Abstract
In Pseudomonas putida, genes for the glucose phosphorylative pathway and the Entner-Doudoroff pathway are organized in two operons; one made up of the zwf, pgl, and eda genes and another consisting of the edd, glk, gltR2, and gltS genes. Divergently with respect to the edd gene is the gap-1 gene. Expression from Pzwf, Pedd, and Pgap is modulated by HexR in response to the availability of glucose in the medium. To study the regulatory process in greater detail we purified HexR and showed that it is a monomer in solution. Electrophoretic mobility shift assays and isothermal titration calorimetry assays were done showing that HexR recognizes the Pedd, Pzwf, and Pgap-1 promoters with affinity in the nanomolar range. DNA footprinting assays identified the binding site between +30 and +1 at Pzwf, between +16 and +41 at Pedd, and between −6 and +18 at Pgap-1. Based on DNA sequence alignment of the target sites and isothermal titration calorimetry data, two monomers of HexR bind to a pseudopalindrome with a consensus sequence of 5′-TTGTN7–8ACAA-3′. Binding of the Entner-Doudoroff pathway intermediate 2-keto-3-deoxy-6-phosphogluconate to HexR released the repressor from its target operators, whereas other chemicals such as glucose, glucose 6-phosphate, and 6-phosphogluconate did not induce complex dissociation. The phosphorylated effector is likely to be recognized by a sugar isomerase domain located at the C-terminal end of HexR, whereas the helix-turn-helix DNA binding domain of HexR exhibits high similarity to proteins of the RpiR family of regulators.
Bacteria of the genus Pseudomonas are ubiquitous inhabitants of soil, water, plant surfaces, and animal tissues. The complete genome sequences of a number of Pseudomonas species and different strains have been deciphered, and their analysis has revealed that Pseudomonas exhibits a limited ability to metabolize sugars; nonetheless, the genomes of all Pseudomonas strains sequenced to date show that they possess the necessary genetic information to metabolize glucose (1–8). In fact, glucose metabolism in Pseudomonas is biochemically rich, as up to three convergent pathways that transform the sugar into 6-phosphogluconate have been described. Subsequently, 6-phosphogluconate is metabolized by Entner-Doudoroff enzymes into central metabolites (9–13).
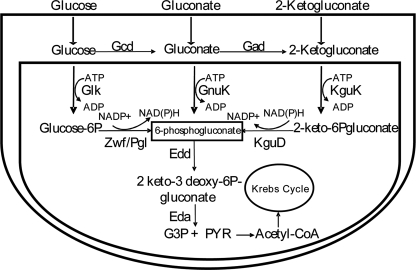
Glucose metabolism is compartmentalized in Pseudomonas in the sense that once glucose passes the outer membrane through the OprB porin and reaches the periplasm (14–16), it can be transported to the cytoplasm, or it can be oxidized by the action of the periplasmic glucose dehydrogenase to yield gluconate, which by the action of gluconate dehydrogenase is transformed into 2-ketogluconate. Gluconate and 2-ketogluconate may also be transported to the cytoplasm through a process mediated by the GnuK and KguP transporters, respectively. In the cytoplasm gluconate is directly phosphorylated to 6-phosphogluconate, whereas two reactions mediated by KguK and KguD are needed to convert 2-ketogluconate into 6-phosphogluconate (Fig. 1). As mentioned above, glucose itself can be transported by an ABC transport system to the cytoplasm (GtsABCD) and then phosphorylated by glucokinase (Glk) and transformed to 6-phosphogluconate by the combined action of glucose 6-phosphate dehydrogenase (Zwf) and 6-phosphogluconolactonase (Pgl). The three pathways co-exist and function simultaneously in Pseudomonas putida KT2440 (11, 12) but not in Pseudomonas aeruginosa (17).
FIGURE 1.
Glucose catabolic pathways in P. putida. The figure is modified from that in del Castillo et al. (11, 13) and del Castillo and Ramos (12) Glk, glucokinase; Zwf, glucose 6-P dehydrogenase; Pgl, 6-phosphogluconolactonase; Edd, 6-phosphogluconate dehydratase; Gcd, glucose dehydrogenase; Gad, gluconate oxidase; KguK, 2-ketogluconate kinase; KguD, 2-ketogluconate-6-phosphate dehydrogenase; G3P, glyceraldehyde 3-phosphate; Pyr, pyruvate; GnuK, gluconate kinase.
The convergent product of glucose metabolism, 6-phosphogluconate, then enters the Entner-Doudoroff pathway, in which it is first converted into 2-keto-3-deoxy-6-phosphogluconate (KDPG)2 by the Edd enzyme (6-phosphogluconate dehydratase) and then hydrolized to produce glyceraldehyde 3-phosphate and pyruvate by action of the Eda enzyme (2-keto-3-deoxy-6-phosphogluconate aldolase). Glyceraldehyde 3-phosphate is further metabolized by the GAP-1 enzyme, whereas pyruvate is decarboxylated to acetyl-CoA and enters the Krebs cycle.
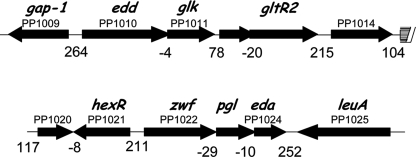
In P. putida KT2440 the zwf, pgl, and eda genes form an operon that encodes, respectively, glucose-6-phosphate dehydrogenase, 6-phosphogluconolactonase (two enzymes of the glucose phosphorylative pathway, and Eda (an enzyme of the Entner-Doudoroff pathway). The edd and glk genes form another operon that encodes, respectively, 6-phosphogluconate dehydratase (the first enzyme of the Entner-Doudoroff pathway) and glucokinase (an enzyme of the glucose phosphorylative pathway) (18–22). Also part of this operon are the gltR2/gltS genes whose gene products are involved in positive transcriptional control of the glucose transport system. Transcribed in the opposite direction to the zwf promoter is an open reading frame (PP1021) that encodes the HexR repressor (Fig. 2), whereas the gap-1 gene is divergently transcribed with respect to the edd promoter.
FIGURE 2.
Organization of the zwf, pgl, eda, edd, and glk genes. Genes zwf/pgl/eda form an operon that is transcribed divergently from hexR gene (12). The genes edd/glk/gltR2/gltS form another operon that is transcribed divergently from the gap-1 gene. The arrows indicate the direction of transcription, and the number underneath indicates the intergenic space.
The two described operons and the gap-1 gene are all induced in cells growing with glucose, gluconate, and 2-ketogluconate, whereas the level of expression of hexR does not vary significantly regardless of the carbon source used for growth (13, 23). It should be noted that due to the physical and transcriptional organization of the two operons that transcribe glucokinase pathway genes (glk, zwf, and pgl), the regulator of the glucose transport system (gltR2), and the Entner-Doudoroff pathway genes (edd, eda), glucokinase pathway genes are co-transcribed with Entner-Doudoroff pathway enzymes. As a result, the glucokinase pathway is induced when bacteria are exposed to gluconate and 2-ketogluconate, whose peripheral pathways are not related to the glucokinase pathway. Another relevant feature of glucose metabolism in Pseudomonas is that the glucose-6-phosphate dehydrogenase enzyme, which is encoded by the zwf-1 gene, is involved in responses to oxidative stress. It has been proposed that the zwf, pgl, eda operon is induced in response to oxygen stress, and Kim et al. (24) have suggested that superoxide-generating chemicals such as menadione and cumene hydroperoxide could act as effectors of HexR.
In this study we report that purified HexR protein is a monomer in solution and that it binds specifically to the promoter regions of the zwf, edd, and gap-1 genes. Two monomers of HexR likely recognize the target operator at a palindromic sequence (5′-TTGTN7–8ACAA-3′). HexR has an HTH DNA binding domain at its N terminus end and a SIS domain at its C terminus, which binds to the Entner-Doudoroff pathway intermediate KDPG. Our results show that the in vitro binding of KDPG to HexR causes its dissociation from target operators.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids Used in This Study
The genotype or the relevant characteristics of the bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown in LB medium or in modified M9 minimal medium with glucose (16 mm) or citrate as the sole C-source (25). When required, antibiotics were added to the culture medium to reach a final concentration of 25 μg/ml kanamycin, 20 μg/ml rifampicin, 50 μg/ml ampicillin, and 30 μg/ml chloramphenicol.
TABLE 1.
Strains and plasmids used in this study
Cmr, Kmr, Rifr, and Apr stand for resistance to chloramphenicol, kanamycin, rifampicin, and ampicillin, respectively.
| Strains or plasmid | Genotype of relevant characteristics | References |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2440 | Wild type, prototroph, Cmr, Rifr | This laboratory |
| M1044a | edd:mini-Tn5-Km; Kmr, Rifr | 34 |
| M1128a | eda:mini-Tn5-Km; Kmr, Rifr | 34 |
| E. coli | ||
| DH5αF′ | F'/hsdR17, recA1, gyrA | 46 |
| BL21 (DE3) | F−, ompI, hsdSB(r−Bm−B)gal, dam, met | 47 |
| Plasmids | ||
| pB-ZWF | Bzwf, TcR, pMP220 bearing a Pzwf ::lacZ fusion | 13 |
| pBEDD | TcR, pMP220 bearing a Pedd ::lacZ fusion | 13 |
| pGAP | TcR, pMP220 bearing a Pgap-1 ::lacZ fusion | 13 |
| pMBLP-ZWF | TcR, pMBL containing zwf promoter | This work |
| pMBL | Vector for cloning fragments, Apr | Dominion |
| pET24b(+) | Kmr, protein expression vector | Novagen |
| pMBLATG | hexR gene in pMBL vector | This work |
| pET24b:HexR | pET24b-containing hexR gene | This work |
| pMBLP-EDD | pMBL-containing edd promoter | This work |
a Collection of KT2440 mutants available at the Consejo Superior de Investigaciones Científicas, Granada, Spain.
Synthesis of KDPG
P. putida eda mutant (Table 1) was grown overnight with shaking at 30 °C in M9 with citrate (5 mm) as a carbon source in the presence of 5 mm glucose. An overnight culture was harvested by centrifugation at 14,000 × g for 10 min. The cell pellet was resuspended, and cell-free extracts were prepared by sonication as previously described (26–28). Subsequently, 6-phosphogluconate (0.5 mm) was added to the whole cell-free extract and transformed into KDPG by incubating for 10 h at 30 °C, which resulted in almost 98% transformation of 6-phosphogluconate. The reaction was stopped by centrifugation of the sample at 14,000 × g for 10 min, and the supernatant was then frozen at −20 °C.
Cloning of hexR and Overproduction in Escherichia coli
The P. putida hexR gene was amplified by PCR using genomic DNA of the KT2440 strain as a template, and the following primers were used: HexRf 5′-GCTAGCATGGACCGCGTGCGAA-3′ (forward) and HexRr 5′-CTCGAGTTGAGGTCGTCGTCCTCGA-3′ (reverse). Upon amplification under standard conditions, the fragment was cloned into the pMBL vector to yield pMBL::hexR. This plasmid was subsequently digested with NheI/XhoI, and the 882-bp fragment bearing hexR was cloned into pET24b(+) digested with the same enzymes. The resulting plasmid, pET24b:HexR, was used to produce HexR protein with a 6 × histidine tag at its C-terminal end. To this end E. coli BL21 (DE3) (pET24b:HexR) was grown in 2-liter conical flasks with 250 ml of LB supplemented with 25 μg/ml kanamycin. Cultures were incubated at 30 °C with shaking until they reached turbidity at 660 nm (A660) of 0.6, at which point 0.5 mm isopropyl-β-d-thiogalactopyranoside was added to induce the expression of the hexR gene. The cultures were then incubated at 18 °C overnight, and cells were harvested by centrifugation (30 min at 20,000 × g) and stored at −80 °C until used for protein purification.
For protein purification, cells were resuspended in 25 ml of buffer A (50 mm Tris-HCl, pH 7.9, 300 mm NaCl, 1 mm dithiothreitol, 10 mm imidazole) supplemented with a tablet of CompleteTM EDTA-free protease inhibitor mixture (Roche Applied Science). Cells were lysed by two passes through a French press at a pressure of 1000 p.s.i. The cell suspension was then centrifuged at 20,000 × g for 1 h. The pellet was discarded, and the supernatant was filtered and loaded onto a 5-ml His-Trap chelating column (GE Healthcare) previously equilibrated with buffer A. His6-HexR was eluted with a 10–500 mm gradient of imidazole in buffer A. The purity of the eluate was determined by using 12% SDS-PAGE gels. Homogeneous protein preparations were dialyzed overnight against buffer B (50 mm Hepes, pH 7.9, 300 mm NaCl, 1 mm dithiothreitol, and 10% (v/v) glycerol). Dialyzed protein at 1.9 mg/ml was separated into 1-ml aliquots and stored at −80 °C.
Analytical Gel Filtration Chromatography
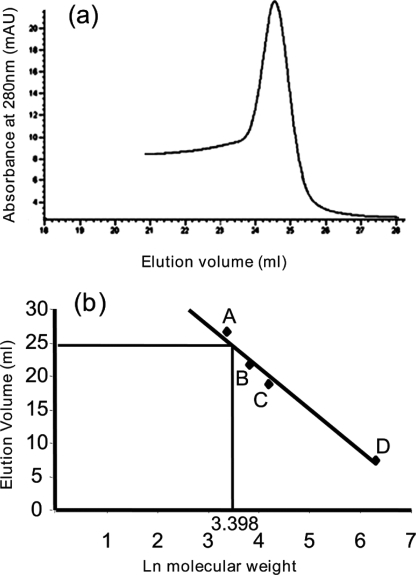
To determine the oligomeric state of HexR in solution, we used analytical gel filtration chromatography using an Åkta fast protein liquid chromatography system (Amersham Biosciences). Purified HexR (32 μm) was loaded onto a Superdex-200 10/300GL column (Amersham Biosciences) that was equilibrated in buffer B. HexR was eluted at a constant flow rate of 0.7 ml/min, and the absorbance of the eluate was monitored at 280 nm. The molecular mass of HexR was estimated from a plot of the elution volume against Ln of the molecular weight of standard calibration proteins, namely, carbonic anhydrase (29 kDa), albumin from chicken egg white (45 kDa), albumin from bovine serum (66 kDa), and urease (545 kDa) (Sigma). HexR was identified by Western blot using a specific anti-His-tag antibody (Fig. 3).
FIGURE 3.
The oligomeric state of HexR was determined by analytical gel filtration. a, elution chromatogram of 32 μm purified HexR. b, calibration curve using protein standards: carbonic anhydrase (A; 29 kDa) and chicken egg albumin (B; 45 kDa), bovine serum albumin (C; 132 kDa), and urease (D; 545 kDa.). The ln of the molecular weight of the protein was plotted versus the corresponding elution volumes, which were fitted by linear regression. The molecular weight of HexR was calculated by extrapolating from the elution volume.
Electrophoresis Mobility Shift Assay
DNA fragments bearing the zwf (280 bp) and edd/gap-1 (340 bp) promoters were generated by PCR amplification from pMBLP-ZWF or pMBLP-EDD using appropriate primers pairs, namely, RHexR2, 5′-GATCTGTTCCAGGAGGTTT-3′, and ZWF1, 5′-GCCAAACAGGGCAAAGGTGC-3′ for the zwf operator and Bedd1, 5′-AGGTCCTGGCGGTAGCCTTG-3, and Bedd2, 5′-GGCTAATTGTAAGGGCGGTT-3′ for the edd/gap-1 operator. Amplification conditions were 90 °C for 5 min followed by 45 cycles at 66 to 60 °C for 1 min and 72 °C for 1 min. Amplified DNA was isolated from agarose gels and end-labeled with deoxy-[γ-32P]ATP using the T4 polynucleotide kinase. A 10-μl sample containing about 2 nm labeled DNA (1.5 × 104 cpm) was incubated with increasing concentrations of purified HexR for 1 h in 10 μl of binding buffer (50 mm Tris-HCl, pH 7.5, 10 mm NaCl, 0.5 m magnesium acetate, 0.1 mm EDTA, 1 mm dithiothreitol, 5% (v/v) glycerol) containing 20 μg/ml of polyd(IC) and 200 μg/ml of bovine serum albumin. The DNA-protein complexes were resolved by electrophoresis in 4% (w/v) nondenaturing polyacrylamide gels in 1 × Tris borate-EDTA using Bio-Rad electrophoresis equipment as described before (29, 30).
DNase I Footprinting
The zwf (280 bp) and edd/gap-1 (340 bp) DNA fragments containing, respectively, the Pzwf and Pgap-1/Pedd promoters, were amplified using the oligonucleotides outlined above. DNA was labeled with deoxy-[γ-32P]ATP. Ten-μl samples containing 2 nm concentrations of probe were mixed with different amounts of HexR (0.1–3 μm) in binding buffer for the formation of the DNA-HexR complex. Samples were incubated for 1 h at 30 °C, and then DNase I (0.4 U; Roche Applied Science) was added to the complexes for 5 min, at which point the reaction was stopped by adding 2 μl of 500 mm EDTA solution. DNA from the footprinting mixtures was phenol-chloroform-extracted, ethanol-precipitated, and dissolved in 12 μl of sequence loading buffer. After 5 min of denaturation at 95 °C, DNA was loaded onto a 6.5% (w/v) DNA-sequencing gel. Appropriate sequencing reactions were loaded onto the gels along with the footprinting samples and used as a size ladder for identification of the sequences of protected sites.
Isothermal Titration Calorimetry
Microcalorimetric experiments were carried out at 25 °C using a VP-microcalorimeter (Microcal, Amherst, MA). Protein and DNA samples were dialyzed into the following buffer: 50 mm Hepes, pH 7.9, 300 mm NaCl, 1 mm dithiothreitol, 10% (v/v) glycerol. For DNA binding studies, oligonucleotides corresponding to both strands of the HexR binding site at the zwf promoter (5′-TTGTCTGTAACACTTGTGTGTAATGTTGTGGTTTTTACTACATTATCCC-3′) were synthesized. Annealing was carried out by mixing 200 μm concentrations of each of the complementary oligonucleotides in 50 mm phosphate buffer, pH 7.0, 0.5 m EDTA, 2.5 m NaCl. The mixture was incubated at 90 °C for 30 min, and then the sample was allowed to cool down to 46 °C to prevent the formation of secondary structures (note that because of secondary structure formation with the chosen oligos for Pedd, it was impossible to achieve the appropriate double-stranded pairs). Typically, reverse titrations (DNA into HexR) involved the injection of aliquots of 17–31.5 μm DNA into 3 μm HexR. All data were corrected using the heat changes arising from injection of the ligand from the syringe into the buffer. The titration data were analyzed using the “one-binding site model” of the MicroCal version of ORIGIN. Titration curves were fitted by a nonlinear least squares method to a function for the binding of a ligand to a macromolecule. The parameters ΔH (reaction enthalpy), KA (binding constant, KA = 1/KD), and n (reaction stoichiometry) were determined from the curve fit. The change in free energy (ΔG) and in entropy (ΔS) was calculated from the values of KA and ΔH with the equation ΔG = −RT ln KA = ΔH-TΔS, where R is the universal molar gas constant, and T is the absolute temperature.
RESULTS
HexR Belongs to the RpiR Family of Transcriptional Regulators
BLAST analysis of the protein sequence of HexR suggests that this 290-residue regulator is very well conserved in all sequenced Pseudomonas genomes exhibiting a high degree of sequence similarity (>90%) and sequence identity (>78%) (supplementalFig. 1). Furthermore, HexR of P. putida exhibits high similarity to members of the RpiR family of transcriptional regulators, which can be repressors or activators. The archetypal regulator of the family is RpiR, which regulates ripB gene expression and whose product, RipB, interconverts ribulose 5-phosphate and ribose 5-phosphate (31). Members of this family possess a helix-turn-helix at the N terminus of the protein and contain a C-terminal SIS domain found in many phosphosugar isomerases and phosphosugar-binding proteins, although at present there is no evidence for catalytic activity of the SIS domain when linked to a HTH DNA binding domain in the same polypeptide (32, 33).
Analysis of HexR with PFAM confirmed that this regulator has two domains, one stretching from residues 20–107 (PFAM 01418) that is predicted to be an HTH DNA binding domain of the RpiR family of regulators (supplementalFig. 1) and another domain involving residues 127–256 (PFAM 01380) that is predicted to be a SIS domain.
HexR Is a Monomer in Solution
With the aim of clarifying the mechanism of action of HexR, we cloned the hexR gene in pET24b and purified hexahistidine-tagged HexR protein from the soluble fraction of E. coli lysates. Upon purification using nickel-trap columns, an apparent homogeneous 30-kDa HexR preparation was obtained with a yield of almost 2.0 mg/liter. To determine the oligomeric state of HexR in solution, the HexR protein was subjected to gel filtration analysis using a Superdex-200 10/300GL column, as described under “Experimental Procedures.” The results indicated that the HexR eluted as a single and symmetric peak (Fig. 3a), and when the elution volume of HexR was extrapolated into the corresponding calibration curve (Fig. 3b) a molecular mass of 29.9 kDa was derived, suggesting that the HexR protein is a monomer in solution.
DNA-HexR Complex Formation and Identification of HexR Binding Sites in the Pzwf, Pgap-1, and Pedd Promoter
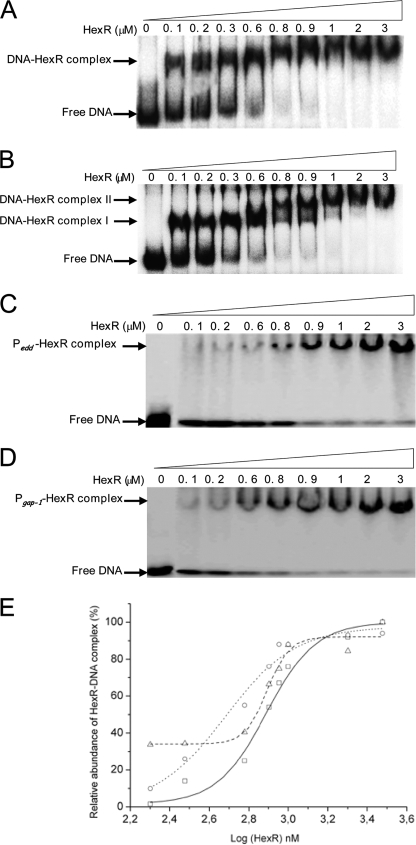
Our previous genetic analysis suggested that HexR modulates expression of the zwf/pgl/edd and eda/glk/gltR2/gltS operons (11, 12, 34) because in a hexR mutant background expression from these promoters was higher than in the absence of glucose. To study the potential interactions of HexR with its target operators in greater detail, we confirmed the previously established transcription start point (tsp) for zwf and mapped the tsp of the edd/gap divergent region. Our primer extension analysis revealed a single tsp for gap-1 and edd (Fig. 4A), and based on the size of the cDNA it appears that their divergent tsp are separated by 28 nucleotides, so that the −10/−35 regions of both promoters overlap (Fig. 4B). Next we tested the ability of HexR to bind to zwf and gap-1/edd promoters using electrophoretic mobility shift assays (EMSA), as described under “Experimental Procedures.” In these assays increasing concentrations of HexR protein (0.1 at 3 μm) were added to 2 nm deoxy-[γ-32P]ATP-labeled DNA (1.5 × 104 cpm) of either a 280-bp fragment bearing the Pzwf promoter or a 340-bp fragment containing the Pedd/Pgap-1 promoters. We found that HexR concentrations as low as 100 nm retarded both the Pzwf and the Pedd/gap-1 DNA fragment (Fig. 5A). Although with Pzwf a major complex was seen in the EMSA, two retard bands were observed with the 340-bp fragment bearing the Pedd/Pgap-1 promoters (Fig. 5A).
FIGURE 4.
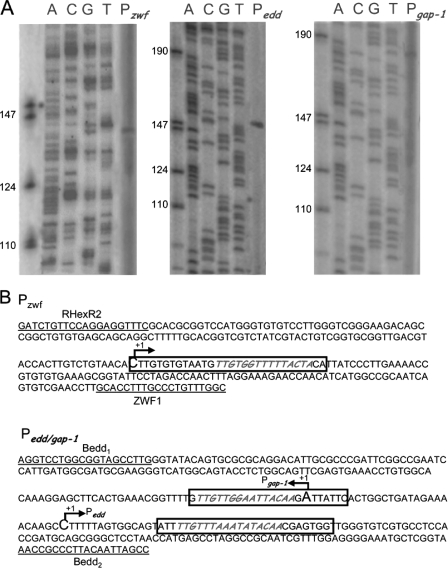
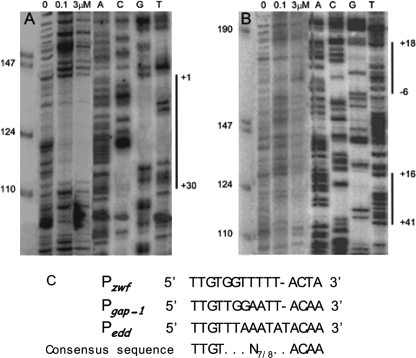
Identification of transcription initiation point in the P zwf, Pedd, and P gap-1 promoters. A, transcription mapping. P. putida cells were grown on M9 minimal medium with glucose, and RNA was extracted as described by Marqués et al. (48). Primer used for extension reactions are given under “Experimental Procedures.” Lanes A, C, G, and T are sequencing ladders. The left lanes show single-stranded DNA markers of the indicated size. B, sequence of the mapped promoters. The tsp is indicated by a letter in larger size and marked +1. Boxed DNA corresponds to protected sequences in footprint (see Fig. 6). Underneath sequences correspond to oligoprimers used for PCR amplification.
FIGURE 5.
Electrophoresis mobility shift assay of the promoter region of P zwf and Pedd/gap-1 by purified HexR. A, the DNA binding assays was done using purified HexR with 2 nm concentrations of the product of PCR (280 bp) of Pzwf end-labeled with 32P and incubated with the concentration of purified HexR indicated at the top of each lane. The position of the free DNA is indicated. B, as in A, but a 340-bp Pedd/gap-1 fragment was used. Complex I and Complex II are indicated. C and D, as in A, but a 280-bp DNA fragment containing the HexR binding site at Pgap-1 (D) or Pedd (C) were used. E, the densitometric analysis of the EMSA gels. Squares, Pzwf promoter fragments; circles, Pedd promoter fragments; triangles, and Pgap-1 promoter fragments.
To identify the HexR binding site(s) in Pzwf, Pgap-1, and Pedd, DNase I footprinting was carried out using 5′ [γ-32P]radiolabeled fragments of 280 bp for the Pzwf promoter and of 340 bp for the Pgap-1/Pedd promoters. DNA was incubated with either 0.1 or 3 μm HexR protein. As shown in Fig. 6A, a protected region was found between base pairs +1 and +30 (5′-CTTGTGTGTAATGTTGTGGTTTTTACTACA-3′) for the Pzwf promoter (Figs. 4 and 6A). Fig. 6B shows two footprints; one from +16 to +41 (5′-ATTTTGTTTAAATATACAACGAGTGG-3′) for the Pedd promoter and another from −6 to +18 at the Pgap-1 promoter (5′-GTTGTTGGAATTACAAGATTATTC-3′. Sequence alignment of the three regions protected by HexR revealed a high degree of identity (65.4%) and a common inverted repeat consensus sequence (5′-TTGTN7/8ACAA-3′), which most likely represents the specific HexR binding motif (Fig. 6C).
FIGURE 6.
DNase I footprinting of promoters regulated by HexR. Assays were done with Pzwf DNA operator (A) and of edd/gap-1 promoter (B). Lane 1 contains a control without HexR, lanes 2 and 3 contain DNA incubated with 0.1 and 3 μm concentrations of purified HexR, and lanes 4-7 show the A C G T sequencing ladder. Vertical lines indicate the target footprinting site. C, alignment of the sequences corresponding to the protected area in the different HexR-regulated promoters and the proposed pseudopalindrome recognized by HexR as consensus sequence.
To estimate the apparent affinity of HexR for the Pzwf promoter, densitometric analysis of three different gels (see Fig. 5, A–D for one gel) was carried out, and the relative amounts of observed HexR-DNA complex were plotted against the logarithm of HexR concentration (Fig. 5E). These data were fitted using the sigmoidal fitting tool of ORIGIN (Fig. 5E). The corresponding apparent KD value was 780 ± 40 nm. To determine the apparent affinity of HexR for the Pedd and Pgap-1 promoter, 280-bp DNA fragments were generated so that only one of the sites identified in the footprint was present. Similar EMSA analyzes as those reported above were carried out (Fig. 5, C and D). In these cases a single retard band was found, in agreement with the presence of a single recognition site for HexR. Based on densitometric analysis (Fig. 5E), apparent affinity of 774 ± 80 nm for edd and 480 ± 50 nm for the gap-1 promoter was estimated. Therefore, HexR exhibits slightly higher affinity for the Pgap-1 promoter than for the Pzwf and Pedd promoters.
To estimate binding stoichiometry, isothermal titration calorimetry assays with the Pzwf binding site and HexR were performed as described under “Experimental Procedures.” Binding stoichiometry was two molecules of HexR per binding site, suggesting that HexR, although a monomer in solution, binds as a dimer (supplemental Fig. 2).
Identification of KDPG as an Effector for HexR
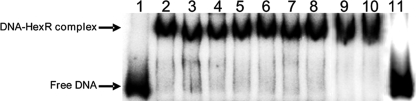
The edd and eda genes must be induced to metabolize glucose, fructose, gluconate, or 2-ketogluconate (Fig. 1). Given the physical organization of the genes for the glucokinase branch and the Entner-Doudoroff pathway (Fig. 2), either HexR recognizes a wide range of sugars or one or more common pathway intermediates, i.e. 6-phosphogluconate or KDPG, could be the true effector of HexR. The above EMSA showed that with 2 nm Pedd/gap-1 promoter and 3 μm HexR 100% of the DNA was retarded. These assays have also been done by incubating for 30 min before running the gel DNA-operator complex with 400 μm concentrations of each of the following compounds: glucose, fructose, gluconate, 2-ketogluconate, glucose 6-phosphate, gluconate 6-phosphate, KDPG, glyceraldehyde 3-phosphate, pyruvate, or acetyl-CoA. Subsequently, samples were electrophoresed. We found that only KDPG was able to release HexR from the HexR-DNA complex (Fig. 7). As such, it appears that KDPG is the specific HexR effector and the chemical responsible of the induction of the genes necessary for glucose metabolism.
FIGURE 7.
EMSA of Pedd/gap-1 with HexR using different effectors. A, mixture of 3 μm concentrations of purified His-tag HexR and DNA promoter and 400 μm concentrations of different effectors, respectively. Lane 1, free DNA; lane 2, purified HexR incubate with 2 nm DNA. Lanes 3-11 are as lane 2, except that 400 μm concentrations of the following chemicals was added: glucose, fructose, gluconate, 2-ketogluconate, glucose 6-phosphate, 6-phosphogluconate, acetyl-CoA, pyruvate, and KDPG.
To corroborate the set of in vitro data, we decided to perform in vivo experiments in which we measured expression from Pedd, Pzwf, and Pgap-1 in wild-type cells and eda mutants growing in M9 minimal medium with citrate in the absence and in the presence of glucose. We found that in the wild type the basal level of expression from these three promoters increased about 2–3-fold in the presence of glucose. In the Δeda background expression levels in the absence of glucose were relatively high and increased again 2–3-fold when glucose was added to the culture medium, in agreement with accumulation of KDPG (Table 2).
TABLE 2.
Expression from Pzwf, Pedd, and Pgap promoters in the wild-type strain and the eda mutant
The indicated host was transformed with a pMP220 derivative (TcR) bearing the indicated fusion to ′lacZ. Cells were grown on M9 minimal medium with citrate (15 mm), overnight cultures were diluted 50-fold in the same medium in the absence or in the presence of glucose (5 mm), and β-galactosidase activity (Miller units) was determined when culture cells had reached a cell density of about 0.7. Data are the average of three independent assays, done in duplicate. ND, not determined.
| Host | Promoter | −Glucose | +Glucose |
|---|---|---|---|
| Wildtype | Pzwf ::lacZ | 500 ± 40 | 1500 ± 100 |
| Wild type | Pedd:lacZ | 100 ± 10 | 200 ± 15 |
| Wild type | Pgap-1:lacZ | 200 ± 25 | 280 ± 10 |
| Δeda | Pzwf ::lacZ | 3700 ± 50 | 10000 ± 1000 |
| Δeda | Pedd:lacZ | 500 ± 30 | 1200 ± 150 |
| Δeda | Pgap-1:lacZ | ND | ND |
DISCUSSION
del Castillo et al. (11, 13) showed that all sequenced Pseudomonas sp. genomes exhibit the same physical organization of the zwf/edd/pgl and eda/glk operons, with the hexR gene being transcribed divergently with respect to the edd promoter. In addition, we show here that all Pseudomonas HexR proteins exhibit highly conserved domains and motifs. This suggests that information derived from studies in P. putida KT2440 can be relevant to understanding the regulation of glucose metabolism in other species of the genus Pseudomonas.
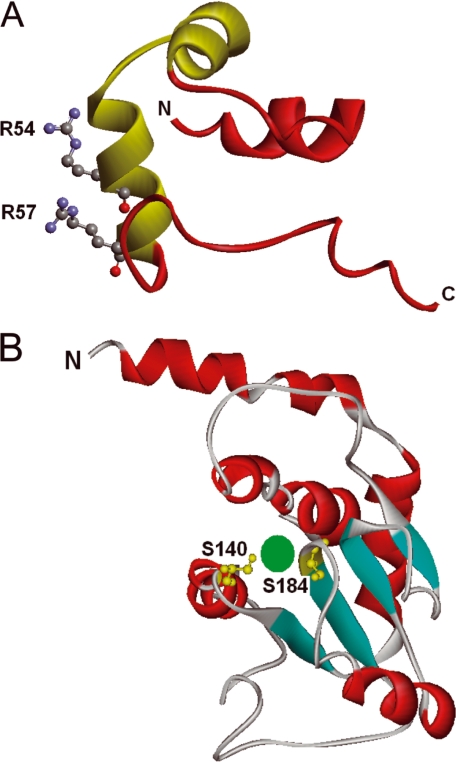
HexR belongs to the RpiR family, and its members are often involved in sugar catabolism control in proteobacteria. RpiR members can be repressors such as RpiR in E. coli (31) and HexR in P. putida or transcriptional activators such as GlvR in Bacillus subtilis that modulates maltose metabolism (35). Regardless of its role as a repressor or activator, the multialignment of HexR with other members of the RpiR family revealed two domains (the HTH binding domain from residues 20 to 66 and the SIS domain from residues 127 to 256) and four blocks of high sequence conservation. These four conserved blocks correspond to residues between 23 and 66, 132 and 153, 183 and 195, and 238 and 250 in HexR of P. putida. Within the first set of residues a potential HTH DNA binding motif was identified, which in turn allowed us to identify the segment of the N terminus of HexR that is likely to directly interact with DNA. The N-terminal region of HexR (PF01418) is 36% identical to the N-terminal region of the putative transcriptional regulator YbbH from B. subtilis, whose three-dimensional structure is known (PDB code 2O3F). We then generated a three-dimensional model of the DNA binding domain of HexR based on the YbbH structure. The analysis of this model shows that the amino acids that appear to be more important for the recognition of DNA are Gln-43, Lys-46, Glu-49, Arg-54, and Arg-57 (Fig. 8A), and the amino acids that may directly interact with DNA are Arg-54 and Arg-57; that is, residues that are particularly well conserved in all of the HexR sequences (supplementalFig. 1). As mentioned above, the C-terminal region of HexR exhibits high homology to SIS domains corresponding to signature PF01380 (36). The C-terminal domain of HexR exhibits 27% identity to the phosphoheptose isomerase of E. coli, whose three-dimensional structure in complex with its substrate sedoheptulose phosphate has been resolved (36). This structure has served as template for the generation of a homology model of the HexR SIS domain (Fig. 8B). Based on an alignment of both sequences and the information obtained from the residues involved in effector recognition in E. coli structure, we suggest that the HexR residues corresponding to serines 140 and 184, which are within two of the conserved blocks of residues, are likely to be involved in effector recognition.
FIGURE 8.
T hree- dimensional homology models of the N-terminal and C-terminal domains of HexR. A, DNA binding domain; the HTH motif is highlighted in green. Amino acids Arg-54 and -57 are likely to interact with the DNA. B, SIS domain, amino acids Ser-140 and -184 are likely to interact with bound effectors, which is schematically depicted in green.
Recognition of KDPG
The HexR SIS domain seems to recognize a phosphorylated molecule, KDPG, as an effector. This indicates that control of the Entner-Doudoroff pathway enzymes, needed for the catabolism of sugars, is modulated by a pathway intermediate rather than initial pathway substrates. In addition, it should be noted that KDPG plays a role as signaling molecule in catabolite repression. Velázquez et al. (37) showed that inactivation of eda gene and the subsequent accumulation of KDPG resulted in catabolite repression of the toluene degradation pathway, whereas this effect is not seen in an edd mutant background in which 6-phosphogluconate is accumulated (11, 12). Furthermore, it has been shown that KDPG is essential to this cross catabolite regulation between the phosphorylative pathway for glucose metabolism and the toluene pathway, so that a fine balance is maintained between the two different carbon sources (11–13). The key position of KDPG is also evident in the Δeda mutant in cells growing in the absence of glucose. As shown in Table 2, citrate-growing cells exhibit high levels of expression from HexR-regulated promoters, probably because of the metabolism of triose phosphate to glucose-6-phosphate and its subsequent metabolism to 6-phosphogluconate. This induction of zwf and other promoters is in agreement with the observation that eda mutants in P. aeruginosa are more resistant to oxidative stress responses, a process in which the zwf gene product, glucose-6-phosphate-ehydrogenase, is involved (24, 38).
The organization of a potential catalytic domain and a DNA binding domain, as seen in HexR, is unusual in regulatory proteins but has been described before for regulators of biosynthesis of amino acids (i.e. Ref. 39). No evidence is available on the catalytic activity of members of the Rip family at present. It is possible that SIS effector binding domain functions as an allosteric pocket that influences the three-dimensional conformation of the HTH DNA binding domain. This type of domain interconnection has been well established for TetR family of repressors in that an α-helix connects the DNA binding domain and the effector pocket so that occupation or not of the pocket influences the angle of the recognition helix in the DNA binding domain (40–43). This in turn dramatically influences the activity of the regulator so that it can bind to target DNA sequences in the absence of effectors rather than in their presence (40–44).
Analysis of DNA Sequences Reveal That HexR May Block Progression of RNA Polymerase
Foot-print analysis revealed that P. putida HexR recognizes an inverted pseudopalindromic sequence, 5′-TTGTN7–8ACAA-3′ that lies between −6 and +18 in Pgap-1, between +16 and +41 in Pedd, and between +1 and +30 in Pzwf, suggesting that regulation of transcription from Pedd, Pgap-1, and Pzwf may involve blockage of RNA polymerase progression. Further in vitro assays are required to determine whether the different locations of the HexR binding sites relative to the tsp in Pedd, Pgap-1, and Pzwf influence repression strength; using EMSA we found that HexR binds with a slightly higher affinity (2-fold) to the Pgap-1 promoter than to the other two promoters. These differences in binding affinity might be because of local differences in DNA sequence or structure, as has been observed for other repressors, e.g. the TtgV repressor binds more tightly to the PttgD promoter than PttgG because of local differences in DNA sequence and the bending degree of DNA, which influences the level of transcription (45).
In summary, HexR modulates the expression of the phosphorylative pathway for glucose metabolism in P. putida through the recognition of KDPG, an intermediate in the Entner-Doudoroff pathway. This pathway is necessary for the catabolism of other sugars such as fructose and gluconate and 2-ketogluconate. Because of the physical organization of the genes encoding Entner-Doudoroff enzymes and the phosphorylative branch, induction by a pathway intermediate leads to the gratuitous induction of the glucose phosphorylative pathway when cells are grown on gluconate. This may not be a “genomic accident” as KDPG plays a relevant role as signaling molecule in catabolite repression and in the response to oxidative stress in Pseudomonas, so that the induction of the phosphorylative pathway may be part of the trade-off cells have to pay to govern levels of the signaling molecules.
Supplementary Material
Acknowledgments
We thank M. Fandila and C. Lorente for secretarial assistance and B. Pakuts for checking the English in this manuscript.
This work was supported by Grant BIO-2006-05668 from the Ministerio de Investigacion y Ciencia of Spain.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- KDPG
- 2-keto-3-deoxy-6-phosphogluconate
- Eda
- 2-keto-3-deoxyphosphogluconate aldolase
- Edd
- 6-phosphogluconate dehydratase
- EMSA
- electrophoretic mobility shift assay
- HTH
- helix-turn-helix
- SIS
- sugar isomerase domain
- TSP
- transcription start point.
REFERENCES
- 1.Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., Brinkman F. S., Hufnagle W. O., Kowalik D. J., Lagrou M., Garber R. L., Goltry L., Tolentino E., Westbrock-Wadman S., Yuan Y., Brody L. L., Coulter S. N., Folger K. R., Kas A., Larbig K., Lim R., Smith K., Spencer D., Wong G. K., Wu Z., Paulsen I. T., Reizer J., Saier M. H., Hancock R. E., Lory S., Olson M. V. (2000) Nature 406, 959–964 [DOI] [PubMed] [Google Scholar]
- 2.Nelson K. E., Weinel C., Paulsen I. T., Dodson R. J., Hilbert H., Martins dos Santos V. A., Fouts D. E., Gill S. R., Pop M., Holmes M., Brinkac L., Beanan M., DeBoy R. T., Daugherty S., Kolonay J., Madupu R., Nelson W., White O., Peterson J., Khouri H., Hance I., Chris Lee P., Holtzapple E., Scanlan D., Tran K., Moazzez A., Utterback T., Rizzo M., Lee K., Kosack D., Moestl D., Wedler H., Lauber J., Stjepandic D., Hoheisel J., Straetz M., Heim S., Kiewitz C., Eisen J. A., Timmis K. N., Düsterhöft A., Tümmler B., Fraser C. M. (2002) Environ. Microbiol. 4, 799–808 [DOI] [PubMed] [Google Scholar]
- 3.Paulsen I. T., Press C. M., Ravel J., Kobayashi D. Y., Myers G. S., Mavrodi D. V., DeBoy R. T., Seshadri R., Ren Q., Madupu R., Dodson R. J., Durkin A. S., Brinkac L. M., Daugherty S. C., Sullivan S. A., Rosovitz M. J., Gwinn M. L., Zhou L., Schneider D. J., Cartinhour S. W., Nelson W. C., Weidman J., Watkins K., Tran K., Khouri H., Pierson E. A., Pierson L. S., 3rd, Thomashow L. S., Loper J. E. (2005) Nat. Biotechnol. 23, 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buell C. R., Joardar V., Lindeberg M., Selengut J., Paulsen I. T., Gwinn M. L., Dodson R. J., Deboy R. T., Durkin A. S., Kolonay J. F., Madupu R., Daugherty S., Brinkac L., Beanan M. J., Haft D. H., Nelson W. C., Davidsen T., Zafar N., Zhou L., Liu J., Yuan Q., Khouri H., Fedorova N., Tran B., Russell D., Berry K., Utterback T., Van Aken S. E., Feldblyum T. V., D'Ascenzo M., Deng W. L., Ramos A. R., Alfano J. R., Cartinhour S., Chatterjee A. K., Delaney T. P., Lazarowitz S. G., Martin G. B., Schneider D. J., Tang X., Bender C. L., White O., Fraser C. M., Collmer A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joardar V., Lindeberg M., Jackson R. W., Selengut J., Dodson R., Brinkac L. M., Daugherty S. C., Deboy R., Durkin A. S., Giglio M. G., Madupu R., Nelson W. C., Rosovitz M. J., Sullivan S., Crabtree J., Creasy T., Davidsen T., Haft D. H., Zafar N., Zhou L., Halpin R., Holley T., Khouri H., Feldblyum T., White O., Fraser C. M., Chatterjee A. K., Cartinhour S., Schneider D. J., Mansfield J., Collmer A., Buell C. R. (2005) J. Bacteriol. 187, 6488–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil H., Feil W. S., Chain P., Larimer F., DiBartolo G., Copeland A., Lykidis A., Trong S., Nolan M., Goltsman E., Thiel J., Malfatti S., Loper J. E., Lapidus A., Detter J. C., Land M., Richardson P. M., Kyrpides N. C., Ivanova N., Lindow S. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler D. L., Church D. M., Edgar R., Federhen S., Helmberg W., Madden T. L., Pontius J. U., Schuler G. D., Schriml L. M., Sequeira E., Suzek T. O., Tatusova T. A., Wagner L. (2004) Nucleic Acids Res. 32, D35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stjepandić D., Weinel C., Hilbert H., Koo H. L., Diehl F., Nelson K. E., Tümmler B., Hoheisel J. D. (2002) Environ. Microbial. 4, 819–823 [DOI] [PubMed] [Google Scholar]
- 9.Entner N., Doudoroff M. (1952) J. Biol. Chem. 196, 853–862 [PubMed] [Google Scholar]
- 10.Fuhrer T., Fischer E., Sauer U. (2005) J. Bacteriol. 187, 1581–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Castillo T., Ramos J. L., Rodríguez-Herva J. J., Fuhrer T., Sauer U., Duque E. (2007) J. Bacteriol. 189, 5142–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Castillo T., Ramos J. L. (2007) J. Bacteriol. 189, 6602–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Castillo T., Duque E., Ramos J. L. (2008) J. Bacteriol. 190, 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wylie J. L., Worobec E. A. (1995) J. Bacteriol. 177, 3021–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saravolac E. G., Taylor N. F., Benz R., Hancock R. E. (1991) J. Bacteriol. 173, 4970–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llamas M. A., Rodríguez-Herva J. J., Hancock R. E., Bitter W., Tommassen J., Ramos J. L. (2003) J. Bacteriol. 185, 4707–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiting P. H., Midgley M., Dawes E. A. (1976) J. Gen. Microbiol. 92, 304–310 [DOI] [PubMed] [Google Scholar]
- 18.Lessie T. G., Phibbs P. V., Jr. (1984) Annu. Rev. Microbiol. 38, 359–388 [DOI] [PubMed] [Google Scholar]
- 19.Detheux M., Vandekerckhove J., Van Schaftingen E. (1993) FEBS Lett. 321, 111–115 [DOI] [PubMed] [Google Scholar]
- 20.Hager P. W., Calfee M. W., Phibbs P. V. (2000) J. Bacteriol. 182, 3934–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J. F., Hager P. W., Howell M. L., Phibbs P. V., Hassett D. J. (1998) J. Bacteriol. 180, 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petruschka L., Adolfa K., Burchhardta G., Derneddea J., Jürgensena J., Hermanna H. (2002) Microbiol. Lett. 215, 89–95 [DOI] [PubMed] [Google Scholar]
- 23.Temple L., Sage A., Christie G. E., Phibbs P. V., Jr. (1994) J. Bacteriol. 176, 4700–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J., Jeon C. O., Park W. (2008) Microbiology 154, 3905–3916 [DOI] [PubMed] [Google Scholar]
- 25.Abril M. A., Michan C., Timmis K. N., Ramos J. L. (1989) J. Bacteriol. 171, 6782–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segura A., Godoy P., van Dillewijn P., Hurtado A., Arroyo N., Santacruz S., Ramos J. L. (2005) J. Bacteriol. 187, 5937–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucio M. I., Cabrera M., Segura E. L., Zenteno E., Salazar-Schettino M. (1999) Immunol. Invest. 28, 257–268 [DOI] [PubMed] [Google Scholar]
- 28.Hoey T. (2001) Curr. Protoc. Mol. Biol., Chapter 16, Unit 16.5 [DOI] [PubMed] [Google Scholar]
- 29.Rojas A., Segura A., Guazzaroni M. E., Terán W., Hurtado A., Gallegos M. T., Ramos J. L. (2003) J. Bacteriol. 185, 4755–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasse J., Gallagher S. R. (2004) Curr. Protoc. Immunol. 8, 8–9 [DOI] [PubMed] [Google Scholar]
- 31.Sørensen K. I., Hove-Jensen B. (1996) J. Bacteriol. 178, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bateman A. (1999) Trends Biochem. Sci. 24, 94–95 [DOI] [PubMed] [Google Scholar]
- 33.Sonnhammer E. L., Eddy S. R., Durbin R. (1997) Proteins 28, 405–420 [DOI] [PubMed] [Google Scholar]
- 34.Duque E., Molina-Henares A. J., de la Torre J., Molina-Henares M. A., del Castillo, et al. ( 2007) Pseudomonas, Vol. V., pp. 227–254, Kluwer Academic Publishers, London [Google Scholar]
- 35.Yamamoto H., Serizawa M., Thompson J., Sekiguchi J. (2001) J. Bacteriol. 183, 5110–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor P. L., Blakely K. M., de Leon G. P., Walker J. R., McArthur F., Evdokimova E., Zhang K., Valvano M. A., Wright G. D., Junop M. S. (2008) J. Biol. Chem. 283, 2835–2845 [DOI] [PubMed] [Google Scholar]
- 37.Velázquez F., di Bartolo I., de Lorenzo V. (2004) J. Bacteriol. 186, 8267–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuskey S. M., Wolff J. A., Phibbs P. V., Jr., Olsen R. H. (1985) J. Bacteriol. 162, 865–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew D. C., Luthey-Schulten Z. (2008) J. Mol. Evol. 66, 519–528 [DOI] [PubMed] [Google Scholar]
- 40.Bertram R., Hillen W. (2008) Microbial. Biotech. 1, 2–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinrichs W., Kisker C., Düvel M., Müller A., Tovar K., Hillen W., Saenger W. (1994) Science 264, 418–420 [DOI] [PubMed] [Google Scholar]
- 42.Schumacher M. A., Miller M. C., Grkovic S., Brown M. H., Skurray R. A., Brennan R. G. (2001) Science 294, 2158–2163 [DOI] [PubMed] [Google Scholar]
- 43.Alguel Y., Meng C., Terán W., Krell T., Ramos J. L., Gallegos M. T., Zhang X. (2007) J. Mol. Biol. 369, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fillet S., Vélez M., Lu D., Zhang X., Gallegos M. T., Ramos J. L. (2009) J. Bacteriol. 191, 1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raleigh E. A., Trimarchi R., Revel H. (1989) Genetics 122, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990) Methods Enzymol. 185, 60–89 [DOI] [PubMed] [Google Scholar]
- 48.Marqués S., Ramos J. L., Timmis K. N. (1993) Biochim. Biophys. Acta 1216, 227–236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.