Abstract
pICln is an essential, highly conserved 26-kDa protein whose functions include binding to Sm proteins in the cytoplasm of human cells and mediating the ordered and regulated assembly of the cell's RNA-splicing machinery by the survival motor neurons complex. pICln also interacts with PRMT5, the enzyme responsible for generating symmetric dimethylarginine modifications on the carboxyl-terminal regions of three of the canonical Sm proteins. To better understand the role of pICln in these cellular processes, we have investigated the properties of pICln and pICln·Sm complexes and the effects that pICln has on the methyltransferase activity of PRMT5. We find that pICln is a monomer in solution, binds with high affinity (Kd ∼ 160 nm) to SmD3-SmB, and forms 1:1 complexes with Sm proteins and Sm protein subcomplexes. The data support an end-capping model of pICln binding that supports current views of how pICln prevents Sm oligomerization on illicit RNA substrates. We have found that by co-expression with pICln, recombinant PRMT5 can be produced in a soluble, active form. PRMT5 alone has promiscuous activity toward a variety of known substrates. In the presence of pICln, however, PRMT5 methylation of Sm proteins is stimulated, but methylation of histones is inhibited. We have also found that mutations in pICln that do not affect Sm protein binding can still have a profound effect on the methyltransferase activity of the PRMT5 complex. Together, the data provide insights into pICln function and represent an important starting point for biochemical analyses of PRMT5.
Although arginine methylation in eukaryotic cells has been known for a number of years, the families of protein arginine methyltransferases (PRMTs)2 responsible for these post-translational modifications have emerged much more recently (1). A diverse set of biological pathways are now thought to be regulated in part by PRMTs, including DNA damage repair, transcriptional regulation, and RNA splicing (2, 3). The majority of PRMTs are type I enzymes which catalyze the formation of monomethylarginine and asymmetric dimethylarginine (4). Human PRMT5 is an example of the less common type II PRMTs that catalyze formation of monomethylarginine and symmetric dimethylarginine (5).
PRMT5 has several known cellular targets, including myelin basic protein (6), histones (7), and the spliceosomal Sm proteins (8, 9). In each case PRMT5 functions in the context of a multiprotein complex where additional proteins contribute to localization and substrate specificity. Methylation of Sm proteins, for example, occurs in a multiprotein complex termed the methylosome or PRMT5 complex, which contains PRMT5, the PRMT5-interacting proteins pICln and MEP50/WD45/IBP42, and Sm protein substrates (8–10).
Seven distinct Sm proteins (SmD3, SmB/B′, SmD1, SmD2, SmE, SmF, SmG) form a protein core that is common to each of the major spliceosomal (U2-type) snRNP particles. In human cells the initial steps of snRNP assembly occur in the cytoplasm, where the seven Sm proteins form a highly stable heptameric ring on the Sm site of the small nuclear RNA to form a small nuclear (snRNP) core (11). The Sm proteins share a small, highly conserved “Sm domain” that is responsible for oligomerization and RNA binding (12). Although formation of the snRNP core can occur spontaneously in vitro, this process is highly regulated and dependent on the survival motor neurons (SMN) complex in vivo (13–15).
Three of the Sm proteins, SmD1, SmD3, and SmB, contain arginine/glycine-rich “RG motifs” in the regions carboxyl-terminal to their Sm domains that are symmetrically dimethylated by the PRMT5 complex (8, 9). Methylation increases the affinity of Sm proteins for the SMN complex, providing a potentially important regulatory mechanism in snRNP assembly (16–18).
The highly conserved 26-kDa protein pICln is involved in both the methylation of Sm proteins by PRMT5 and the assembly of spliceosomal snRNPs. pICln was originally cloned by screening a canine cDNA library in Xenopus oocytes for induction of a novel nucleotide-sensitive chloride current (19). Although it was originally proposed that pICln acts as an ion channel in vivo (19), subsequent work has led to considerable doubt that this is a physiological function of the protein (20, 21). Indeed, pICln lacks transmembrane motifs and is water-soluble, and the NMR structure of an amino-terminal fragment of canine pICln revealed a pleckstrin homology (PH) domain fold (22). It is clear, however, that pICln participates in critical cellular pathways as disruption of the ICln gene causes embryonic lethality in the mouse (23). In addition, reduced expression of pICln causes motor neuron outgrowth deficiencies that mimic the effects of a reduction of SMN in a zebrafish model of spinal muscular atrophy (24).
pICln has been shown to bind directly to both spliceosomal Sm proteins and to PRMT5 in vitro and in vivo (8, 9, 21). Pu et al. (21) made the important observation that pICln-bound Sm proteins are strongly inhibited from assembling into snRNP cores in vitro, leading to a model where pICln plays a role in the stepwise snRNP assembly process by acting as a Sm chaperone. In this model pICln·Sm complexes associate with PRMT5 for methylation of the RG tails of SmD1, D3, and B but are otherwise sequestered and prevented from associating with RNA or with one another (25). This report focuses on the roles of pICln in mediating Sm protein methylation and in the early steps in snRNP assembly. Although several of the interacting partners in these processes have been identified, there are a number of important mechanistic questions that have not yet been addressed. For example, the oligomeric state of pICln, the affinity and stoichiometry of complexes formed between pICln and Sm proteins, and the properties of pICln bound to Sm subcomplexes are all directly relevant to understanding the methylation reaction and potential models for SMN-mediated snRNP assembly. The methyltransferase activities of PRMT5 complexes purified from cultured cells and from tissues have been reported (5, 8, 9, 26–28); however, it has not yet been possible to study PRMT5 in isolation or to study the effects of individual components of any PRMT5 complex because of the difficulties encountered in producing active, recombinant PRMT5. In this report we describe progress on each of these fronts, including the initial characterization of active PRMT5 produced in bacteria.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Proteins were overexpressed in BL21(DE3) cells at 37 °C for 3 h after induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were lysed by sonication in lysis buffer (50 mm phosphate, pH 7.5, 0.15 m NaCl, 0.1% Triton X-100, 100 μm phenylmethylsulfonyl fluoride, 50 μm pepstatin A (Sigma), 0.25 trypsin inhibitory units of aprotinin (Sigma), and 1 Complete Protease Inhibitor tablet (Roche Applied Science)). pICln was cloned into pACYCDuet (Novagen) with an amino-terminal His6 tag that was cleavable with tobacco etch virus (TEV) protease. For pICln·SmD3 co-expression, the SmD3 coding region was cloned into the second multiple cloning site of pACYCDuet. The pICln·SmD3 complex was purified using Talon resin (Clontech) according to the manufacturer's protocol for imidazole elution. The His6 tag was removed by overnight incubation with TEV protease (29). The cleaved mixture was subsequently purified on Talon resin, hydroxyapatite (Bio-Rad) with phosphate elution and gel filtration using a Superdex-75 column (GE Healthcare) equilibrated with PBS. Free pICln was purified from the pICln·SmD3 complex by anion exchange using a Mono Q column (GE Healthcare) and elution with a NaCl gradient.
For co-expression of pICln with PRMT5, His6-TEV-pICln was expressed from pACYCDuet and PRMT5 or PRMT5ΔSAM (G367A/R368A) was expressed from pCDFDuet (Novagen). When SmD3 was included in the co-expression, it was expressed from pETDuet (Novagen). PRMT5·pICln·SmD3 and PRMT5·pICln complexes were purified on Talon resin followed by gel filtration on a Superdex-200 column. Free PRMT5 was obtained by anion exchange chromatography of the PRMT5·pICln complex or the PRMT5·pICln·SmD3 complex using a Mono Q column and sodium chloride gradient.
GST fusions of PRMT5 were expressed from a pETDuet derivative containing the GST coding sequence. For full-length PRMT5 and the PRMT5 regulatory domain (residues 1–290), the GST fusions were co-expressed with pICln to promote the expression of soluble protein followed by affinity chromatography using glutathione-agarose beads (Sigma). A high salt wash (0.5 m NaCl) was sufficient to remove pICln before elution with glutathione. The PRMT5 catalytic domain (residues 322–637) was expressed in soluble form in the absence of pICln. Yeast HMT1p was amplified from Saccharomyces cerevisiae genomic DNA, expressed as an amino-terminal His6 fusion, and purified using Talon beads. Lsm4 and a GST fusion of fibrillarin were generated and purified as previously described (22, 30). Myelin basic protein and histones were purchased from Sigma and Worthington, respectively. SmD3-SmB and SmD1-SmD2 constructs were prepared, and the protein heterodimers were purified essentially as described (12) except that we used full-length SmD3. All proteins were stored in 0.02 m phosphate buffer, pH 7.5, 0.1 m NaCl, 5 mm dithiothreitol at −80 °C and retained full activity after multiple freeze-thaws.
Methyltransferase Assays
Methylation reactions were performed as previously described (31) with a few modifications. Reactions containing the indicated amounts of PRMT5 and substrate were incubated in the presence of 40–80 μm S-adenosylmethionine (AdoMet; Sigma) and 0.5–1 μCi of [methyl-3H]AdoMet (GE Healthcare) in PBS, pH 7.6, 1 mm dithiothreitol. For SDS-PAGE analysis, reactions were first boiled in SDS sample loading buffer. For native PAGE analysis, reactions were separated on a 10–20% polyacrylamide gradient gel (Cambrex) with a Tris-glycine buffer system (25 mm tris, 0.2 m glycine, pH 8.5). Gels were fixed for 30 min in 40% methanol, 10% acetic acid, soaked in Amplify (GE Healthcare), dried, and exposed to film for 24 h at −80 °C. Relative protein concentrations in methylation reactions were visualized by Coomassie staining of a duplicate gel. Reactions assessed by scintillation counting were first precipitated with 25% trichloroacetic acid. Precipitated proteins were washed five times with 500 μl of 25% trichloroacetic acid, once with 500 μl of cold acetone, and resuspended in 0.1% SDS, 0.1 m Tris, pH 8.0, followed by scintillation counting.
Ultracentrifugation
Ultracentrifugation experiments were performed at 20 °C with an XL-A analytical ultracentrifuge (Beckman) and a TiAn60 rotor with charcoal-filled Epon centerpieces and quartz windows. Sedimentation equilibrium experiments were performed at three concentrations in PBS, pH 7.4, and three rotor speeds. Radial absorption data for each experiment were globally fit well, with single species models and good model statistics (root mean square deviation < 0.007) using the program SEDPHAT (32). Complete sedimentation velocity profiles were collected every 30 s at 55,000 rpm in PBS, pH 7.4, followed by data analysis using the program SEDFIT (33).
Protein-Protein Interaction
The binding affinity between pICn and the SmD3-SmB heterodimer was measured using a fluorescence anisotropy assay. pICln was labeled with amine reactive Oregon Green-488 (OG488; Invitrogen) under conditions that favor labeling of the amino terminus as described by the vendor. Binding reactions (100 μl) containing 10 nm OG488-labeled pICln and increasing concentrations of the SmD3-SmB heterodimer (0–1 μm) were analyzed using a Beacon 2000 (Pan Vera) instrument in PBS, pH 7.4. Data were fit to a simple binding isotherm. Qualitative binding of GST-pICln deletion mutants to FLAG-SmD3 was assessed by co-expression of the proteins and partial purification of stable complexes that formed on glutathione-agarose (Sigma). For these co-expressions, GST fusions of pICln and pICln deletion mutants were expressed from pACYCDuet, and FLAG-tagged SmD3 was expressed from pCDFDuet. After 5× washes with PBS, proteins bound to the beads were stripped with SDS, run on SDS-PAGE, and analyzed by anti-FLAG Western blot and Coomassie staining. PRMT5·pICln interactions were assayed using a standard GST pulldown approach (34). 1 μg of purified pICln was incubated with 5 μg of GST, GST-PRMT5, GST-PRMT5reg, or GST-PRMT5cat in RSB-100 buffer (10 mm Tris, pH 8, 0.1 m NaCl, 0.1% Nonidet P-40). After incubating 1 h at 4 °C, the beads were washed and stripped as described followed by SDS-PAGE and Western blotting with anti-pICln antibody (Millipore). [35S]Methionine-labeled deletion mutants of pICln were produced by in vitro translation in Escherichia coli S30 extract (Promega) followed by incubation with 5 μg of GST-PRMT5 bound to glutathione-agarose beads. Binding reactions were washed, eluted with SDS, and visualized after SDS-PAGE and PhosphorImager analysis of dried gels (GE Healthcare).
RESULTS
pICln Is an Elongated Monomer in Solution
Based on size-exclusion chromatography profiles, pICln appears to be oligomeric in solution (35). If this interpretation is correct, then an oligomeric pICln could provide multiple independent binding surfaces, each capable of interacting with a Sm protein, an Sm protein hetero-oligomer, or a PRMT5 oligomer. To determine whether higher order pICln·Sm assemblies containing multimers of pICln are likely to exist in vivo, we investigated the oligomeric state of pICln using analytical ultracentrifugation techniques.
Using sedimentation equilibrium ultracentrifugation, we found that even at the relatively high concentration of 20 μm, pICln exists as a monomer in solution, with no indication that dimeric or higher order species are formed (Fig. 1A). One explanation for the anomalous behavior of pICln on size-exclusion chromatography columns is that the protein has an elongated shape that gives rise to hydrodynamic properties consistent with a larger globular protein. To test this idea, we determined the sedimentation coefficient of pICln by sedimentation velocity (Fig. 1B). Indeed, pICln does not sediment as expected for a 27-kDa spherical-shaped protein but rather as an elongated molecule that can be modeled as an ellipsoid with an ∼7:1 axial ratio. Because the amino-terminal half of pICln adopts a PH domain fold, the extended nature of the protein overall shape must be because of the carboxyl-terminal half of the protein and/or the large acidic insertion located between the β6 and β7 strands of the PH domain (22).
FIGURE 1.
pICln is monomeric and adopts an extended conformation. A, sedimentation equilibrium analysis of pICln at 25 μm. Radial absorbance distributions fit well to a single 26.5-kDa species, indicating that pICln is a monomer in solution. B, sedimentation velocity c(s) distribution of pICln (solid line) compared with that expected if pICln had a spherical shape (dotted line). The axial ratio of elongated pICln in solution can be estimated as ∼7:1, assuming a prolate ellipsoidal shape.
pICln Forms Tight 1:1 Complexes with Sm Proteins
Several groups have shown that pICln interacts with spliceosomal Sm proteins (8, 9, 21), yet the affinity and stoichiometry of these interactions has not been reported. One of the difficulties involved with biochemical and biophysical characterization of pICln·Sm complexes is the poor solubility properties of Sm proteins expressed and purified in isolation (36). However, the SmD3-SmB and SmD1-SmD2 heterodimers as well as the SmF-SmE-SmG heterotrimer can be produced in a soluble form that is amenable to biochemical analysis by co-expression in bacteria. These hetero-oligomers of Sm proteins have been shown to be intermediates in the assembly of spliceosomal snRNPs (13, 37). To test whether pICln can stabilize single Sm proteins in the absence of cognate Sm oligomeric partners, we separately co-expressed and co-purified pICln with each Sm protein. We found that pICln can be co-expressed and co-purified with each of the individual Sm proteins, with the exception of SmE (supplemental Fig. 1). By employing these co-expression approaches, we have been able to produce soluble pICln·Sm and Sm hetero-oligomers for biochemical and biophysical studies.
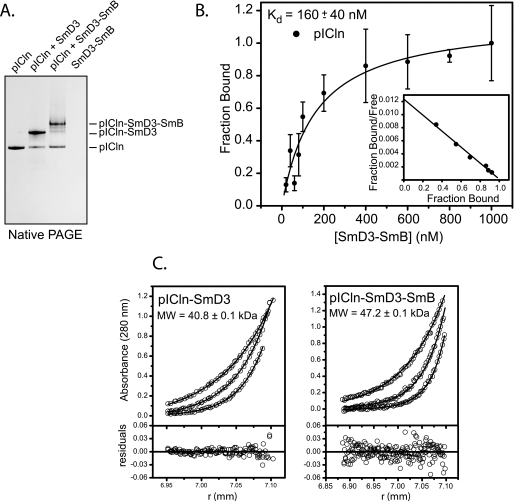
To investigate the interaction between pICln and Sm proteins, we chose SmD3 and the SmD3/SmBΔC heterodimer to use as model systems (SmBΔC is the Sm domain of SmB). Because pICln is quite acidic, pICln and complexes of pICln with Sm proteins can be readily separated and visualized by native PAGE (Fig. 2A). When equilibrium binding mixtures of pICln and SmD3/SmBΔC heterodimer are analyzed, the primary shifted species is the pICln·SmD3/SmBΔC complex, with only minor formation of pICln·SmD3 and pICln·SmBΔC. This indicates that the SmD3-SmB interaction is stable and does not readily dissociate into pICln·SmD3 and pICln·SmBΔC complexes, an observation that is consistent with a role for SmD3/B (and SmD1/D2 and SmF/E/G) as snRNP building blocks (13, 14, 38).
FIGURE 2.
Interaction of pICln with Sm proteins. A, native PAGE of purified pICln, pICln·SmD3 complex, and a 2:1 mixture of pICln with SmD3/SmBΔC heterodimer. The SmD3/SmBΔC dimer is positively charged and, therefore, does not enter the polyacrylamide gel unless it is bound to pICln. B, pICln binding to SmD3/SmBΔC monitored by fluorescence polarization of labeled pICln. pICln binds with a Kd of 160 nm and a 1:1 stoichiometry. Error bars represent 1 S.D. derived from three replicate experiments. C, sedimentation equilibrium ultracentrifugation of pICln·SmD3 complex at 25 μm and pICln·SmD3/SmBΔC complex at 12.5 μm concentrations. pICln·SmD3 data fit well to a 1:1 heterodimer, and pICln·SmD3/SmBDC data fit well to a 1:1:1 heterotrimer with the experimental molecular weights shown.
Interestingly, co-expression of pICln in the presence of both SmD3 and SmBΔC primarily yields the pICln·SmD3 and pICln·SmBΔC complexes (data not shown). Under these conditions, pICln is present in excess of the Sm proteins. These observations support the idea that monomeric Sm proteins (e.g. SmD3) can form stable complexes with pICln in the presence of cognate binding partners (e.g. SmBΔC).
To estimate the binding affinity of pICln for a Sm protein, we titrated fluorescently labeled pICln with the SmD3/SmBΔC heterodimer and measured changes in fluorescence polarization that occurs when the complex is formed (Fig. 2B). For this Sm heterodimer, the apparent dissociation constant is 160 nm, with a 1:1 stoichiometry of binding. We were not able to perform the analogous experiment with SmD3 alone because of the low solubility of the isolated Sm protein.
We further explored the stoichiometry and oligomeric state of the pICln·SmD3 and pICln·SmD3/SmBΔC complexes using sedimentation equilibrium analysis (Fig. 2C). Good fits of the radial absorbance distributions to a single-species model indicate that the pICln·SmD3 complex is a 1:1 heterodimer in solution, with experimental and calculated molecular masses of 40.8 and 40.1 kDa, respectively. Similarly, the pICln·SmD3/SmBΔC complex is a 1:1:1 heterotrimer in solution, with experimental and calculated molecular masses of 47 and 50.2 kDa, respectively. These results, together with structural (12) and mutagenesis (9) data are consistent with a model in which pICln interacts specifically with one of the two surfaces used in forming Sm oligomers but not both. An alternative model, where pICln binds to a Sm domain surface that is not used in formation of a Sm-Sm interface, would require one pICln bound per Sm protein or a 2:1:1 stoichiometry for the pICln·SmD3/SmBΔC complex.
The concentration of pICln in Madin-Darby canine kidney cells has been estimated at 200–300 nm (39). That estimate together with the relatively high affinity (Kd ∼ 160 nm) between pICln and SmD3-SmB suggests that under physiological conditions newly synthesized Sm proteins are all likely to exist as some type of complex with pICln. Although we have not been able to directly measure the affinity of the pICln·SmD3 interaction, sedimentation equilibrium results indicate that 300 nm pICln·SmD3 complex exists as a 1:1 heterodimer. This indicates that the pICln·SmD3 interaction is at least as strong as that measured for pICln·SmD3/SmBΔC. The results summarized in Fig. 2 explain why pICln is able to effectively inhibit snRNP assembly in vitro (21). High affinity binding to one of the oligomerization surfaces of a Sm protein could effectively block the ability of that protein to participate in formation of a heptameric ring on Sm site-containing RNA substrates.
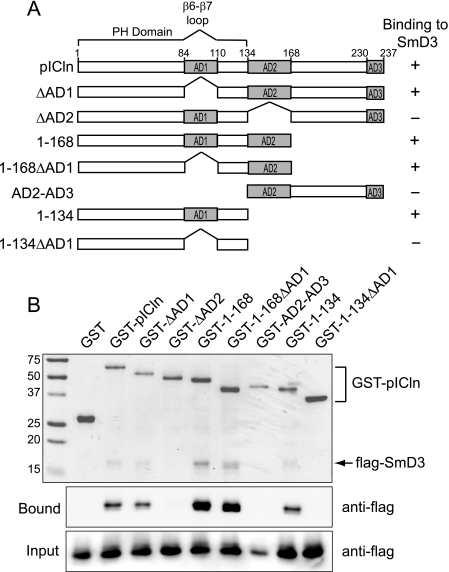
A report showing that the second acidic region (AD2; see Fig. 3) of pICln is responsible for its interaction with the Sm-like protein LSm4 (22) prompted us to ask whether pICln interacts similarly with SmD3. To establish the region(s) in pICln responsible for interacting with Sm proteins, we co-expressed FLAG-tagged SmD3 with a series of pICln deletion mutants fused to GST and asked which mutants were able to retain SmD3 when co-purified on glutathione-agarose. With this approach, binding takes place inside the cell (albeit bacterial cells) in the presence of a variety of competitor macromolecules, and we were able to evaluate binding to individual Sm proteins in the absence of eukaryotic factors that could potentially form bridging interactions.
FIGURE 3.
Participation of pICln PH domain and acidic regions in binding to Sm proteins. A, schematic of pICln constructs used. AD1, AD2, and AD3 refer to the three acidic regions previously described (45). B, interaction assay for pICln and SmD3. FLAG-tagged SmD3 was co-expressed with GST alone or with the indicated GST-pICln fusion. The complexes were purified on glutathione-agarose and analyzed by SDS-PAGE (top panel). SmD3 in the input and bound fractions was identified by anti-FLAG immunoblot (bottom panels).
The results summarized in Fig. 3 reveal that the deletion of AD2 abolishes SmD3 binding to pICln. However, the AD2-AD3 construct alone is not sufficient for a high affinity interaction, indicating that multiple regions of pICln are involved in binding to Sm proteins. Surprisingly, pICln constructs containing the PH domain and either AD1 (1–134) or AD2 (1–168ΔAD1) are able to efficiently bind SmD3, but the PH domain lacking the AD1 loop (1–134ΔAD1) does not bind. Together, these observations are consistent with a model in which the pICln PH domain, the adjacent AD2 region, and the AD1 loop contribute to form an interaction surface for binding Sm proteins.
Interestingly, the AD1 loop is able to compensate for the loss of AD2 in a pICln mutant truncated after the PH domain but not in the context of the internal AD2 deletion (1–237ΔAD2). One explanation could be that replacement of the AD2 sequence by downstream residues in the ΔAD2 construct introduces a steric clash that is incompatible with Sm protein binding, whereas simple truncation of the protein after the PH domain still allows Sm proteins to interact. A structural model of the pICln·Sm complex will no doubt provide valuable insight into interpretation of these results.
Active PRMT5 Can Be Produced by Co-expression with pICln
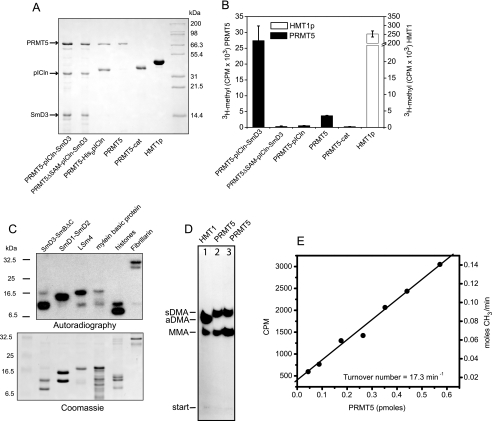
Our initial attempts to produce PRMT5 in bacteria resulted in an inactive enzyme. Although one group has reported that they have produced bacterial GST-PRMT5 that methylates myelin basic protein (27), others have noted that the enzyme is inactive when expressed in E. coli (8, 40). We found that when co-expressed with pICln, PRMT5 can be produced at reasonable levels in a soluble and active form (Fig. 4, A and B). The two proteins co-purify, resulting in a stable PRMT5·pICln complex. When SmD3 is included in the co-expression, a ternary PRMT5·pICln·SmD3 complex can be readily purified (Fig. 4A). As controls for methyltransferase assays, we also produced a catalytically inactive PRMT5 that does not bind S-adenosylmethionine (PRMT5ΔSAM), the PRMT5 catalytic domain (PRMT5cat), and yeast HMT1, a well characterized type I PRMT (41, 42).
FIGURE 4.
Active PRMT5 produced by co-expression with pICln. A, SDS-PAGE of PRMT5 complexes produced by co-expression with pICln in E. coli. PRMT5ΔSAM is a catalytically defective PRMT5 mutant. PRMT5 alone was obtained by separation of the purified PRMT5·pICln complex. The PRMT5 catalytic domain (296–637) and yeast HMT1 were expressed independently. B, arginine methylation activity of PRMT5 complexes. Reactions containing 20 μm SmD3/SmBΔC substrate and 40 μm [3H]SAM were separated after 1 h, and the amount of substrate methylation was determined by scintillation counting. Error bars represent the S.D. based on three replicates. C, methylation of RG-containing substrates by PRMT5. Reactions containing 1 μm PRMT5, 40 μm [3H]SAM (1 μCi/reaction) and the indicated substrates were incubated for 2 h followed by SDS-PAGE and autoradiography. D, recombinant PRMT5 produces monomethylarginine (MMA) and symmetric dimethylarginine (sDMA) but not asymmetric dimethylarginine (aDMA). Methylation reactions containing 5 μm HMT1 + 5 μm pICln·SmD3 (lane 1), 5 μm PRMT5·pICln·SmD3 complex (lane 2), or 5 μm PRMT5·pICln·SmD3 + 5 μm pICln·SmD3 (lane 3) were incubated with 80 μm [3H]SAM (20 μCi/reaction), and the resulting modified arginine residues were analyzed by thin layer chromatography and autoradiography as described (8). E, PRMT5 specific activity was determined from methylation reactions containing varying amounts of PRMT5 and saturating levels (20 μm) of pICln·SmD3 and [3H]SAM (80 μm SAM, 1 μCi/ reaction).
To test recombinant PRMT5 for methyltransferase activity, we incubated purified PRMT5 or PRMT5 complexes with SmD3/SmBΔC substrate in the presence of S-[methyl-3H]adenosylmethionine ([3H]SAM). SmD3 contains multiple RG repeats in its carboxyl terminus that are methylated by PRMT5 in vivo (5, 8, 9). SmBΔC contains only the Sm domain of SmB, so it is not methylated by PRMT5. The highest PRMT5 activity was observed for a co-purified complex containing PRMT5, pICln, and SmD3, which has roughly 10% that of the activity of an equivalent amount of HMT1 on the same substrate (Fig. 4B). Interestingly, the PRMT5·pICln complex showed only background levels of methyltransferase activity, suggesting that pICln alone inhibits PRMT5 when it is pre-bound to the enzyme. By chromatographically separating PRMT5 from the PRMT5·pICln complex we were able to recover methyltransferase activity, but the activity was less than that observed for the ternary complex. PRMT5 that was chromatographically separated from the PRMT5·pICln·SmD3 complex showed a similar level of methyltransferase activity (data not shown).
The PRMT5 catalytic domain does not possess measurable methyltransferase activity (Fig. 4B), in agreement with the results of Rho et al. (27). Because dimerization of other PRMTs has been shown to be essential for catalytic activity (31, 41), we analyzed PRMT5cat by sedimentation equilibrium ultracentrifugation to determine whether the isolated domain exists as a dimer. We found that PRMT5cat dimerizes only weakly (Kd ∼ 20 μm), indicating that under our assay conditions, the isolated catalytic domain is 95% monomeric (supplemental Fig. 2A). When we assayed PRMTcat at 30 μm enzyme (where [dimer] > 15 μm), we saw only a 4-fold increase in activity above background, despite the ∼400-fold increase in dimer concentration (supplemental Fig. 2B). We conclude that even when dimeric, the PRMT5 catalytic domain lacks significant levels of methyltransferase activity.
Although we have not yet established the oligomeric states of full-length PRMT5 and PRMT5-containing complexes, it is clear from preliminary size-exclusion chromatography and ultracentrifugation studies that they do form stable oligomers at low concentration (data not shown), indicating that the amino-terminal domain of PRMT5 is important for oligomerization and methyltransferase activity. This observation is consistent with results obtained from PRMT5-containing complexes purified from mammalian sources (28).
To further characterize full-length PRMT5 methyltransferase activity, we incubated the free enzyme with several known substrates and visualized the products by SDS-PAGE and autoradiography (Fig. 4C). Recombinant PRMT5 methylates SmD3, SmD1, LSm4, myelin basic protein, histones H3 and H4, and fibrillarin. PRMT5 does not methylate itself or pICln. We also confirmed that recombinant PRMT5 generates symmetric dimethylarginine by hydrolyzing methylated SmD3 and analyzing the resulting 3H-labeled arginine residues by thin layer chromatography (Fig. 4D). As noted previously for PRMT7, a larger fraction of monomethylarginine is produced when the substrate:enzyme ratio is increased (43). A typical activity assay for a preparation of PRMT5 (purified from pICln-bound complex) is shown in Fig. 4E.
pICln Stimulates Methylation of SmD3
The results summarized in Fig. 4B indicate that pICln is able to affect PRMT5 methyltransferase activity for SmD3. One possibility is that pICln may act as a co-substrate when bound to Sm proteins, presenting its Sm partner for more efficient methylation by PRMT5 (8). In this case one might predict that pICln-mediated PRMT5 activation should be specific for Sm proteins and that a direct interaction between pICln and the Sm substrate is required. To test this idea we performed methylation reactions in which SmD3/SmBΔC or histone H3/H4 substrates were preincubated with increasing concentrations of pICln (Fig. 5A).
FIGURE 5.
pICln stimulates methylation of SmD3 but not histones. A, methylation reactions containing 1 μm PRMT5, 40 μm SAM (0.5 μCi/reaction [3H]SAM), and either SmD3/SmBΔC (SmD3, 4 μm) or histones (H, 4 μm) were performed with increasing concentrations of pICln and analyzed by SDS-PAGE and autoradiography (top panels) or Coomassie staining (bottom panels). B, effect of pICln on the initial PRMT5 methylation rate of the SmD3 RG tail (D3C32). Methylation of the isolated peptide in the absence of pICln is slowest. The addition of pICln stimulates methylation in the absence of direct interaction with the RG substrate. Methylation is fastest for full-length SmD3 in the presence of pICln.
For the SmD3-SmB heterodimer we found that pICln has a strong stimulatory effect on methylation that increases up to about a 2:1 pICln:substrate ratio. In contrast, pICln inhibits the methylation of histones by PRMT5. pICln does not inhibit methylation of histones by HMT1, indicating that the effect is specific for PRMT5 and is not a result of pICln nonspecifically binding the histone substrates (data not shown). Thus, pICln stimulation of PRMT5 methyltransferase activity appears to be specific for Sm proteins.
When we looked at time courses for methylation of just the 32-residue carboxyl-terminal tail of SmD3 (D3C32), we found that pICln still stimulates PRMT5 methylation, although to a lesser extent than for full-length SmD3 (Fig. 5B). In this case, pICln does not bind to the D3C32 peptide (8) yet is still able to stimulate PRMT5. These results are consistent with a model where pICln binding to PRMT5 leads to an increase in activity that is not simply the result of enhanced substrate binding.
It is important to note that when pICln is pre-bound or binds independently to PRMT5, we only observe stimulation of methyltransferase activity for peptide substrates, as shown in Fig. 5B. Methylation of histone (Fig. 5A) and SmD3/SmBΔC (Fig. 4B) substrates are both inhibited under these conditions. One explanation could be that substrates have limited access to the PRMT5 active site when pICln is bound, and free peptide substrates may be able to circumvent this steric filter (see “Discussion”).
Interaction of pICln and SmD3 with PRMT5
To extend our understanding of how pICln affects PRMT5 activity, we examined which regions of pICln are required for interaction with PRMT5. A series of pICln deletion mutants were generated by in vitro transcription and translation in the presence of [35S]methionine and tested for binding to immobilized GST-PRMT5 (Fig. 6A). Deletion of the first two acidic regions in pICln (AD1, AD2, or both) had no effect on PRMT5 binding. However, deletion of the third acidic region (AD3) resulted in a loss of detectable binding, consistent with previous reports (44, 45). Interestingly, neither the 1–168 nor the 169–237 pICln constructs retained the ability to bind PRMT5 in this assay, suggesting that both are required, but neither is sufficient on their own for a stable interaction. Thus, the PH domain-AD2 region and AD3, both, participate in PRMT5 binding. To determine which PRMT5 domain(s) mediates the interaction with pICln, we incubated pICln with GST fusions of full-length PRMT5, the amino-terminal regulatory domain (1–290), and the carboxyl-terminal catalytic domain (311–637) and partially purified the resulting complexes on glutathione-agarose. Anti-pICln Western blots of the GST pulldown assays revealed that the PRMT5 regulatory domain binds independently to pICln, although the strength of the interaction is reproducibly less than that of the full-length enzyme (Fig. 6C). The isolated catalytic domain shows no significant binding above that of the GST control. Together, the data are consistent with a model in which pICln interacts primarily with the regulatory domain of PRMT5. Because efficient oligomerization of PRMT5 requires both the amino-terminal and catalytic domains (discussed above), it is also possible that pICln interacts with a PRMT5 surface that is generated by oligomerization and would not be present in a binding assay with either of the isolated domains.
FIGURE 6.
Interaction of pICln with PRMT5. A, pICln-PRMT5 interaction assay. pICln or pICln deletion mutants were synthesized by in vitro transcription and translation in the presence of [35S]methionine (top panel) and added to GST or GST-PRMT5 bound to glutathione-agarose beads. Bound pICln was visualized by SDS-PAGE and autoradiography (bottom panel) after extensive washing of the beads. B, domain structure of pICln. The pICln deletion mutants are shown schematically in Fig. 3A. C, purified pICln was incubated with GST, GST-PRMT5, and GST fused to the PRMT5 regulatory domain (GST-PRMT5-reg) or GST fused to the PRMT5 catalytic domain (GST-PRMT5-cat). The complexes were bound to glutathione-agarose beads, washed, eluted in SDS gel loading buffer, and analyzed by anti-pICln immunoblotting.
A third point of interaction with PRMT5 in the PRMT5·pICln·SmD3 complex involves the carboxyl-terminal RG tail of SmD3. Indeed, Friesen et al. (8) originally purified PRMT5 from HeLa cell extracts using a RG-peptide affinity column. We have verified a tight interaction between the D3C32 peptide (the carboxyl-terminal tail of SmD3) and PRMT5cat with Kd ∼ 1 μm (supplemental Fig. 3). This interaction is expected to make a significant contribution to overall binding affinity in the context of a PRMT5·pICln·Sm assembly. Thus, pICln·SmD3 interacts with PRMT5 via the PH and AD3 regions of pICln as well as via the RG tail of SmD3. The Sm motif alone does not interact substantially with PRMT5 (8).
Mutations in pICln That Block PRMT5 Methylation
Because the AD3 region of pICln is required for the interaction with PRMT5, we reasoned that AD3 might also play a role in regulating methyltransferase activity. First, we attempted to purify a recombinant PRMT5 complex containing pICln-(1–168) and SmD3. As shown in Fig. 7A, the PRMT5·pICln-(1–168)·SmD3 complex can be co-expressed and co-purified similarly to the complex containing full-length pICln. This result appears to contradict the requirement of AD3 for a stable interaction with PRMT5 (Fig. 6). However, the presence of SmD3 and the PH domain of pICln compensate for the loss of AD3 such that a stable PRMT5·pICln-(1–168)·SmD3 complex is formed. As expected, we were not able to purify a stable PRMT5·pICln-(1–168) complex in the absence of SmD3, as the interaction between the RG tail of SmD3 and PRMT5 is not present.
FIGURE 7.
The carboxyl terminus of pICln stimulates PRMT5 activity. A, silver-stained SDS-PAGE of purified PRMT5·pICln·SmD3 complexes (top). The addition of 40 μm SAM (0.5 μCi of [3H]SAM) to the complexes followed by SDS-PAGE and autoradiography (bottom) shows that full-length pICln, but not pICln 1–168, produces a high level of methyltransferase activity. B, attempts to rescue the catalytic activity of PRMT5·pICln-(1–168)·SmD3 by adding alternative substrate or pICln-substrate complexes. C, products of PRMT5 methylation reactions. Methyltransferase reactions containing PRMT5·pICln·SmD3 and additional SmD3/SmBΔC, pICln·SmD3, or pICln were analyzed by Native-PAGE (Coomassie and autoradiography) and by anti-PRMT5 immunoblot. Methylated pICln·SmD3 and pICln·SmD3/SmBΔC are the primary products of the reaction. The faster migrating band containing PRMT5 has not yet been characterized but could contain an alternative oligomeric form of PRMT5.
To assess the effect of the pICln AD3 deletion on methyltransferase activity, [3H]SAM was incubated with PRMT5·pICln-(1–168)·SmD3, and the level of [3H]methyl was incorporated into the SmD3 RG tail was determined by SDS-PAGE and autoradiography (Fig. 7, A and B). Surprisingly, we found that deletion of the carboxyl terminus of pICln (residues 169–237) led to strongly diminished levels of methyltransferase activity for the co-expressed PRMT5 complex. The activity of this complex was less than that observed for isolated PRMT5, purified from either the same complex or from a complex with full-length pICln. These observations indicate that the carboxyl terminus of pICln is required for optimal methylation of SmD3 and that the role of pICln extends beyond simply binding and escorting SmD3 to PRMT5.
To further investigate the properties of the PRMT5·pICln-(1–168)·SmD3 complex, we attempted to rescue activity by the addition of pICln·SmD3 heterodimer, D3C32 peptide, or pICln(169–237). As shown in Fig. 7B, only minor increases in methyltransferase activity resulted from the addition of pICln·SmD3 or the carboxyl-terminal fragment of pICln. A higher level of activity was observed after the addition of D3C32 peptide substrate, but the amount of methylated peptide still fell below that observed for methylation of the same substrate by PRMT5 alone (Fig. 5B). We do not yet know the basis of pICln-(1–168) inhibition of PRMT5 when the SmD3 substrate is bound. One possibility is that the tightly bound SmD3 is methylated so slowly that there is very little product release. Under these conditions a slow rate of exchange of substrates may limit the ability of PRMT5 to methylate the pICln·SmD3 complex.
pICln·SmD3-methylated Product Complexes
The tight interaction of unmethylated pICln·SmD3 and pICln-(1–168)-SmD3 with PRMT5 prompted us to consider the nature of the product(s) of methylation. It has been noted that Sm proteins have much reduced affinity for the PRMT5 complex once they have been methylated (8), consistent with tighter binding of the enzyme to substrate relative to product. In principle, methylated SmD3 could dissociate from the PRMT5 complex as an isolated product. However, this seems unlikely given the high affinity of the pICln·Sm interaction, and previous observations that the RG-tail does not play a significant role in binding to pICln. A more likely model is that methylated SmD3 dissociates from PRMT5 as a pICln·SmD3 complex. Because methylation of the RG tail of SmD3 lowers the affinity of the tail for PRMT5, unmethylated pICln·SmD3 would be an effective competitor for binding to the enzyme and displacing methylated pICln·SmD3. To address this question we carried out methylation reactions using [3H]SAM, co-purified PRMT5·pICln·SmD3 as the source of enzyme, and either pICln·SmD3 or pICln·SmD3/SmBΔC as the methylation substrate. The reaction products were then separated by native PAGE and analyzed by Coomassie stain, autoradiography, and anti-PRMT5 Western blot (Fig. 7C).
As anticipated, most of the [3H]methyl label is found in pICln·SmD3 and pICln·SmD3/SmBΔC complexes. Some of the label is also found within the PRMT5 complex, consistent with formation of a PRMT5·pICln·Sm product complex where SmD3 has been at least partially methylated. Because free SmD3 and SmD3/SmBΔC do not migrate into the non-denaturing polyacrylamide gel, we cannot rule out the possibility that a small fraction of free methylated SmD3 or SmD3/SmBΔC is also present. However, a comparison to similar reactions analyzed by SDS-PAGE (to determine total product label) indicates that most if not all of the labeled products is present as pICln complexes. Together, these observations are consistent with a model where pICln escorts Sm proteins both to and from the PRMT5 complex.
Interestingly, native PAGE analyses of PRMT5 methylation reactions indicate that PRMT5 forms two distinct complexes with different mobilities (Fig. 7C). Based on Western blots of purified complexes run on SDS-PAGE, both species on native PAGE contain full-length PRMT5. The slower migrating complex contains methylated SmD3 and, therefore, represents an active species. The faster moving complex does not contain radioactive label. We suggest that these PRMT5 species may be dimeric and monomeric forms of PRMT5 complexes, respectively, although further work will be required to confirm this. The requirement for dimerization in other PRMT systems would be consistent with this interpretation.
DISCUSSION
Since the initial report that pICln interacts with Sm proteins (21), several groups have confirmed and extended these observations (8, 9, 25). Here, we have used the SmD3 protein and a heterodimeric complex containing SmD3 bound to the Sm domain of SmB as models to study the interaction of pICln with Sm proteins.
A binding stoichiometry of one pICln per Sm protein complex together with observations that Sm ring formation is inhibited by pICln (21) suggest an “end-capping” model in which pICln interacts primarily with one of the two surfaces used by Sm proteins to oligomerize (Fig. 8). These have been described as the β4 and β5 surfaces, based on the exposed β-strands that mediate interactions with neighboring Sm domains (12). Although the experiments described here do not distinguish between pICln binding to the β4 versus the β5 surface of the Sm domain, Meister et al. (9) have shown that mutations in β4 of SmD1 disrupt the interaction with pICln. We, therefore, favor the configuration illustrated in Fig. 8, where pICln binds to the β4 surface. If this is indeed the case, then it follows that pICln interacts with SmD3 when bound to the SmD3-SmB heterodimer and with SmD1 when bound to SmD1-SmD2. Similarly, pICln would be expected to bind to SmF in the SmF-SmE-SmG heterotrimer.
FIGURE 8.
pICln complexes with Sm proteins and PRMT5. A, schematic illustration of pICln bound to a Sm monomer and to Sm heterooligomers. pICln is shown here interacting with the β4-surfaces of SmD3, SmD1, and SmF. B, schematic drawing of a PRMT5·pICln·Sm complex. The oligomeric state of the PRMT5 complex is likely to be at least a dimer but is illustrated here as a monomeric complex for simplicity. pICln is expected to have multiple points of contact with PRMT5, with AD3 and the PH domain regions being the most important. SmD1, SmD3, and SmB can interact with PRMT5 via their RG-tail substrates.
When cytosolic extracts of mammalian cells are fractionated by density gradient ultracentrifugation or size-exclusion chromatography, pICln is found in two major fractions with approximate sedimentation coefficients of 6 S and 20 S (8). The 6 S fraction is enriched in Sm proteins D1, D2, E, F, and G, and the 20 S PRMT5-containing fraction has a higher percentage of SmD1, SmB/B′, and SmD3 (8, 25). The broad nature of the 6 S peak suggests that there may be several pICln·Sm complex constituents. Data from previous experiments do not exclude the idea that pICln could be bound to Sm protein monomers, heterodimers, heterotrimers, or even the SmD1/D2/F/E/G snRNP assembly precursor (13). Indeed, Chari et al. (25) have recently argued that pICln bound to the SmD1/D2/F/E/G pentamer is a major constituent of the 6 S complex. Given the high concentration of pICln in human cells (39), one might expect that Sm proteins should be rapidly captured as pICln complexes after their synthesis and folding. The role of pICln in this context could be to prevent illicit oligomerization and/or aggregation as well as to assist in the methylation of the RG-tails of SmD1, D3, and B/B′. If this is the case, then an early assembly step in snRNP biogenesis would be the formation of pICln bound Sm oligomers (such as SmD3-SmB) from the monomeric complexes. A similar type of regulated exchange of Sm subunits has been implicated in the transfer of oligomeric Sm precursors to the SMN complex (14, 25, 38). The composition of the 6 S cytosolic fraction may, therefore, represent a steady-state pool of pICln-bound Sm intermediates that are themselves the result of a regulated assembly or redistribution of oligomeric states.
Little is currently known about the effect of pICln on PRMT5 methyltransferase activity. By co-expressing pICln with PRMT5 in E. coli, we were able to produce soluble, active PRMT5 and study the role of pICln in PRMT5 methylation activity on Sm substrates. We found that SmD3 methylation is stimulated by pICln in a dose-dependent manner relative to methylation of SmD3 by PRMT5 alone. This stimulation is dependent on the carboxyl terminus of pICln, a region previously shown to participate in binding to PRMT5 (44, 45). When pICln was added to histone methylation reactions, we observed a dose-dependent inhibition of methylation that was not observed in control reactions with HMT1. Thus, pICln acts as a co-substrate for Sm protein methylation by PRMT5, providing both specificity and stimulation of activity.
We attempted to compare the activity of our recombinant PRMT5 with the activities of PRMT5 complexes purified from cultured cells and mammalian tissues (5, 8, 9, 26–28). For example, Rho et al. (27) purified FLAG-PRMT5 from HEK-293 cells and reported activities as a function of PRMT5 concentration using a GST-RG peptide fusion as substrate. In this case, our most active recombinant PRMT5 appears to have a higher specific activity than that described by Rho et al. (27), although this comparison assumes a similar normalization of tritium counting statistics in the methylation assays. Qualitatively, the HeLa cell-derived PRMT5 reported by Friesen et al. (8) appears comparable with our recombinant PRMT5, taking differences in enzyme concentration and amount of radiolabel used into account.
The PRMT5 complexes that have been affinity-purified from mammalian cells range in size from 250 to 500 kDa, based on reports from gradient sedimentation and size-exclusion chromatography (8, 9, 27, 28). These complexes are known to contain at a minimum PRMT5, pICln, Mep50, and Sm proteins, although the stoichiometry and degree of homogeneity within these purified fractions has not been established. Rho et al. (27) have reported that PRMT5 exists in an equilibrium between dimers and tetramers, based partly on peaks of activity corresponding to sedimentation markers of 68–158 and ∼240 kDa. Although these sizes could correspond to a dimer and tetramer of 72-kDa PRMT5, this study was conducted before the identification of Mep50 and pICln as tight binding interactors of cytoplasmic PRMT5. If these proteins are included stoichiometrically, then the data could be reinterpreted as a monomer-dimer equilibrium, a result that would not be surprising in light of the oligomeric properties of model PRMT enzymes (31, 41).
The recombinant PRMT5 that we have used in our studies thus far lacks the Mep50 protein that has been identified in 20 S PRMT5-containing cytosolic fractions and which is efficiently immunoprecipitated with both anti-PRMT5 and anti-pICln antibodies (9, 10). The function of Mep50 in the context of the 20 S complex is not yet known. Although the work described here demonstrates that it is not required for PRMT5 methyltransferase activity, it is possible that Mep50 plays a regulatory role by conferring additional or alternative specificity, modulating oligomerization state, or affecting localization. Friesen et al. (10) have demonstrated that anti-Mep50 antibodies can interfere with PRMT5 complex methylation of Sm proteins, suggesting an involvement at some level. Our attempts to express and purify Mep50 alone, PRMT5·Mep50, PRMT5·pICln-Mep50, and the PRMT5·pICln·Mep50·SmD3 complexes have so far been unsuccessful.
Although we have shown that PRMT5 has promiscuous methyltransferase activity for RG-containing substrates on its own, the theme that has emerged from studies of PRMT5 complexes purified from mammalian cells is that the PRMT5 binding partners are responsible for providing substrate specificity (2). Indeed, histone H3R8 methylation by PRMT5 is associated with the SWI/SNF proteins Brg1-Brm1 and has emerged as a component of regulatory networks governing transcription of some target genes (7, 46–48). In another example, the human transcription factor COPR5 protein has been demonstrated to interact directly with PRMT5 and to mediate nuclear methyltransferase activity on histone H4R3 (49). It is interesting to note that, like pICln, the carboxyl terminus of COPR5 is required for PRMT5 binding, and the carboxyl-terminal 8–10 residues of the two proteins share very similar acidic sequences. In yet another example of a PRMT5-binding protein, the tumor suppressor DAL-1/4.1B has been shown to interact with PRMT5 in promoting methylation of myelin basic protein over that of Sm proteins (50). Interestingly, Mep50 has been observed in several different PRMT5 complexes (10, 48, 51), suggesting that it may play a role in methylation that is distinct from target selection.
The work described here provides biochemical groundwork for understanding how pICln functions in the early steps of snRNP assembly. An important issue that we have not addressed involves the role of phosphorylation in mediating Sm protein methylation and delivery to the SMN complex. pICln is phosphorylated in vivo, and there is strong evidence that at least some of the phosphorylation sites lie in the acidic regions of the protein (22, 52, 53). Because phosphorylation could be used to regulate the binding and/or release of Sm proteins from pICln complexes, it will be important to understand this aspect of pICln biochemistry. These questions along with the role of Mep50 in the PRMT5 complex are the subjects of ongoing investigations.
Supplementary Material
Acknowledgments
We are grateful to Gideon Dreyfuss, Wes Friesen, Jeongsik Yong, and the Van Duyne laboratory members for helpful discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- PRMT
- protein arginine methyltransferase
- snRNP
- small nuclear ribonucleoprotein
- SMN
- survival motor neuron
- PH
- pleckstrin homology
- TEV
- tobacco etch virus
- SAM
- S-adenosylmethionine
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase.
REFERENCES
- 1.Bachand F. (2007) Eukaryot. Cell 6, 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford M. T., Richard S. (2005) Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 4.Gary J. D., Clarke S. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 5.Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) J. Biol. Chem. 276, 32971–32976 [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S. K., Paik W. K., Kim S. (1988) J. Biol. Chem. 263, 19024–19033 [PubMed] [Google Scholar]
- 7.Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. (2004) Mol. Cell. Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001) Mol. Cell. Biol. 21, 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G., Eggert C., Bühler D., Brahms H., Kambach C., Fischer U. (2001) Curr. Biol. 11, 1990–1994 [DOI] [PubMed] [Google Scholar]
- 10.Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 11.Will C. L., Lührmann R. (2001) Curr. Opin. Cell Biol. 13, 290–301 [DOI] [PubMed] [Google Scholar]
- 12.Kambach C., Walke S., Young R., Avis J. M., de la Fortelle E., Raker V. A., Lührmann R., Li J., Nagai K. (1999) Cell 96, 375–387 [DOI] [PubMed] [Google Scholar]
- 13.Raker V. A., Plessel G., Lührmann R. (1996) EMBO J. 15, 2256–2269 [PMC free article] [PubMed] [Google Scholar]
- 14.Pellizzoni L., Yong J., Dreyfuss G. (2002) Science 298, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 15.Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. (2001) Nat. Cell Biol. 3, 945–949 [DOI] [PubMed] [Google Scholar]
- 16.Friesen W. J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. (2001) Mol. Cell 7, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 17.Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. (2001) Nat. Struct. Biol. 8, 27–31 [DOI] [PubMed] [Google Scholar]
- 18.Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001) RNA 7, 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulmichl M., Li Y., Wickman K., Ackerman M., Peralta E., Clapham D. (1992) Nature 356, 238–241 [DOI] [PubMed] [Google Scholar]
- 20.Strange K. (1998) J. Gen. Physiol. 111, 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu W. T., Krapivinsky G. B., Krapivinsky L., Clapham D. E. (1999) Mol. Cell. Biol. 19, 4113–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fürst J., Schedlbauer A., Gandini R., Garavaglia M. L., Saino S., Gschwentner M., Sarg B., Lindner H., Jakab M., Ritter M., Bazzini C., Botta G., Meyer G., Kontaxis G., Tilly B. C., Konrat R., Paulmichl M. (2005) J. Biol. Chem. 280, 31276–31282 [DOI] [PubMed] [Google Scholar]
- 23.Pu W. T., Wickman K., Clapham D. E. (2000) J. Biol. Chem. 275, 12363–12366 [DOI] [PubMed] [Google Scholar]
- 24.Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U. (2005) Genes Dev. 19, 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 26.Pollack B. P., Kotenko S. V., He W., Izotova L. S., Barnoski B. L., Pestka S. (1999) J. Biol. Chem. 274, 31531–31542 [DOI] [PubMed] [Google Scholar]
- 27.Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., Im D. S. (2001) J. Biol. Chem. 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 28.Lim Y., Kwon Y. H., Won N. H., Min B. H., Park I. S., Paik W. K., Kim S. (2005) Biochim. Biophys. Acta 1723, 240–247 [DOI] [PubMed] [Google Scholar]
- 29.Parks T. D., Leuther K. K., Howard E. D., Johnston S. A., Dougherty W. G. (1994) Anal. Biochem. 216, 413–417 [DOI] [PubMed] [Google Scholar]
- 30.Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. (2001) Curr. Biol. 11, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., Zhou L., Cheng X. (2000) EMBO J. 19, 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., Schuck P. (2004) Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 33.Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Einarson M. B., Pugacheva E. N., Orlinick J. R. (2007) Cold Spring Harbor Protocols 2007 pdb.prot4757, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
- 35.Buyse G., de Greef C., Raeymaekers L., Droogmans G., Nilius B., Eggermont J. (1996) Biochem. Biophys. Res. Commun. 218, 822–827 [DOI] [PubMed] [Google Scholar]
- 36.Zaric B., Chami M., Rémigy H., Engel A., Ballmer-Hofer K., Winkler F. K., Kambach C. (2005) J. Biol. Chem. 280, 16066–16075 [DOI] [PubMed] [Google Scholar]
- 37.Raker V. A., Hartmuth K., Kastner B., Lührmann R. (1999) Mol. Cell. Biol. 19, 6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meister G., Fischer U. (2002) EMBO J. 21, 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krapivinsky G. B., Ackerman M. J., Gordon E. A., Krapivinsky L. D., Clapham D. E. (1994) Cell 76, 439–448 [DOI] [PubMed] [Google Scholar]
- 40.Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. (2003) Mol. Cell. Biol. 23, 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss V. H., McBride A. E., Soriano M. A., Filman D. J., Silver P. A., Hogle J. M. (2000) Nat. Struct. Biol. 7, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 42.McBride A. E., Weiss V. H., Kim H. K., Hogle J. M., Silver P. A. (2000) J. Biol. Chem. 275, 3128–3136 [DOI] [PubMed] [Google Scholar]
- 43.Lee J. H., Cook J. R., Yang Z. H., Mirochnitchenko O., Gunderson S. I., Felix A. M., Herth N., Hoffmann R., Pestka S. (2005) J. Biol. Chem. 280, 3656–3664 [DOI] [PubMed] [Google Scholar]
- 44.Krapivinsky G., Pu W., Wickman K., Krapivinsky L., Clapham D. E. (1998) J. Biol. Chem. 273, 10811–10814 [DOI] [PubMed] [Google Scholar]
- 45.Emma F., Sanchez-Olea R., Strange K. (1998) Biochim. Biophys. Acta 1404, 321–328 [DOI] [PubMed] [Google Scholar]
- 46.Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. (2007) Mol. Cell. Biol. 27, 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J. H., Negre V., Rousset M., Pestka S., Le Cam A., Sardet C. (2002) EMBO Rep. 3, 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Guezennec X., Vermeulen M., Brinkman A. B., Hoeijmakers W. A., Cohen A., Lasonder E., Stunnenberg H. G. (2006) Mol. Cell. Biol. 26, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacroix M., Messaoudi S. E., Rodier G., Le Cam A., Sardet C., Fabbrizio E. (2008) EMBO Rep. 9, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang W., Roemer M. E., Newsham I. F. (2005) Biochem. Biophys. Res. Commun. 329, 522–530 [DOI] [PubMed] [Google Scholar]
- 51.Gonsalvez G. B., Rajendra T. K., Tian L., Matera A. G. (2006) Curr. Biol. 16, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 52.Grimmler M., Bauer L., Nousiainen M., Körner R., Meister G., Fischer U. (2005) EMBO Rep. 6, 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-Olea R., Emma F., Coghlan M., Strange K. (1998) Biochim. Biophys. Acta 1381, 49–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.