70 were mapped and 41 and 34 were network and function/pathway/list eligible, respectively. The top four functional categories were cardiovascular system development and function (8 molecules; p value 1.49 × 10−6 to 1.27 × 10−2), cellular movement (14 molecules; p value 1.49 × 10−6 to 1.27 × 10−2), skeletal and muscular system development and function (9 molecules; p value 1.49 × 10−6 to 1.27 × 10−2), and cardiovascular disease (6 molecules; p value 9.73 × 10−6 to 1.27 × 10−2). Overall, the microarray data suggest a role of PRMT6 in the regulation of genes involved in cell movement and cardiovascular-associated diseases. These latter changes would best be analyzed in PRMT6 mice models that are currently not available. Therefore, we focused our studies on the role of PRMT6 in cell movement.
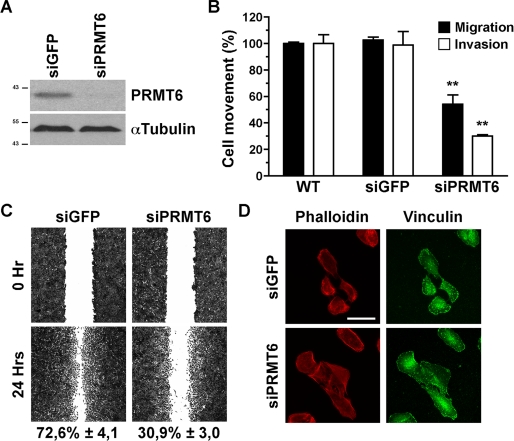
FIGURE 1.
PRMT6-deficient cells exhibit impaired cell movement. A, cell lysates from control and PRMT6 siRNA-treated U2OS cells were immunoblotted with α-PRMT6 and α-tubulin as a loading control. The molecular mass markers are shown on the left in kDa. B, U2OS cell migration and invasion assays were performed following PRMT6 siRNA-mediated transient knockdown. Cell migration (black bars) and cell invasion (white bars) were measured using modified Boyden chambers with filters coated with gelatin or with air-dried Matrigel, respectively. Cells that had moved to the lower surface of the filters were fixed, stained with crystal violet, and quantified as described under “Experimental Procedures.” Results are expressed as a percentage of cell movement seen in PRMT6 knockdown cells compared with control cells. Data represent the means ± S.D. of two independent experiments performed in triplicate. Statistically significant differences, as compared with respective control conditions, are indicated by **, p < 0.01 (Student's t test). WT, wild type. C, monolayer of confluent siGFP- or siPRMT6-treated U2OS cells were wounded with pipette tips, and the cells were incubated with fresh culture medium for 48 h as described under “Experimental Procedures.” The wounded areas were photographed at the beginning (0 h) and at the end (24 h) of the assay. Photographs (original magnification, ×25) obtained from a representative experiment are shown. Results are expressed as the percentage of wound healing after 24 h. Data represent the means ± S.D. of two independent experiments performed in triplicate. D, U2OS cell morphology was monitored following PRMT6 siRNA-mediated transient knockdown. siRNA-treated U2OS cells were plated on coverslips, and stress fibers and focal adhesions were visualized by phalloidin staining and α-vinculin antibodies, respectively, followed by fluorescence microscopy as described under “Experimental Procedures.” Scale bar represents 50 μm.
PRMT6 Knockdown Inhibits Cell Migration and Invasion
Because the PRMT6 knockdown affects several genes involved in cell migration, we tested whether the loss of PRMT6 affected cell movement and invasion (Fig. 1B). The migration capacity of control and PRMT6-deficient U2OS cells was assessed using a Boyden chamber where the Transwell filter was coated with gelatin (Fig. 1B, black bars). PRMT6 knockdown cells had an ∼50% reduction in serum-induced migration as compared with control U2OS cells (Fig. 1B, black bars). To further measure the impact of PRMT6 knockdown on the cell movement, the invasive capacity of U2OS was assessed using Transwell filters coated with Matrigel matrix following siRNA-mediated down-regulation of PRMT6 (Fig. 1B, white bars). The knockdown of PRMT6 also significantly inhibited U2OS cell invasion by ∼70% compared with their siGFP control counterpart (Fig. 1B, white bars). Similar findings were also observed with HeLa cells (data not shown). A wound healing assay was performed to further examine the migratory capabilities of the PRMT6 knockdown U2OS cells. Although control siGFP-treated U2OS cells closed the wound by ∼72.6% after 24 h, the migration of PRMT6-deficient U2OS cells was significantly impaired (∼30.9%, see Fig. 1C), further supporting that the directional migration of U2OS cells requires PRMT6. Taken together, the different cell movement assays show that PRMT6 regulates cell migration and invasion. We next examined whether the PRMT6-deficient cells had morphological defects, as assessed by stress fiber organization and focal adhesion organization. The depletion of PRMT6 in U2OS cells did not induce noticeable morphological alterations (Fig. 1D). Moreover, the stress fibers and focal adhesions of PRMT6-deficient U2OS cells displayed normal cellular organization compared with their siGFP control counterpart, as stained with phalloidin and α-vinculin antibodies, respectively (Fig. 1D).
PRMT6 Knockdown-mediated U2OS Cell Migration Inhibition Is Associated with Elevated TSP-1 Protein Levels
PRMT6-deficient U2OS cells exhibit a statistical increase in mRNAs of proteins implicated in cell movement, including α1-antitrypsin, urokinase-type plasminogen activator, tumor necrosis factor receptor 11b (osteoprotegerin), matrix metalloproteinase-7 and -9, Rho GDP dissociation inhibitor b, interleukin 8, KISS-1 metastasis-suppressor, plasminogen activator inhibitor-1 and -2, colony-stimulating factor 2, α2-integrin, procollagen lysine, 2-oxoglutarate 5-dioxygenase 2, and TSP-1. To confirm that the alteration of the TSP-1 mRNA was also observed at the TSP-1 protein levels, we verified TSP-1 expression by immunoblotting (Fig. 2A). Because TSP-1 is a known secreted protein (38), we probed cell extracts and tissue culture supernatant from siGFP- and siPRMT6-treated U2OS cells with α-TSP-1 antibodies. PRMT6-depleted cells had elevated levels of cellular and secreted TSP-1 compared with control siGFP-treated U2OS cells (Fig. 2A), suggesting that PRMT6 also regulates TSP-1 protein expression.
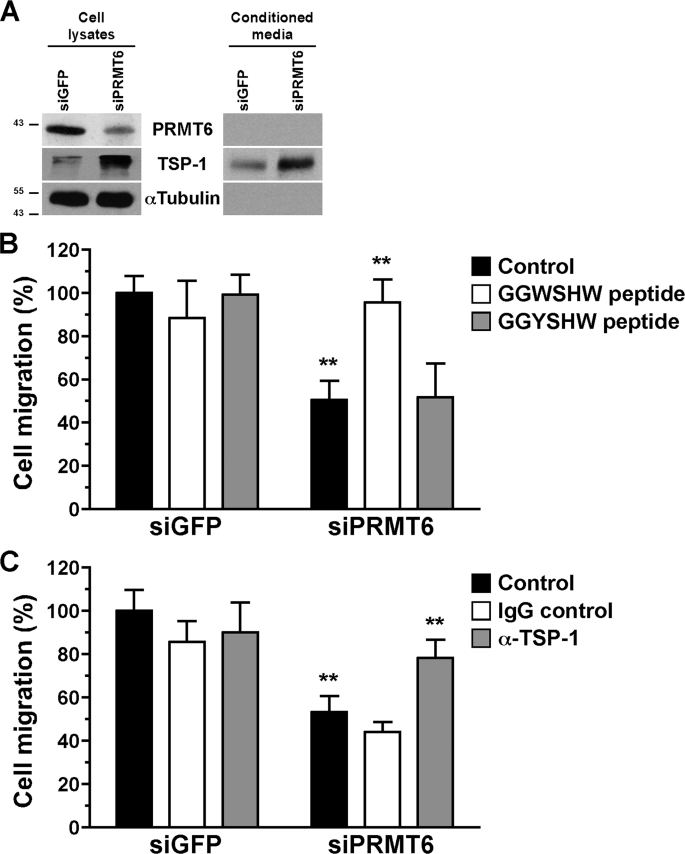
FIGURE 2.
Interference with the activity of TSP-1 restores cell migration properties to PRMT6-deficient U2OS cells. A, cell lysates or the conditioned media from control and PRMT6 siRNA-treated U2OS cells were immunoblotted with α-PRMT6, α-TSP-1, and α-tubulin as a loading control. The molecular mass markers are shown on the left in kDa. B, cell migration of siGFP- and siPRMT6-treated U2OS cells was assayed in the presence of either 20 μm neutralizing TSP-1 (GGWSHW) or the control (GGYSHW) peptide in the conditioned media. The data represent the means ± S.D. of two independent experiments performed in triplicate. Statistically significant differences, as compared with respective control conditions, are indicated by **, p < 0.01 (Student's t test). C, cell migration assays were also performed with the presence of either 10 μg/ml blocking α-TSP-1 or immunoglobulin G in the conditioned media. The data represent the means ± S.D. of two independent experiments performed in triplicate. Statistically significant differences, as compared with respective control conditions, are indicated by **, p < 0.01 (Student's t test).
Neutralizing Peptide and Antibody against TSP-1 Rescue the PRMT6 Knockdown Inhibition of Cell Movement
TSP-1 is a known inhibitor of cell migration (42), and thus we reasoned that blocking TSP-1 with neutralizing peptides and antibodies might reverse the cellular defect of PRMT6-deficient U2OS cells. Cell migration assays were performed with control and PRMT6-deficient cells in the presence of a TSP-1 neutralizing peptide (GGWSHW) or a control peptide (GGYSHW; Fig. 2B). The GGWSHW peptide mimics the WXXW motif in the type 1 repeats of TSP-1 and blocks its interaction with known growth factors (54). The presence of the GGWSHW peptide completely reversed the U2OS cell migration inhibition observed in PRMT6 knockdown cells (Fig. 2B). Moreover, the GGWSHW peptide had no effect on the migration of siGFP control cells, and the related inactive control GGYSHW peptide had no effect on either siGFP- or siPRMT6-transfected U2OS cells (Fig. 2B). These results suggest that the cell migration inhibition observed with the PRMT6-deficient cells is mainly caused by an increase in TSP-1 expression. Moreover, we performed cell migration assays in the presence of either a blocking mouse monoclonal antibody against TSP-1 or a nonspecific immunoglobulin G as control (Fig. 2C). The α-TSP-1 antibody reversed, albeit partially, the inhibition of cell migration of PRMT6-deficient U2OS cells (Fig. 2C). These data suggest that the reduced cell migration phenotype observed in PRMT6-deficient U2OS cells is mainly the result of the up-regulation of TSP-1 expression.
PRMT6 Knockdown Increases TSP-1 Gene Promoter Expression in U2OS Cells
PRMT6 has been shown to negatively regulate transcription of genes known to be modified by epigenetic changes (32, 33). Our microarray analysis provides an unbiased approach to identify the genes regulated by PRMT6. The ability of PRMT6 to regulate the transcriptional expression of the TSP-1 gene promoter was examined by using TSP-1 promoter driving a luciferase reporter gene (Fig. 3, A and B). We cotransfected siGFP or siPRMT6 in the presence of both the TSP-1 luciferase reporter plasmid in U2OS cells (Fig. 3A). We observed that siRNA-mediated PRMT6 knockdown in U2OS cells led to the activation of the TSP-1 gene by a significant 1.7-fold (Fig. 3A). The converse was also performed using overexpressed active and inactive PRMT6. We observed that wild-type PRMT6 significantly inhibited the activation of the TSP-1 promoter by ∼50% in U2OS cells, whereas the methyltransferase-inactive PRMT6 (VLD-KLA) did not significantly inhibit the TSP-1 promoter expression (Fig. 3B). These results show that PRMT6 arginine methylation directly regulates the activity of the TSP-1 promoter.
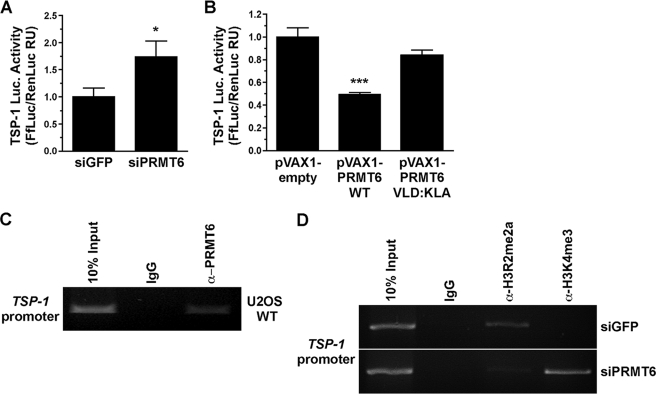
FIGURE 3.
PRMT6 regulates the activity of the TSP-1 promoter by methylating H3R2 in U2OS cells. A, activity of the TSP-1 promoter luciferase (Luc.) reporter gene was assessed in U2OS cells transfected with either siGFP or siPRMT6. The luciferase activity was normalized to Renilla, and the latter was used to assess transfection efficiency. The transfected cells were lysed and assayed for luciferase activity as described under “Experimental Procedures.” The results are expressed as a ratio of firefly luciferase (FfLuc) on Renilla luciferase (RenLuc) fold induction activities. Data represent the means ± S.D. of two independent experiments performed in triplicate. Statistically significant differences as compared with control conditions are as follows: *, p < 0.05; ***, p < 0.001 (Student's t test). B, activity of the TSP-1 promoter luciferase reporter gene was assessed in U2OS cells transfected with either empty vector (pVAX1), wild-type PRMT6 (pVAX1 PRMT6 WT), and catalytically inactive PRMT6 (pVAX1 PRMT6 VLD:KLA). The activity of a TSP-1 promoter luciferase reporter gene was assessed as in A. C, chromatin immunoprecipitation assays were performed with control IgG and α-PRMT6 antibodies. DNA oligonucleotides from the TSP-1 promoter were used in a PCR assay to amplify a DNA fragment. The latter was visualized by ethidium bromide-stained agarose gel electrophoresis. D, analysis of the occupancy of H3R2me2a and H3K4me3 histone marks at the TSP-1 gene promoter by ChIP analysis. ChIP assays were performed with chromatin prepared from U2OS cells transfected or not with control siGFP or siPRMT6. After 72 h, the cells were harvested for ChIP analysis using the indicated antibodies and a primer pair located in the promoter region of TSP-1. ChIPs experiments were performed in duplicate, and the results shown are representative of the duplicates.
PRMT6 Methylation of H3R2me2a Regulates TSP-1 Gene Promoter Expression
We next examined whether the effect of PRMT6 is mediated by epigenetic changes at the TSP-1 promoter (Fig. 3, C and D). Because the expression of TSP-1 is elevated in PRMT6-deficient cells, we investigated whether the H3R2me2a mark was lost at the TSP-1 promoter. By ChIP analysis, we first observed that PRMT6 was indeed recruited to the TSP-1 promoter of U2OS cells (Fig. 3C). In PRMT6-deficient cells, we observed the loss of H3R2me2a at the TSP-1 promoter and a corresponding gain of H3K4me3 (Fig. 3D). These histone marks are consistent with the increase TSP-1 gene activation seen in PRMT6-deficient cells. These findings suggest that PRMT6 regulates TSP-1 expression by regulating epigenetic marks at the level of the TSP-1 promoter.
DISCUSSION
In this study, we identified a set genes involved in cardiovascular and cellular migration regulated by PRMT6. The knockdown by ∼90% of PRMT6 in U2OS cells led to the identification of 51 genes that were up-regulated and 28 genes that were repressed by >2-fold using Illumina microarrays. One of the genes that is repressed by PRMT6 is the secreted glycoprotein TSP-1, a potent inhibitor of angiogenesis and cell migration. We show that PRMT6-deficient U2OS cells exhibited cell migration defects that were rescued by blocking the secreted TSP-1 with a neutralizing peptide or blocking α-TSP-1 antibodies. PRMT6 was associated with the TSP-1 promoter and regulated the balance of methylation of H3R2 and H3K4, such that in PRMT6-deficient cells H3R2 was hypomethylated and H3K4 was trimethylated at the TSP-1 promoter consistent with gene activation. Using a TSP-1 promoter reporter gene, we further show that PRMT6 directly regulates the TSP-1 promoter. These findings show that TSP-1 is a transcriptional repression target of PRMT6 and that epigenetic changes of H3R2 regulate TSP-1 expression.
Arginine methylation and PRMTs have not been shown to regulate cell migration. However, cytoskeleton proteins and signaling proteins have been identified as targets of PRMTs using proteomic analyses (3, 55). These findings suggest that arginine methylation is likely to play a role in regulating key protein-protein interactions during cell migration. Indeed, the estrogen receptor α was recently shown to be arginine-methylated by PRMT1, and this regulates complex formation with Src kinase, phosphoinositide 3-kinases, and focal adhesion kinase (56). However, the role of PRMT1, or other PRMTs, in cell migration remains elusive. Further evidence for a potential role of PRMTs in signaling or cell migration is suggested by the identification of PRMT8 as a membrane-bound PRMT (57). Our findings show that the nuclear PRMT6 influences cell migration by regulating genes, including TSP-1, involved in cellular migration.
The presence of the asymmetrically dimethylated H3R2 (H3R2me2a) mark is associated with gene repression and infrequently linked with active gene promoters (32). The asymmetrical dimethylation of H3R2 interferes with key contacts with plant homology domain (30, 31) and WD40 domains (32–34) such that in mammals the mixed lineage leukemia lysine methyltransferase complex is not recruited to promoter regions that harbor H3R2me2a. Therefore, the knockdown of PRMT6 would be predicted to activate H3R2me2a-repressed genes, and thus the genes that are up-regulated in U2OS PRMT6-deficient cells (supplemental Table 1) likely represent genes that are normally repressed by PRMT6 via H3R2me2a. It is also possible that the monomethyl H3R2, generated by other PRMTs, itself may be an activating mark, as recently demonstrated in yeast (30). The hox and myc regulated genes are genes known to be regulated by H3K4 trimethylation (32, 58, 59), and these should be increased in PRMT6 knockdown cells. We did not identify them in our microarray analysis suggesting that these genes are active in U2OS and that the knockdown of PRMT6 does not further change the H3R2 mark that is likely hypomethylated in these cells. The genes that are down-regulated in the PRMT6-deficient cells represent a new category of regulated genes by PRMT6 and suggest that PRMT6 may serve as a coactivator for certain genes via H3R2me2a or via other methyl marks.
We reported previously that the hypomethylation of H3R2 in the presence of DNA damage reduced cyclin D1 expression (34). DNA damage was required to induce ING2 expression (60), which then binds H3K4me3 and represses gene expression (61). It has been reported that cyclin D1-deficient cells up-regulate TSP-1 leading to defects in cellular migration (53). The mechanism by which cyclin D1 via p27KIP1 leads to TSP-1 gene regulation is unknown. We did not identify cyclin D1 in our microarray analysis (supplemental Table 1) consistent with our previous findings that PRMT6 knockdown alone was insufficient to reduce cyclin D1 expression (34).
TSP-1 is known to be regulated by serum (46), p53 (47), as well as DNA methylation (48), and we now extend this to histone arginine modification by PRMT6. Our data show that blocking secreted TSP-1 restored cell migration in PRMT6-deficient U2OS cells. However, a total of 14 genes was identified in the cellular migration category in our microarray analysis of PRMT6-deficient cells, suggesting that the other 13 genes may also contribute to the phenotype of PRMT6-deficient cells. In conclusion, our results show that PRMT6 regulates cell migration and invasion by regulating epigenetic changes in the TSP-1 promoter. Our findings suggest that specific PRMT6 inhibitors may be useful to suppress cancer cell invasion and malignant development and/or progression.
Supplementary Material
Acknowledgment
We thank Amber Zabarauskas for assistance with the microarray data analysis.
This work was supported in part by Grant MOP-93811 from the Canadian Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- PRMT
- protein arginine methyltransferase
- ChIP
- chromatin immunoprecipitation
- PBS
- phosphate-buffered saline
- TSP-1
- thrombospondin-1
- WD40 repeat (also known as WD or β-transducin repeat)
- tryptophan-aspartic acid (W-D) dipeptide repeat
- siRNA
- small interfering RNA
- HIV
- human immunodeficiency virus
- GFP
- green fluorescent protein.
REFERENCES
- 1.Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y. H., Stallcup M. R. (2009) Mol. Endocrinol. 23, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert F. M., Côté J., Boulanger M. C., Richard S. (2003) Mol. Cell. Proteomics 2, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 4.Shen E. C., Henry M. F., Weiss V. H., Valentini S. R., Silver P. A., Lee M. S. (1998) Genes Dev. 12, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedford M. T., Frankel A., Yaffe M. B., Clarke S., Leder P., Richard S. (2000) J. Biol. Chem. 275, 16030–16036 [DOI] [PubMed] [Google Scholar]
- 6.Friesen W. J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. (2001) Mol. Cell 7, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 7.Bachand F., Silver P. A. (2004) EMBO J. 23, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swiercz R., Person M. D., Bedford M. T. (2005) Biochem. J. 386, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisvert F. M., Déry U., Masson J. Y., Richard S. (2005) Genes Dev. 19, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Andaloussi N., Valovka T., Toueille M., Steinacher R., Focke F., Gehrig P., Covic M., Hassa P. O., Schär P., Hübscher U., Hottiger M. O. (2006) Mol. Cell 22, 51–62 [DOI] [PubMed] [Google Scholar]
- 11.Yu Z., Chen T., Hebert J., Li E., Richard S. (2009) Mol. Cell. Biol. 29, 2982–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001) Mol. Cell. Biol. 21, 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boisvert F. M., Cote J., Boulanger M. C., Cleroux P., Bachand F., Autexier C., Richard S. (2002) J. Cell Biol. 159, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brahms H., Raymackers J., Union A., de Keyser F., Meheus L., Lührmann R. (2000) J. Biol. Chem. 275, 17122–17129 [DOI] [PubMed] [Google Scholar]
- 15.Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. (2001) Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 17.Strahl B. D., Briggs S. D., Brame C. J., Caldwell J. A., Koh S. S., Ma H., Cook R. G., Shabanowitz J., Hunt D. F., Stallcup M. R., Allis C. D. (2001) Curr. Biol. 11, 996–1000 [DOI] [PubMed] [Google Scholar]
- 18.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 19.An W., Kim J., Roeder R. G. (2004) Cell 117, 735–748 [DOI] [PubMed] [Google Scholar]
- 20.Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. (2002) J. Biol. Chem. 277, 3537–3543 [DOI] [PubMed] [Google Scholar]
- 21.Boulanger M. C., Liang C., Russell R. S., Lin R., Bedford M. T., Wainberg M. A., Richard S. (2005) J. Virol. 79, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakowski T. M., Frankel A. (2008) J. Biol. Chem. 283, 10015–10025 [DOI] [PubMed] [Google Scholar]
- 23.Xie B., Invernizzi C. F., Richard S., Wainberg M. A. (2007) J. Virol. 81, 4226–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Invernizzi C. F., Xie B., Frankel F. A., Feldhammer M., Roy B. B., Richard S., Wainberg M. A. (2007) AIDS 21, 795–805 [DOI] [PubMed] [Google Scholar]
- 25.Invernizzi C. F., Xie B., Richard S., Wainberg M. A. (2006) Retrovirology 3, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miranda T. B., Webb K. J., Edberg D. D., Reeves R., Clarke S. (2005) Biochem. Biophys. Res. Commun. 336, 831–835 [DOI] [PubMed] [Google Scholar]
- 27.Zou Y., Webb K., Perna A. D., Zhang Q., Clarke S., Wang Y. (2007) Biochemistry 46, 7896–7906 [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 29.Ruthenburg A. J., Allis C. D., Wysocka J. (2007) Mol. Cell 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 30.Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Vermeulen M., Mann M., Bähler J., Green R. D., Kouzarides T. (2007) Nature 449, 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. (2007) Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 32.Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Lüscher B., Amati B. (2007) Nature 449, 933–937 [DOI] [PubMed] [Google Scholar]
- 33.Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U. M. (2007) Genes Dev. 21, 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iberg A. N., Espejo A., Cheng D., Kim D., Michaud-Levesque J., Richard S., Bedford M. T. (2008) J. Biol. Chem. 283, 3006–3010 [DOI] [PubMed] [Google Scholar]
- 35.Bornstein P. (1992) FASEB J. 6, 3290–3299 [DOI] [PubMed] [Google Scholar]
- 36.Lawler J. (2008) Curr. Drug Targets 9, 820–821 [DOI] [PubMed] [Google Scholar]
- 37.Mosher D. F., Doyle M. J., Jaffe E. A. (1982) J. Cell Biol. 93, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baenziger N. L., Brodie G. N., Majerus P. W. (1972) J. Biol. Chem. 247, 2723–2731 [PubMed] [Google Scholar]
- 39.Mosher D. F. (1990) Annu Rev. Med. 41, 85–97 [DOI] [PubMed] [Google Scholar]
- 40.Varani J., Riser B. L., Hughes L. A., Carey T. E., Fligiel S. E., Dixit V. M. (1989) Clin. Exp. Metastasis 7, 265–276 [DOI] [PubMed] [Google Scholar]
- 41.Jaffe E. A., Ruggiero J. T., Leung L. K., Doyle M. J., McKeown-Longo P. J., Mosher D. F. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 998–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson D. W., Pearce S. F., Zhong R., Silverstein R. L., Frazier W. A., Bouck N. P. (1997) J. Cell Biol. 138, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson D. W., Volpert O. V., Pearce S. F., Schneider A. J., Silverstein R. L., Henkin J., Bouck N. P. (1999) Mol. Pharmacol. 55, 332–338 [DOI] [PubMed] [Google Scholar]
- 44.Asch A. S., Silbiger S., Heimer E., Nachman R. L. (1992) Biochem. Biophys. Res. Commun. 182, 1208–1217 [DOI] [PubMed] [Google Scholar]
- 45.Tuszynski G. P., Rothman V. L., Papale M., Hamilton B. K., Eyal J. (1993) J. Cell Biol. 120, 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Framson P., Bornstein P. (1993) J. Biol. Chem. 268, 4989–4996 [PubMed] [Google Scholar]
- 47.Dameron K. M., Volpert O. V., Tainsky M. A., Bouck N. (1994) Science 265, 1582–1584 [DOI] [PubMed] [Google Scholar]
- 48.Rojas A., Meherem S., Kim Y. H., Washington M. K., Willis J. E., Markowitz S. D., Grady W. M. (2008) Int. J. Cancer 123, 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shingu T., Bornstein P. (1994) J. Biol. Chem. 269, 32551–32557 [PubMed] [Google Scholar]
- 50.Zhao H. Y., Ooyama A., Yamamoto M., Ikeda R., Haraguchi M., Tabata S., Furukawa T., Che X. F., Zhang S., Oka T., Fukushima M., Nakagawa M., Ono M., Kuwano M., Akiyama S. (2008) Cancer Res. 68, 7035–7041 [DOI] [PubMed] [Google Scholar]
- 51.Kastan M. B., Zhan Q., el-Deiry W. S., Carrier F., Jacks T., Walsh W. V., Plunkett B. S., Vogelstein B., Fornace A. J., Jr. (1992) Cell 71, 587–597 [DOI] [PubMed] [Google Scholar]
- 52.Lukas J., Parry D., Aagaard L., Mann D. J., Bartkova J., Strauss M., Peters G., Bartek J. (1995) Nature 375, 503–506 [DOI] [PubMed] [Google Scholar]
- 53.Li Z., Wang C., Jiao X., Lu Y., Fu M., Quong A. A., Dye C., Yang J., Dai M., Ju X., Zhang X., Li A., Burbelo P., Stanley E. R., Pestell R. G. (2006) Mol. Cell. Biol. 26, 4240–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz-Cherry S., Chen H., Mosher D. F., Misenheimer T. M., Krutzsch H. C., Roberts D. D., Murphy-Ullrich J. E. (1995) J. Biol. Chem. 270, 7304–7310 [DOI] [PubMed] [Google Scholar]
- 55.Wu C. C., MacCoss M. J., Mardones G., Finnigan C., Mogelsvang S., Yates J. R., 3rd, Howell K. E. (2004) Mol. Biol. Cell 15, 2907–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Romancer M., Treilleux I., Leconte N., Robin-Lespinasse Y., Sentis S., Bouchekioua-Bouzaghou K., Goddard S., Gobert-Gosse S., Corbo L. (2008) Mol. Cell 31, 212–221 [DOI] [PubMed] [Google Scholar]
- 57.Lee J., Sayegh J., Daniel J., Clarke S., Bedford M. T. (2005) J. Biol. Chem. 280, 32890–32896 [DOI] [PubMed] [Google Scholar]
- 58.Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C. M., Canaani E. (2002) Mol. Cell 10, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 59.Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. (2002) Mol. Cell 10, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 60.Nagashima M., Shiseki M., Miura K., Hagiwara K., Linke S. P., Pedeux R., Wang X. W., Yokota J., Riabowol K., Harris C. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.