Abstract
Autophagy-essential proteins are the molecular basis of protective or destructive autophagy machinery. However, little is known about the signaling mechanisms governing these proteins and the opposing consequences of autophagy in mammals. Here we report that a non-canonical MEK/ERK module, which is positioned downstream of AMP-activated protein kinase (AMPK) and upstream of tuberous sclerosis complex (TSC), regulates autophagy by regulating Beclin 1. Depletion of ERK partially inhibited autophagy, whereas specific inhibition on MEK completely inhibited autophagy. MEK could bypass ERK to promote autophagy. Basal MEK/ERK activity conferred basal Beclin 1 by preventing disassembly of mammalian target of rapamycin complex 1 (mTORC1) and mTORC2. Activation of MEK/ERK by AMPK upon autophagy stimuli disassembled mTORC1 via binding to and activating TSC but disassembled mTORC2 independently of TSC. Inhibition of mTORC1 or mTORC2 by transiently or moderately activated MEK/ERK caused moderately enhanced Beclin 1 resulting in cytoprotective autophagy, whereas inhibition of both mTORC1 and mTORC2 by sustained MEK/ERK activation caused strongly pronounced Beclin 1 leading to cytodestructive autophagy. Our findings thus propose that the AMPK-MEK/ERK-TSC-mTOR pathway regulation of Beclin 1 represents different thresholds responsible for a protective or destructive autophagy.
Autophagy is an evolutionally conserved machinery involving the degradation and turnover of cytoplasmic material in lysosomes. Autophagy plays a role in cellular homeostasis (1), antiaging (2–4), development (1, 5), protection of the genome (6), and regulation of cell size (7). Autophagy may act as a means of defense against bacterium and virus invasion and be linked to various diseases including cancer (8–10), cardiomyopathy (11), and neurodegenerative disorders (12).
Autophagy starts with the formation of an autophagosome, enclosed within a double membrane that engulfs part of the cytoplasm. During periods of autophagy stimuli, cells respond to either maintain the metabolism essential for survival or execute cell death. Autophagy-essential proteins (Atg)2 are the molecular basis of autophagy machinery. About 30 Atg proteins in yeast and 10 in mammals have been identified. In yeast, the protein kinase target of rapamycin (TOR) mediates autophagy via Atg1-Atg13 kinase complex. Atg1 interacts with multiple components of the autophagic machinery through direct association, phosphorylation, and/or intracellular localization (13, 14).
In mammalian systems, autophagosomes fuse with lysosomes to generate autophagolysosomes, which undergo a maturation process by fusing with endocytic compartments and lysosomes (15). Because it is not known how the Atg1 homolog acts in mammals, a different mechanism may be involved in regulating autophagy. Beclin 1/Atg6, microtubule-associated protein 1 light chain 3 (LC3)/Atg8, Atg5, Atg12, and Atg13 are essential for autophagosome formation in mammalian species (5, 16–20). Atg7 and Atg3 are required in the conjugation reaction between Atg12 and Atg5 and in the lipidation of LC3. During the formation of autophagosomes in mammalian cells, LC3 is lipidated via a ubiquitylation-like system (17, 21), generating a soluble form, LC3-I. LC3-I is further modified to a membrane-bound form, LC3-II, which is subsequently localized to autophagosomes and autolysosomes until being degraded by the lysosome.
Beclin 1 was initially isolated as a B-cell lymphoma-2 (Bcl2)-interacting tumor suppressor in mammalian cells (22). Overexpression of Bcl2 attenuates the formation of the kinase complex Beclin 1-class III phosphatidylinositol 3-kinase (PI3KC3) essential for the formation of autophagosomes (23). The UV radiation resistance-associated gene tumor suppressor and the activating molecule in Beclin 1-regulated autophagy protein 1 (Ambra 1) were identified as new Beclin 1-binding partners that also regulate autophagy by regulating the Beclin 1-PI3KC3 kinase complex. Association of Beclin 1 with PI3KC3 is negatively regulated by Bcl2 (22) and positively regulated by UV radiation resistance-associated gene tumor suppressor and Ambra 1 (24, 25). Beclin 1 is homoallelically deleted in many human tumors. A decreased Beclin 1 level causes defective autophagy and breast cancer, but restoration of Beclin 1 induces autophagy and inhibits tumorigenicity of human breast cancer cells (18). These reports evidence the dependence on Beclin 1 for a functional autophagy mechanism.
Diverse signaling pathways have been reported in the regulation of autophagy in mammalian cells (26, 27). In contrast to yeast, mammalian cells regulate autophagy via both class I and class III PI3K. Class I PI3K plays an inhibitory role, whereas class III PI3K kinase complex, which includes Beclin 1, plays a stimulatory role in autophagy by promoting the nucleation of autophagic vesicles (28, 29). A recent study also indicates that hVps15 is required in regulation of class III PI3K in mammalian cells (30). However, the signaling mechanisms controlling autophagy-essential proteins, in particular Beclin 1, and the opposing consequences of autophagy remain to be resolved.
Our present studies identified and positioned a non-canonical MEK/ERK pathway downstream of AMPK and upstream of TSC and mTOR. This MEK/ERK module regulated autophagy via regulating the Beclin 1 level through the AMPK-MEK/ERK-TSC-mTOR pathway. Moderately enhanced Beclin 1 by transient or moderate activation of MEK/ERK and subsequent inhibition on mTORC1 and mTORC2 individually caused protective autophagy. Strongly pronounced Beclin 1 by sustained or strong activation of MEK/ERK followed by dual inhibition on mTORC1 and mTORC2 caused destructive autophagy. Our results thus reveal interesting Beclin 1 thresholds in regulating autophagy.
EXPERIMENTAL PROCEDURES
Reagents
Antibody and reagent sources were as follows: LC3, Beclin 1, PI3KC3 (Abgent), Akt, Raptor, Rictor, GβL, β-actin, Raf1, non-phospho- or phospho-MEK1/2, ERK1/2, p90RSK, Elk, AMPKα, TSC1, TSC2, mTOR, 4EBP1, and S6K (Cell Signaling Technology), vitamin D3 (Cayman Chemical), rapamycin and bafilomycin A1 (Calbiochem), pepstatin A (Sigma), PD98059, 5-aminoimidazole-4-carboxamide riboside (AICAR) (Toronto Research Chemicals), and compound C (Merck).
Cell Culture
Human erythroleukemia K562 cells were grown in Iscove's modified Eagle's medium, and rat hepatoma H4IIE cells were grown in Dulbecco's modified Eagle's medium. The cells were starved for amino acid by culturing in Dulbecco's phosphate-buffered saline with 10% of fetal calf serum or starved for serum by culturing in Iscove's modified Eagle's medium (K562) or Dulbecco's modified Eagle's medium (H4IIE).
Mice
C57BL/6 mice were used to test physiological response of the autophagic signaling. Heart and liver tissues were surgically excised, immediately frozen in liquid nitrogen, ground in liquid nitrogen with a mortar and pestle, and homogenized briefly with an electric blender in 5 m guanidinium thiocyanate, 0.5% Sarkosyl, 0.1 m 2-mercaptoethanol, and 25 mm sodium citrate.
DNA Constructs and Transfection
Constitutively active MEK(S218D/S222D) was ordered from Addgene (31), constitutively active MEK1 mutant with disrupted ERK docking site motif (L11A/L40A/I99A/I204A) was created from the MEK1(S218D/S222D) by mutagenesis, wild-type ERK1 and dominant negative ERK1(T202A/Y204F) was provided by Melanie Cobb (32), constitutively active ERK1(T217D/Y221D) was created from the wild-type ERK1 by mutagenesis, and constitutively active Akt was created as described previously (33). Rat AMPKα2 kinase-dead construct was made by mutating its cDNA at the lysine residue critical for ATP binding and hydrolysis with lysine 45 changed to arginine, resulting in a cDNA encoding a kinase-dead protein (34). The mutated cDNA was subcloned into a mammalian expression vector with green fluorescent protein (GFP) marker. GFP-LC3 plasmid was from Addgene. The above DNA constructs were transfected into H4IIE cells with DEAE-dextran or K562 cells with electroporation followed by selection against G418. Transfectants with a GFP marker were further sorted by flow cytometry.
RNA Interference
RNA interference was performed by transfection of the cells with 20 nm desalted, purified, and annealed double-stranded siRNA against the designated genes using DEAE-dextran transfection for H4IIE cells or electroporation for K562 cells. Human siRNA sources were as follows: MEK1/2, Raf1, ERK1/2, mTOR, TSC2 (Cell Signaling Technology), Beclin 1, p90RSK, Elk, Raptor, and Rictor (Santa Cruz Biotechnology).
Kinase Assay
mTOR kinase assay was performed as described previously (35). Radioactivity in GST-4EBP1 was quantitated by GE Healthcare PhosphorImager and ImageQuant software (35).
Autophagy Assay
Autophagy was determined by detection of the processing of autophagy marker LC3 by autophagic flux assay (36) and fluorescence microscopic detection of the formation of the autophagosomes in the cells transfected with GFP-LC3 as described previously (37).
Immunoblotting and Co-immunoprecipitation
Western blotting and co-immunoprecipitation were performed as described previously (38).
RESULTS
Autophagy Response Is MEK/ERK-dependent
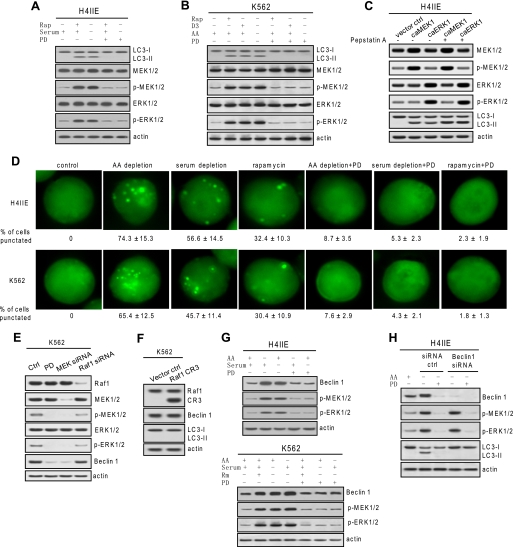
To explore the role of MEK/ERK in the induction of autophagy, we examined whether MEK/ERK is activated in response to autophagy stimuli using rat hepatoma H4IIE and human erythroleukemia K562 cells. Autophagic response involves the cleavage of cytosolic LC3-I into a lipidated LC3-II, which is then recruited to the autophagosomal membrane (19, 21, 39). Thus, processing of LC3 and a punctate GFP-labeled LC3 pattern represents autophagosomes or autophagic responses. Autophagy stimuli including rapamycin or vitamin D3 treatment and amino acid or serum starvation caused autophagic responses as shown by the processing of LC3-I into LC3-II. These autophagic activities coincided with MEK/ERK activation in both H4IIE (Fig. 1A) and K562 cells (Fig. 1B). To test whether the above coincidence is a causal event, the rat and human cell lines were treated with autophagy stimuli together with MEK inhibitor PD98059. Inhibiting MEK/ERK by PD98059 abolished the autophagic response (Fig. 1, A and B), indicating that induction of autophagy depends on MEK/ERK activation. Similar results were observed in human myeloid leukemia HL-60 cells and human breast cancer MCF-7 cells (data not shown). To test whether activation of MEK/ERK is sufficient to trigger autophagy, H4IIE cells were transfected with constitutively active MEK or ERK. The results show that constitutive expression of active MEK or ERK triggered autophagic responses indicated by autophagic flux assay (Fig. 1C).
FIGURE 1.
Autophagic activity and Beclin 1 expression is MEK/ERK-dependent. A and B, autophagic response coincided with MEK/ERK signaling. H4IIE and K562 cells were treated with various autophagy stimuli (10 μm rapamycin (Rap) for 48 h, serum depletion for 48 h, amino acid (AA) depletion for 6 h, or 50 nm vitamin D3 (D3) for 72 h) with or without 2 μm PD98059 (PD). C, constitutive expression of active MEK or ERK triggered autophagic responses. H4IIE cells were transfected with constitutively active (ca) MEK or active ERK, and LC3 processing was determined in the presence of lysosomal protease inhibitor pepstatin A. D, fluorescence microscopy of GFP-LC3 transfectant cells undergoing the designated treatments. The percentage of cells with a punctate pattern is indicated as means ± S.D. (n = 3). E, basal MEK/ERK activity confers constitutive Beclin 1 expression. Depleting basal phospho (p)-MEK1/2 but not Raf1 by siRNA depleted basal Beclin 1. F, constitutive expression of CR3 domain of Raf1 did not trigger autophagic responses. G, autophagy stimuli-enhanced Beclin 1 was diminished by inhibition of MEK/ERK. H4IIE and K562 cells were starved for amino acids for 6 h or serum for 48 h or treated with 10 μm rapamycin (Rm) together with or without 2 μm PD98059. H, depleting Beclin 1 abolished autophagy in response to MEK/ERK activation. Beclin 1 knockdown H4IIE cells were starved for amino acids with or without 2 μm PD98059. Shown in this figure from A to G are representative Western blots or fluorescence microscopy of each of three independent experiments. ctrl, control.
To visually observe the dependence of autophagy on an MEK/ERK signal, we transfected H4IIE or K562 cells with a GFP-LC3 construct followed by autophagy stimuli with or without PD98059 treatment. The fluorescence microscopic results show that inhibiting MEK/ERK inhibited the punctate GFP-LC3 pattern caused by autophagy stimuli (Fig. 1D), further indicating that the induction of autophagy requires MEK/ERK signals. These data suggest that induction of autophagic responses may ubiquitously depend on MEK/ERK activation.
Expression of Beclin 1 Is MEK/ERK-dependent
The immediate question is how autophagy is regulated by MEK/ERK signaling. Beclin 1 is ubiquitously and constitutively expressed at basal level in mammalian cells despite its frequent down-regulation in some cancer cells. Beclin 1 expression level coincides with autophagy activity and tumorigenesis suppression (8, 18, 40). Defective Beclin 1 expression or depleting Beclin 1 cripples autophagy (18, 37, 41), but its overexpression triggers autophagy (18, 42). Because autophagy response depends on both Beclin 1 and activation of MEK/ERK, we asked whether constitutive Beclin 1 is regulated by MEK/ERK. Inhibiting the basal MEK/ERK activity by PD98059 or knockdown of MEK by RNA interference resulted in the depletion of constitutive Beclin 1 (Fig. 1E), indicating that expressing or maintaining the constitutive Beclin 1 depends on the basal activity of MEK/ERK. However, knockdown of Raf1 by RNA interference or constitutive expression of the Raf1 CR3 domain, the active Raf1, which was proven functional previously (38), hardly affected the basal activity of MEK/ERK or the constitutive Beclin 1 (Fig. 1, E and F), suggesting that basal MEK/ERK activity conferring the constitutive Beclin 1 may largely be independent of Raf1. We then asked whether MEK/ERK activation in response to autophagy stimuli up-regulates Beclin 1. Indeed the increase in Beclin 1 by autophagy stimuli coincided with MEK/ERK activation, and inhibiting MEK/ERK activation abolished Beclin 1 up-regulation in response to autophagy stimuli in both HII4E and K562 cells (Fig. 1G). Despite MEK/ERK activation by autophagy stimuli, depleting Beclin 1 by RNA interference abolished autophagic responses (Fig. 1H). These data thus suggest that both basal Beclin 1 and up-regulation of Beclin 1 are MEK/ERK-dependent and that MEK/ERK regulates autophagy through regulating the Beclin 1 level.
MEK Can Bypass ERK to Trigger Autophagy
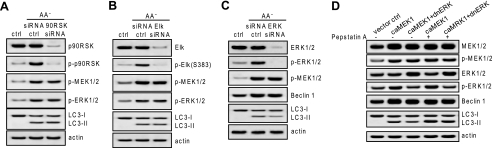
Previous studies suggested that ERK is required in autophagy; however, the autophagy dependence on ERK was largely concluded from pharmacological inhibition with an MEK inhibitor (43, 44). To better understand the role of ERK in regulating autophagy, we examined the consequences on autophagy by depleting the ERK1/2 canonical downstream effectors, p90RSK and Elk, by RNA interference in K562 cells. MEK1/2 and ERK1/2 activation triggered by amino acid starvation coincided with the phosphorylation of p90RSK (Fig. 2A) and Elk (Fig. 2B). Knockdown of either p90RSK or Elk did not affect the autophagic response to amino acid starvation (Fig. 2, A and B), suggesting that ERK may cross-talk with other pathways or use non-canonical downstream effectors to regulate autophagy or that MEK may bypass ERK to regulate autophagy. To answer this question, ERK1/2 was knocked down by RNA interference in K562 cells, and the cells were starved for amino acids. Depleting ERK1/2 partially inhibited autophagic responses (Fig. 2C), suggesting that although ERK is involved in the regulation of autophagy MEK may play a more critical role in regulating this cellular cascade by bypassing ERK. To confirm this, K562 cells were transfected with constitutively active MEK1 or co-transfected with both constitutively active MEK1 and dominant negative ERK1. MEK activation up-regulated Beclin 1 and triggered LC3 processing, and inhibition on ERK by dominant negative ERK expression attenuated, but did not block, the autophagic response by constitutively active MEK as shown by the autophagic flux assay (Fig. 2D). The above data propose that despite the role of ERK in regulating autophagy to some extent MEK can bypass ERK to induce autophagy.
FIGURE 2.
MEK can bypass ERK to trigger autophagy. A and B, knockdown of p90RSK or Elk did not abolish autophagy. K562 cells were transfected with siRNA against p90RSK or Elk, and LC3 processing was determined in response to amino acid (AA) starvation. C, knockdown of ERK did not abolish autophagy. K562 cells were transfected with siRNA against ERK1/2, and LC3 processing was determined in response to starvation. D, MEK can trigger autophagic response independently of ERK. K562 cells were transfected with constitutively active MEK (caMEK) or constitutively active MEK together with dominant negative ERK (dnERK), and LC3 processing was determined by autophagic flux assay. Shown in this figure from A to D are the representative Western blots of each of three independent experiments. ctrl, control; p-, phospho-.
AMPK Is an Upstream Regulator of MEK in Regulating Autophagy
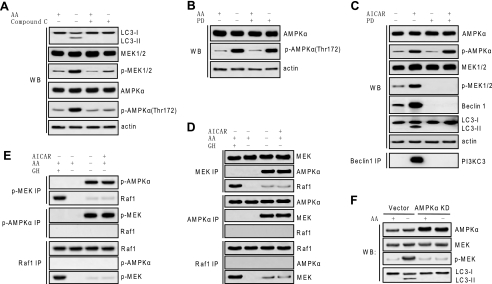
An early study reported that activation of AMPK by addition of the cell-permeable nucleotide analogue AICAR in hepatocytes inhibits autophagy (45). But a recent study using different mammalian cell types and compound C, a specific AMPK inhibitor, showed that AMPK is required for autophagy (46). Our results show that starvation-caused MEK activation as well as AMPK activation and autophagic response were inhibited by compound C in H4IIE cells (Fig. 3A), whereas inhibition of MEK by PD98059 did not inhibit AMPK activation in response to the starvation (Fig. 3B), suggesting that AMPK may be an upstream regulator of MEK in modulating autophagy.
FIGURE 3.
MEK/ERK is positioned downstream of AMPK in autophagic signaling. A, inhibiting AMPK inhibited MEK activation in response to autophagy stimuli. H4IIE cells were starved for amino acids (AA) and treated with 10 μm compound C. B, inhibition of MEK did not inhibit AMPK activation in response to autophagy stimuli. H4IIE cells were starved for amino acids and treated with or without 2 μm PD98059 (PD), and AMPK activation was determined. C, up-regulation of Beclin 1 and induction of autophagy by AMPK activation was dependent on MEK. H4IIE cells were treated with 1 mm AICAR with or without 2 μm PD98059, and activation of AMPK and MEK, expression of Beclin 1, LC3 processing, and Beclin 1 binding to PI3KC3 were determined. D and E, MEK interacted with AMPK but not Raf1 in response to autophagy stimuli. H4IIE cells were starved for amino acids or treated with 1 mm AICAR, and physical interaction between MEK and AMPK, between Raf1 and MEK, or between AMPK and Raf1 was determined by using total (D) or phospho antibody (E). Growth hormone (GH) treatment served as a positive control for canonical Raf/MEK/ERK activation and interaction in H4IIE cells. F, specific inhibition on AMPK by kinase-dead AMPK crippled activation of MEK and autophagic activity in response to autophagy stimuli. H4IIE cells stably transfected with either rat AMPKα2 kinase-dead (KD) construct or vector control were starved for amino acids for 6 h. Expression of total AMPKα and total and phospho (p)-MEK and processing of LC3 were determined. Shown in this figure from A to F are the representative Western blots (WB) or co-immunoprecipitate (IP) blots of each of three independent experiments.
To understand how MEK is linked to AMPK, H4IIE cells were treated with AICAR to trigger autophagic responses. Inhibiting MEK did not inhibit AMPK activation by AICAR. AICAR activated MEK and triggered autophagic responses as shown by an elevated Beclin 1 level and processing of LC3. Inhibiting MEK blocked the autophagic response by AICAR (Fig. 3C). This result suggests that enhancing Beclin 1 and triggering autophagic responses by AMPK activation depends on MEK activation. Furthermore co-immunoprecipitation results revealed that MEK interacts with AMPK but not Raf1 in response to autophagy stimuli (Fig. 3D), and this interaction is apparently caused by activation of AMPK and MEK signaling because activated MEK bound to activated AMPK but hardly bound to Raf1 upon exposure to autophagy stimuli (Fig. 3E). In contrast, growth hormone treatment, which is known to activate the canonical Raf/MEK/ERK pathway in H4IIE cells (47), caused strong Raf1 binding to MEK (Fig. 3, D and E). The results thus propose that MEK interacts with its upstream regulator AMPK, but not Raf1, in response to autophagy stimuli.
To confirm the role of AMPK as an upstream regulator of MEK in regulating autophagy proposed by the pharmacological data, we generated a kinase-dead AMPKα2 H4IIE cell line by mutating rat AMPKα2 cDNA at the lysine residue critical for ATP binding and hydrolysis that was reported by an early study (34). The results show that in kinase-dead AMPKα2 cells, amino acid starvation, which is a strong autophagy stimulus, failed to activate MEK and cause autophagic processing of LC3 (Fig. 3F), further supporting the notion that AMPK regulates autophagy via regulating its downstream effector MEK.
MEK/ERK Binds to and Activates TSC2 in Regulating Autophagy
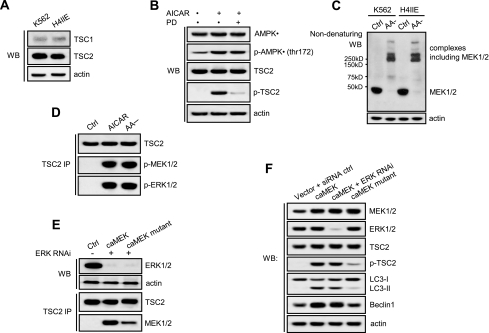
AMPK activates TSC2 by phosphorylation (48), whereas activation of TSC2 suppresses mTOR signaling (49), and activation of AMPK down-regulates mTOR activity (50), supporting a positive role of AMPK in regulating autophagy. Both H4IIE and K562 cells expressed a dominant TSC2 and a weak TSC1 (Fig. 4A). To examine whether MEK/ERK plays a role in TSC activation by AMPK, H4IIE cells were stimulated with AICAR with or without PD98059. Activation of AMPK activated TSC2, but inhibiting MEK inhibited TSC2 activation by AMPK as shown by the crippled phosphorylation of TSC2 (Fig. 4B), suggesting that activation of TSC2 by AMPK is MEK/ERK-dependent.
FIGURE 4.
MEK/ERK is positioned upstream of TSC in autophagic signaling. A, TSC2 was the dominant form of TSC1/2 in both H4IIE cells and K562 cells. B, AMPK activation of TSC2 was MEK/ERK-dependent. H4IIE cells were stimulated by 1 mm AICAR with or without 2 μm PD98059 (PD), and activation of AMPK or TSC2 was determined. C, activated MEK formed multiple complexes in response to autophagy stimuli. K562 and H4IIE cells were starved for amino acids (AA), and formation of the complexes including MEK were determined by non-denaturing Western blotting (WB). D, MEK and ERK associated with TSC2 in response to autophagy stimuli. H4IIE cells were starved for amino acids or treated with AICAR, and TSC2 binding to MEK or ERK was determined. E, MEK binding to TSC2 required ERK D domain. H4IIE cells were transfected with constitutively active MEK (caMEK) or constitutively active MEK mutant with the ERK D domain disrupted, and TSC2 binding to active MEK or MEK mutant without ERK D domain was determined. F, disruption of the association between MEK and TSC2 crippled TSC2 activation and the autophagic response. H4IIE cells transfected with constitutively active MEK, constitutively active MEK together with ERK siRNA, or constitutively active MEK mutant with the ERK D domain disrupted. Shown in this figure from A to F are the representative Western blots or co-immunoprecipitate (IP) blots of each of three independent experiments. p-, phospho-; Ctrl, control.
Upon activation by autophagy stimuli, MEK formed multiple complexes (Fig. 4C). TSC2 bound to the activated MEK and activated ERK (Fig. 4D), suggesting that TSC2 is the downstream effector of MEK/ERK in regulation of autophagy.
Physical interaction between TSC2 and ERK was reported previously (51). TSC2 binds to ERK through the ERK docking site (ERK D domain), which consists of a conserved (L/I)X(L/I) motif located three to five residues downstream of a cluster of basic amino acids (52). Because MEK activation can induce autophagy when ERK activation is inhibited (Fig. 2D), we asked whether MEK can directly activate TSC through physical interaction. To test whether MEK interacts with TSC2 through the ERK D domain, the four ERK D domains were disrupted by mutating the four amino acids in the ERK docking site motif (L11A/L40A/I99A/I204A) of the active MEK1. In the absence of ERK by RNA interference, constitutively active wild-type MEK interacted with TSC2, but disruption of the ERK D domains of MEK1 attenuated this interaction (Fig. 4E), suggesting that MEK interacted with TSC2 through the ERK D domains. Constitutively active MEK phosphorylated TSC2 and caused LC3 processing, which was not affected by depleting ERK with RNA interference, but disrupting these docking sites of MEK1 crippled the phosphorylation of TSC2 and the processing of LC3 in the cells that express ERK (Fig. 4F). These results indicate that MEK binds to and activates TSC2 independently of ERK in regulating autophagy.
AMPK-MEK/ERK-TSC Signaling Functions in Physiological Response of Autophagy
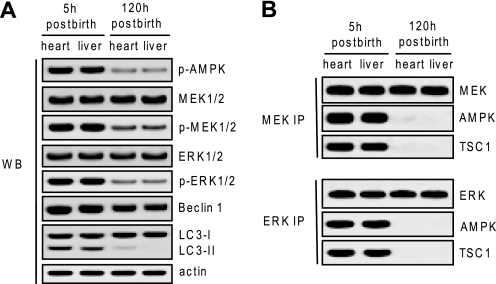
To test whether this non-canonical MEK/ERK module physiologically regulates autophagy, we detected MEK/ERK signaling in neonatal mice, which adapt to sudden interruption of trans-placental nutrient by inducing autophagy until supply can be restored through milking (5). At 5 h after birth without milking, the heart and liver samples displayed AMPK and MEK/ERK activation, Beclin 1 up-regulation, and LC3 processing as well as MEK/ERK association with AMPK or TSC1 due to nutrient interruption. By contrast, at 120 h after birth when the mice began suckling for several days, activation of AMPK or MEK/ERK or physical interaction between MEK/ERK and AMPK or TSC1 was not detected (Fig. 5, A and B). These results suggest that the non-canonical MEK/ERK signaling plays a physiological role in the regulation of mammalian autophagy.
FIGURE 5.
AMPK-MEK/ERK-TSC signaling functions in physiological response of autophagy in mice. A, AMPK-MEK/ERK signaling responded to autophagy stimuli in neonatal mice. Western blotting was performed to determine activation of AMPK, MEK, and ERK; expression of Beclin 1; and processing of LC3 using cell lysates prepared from the hearts and livers of mice at 5 or 120 h after birth. B, MEK/ERK interacted with AMPK or TSC1 in response to autophagy stimuli in neonatal mice. MEK or ERK co-immunoprecipitation was performed to examine their interactions with upstream regulator or downstream effector in response to autophagy or non-autophagy stimuli using the cell lysates from the hearts and livers of the mice 5 or 120 h after birth. Shown in this figure are the representative Western blots (WB) (A) or co-immunoprecipitate (IP) blots (B) of each of three independent experiments. p-, phospho-.
MEK/ERK Regulates Beclin 1 through Negatively Regulating mTORC1 and mTORC2
Unlike positive regulation of autophagy by MEK/ERK, mTOR is a major negative regulator of autophagy. Activation of AMPK and TSC2 up-regulated Beclin 1 (Figs. 3C, 4F, and 5A) and negatively regulates mTOR activity (48, 49). Furthermore, AMPK activation of TSC2 was dependent on MEK/ERK (Fig. 4B). We thus hypothesize that MEK/ERK may up-regulate Beclin 1 and autophagy through down-regulation of mTOR. To test this, we made transfectant H4IIE cells constitutively expressing active Akt, which is a known upstream positive regulator of mTOR (53). Although overexpressing active Akt did not inhibit MEK/ERK activation in response to autophagy stimulus AICAR, it activated mTOR and crippled LC3 processing (Fig. 6A). Activation of AMPK by AICAR enhanced the Beclin 1 level and caused LC3 processing (Fig. 6A) and punctate GFP-LC3 distribution representing autophagy, but active Akt overexpression inhibited them (Fig. 6, A and B). Thus, MEK/ERK activation failed to up-regulate Beclin 1 and trigger autophagy when mTOR was activated. These data suggest that MEK/ERK regulates Beclin 1 and autophagy through down-regulation of mTOR activity.
FIGURE 6.
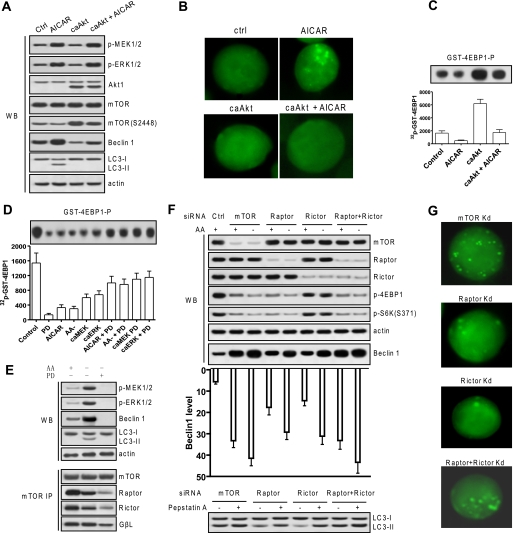
MEK/ERK up-regulates Beclin 1 and autophagy by down-regulating mTOR activity. A, MEK/ERK failed to up-regulate Beclin 1 and autophagy when mTOR was activated. H4IIE cells were transfected with constitutively active Akt with or without 1 mm AICAR, and MEK/ERK or mTOR activation, Beclin 1 expression, and LC3 processing were determined. B, fluorescence microscopy of the GFP-LC3 cells transfected with constitutively active Akt (caAkt) with or without autophagy stimuli. C, overexpressing active Akt enhanced mTORC1 activity using GST-4EBP1 as a substrate. The 32P incorporated into GST-4EBP1 (GST-4EBP1-P) is shown in a bar graph. D, MEK/ERK activation inhibits mTORC1 activity. H4IIE cells were stimulated with the designated autophagy stimuli or transfected with constitutively active MEK (caMEK) or constitutively active ERK (caERK) with or without 2 μm PD98059 (PD), and mTORC1 activity was determined as described in C. E, inhibiting MEK/ERK disassociated both mTORC1 and mTORC2. H4IIE cells were starved for amino acids (AA) with or without 2 μm PD98059, and Beclin 1 expression and LC3 processing as well as mTOR binding to Raptor, Rictor, or GβL were determined. F, inhibiting mTORC1 or mTORC2 individually caused a mildly increased Beclin 1 and autophagic response, but inhibiting both mTORC1 and mTORC2 caused strongly pronounced Beclin 1 levels and autophagic responses. H4IIE cells were transfected with siRNA against mTOR or against Raptor or Rictor individually or in combination, and activation of mTOR effectors, Beclin 1 expression, and LC3 processing was determined by autophagic flux assay. The Beclin 1 levels were quantified by density scanning. G, fluorescence microscopy of GFP-LC3 transfectant H4IIE cells with Raptor or Rictor knockdown individually or in combination. Shown in this figure from A to G are the representative Western blots (WB) or co-immunoprecipitate (IP) blots or quantified data (means ± S.D.) of each of three independent experiments. Ctrl, control; Kd, kinase-dead; p-, phospho-.
The role of MEK/ERK in regulating mTOR activity was further assessed by measuring mTORC1 activity using GST-4EBP1 as substrate. Overexpressing active Akt enhanced mTORC1 activity (Fig. 6C). Various autophagy stimuli inhibited mTORC1 activity (Fig. 6D). However, PD98059 released the suppression of mTORC1 activity by MEK/ERK activation (Fig. 6D), indicating that MEK/ERK up-regulates autophagy possibly through down-regulating mTORC1 activity. Surprisingly, depleting basal MEK/ERK activity by PD98059 almost completely abolished mTORC1 activity (Fig. 6D). These data suggest that basal MEK/ERK activity is needed for constitutive mTORC1 activity, but pronounced MEK/ERK activation inhibits mTORC1 activity and thus activates autophagy.
Amino acid starvation-caused MEK/ERK activation, ultimately leading to an autophagic response, attenuated the binding of mTOR to Raptor or Rictor but not GβL (Fig. 6E), suggesting that assembly of mTOR complexes mTORC1 and mTORC2 is sensitive to MEK/ERK activation and that up-regulation of Beclin 1 by MEK/ERK activation, which triggers autophagy, may require inactivation of both mTORC1 and mTORC2. Surprisingly, depleting MEK/ERK activity by PD98059, which depleted basal Beclin 1 (Figs. 1E, 3C, and 6E), disassembled both mTORC1 and mTORC2 (Fig. 6E), suggesting that assembly of mTORC1 and mTORC2 requires basal MEK/ERK activity. It thus appears that basal MEK/ERK activity confers basal mTOR activity by conferring the assembly of complexes mTORC1 and mTORC2, whereas activation of MEK/ERK inactivates mTOR by disassembling both mTORC1 and mTORC2.
To test whether down-regulation of both mTORC1 and mTORC2 is required to up-regulate Beclin 1 and trigger autophagy in response to MEK/ERK activation, mTOR, Raptor, or Rictor was knocked down individually or in combination by RNA interference using the K562 cells stably transfected with GFP-LC3. The mTOR was activated in the control cells in rich medium due to basal MEK/ERK activity (Fig. 6D). The mTOR activity phosphorylated p70S6K and the translation repressor protein 4EBP1 in control cells (Fig. 6F). Efficient knockdown of mTOR caused hypophosphorylation of S6K and 4EBP1, enhanced Beclin 1 (Fig. 6F), and punctate distribution of the GFP-LC3 (Fig. 6G). Knockdown of mTOR plus amino acid starvation caused more pronounced Beclin 1 and autophagic response. By contrast, knockdown of Raptor or Rictor individually only caused a moderate enhancement of Beclin 1 and weak autophagic responses as shown by the LC3 processing determined with autophagic flux, but knockdown of Raptor and Rictor together caused a pronounced Beclin 1 and autophagic response comparable to that caused by global mTOR knockdown (Fig. 6, F and G). These data suggest that efficient up-regulation of Beclin 1 attributed to the induction of autophagy requires inactivation of both mTORC1 and mTORC2 activities.
MEK/ERK Activation Inhibits mTORC1 via TSC2 but Inhibits mTORC2 Independently of TSC2
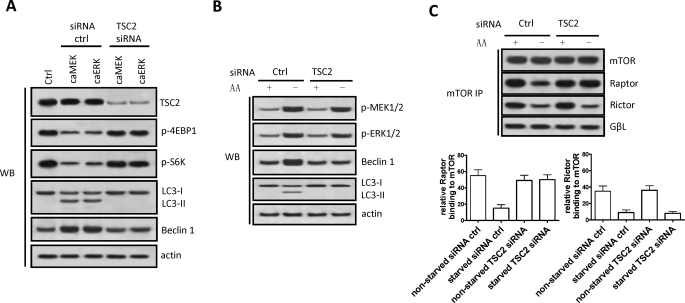
TSC2 mediates cellular energy response through regulating mTOR (49). To determine whether MEK/ERK down-regulation of mTOR kinase activity and up-regulation of Beclin 1 and autophagy involves TSC1/2, the dominant form of TSC, TSC2, was knocked down by RNA interference using the H4IIE cells transfected with constitutively active MEK or ERK. Constitutive expression of active MEK or active ERK caused dephosphorylation of both 4EBP1 and p70S6K, the two direct downstream effectors of mTORC1, that coincided with Beclin 1 up-regulation and LC3 processing, whereas knockdown of TSC2 crippled the hypophosphorylation of 4EBP1 and S6K as well as the up-regulation of Beclin 1 and processing of LC3 (Fig. 7A). These data suggest that the down-regulation of mTORC1 activity by MEK/ERK, up-regulation of Beclin 1, and induction of LC3 processing requires TSC2. Knockdown of TSC2 did not cripple activation of MEK/ERK in response to amino acid starvation but crippled the autophagic response (Fig. 7B), further confirming that TSC2 is downstream of MEK/ERK in the signaling pathway that regulates autophagy.
FIGURE 7.
MEK/ERK inhibition on mTORC1 is TSC2-dependent, but its inhibition on mTORC2 is TSC2-independent. A, depleting TSC2 increased mTOR activity and crippled Beclin 1 up-regulation and LC3 processing by MEK/ERK activation. H4IIE cells stably transfected with constitutively active (ca) MEK or ERK were transiently transfected with siRNA against TSC2. B, depleting TSC2 did not affect MEK/ERK activation but crippled Beclin 1 up-regulation and LC3 processing in response to autophagy stimuli. H4IIE cells were transfected with siRNA against TSC2 and starved for amino acids (AA). C, depleting TSC2 crippled the disassembly of mTORC1 but not mTORC2 in response to autophagy stimuli. H4IIE cells were transfected with siRNA against TSC2 and starved for amino acids, and mTOR binding to Raptor, Rictor, or GβL was determined. The binding was quantified by density scanning. Shown in this figure from A to C are the representative Western blots (WB) or co-immunoprecipitate (IP) blots or quantified data (means ± S.D.) of each of three independent experiments. Ctrl, control; p-, phospho-.
Autophagy stimuli caused MEK/ERK activation (Fig. 1) and attenuated mTOR binding to Raptor or Rictor (Fig. 7C). Knockdown of TSC2 crippled Raptor but not Rictor and caused disassociation from mTOR in response to autophagy stimuli that trigger MEK/ERK activation (Fig. 7C). These data suggest that MEK/ERK inhibition of mTORC1 activity is dependent on TSC2, but MEK/ERK inhibition of mTORC2 is independent of TSC2.
Beclin 1 Modulates a Protective or Destructive Autophagy
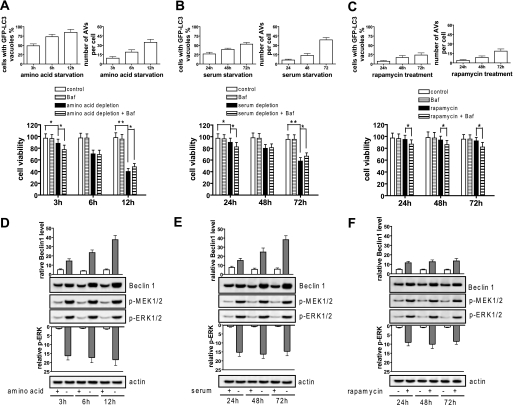
To explore the signaling mechanisms by which a protective or destructive autophagy is modulated, H4IIE cells were stimulated with amino acid or serum starvation or treated with rapamycin either in combination with or without autophagy inhibitor bafilomycin A1. Autophagic response increased from 3 to 12 h of amino acid starvation as shown by the number of autophagic vacuoles per cell or by the percentage of the cells with GFP-LC3 vacuoles (Fig. 8A, upper panel). At 3 h, the amino acid-starved cells had a decreased cell viability of 88.3% compared with the control cells, which had 97.2% viability. Inhibiting autophagy by bafilomycin A1 further reduced cell viability to 77.5%, suggesting that autophagy played a protective role at 3 h of amino acid starvation. At 6 h, cell viability dropped to 70.3% in the starved cells and 69.2% in the starved cells treated with bafilomycin A1, suggesting that a balance exists between protective and destructive autophagy. At 12 h, cell viability further dropped to 40.3%, whereas treatment with bafilomycin A1 increased the cell viability to 48.3%, suggesting that destructive autophagy occurs in the late stage of the starvation (Fig. 8A, lower panel). Compared with amino acid starvation, serum starvation caused slower and less intense autophagy (Fig. 8B, upper panel). Inhibiting autophagy by bafilomycin A1 reduced cell viability at 24 h and did not change the viability at 48 h but increased the viability at 72 h compared with the control cells (Fig. 8B, lower panel), suggesting that autophagy by serum starvation is protective at 24 h, neutral at 48 h, and destructive at 72 h. By contrast, rapamycin caused weak autophagy (Fig. 8C, upper panel). Inhibiting autophagy on the rapamycin-treated cells decreased cell viability at 24, 48, and 72 h, suggesting that rapamycin causes protective autophagy (Fig. 8C, lower panel). These data suggest that transient or less intense autophagy stimuli cause protective autophagy, but sustained or intense autophagy stimuli cause destructive autophagy.
FIGURE 8.
Moderately enhanced Beclin 1 causes cytoprotective autophagy, but prominently enhanced Beclin 1 causes cytodestructive autophagy. A, amino acid starvation caused a protective autophagy at early stage and a destructive autophagy at late stage. H4IIE cells were starved for amino acids, and autophagic activity represented by LC3 dots (upper panel) and cell viability (lower panel) were determined at the designated time points. B, serum starvation caused a protective autophagy at early stage and a destructive autophagy at late stage determined at the designated times as in A. C, rapamycin (10 μm) caused a protective autophagy, determined at the designated times as described in A, throughout. Data shown in A, B, and C are means ± S.D. (n = 3) (*, p < 0.05, **, p < 0.01). D, amino acid starvation caused a sustained high MEK/ERK activation and an increasingly pronounced Beclin 1 expression. Beclin 1 expression and ERK activation were quantitated by density scanning. E, serum starvation caused a sustained high MEK/ERK activation and significantly pronounced Beclin 1 expression. F, rapamycin causes a sustained moderate MEK/ERK activation and moderately enhanced Beclin 1 determined as in D. Shown in this figure from D to F are the representative Western blots and quantified data of each of three independent experiments. AVs, autophagic vacuoles; Baf, bafilomycin A1; p-, phospho-.
Because we observed that MEK/ERK activation triggers autophagy by regulating Beclin 1, we asked whether Beclin 1 regulates both protective and destructive autophagy and if so how. The results show that MEK/ERK activation at the three designated time points remained unchanged, but the Beclin 1 level was increased along with the starvation, suggesting that less enhanced Beclin 1 causes protective autophagy, moderately enhanced Beclin 1 causes neutral autophagy, and strongly pronounced Beclin 1 causes destructive autophagy (Fig. 8D). A similar pattern that the Beclin 1 level determines autophagic consequences was also revealed by serum starvation; however, serum starvation triggered a slower and less intense autophagy than amino acid starvation (Fig. 8E). By contrast, rapamycin caused much less enhanced Beclin 1, which was unchanged throughout the protective autophagy response (Fig. 8F). These data thus suggest that the cellular consequences of autophagy may depend on the longevity of MEK/ERK activation and the enhanced Beclin 1 level.
DISCUSSION
The results presented here demonstrate that a non-canonical MEK/ERK module regulates autophagy by regulating Beclin 1 level through the AMPK-MEK/ERK-TSC-mTOR signaling pathway (Fig. 9). Basal MEK/ERK activity conferred constitutive Beclin 1 that was insufficient to trigger the autophagic response. MEK/ERK activation by AMPK upon various autophagy stimuli inactivated mTOR by disassembling the functional complexes mTORC1 and/or mTORC2, resulting in the up-regulation of Beclin 1 to different thresholds leading to autophagy of opposing consequences, suggesting that Beclin 1 may play a central role in integrating signals downstream of mTOR. Our studies thus provide the first mechanistic link between a signaling pathway and autophagy-essential proteins in the mammalian system.
FIGURE 9.
A diagram for the non-canonical MEK/ERK signaling pathway regulating autophagy via regulating Beclin 1. Basal MEK/ERK activity maintains the activity of mTORC1 and mTORC2 by protecting them from being disassembled and thus maintains a basal Beclin 1 level incapable of triggering autophagic response. The non-canonical MEK/ERK module is activated by upstream regulator AMPK upon autophagy stimuli. Activation of MEK/ERK up-regulates the Beclin 1 level through disassembling mTORC1 and/or mTORC2 with a significantly sustained and elevated Beclin 1 level causing cytodestructive autophagy and moderately elevated Beclin 1 causing cytoprotective autophagy. Disassembling mTORC1 by MEK/ERK depends on TSC, whereas disassembling mTORC2 does not depend on TSC2. Although ERK is implicated in the regulation of autophagy, MEK can bypass ERK to trigger autophagic response.
Mechanism of the Non-canonical MEK/ERK Module Regulating Autophagy
ERKs are a widely conserved family of serine/threonine protein kinases implicated in many cellular programs such as cell proliferation, differentiation, and apoptosis. ERK can be activated by a wide variety of oncogenes and extracellular stimuli including mitogens, growth factors, cytokines, and chemokines. An increasing number of studies have suggested that ERK plays a role in modulating autophagy (27, 43, 44, 54–57). However, the role of ERK in autophagy was primarily concluded from the pharmacological blockage of autophagy by compound PD98059, an MEK-specific inhibitor. Because this chemical indirectly blocks ERK via inhibiting its canonical upstream regulator, MEK, it is not surprising that ERK was exclusively credited in regulating autophagy because ERK has been believed to be the only known substrate of MEK.
We demonstrated in this study that basal MEK/ERK activity maintains the activity of mTORC1 and mTORC2 by protecting them from being disassembled (Fig. 6E, lower panel) and thus maintains a basal Beclin 1 level incapable of triggering autophagic response (Figs. 1E, 3C, and 6E, upper panel). Although we found that ERK is involved in the regulation of autophagy because constitutive expression of active ERK caused autophagy response (Figs. 1C and 7A) whereas specific inhibition on ERK by RNA interference (Fig. 2C) or dominant negative ERK (Fig. 2D) partially inhibited autophagy, knockdown of the canonical effectors ERK, p90RSK, and Elk by RNA interference did not cripple autophagic response (Fig. 2, A and B). This unusual observation suggests that ERK may use non-canonical downstream effectors in the context of autophagic signaling. Coincidentally MEK could bypass ERK (Fig. 2D) and interact with a downstream effector other than ERK in the context of autophagy stimuli (Figs. 4D and 5B), indicating that ERK is not the sole downstream effector of MEK and that MEK plays a more important role than ERK in regulating autophagy. Interestingly MEK regulates Golgi fragmentation in mitosis independently of ERK1/2 (58, 59), and the nucleation of autophagy by Beclin 1-PI3KC3 kinase complex is executed at the trans-Golgi network in mammalian cells (60), both of which support the unusual role and the non-canonical module of MEK/ERK in regulating Beclin 1 and autophagy.
Recent studies show that ERK up-regulates starvation-caused autophagy by down-regulating Akt/mTOR/S6K (27), but the molecular link between this pathway and the basis of autophagy machinery remains largely unclear. We observed here that activation of MEK/ERK coincided with up-regulation of Beclin 1 and autophagic responses, and MEK/ERK was activated by AMPK but not Raf1 in response to autophagy stimuli. AMPK has been proposed as a physiological cellular energy sensor because AMP is the most sensitive indicator for a cellular energy status. The cellular ATP concentration is much higher than that of AMP, and an insignificant decrease in ATP levels can result in an extraordinary increase in AMP levels that are sensed by and stimulate AMPK (24). MEK/ERK activation by AMPK thus confers the sensitivity and accuracy of MEK/ERK in regulating autophagic activity. TSC is known to be regulated by cellular energy levels. Activation of AMPK by energy starvation results in direct phosphorylation of TSC (49). Our study showed that AMPK activation of TSC depends on MEK/ERK. Furthermore, activated MEK interacted with TSC, and the physical interaction between activated MEK and TSC required the ERK D docking site sequence motif contained in both MEK and TSC, consistent with the notion that stable binding of substrates to mitogen-activated protein kinase (MAPK) kinase involves recognition of a docking site on the substrate (2). This non-canonical MEK/ERK signaling pathway functions not only in human and rat cell lines but also in neonatal mice, indicating a physiological significance. Our findings thus position this unusual MEK/ERK module downstream of AMPK and upstream of TSC in response to autophagy stimuli.
Our present study showed that mTOR regulates autophagy caused both by starvation and non-starvation stimuli that activate MEK/ERK, consistent with the report that AMPK regulates autophagy through mTOR also in non-starved conditions (61), suggesting a possible universal mechanism in the regulation of autophagy through mTOR. Because constitutive expression of Beclin 1 is due to the basal activity of MEK/ERK and enhanced Beclin 1 is attributed to the enhanced MEK/ERK activation in both human and rat cells, we postulate that the dependence of Beclin 1 on MEK/ERK signals may also be a universal mechanism in regulating autophagy.
Beclin 1 Plays a Central Role in the Mechanisms Determining a Protective or Destructive Autophagy
Our previous study shows that autophagic activity depends on the Beclin 1 level (41). The present study found that the non-canonical MEK/ERK module regulates mutually antagonistic processes by regulating Beclin 1 levels. Basal activity of MEK/ERK protected mTORC1 and mTORC2 from being disassembled, thus conferring the basal mTOR activity and basal Beclin 1 level, which are incapable of triggering autophagy. MEK/ERK activation by AMPK up-regulated Beclin 1 through down-regulation of mTOR activity. Transient or moderate activation of MEK/ERK caused inhibition of mTORC1 or mTORC2, resulting in a moderate Beclin 1 increase and an ultimate cytoprotective autophagy, but sustained or strong MEK/ERK activation completely inhibited both mTORC1 and mTORC2, resulting in a strongly pronounced Beclin 1 and an ultimate cytodestructive autophagy. The requirement of an enhanced Beclin 1 level by MEK/ERK activation for autophagy thus appears to represent interesting threshold effects. The degree of enhanced Beclin 1 level may determine the level of autophagic activity, and the degree of autophagy elicited may play a role in cell fate decisions with low levels serving a homeostatic role and high levels promoting cell death. Our findings thus establish the molecular connection between MEK/ERK and mTOR in the regulation of mammalian autophagy-essential proteins and the opposing consequences of autophagy.
Mammalian cells undergo autophagy resulting from exposure to nutritional, chemical, or physical autophagy stimuli or extreme physiological conditions such as birth. Despite the ubiquitous and constitutive expression of Beclin 1 in mammalian cells, our data demonstrate that autophagy is not evident until Beclin 1 is elevated, and autophagic activity is not detectable at a constitutive Beclin 1 level under normal growth conditions in cancer cells. In the brain, autophagic activity is very low even under starvation conditions (20). These data suggest that either there is no constitutive autophagy or autophagy can only occur at extremely low levels under normal growth conditions when Beclin 1 is maintained at a basal level. Therefore, we doubt that autophagy plays a housekeeping role in all types of mammalian cells. Constitutive or basal Beclin 1 may be implicated in other cellular processes. Our recent studies show that constitutive Beclin 1 inhibits activation of apoptosis and confers differentiation capability in leukemia HL-60 cells (37, 41). Because autophagy can be regulated by several proteins capable of binding Beclin 1, including Bcl2, UV radiation resistance-associated gene tumor suppressor, and Ambra 1 (22, 24, 25), Beclin 1 may play a central role in integrating signals by various stimuli.
Studies on Beclin 1 demonstrate its increasing importance in the mammalian system because mammalian species fail to survive in the absence of Beclin 1, develop cancers with low Beclin 1 levels, suppress tumorigenesis with normal Beclin 1 levels, secure a protective autophagy at moderately enhanced Beclin 1 levels, and suffer from a destructive autophagy when too much Beclin 1 is induced. Thus, our present study adds new understanding of Beclin 1 in the regulation of autophagy.
Acknowledgments
We thank Samantha Spindel and Ariel A. Kauss for comments on this manuscript.
This work was supported by grants from Shantou University and the Unimed Foundation.
- Atg
- autophagy-essential protein
- AMPK
- AMP-activated protein kinase
- TSC
- tuberous sclerosis complex
- mTOR
- mammalian target of rapamycin
- LC3
- light chain 3
- Bcl2
- B-cell lymphoma-2
- PI3K
- phosphatidylinositol 3-kinase
- PI3KC3
- class III phosphatidylinositol 3-kinase
- Ambra 1
- activating molecule in Beclin 1-regulated autophagy protein 1
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- ERK
- extracellular signal-regulated kinase
- mTORC
- mTOR complex
- GβL
- G-protein β-subunit-like
- AICAR
- 5-aminoimidazole-4-carboxamide riboside
- GFP
- green fluorescent protein
- siRNA
- short interfering RNA
- GST
- glutathione S-transferase.
REFERENCES
- 1.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 2.Longo V. D., Finch C. E. (2003) Science 299, 1342–1346 [DOI] [PubMed] [Google Scholar]
- 3.Meléndez A., Tallóczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. (2003) Science 301, 1387–1391 [DOI] [PubMed] [Google Scholar]
- 4.Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A., Orosz L., Müller F. (2003) Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- 5.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 6.Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. (2007) Genes Dev. 21, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vellai T., Bicsák B., Tóth M. L., Takács-Vellai K., Kovács A. L. (2008) Autophagy 4, 507–509 [DOI] [PubMed] [Google Scholar]
- 8.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) J. Clin. Investig. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lüllmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. (2000) Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- 12.Yuan J., Lipinski M., Degterev A. (2003) Neuron 40, 401–413 [DOI] [PubMed] [Google Scholar]
- 13.Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. (2004) Dev. Cell 6, 79–90 [DOI] [PubMed] [Google Scholar]
- 15.Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 16.Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Mol. Cell. Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. (2001) J. Cell Biol. 152, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanida I., Ueno T., Kominami E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X. H., Kleeman L. K., Jiang H. H., Gordon G., Goldman J. E., Berry G., Herman B., Levine B. (1998) J. Virol. 72, 8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 24.Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P., Piacentini M., Chowdhury K., Cecconi F. (2007) Nature 447, 1121–1125 [DOI] [PubMed] [Google Scholar]
- 25.Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. (2006) Nat. Cell Biol. 8, 688–699 [DOI] [PubMed] [Google Scholar]
- 26.Meijer A. J., Codogno P. (2004) Int. J. Biochem. Cell Biol. 36, 2445–2462 [DOI] [PubMed] [Google Scholar]
- 27.Shinojima N., Yokoyama T., Kondo Y., Kondo S. (2007) Autophagy 3, 635–637 [DOI] [PubMed] [Google Scholar]
- 28.Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 30.Yan Y., Flinn R. J., Wu H., Schnur R. S., Backer J. M. (2009) Biochem. J. 417, 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehm J. S., Zhao J. J., Yao J., Kim S. Y., Firestein R., Dunn I. F., Sjostrom S. K., Garraway L. A., Weremowicz S., Richardson A. L., Greulich H., Stewart C. J., Mulvey L. A., Shen R. R., Ambrogio L., Hirozane-Kishikawa T., Hill D. E., Vidal M., Meyerson M., Grenier J. K., Hinkle G., Root D. E., Roberts T. M., Lander E. S., Polyak K., Hahn W. C. (2007) Cell 129, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 32.Boulton T. G., Yancopoulos G. D., Gregory J. S., Slaughter C., Moomaw C., Hsu J., Cobb M. H. (1990) Science 249, 64–67 [DOI] [PubMed] [Google Scholar]
- 33.Kohn A. D., Takeuchi F., Roth R. A. (1996) J. Biol. Chem. 271, 21920–21926 [DOI] [PubMed] [Google Scholar]
- 34.Mu J., Brozinick J. T., Jr., Valladares O., Bucan M., Birnbaum M. J. (2001) Mol. Cell 7, 1085–1094 [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Rhodes C. J., Lawrence J. C., Jr. (2006) J. Biol. Chem. 281, 24293–24303 [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Lian H., Zhao Y., Kauss M. A., Spindel S. (2008) J. Biol. Chem. 283, 25596–25605 [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Yen A. (2008) J. Biol. Chem. 283, 4375–4386 [DOI] [PubMed] [Google Scholar]
- 39.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 40.Aita V. M., Liang X. H., Murty V. V., Pincus D. L., Yu W., Cayanis E., Kalachikov S., Gilliam T. C., Levine B. (1999) Genomics 59, 59–65 [DOI] [PubMed] [Google Scholar]
- 41.Wang J. (2008) Autophagy 4, 947–948 [DOI] [PubMed] [Google Scholar]
- 42.Scott R. C., Juhász G., Neufeld T. P. (2007) Curr. Biol. 17, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki H., Takada Y., Kondo S., Sawaya R., Aggarwal B. B., Kondo Y. (2007) Mol. Pharmacol. 72, 29–39 [DOI] [PubMed] [Google Scholar]
- 44.Zhu J. H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C. T. (2007) Am. J. Pathol. 170, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samari H. R., Seglen P. O. (1998) J. Biol. Chem. 273, 23758–23763 [DOI] [PubMed] [Google Scholar]
- 46.Meley D., Bauvy C., Houben-Weerts J. H., Dubbelhuis P. F., Helmond M. T., Codogno P., Meijer A. J. (2006) J. Biol. Chem. 281, 34870–34879 [DOI] [PubMed] [Google Scholar]
- 47.Xu J., Keeton A. B., Franklin J. L., Li X., Venable D. Y., Frank S. J., Messina J. L. (2006) J. Biol. Chem. 281, 982–992 [DOI] [PubMed] [Google Scholar]
- 48.Corradetti M. N., Inoki K., Bardeesy N., DePinho R. A., Guan K. L. (2004) Genes Dev. 18, 1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 50.Shaw R. J., Bardeesy N., Manning B. D., Lopez L., Kosmatka M., DePinho R. A., Cantley L. C. (2004) Cancer Cell 6, 91–99 [DOI] [PubMed] [Google Scholar]
- 51.Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 52.Sharrocks A. D., Yang S. H., Galanis A. (2000) Trends Biochem. Sci. 25, 448–453 [DOI] [PubMed] [Google Scholar]
- 53.Navé B. T., Ouwens M, Withers D. J., Alessi D. R., Shepherd P. R. (1999) Biochem. J. 344, 427–431 [PMC free article] [PubMed] [Google Scholar]
- 54.Corcelle E., Nebout M., Bekri S., Gauthier N., Hofman P., Poujeol P., Fénichel P., Mograbi B. (2006) Cancer Res. 66, 6861–6870 [DOI] [PubMed] [Google Scholar]
- 55.Ogier-Denis E., Pattingre S., El Benna J., Codogno P. (2000) J. Biol. Chem. 275, 39090–39095 [DOI] [PubMed] [Google Scholar]
- 56.Pattingre S., Bauvy C., Codogno P. (2003) J. Biol. Chem. 278, 16667–16674 [DOI] [PubMed] [Google Scholar]
- 57.Subramaniam S., Unsicker K. (2006) Neuroscience 138, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 58.Colanzi A., Deerinck T. J., Ellisman M. H., Malhotra V. (2000) J. Cell Biol. 149, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colanzi A., Suetterlin C., Malhotra V. (2003) Curr. Opin. Cell Biol. 15, 462–467 [DOI] [PubMed] [Google Scholar]
- 60.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. (2001) EMBO Rep. 2, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Høyer-Hansen M., Jäättelä M. (2007) Autophagy 3, 381–383 [DOI] [PubMed] [Google Scholar]