Abstract
The Bcl-x pre-mRNA is alternatively spliced to produce the anti-apoptotic Bcl-xL and the pro-apoptotic Bcl-xS isoforms. By performing deletion mutagenesis on a human Bcl-x minigene, we have identified a novel exonic element that controls the use of the 5′ splice site of Bcl-xS. The proximal portion of this element acts as a repressor and is located downstream of an enhancer. Further mutational analysis provided a detailed topological map of the regulatory activities revealing a sharp transition between enhancer and repressor sequences. Portions of the enhancer can function when transplanted in another alternative splicing unit. Chromatography and immunoprecipitation assays indicate that the silencer element interacts with heterogeneous ribonucleoprotein particle (hnRNP) K, consistent with the presence of putative high affinity sites for this protein. Finally, down-regulation of hnRNP K by RNA interference enhanced splicing to Bcl-xS, an effect seen only when the sequences bound by hnRNP K are present. Our results therefore document a clear role for hnRNP K in preventing the production of the pro-apoptotic Bcl-xS splice isoform.
Alternative splicing is a major mechanism used to augment the number of proteins encoded by the genome. It is estimated that as many as 97% of multiple exon pre-mRNAs undergo alternative splicing (1, 2). Disruption of alternative splicing by mutating important regulatory sequences or by altering the expression or activity of proteins controlling splice site selection has been linked with different diseases, including cancer (3–7). Apoptosis is an important and complex cellular program involved in development and differentiation in higher organisms (8, 9). However, its aberrant control often contributes to cancer development and the resistance of cancer cells to drug therapy (10–13).
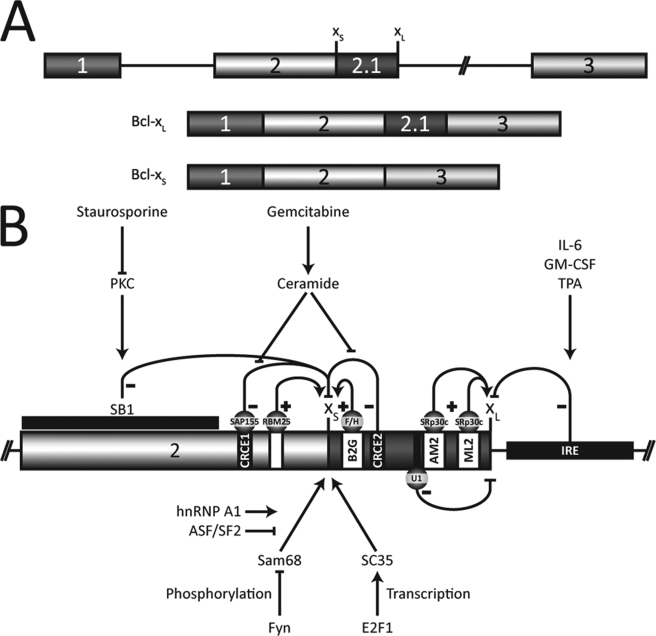
Genes implicated in the apoptotic pathway are alternatively spliced often to produce protein isoforms with distinct or even antagonistic activities (14, 15). A good example is the apoptotic regulator Bcl-x, which is alternatively spliced to produce two major isoforms, the anti-apoptotic Bcl-xL protein and the shorter pro-apoptotic Bcl-xS isoform (16). This alternative splicing decision involves a competition between two 5′ splice sites; the use of the downstream site creates Bcl-xL, and the use of the upstream one produces Bcl-xS (Fig. 1A). Bcl-xL is always the predominant form in cancer cells, and overexpressing it can confer resistance to chemotherapeutic agents (17–22). On the other hand, overexpression of the pro-apoptotic Bcl-xS isoform enhances sensitivity to the topoisomerase inhibitor etoposide and to taxol in a breast cancer cell line, while triggering apoptosis in melanoma cell lines (23, 24). Using antisense technologies to improve the production of the Bcl-xS splice variant can also induce apoptosis in cancer cells (25–27).
FIGURE 1.
A, alternative splicing of Bcl-x produces two major isoforms, Bcl-xL and Bcl-xS. B, regulation of Bcl-x alternative splicing. The enhancer elements are shown as white boxes, and the repressors are black. The pointed and flat arrows indicate positive and negative regulation, respectively. Protein kinase C inhibition relieves repression caused by the SB1 element on the Bcl-xS splice site (36). The repressor elements CRCE1, recognized by SAP155, and CRCE2 mediate the production of Bcl-xS by ceramide as when induced by gemcitabine in A549 cells (38, 39). hnRNP F/H binds to the B2G element to enhance the production of the Bcl-xS isoform (41). RBM25, through an element located upstream of the Bcl-xS splice site, can also augment its use (44). A large intronic region (IRE) mediates the Bcl-xL increase caused by interleukin-6 (IL-6), granulocyte-macrophage colony-stimulating factor (GM-CSF), and 12-O-tetradecanoylphorbol-13-acetate (TPA) (35). Finally, the B3 region also enhances Bcl-xL formation through the binding of SRp30c to AM2 and ML2 and the U1 snRNP to two cryptic 5′ splice sites (42).
Alternative splicing is regulated by different proteins bound to sequence elements near splice sites. A variety of mechanisms is used to achieve regulation. Some splicing factors act by recruiting or inhibiting the binding of different components of the spliceosome. Others may change the conformation of the pre-mRNA to mask a splice site or to bring a pair of splice sites into closer proximity (28, 29).
Although individual factors can have a strong and specific effect on splicing decisions, alternative splicing often relies on a combination of factors to determine the appropriate levels of isoforms. The implication of multiple proteins likely provides additional levels of regulation that helps attuned splicing control to a variety of stresses, environmental cues, and growth conditions. In several cases, the interaction of regulatory factors can be antagonistic. For example, in the Drosophila male-specific-lethal-2 (msl-2) pre-mRNA, recruitment of SXL to a uridine-rich region interferes with the binding of TIA-1 that is necessary for efficient U1 snRNP2 recruitment at the 5′ splice site (30). On the same pre-mRNA, SXL also diminishes U2AF recognition of the polypyrimidine tract at the 3′ splice site. TIA proteins bound to a U-rich element on the avian myosin phosphatase targeting subunit-1 (MYPT1) pre-mRNA repress the binding of PTB (31). PTB can also reduce the recruitment of ETR-3 to intronic elements near exon 5 of cardiac troponin T (32). In neurons, the binding of PTB to the introns surrounding the N1 exon of c-src is antagonized by nPTB protein, promoting exon inclusion. On the hnRNP A1 pre-mRNA, PTB diminishes the binding of SRp30c to the intronic CE9 element, reducing the inhibition by this protein on the use of the downstream 3′ splice site (33). SC35 and hnRNP A1 have partially overlapping binding sites on the human immunodeficiency virus 1 (HIV-1) tat exon 2. Preferential binding of SC35 enhances the inclusion of the exon, whereas hnRNP A1, by reducing SC35 binding, increases exclusion (34). Thus, the competition provided by an overlapping or a closely abutting pair of enhancer/ silencer represents a simple and frequent mechanism of splicing control.
The regulation of Bcl-x alternative splicing has received some attention in recent years leading to the discovery of several cis-acting elements and a few trans-acting control factors (Fig. 1B). Intronic regions downstream from the Bcl-xL 5′ splice site have been implicated as mediating signals from cytokines such as interleukin-6 and granulocyte-macrophage colony-stimulating factor (35). In addition, we have reported that an element located 187 nt upstream of the Bcl-xS splice site mediates a protein kinase C-dependent signal that represses splicing to the Bcl-xS donor site (36). On the other hand, ceramide enhances the use of the Bcl-xS 5′ splice site by lifting the repression mediated by two other elements (37, 38). The activity of one of these apparently involves SAP155 (39). The RNA-binding protein Sam68, under the control of the tyrosine kinase Fyn, can also increase the production of Bcl-xS in cooperation with hnRNP A1 (40), and this effect is inhibited by overexpression of ASF/SF2. The Bcl-x sequences bound by the above factors remain to be identified. We also uncovered enhancer elements for Bcl-xS and Bcl-xL. hnRNP F and H bind downstream of the Bcl-xS 5′ splice site to stimulate splicing to that site (41). Enhancement of Bcl-xL is conferred by SRp30c, which binds upstream of the 5′ splice site to antagonize the repressor activity of pseudo 5′ splice sites (42). Recently, the SR protein SC35 was shown to increase the production of Bcl-xS (43). Finally, the binding of RBM25 to a sequence element upstream of the Bcl-xS 5′ splice site stimulated its use, possibly by recruiting U1 snRNP through its interaction with the U1-associated protein hLuc7A (44). Thus, the region located between the two competing 5′ splice sites of Bcl-x is densely populated by splicing control elements.
In this study, we have pursued our characterization of Bcl-x splicing control by examining the contribution of sequences directly upstream of the Bcl-xS donor site. Our mutational approach identified a region containing flanking enhancer and silencer activities. The activity of the repressor portion is mediated by hnRNP K, which makes this protein an anti-apoptotic regulator.
EXPERIMENTAL PROCEDURES
Cell Culture
The 293 cells used in this study were the EcR-293 cell line (Invitrogen). EcR-293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% glutamine. PC-3 cells were maintained in Ham's F-12 medium containing 10% fetal bovine serum and 1% glutamine.
Plasmid Construction
The Bcl-x minigene X2.13 was constructed as described previously (41). The ΔB1 deletion was created by amplifying the X2.13 with primers XS + BsmI (ATATAGGGCATTCCTTTGAACAGGT) and Bcl-x AccI (ATCTCCTTGTCTACGCTT) using Pfu polymerase. The resulting insert was digested with BsmI and AccI and inserted in the S2.13 minigene digested with the same enzymes. The resulting construction was inserted in pcDNA3.1+ as described previously (41). To construct ΔB1d, a first fragment of X2.13 was amplified with Pfu using oligos B1down25 (TACCGGCGGGCATTCTCACCCCAGGGACAG) and Human-4 (ATGCCTGATCTCTGAAGCACAG). A second fragment was amplified using B1down25-B (GAATGCCCGCCGGTACCGCAG) and RT-1 (GAACCCACTGCTTACTGGCT). Both fragments were purified on gel and extracted (Qiagen) and then mixed in equimolar quantities and amplified with Pfu using oligos RT-1 and Human-4. The resulting product was digested with NheI and HpaI, purified on agarose gel, and ligated in X2.13 previously digested with the same enzymes. The same technique was used to obtain ΔB1u (oligos B1up25 (ACTACCTGTTCAAAGTGTGGAGCTGGGATG) with Human-4 and B1up25-B (CTTTGAACAGGTAGTGAATGA) with RT-1), ΔB1AG (oligos B1AGdown25 (GGAGGCAGGCGAAGTGACCTGACA) with Human-4 and B1ACGdown25-B (TCGCCTGCCTCCCTC) with RT-1), and ΔB1AC (oligos B1ACdown25 (GGAGGCAGGCGATCACCCCAGGGA) with Human-4 and B1ACGdown25-B with RT-1). Point mutations were obtained by Pfu amplification using the same overlapping primer technique. To insert the elements in minigene 45, we hybridized oligos representing the elements using an equimolar ratio of DNA oligomers for the sense and antisense strands, incubated in 1× One-Phor-All buffer (GE Healthcare) at 100 °C for 10 min, and then slowly cooling down to 21 °C during 2 h. These mixtures were then ligated in minigene 45 previously cut with BseRI and blunted with Klenow. All constructs were verified by digestion and sequencing.
Transfection of Plasmids or siRNA and RNA Extraction
All transfections of plasmids, RNA extractions, and consequent RT-PCR analysis were done as described previously (36) as were treatments with RNAi (41). The target sequences for siK1 and siK2 were GAGCGCAUAUUGAGUAUCA and UCUAGCAGGAGGAAUUAUU, respectively.
Transcription and Splicing Assays
In vitro transcription and splicing of minigene 45 and derivatives, as well as RT-PCR analysis of the products, were done as described previously (45). Templates for the RNA transcripts used in the immunoprecipitation were amplified by PCR (PfuTurbo, Stratagene) and gel-purified (Qiagen). Transcription was done using T3 RNA polymerase (Promega) and [α-32P]CTP (PerkinElmer Life Sciences).
RNA Chromatography
Coupling the RNA (IDT) to the beads and incubation of nuclear extracts in splicing conditions were done as described previously (33). After incubation, the beads were washed twice with 8 volumes of KCl-free buffer D (60 mm HEPES, pH 7.9, 0.2 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, and 20% glycerol) and then eluted twice with 4 volumes of buffer D containing 100 mm KCl. These were pooled and then precipitated by adding 1 volume of trichloroacetic acid, incubating for 20 min on ice, and spinning for 5 min at 10,000 × g. The pellets were resuspended in 0.1 n NaOH. These steps, starting with washing with the previous eluting KCl concentration, were repeated for buffer D containing 250, 500, and 1000 mm KCl. The various eluates were loaded on a 10% polyacrylamide gel and stained with silver nitrate (Invitrogen), and the bands of interest were analyzed by mass spectroscopy.
RNA Immunoprecipitation
Sepharose-protein A beads (GE Healthcare) were incubated in buffer A (10 mm Tris-HCl, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2) containing 0.5% Triton X-100 for 1 h at 25 °C, washed three times in the same buffer, and then resuspended for a final volume of 50% beads. One μl of 12G4 anti-hnRNP K antibodies (kindly provided by G. Dreyfuss) was added per 50 μl of 50% slurry and incubated for 1 h at 4 °C. The beads were washed three times with cold buffer A. Equivalent quantities of transcripts were incubated in 12.5 μl of splicing mix (45) for 30 min at 4 °C and then washed five times with 1 ml of buffer A. Proteinase K and SDS were added to the beads and then incubated at 37 °C for 15 min. Following phenol extraction and ethanol precipitation, the RNA was resuspended in formamide dye and loaded onto a 6.5% denaturing acrylamide gel. The gels were then analyzed on a PhosphorImager and the results adjusted for relative amounts of radioactivity.
RESULTS
Mapping of Enhancer and Silencer Elements Upstream of the 5′ Splice Site of Bcl-xS
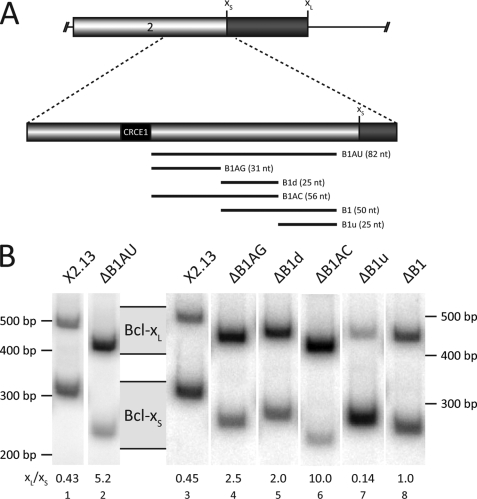
Previous deletion mutagenesis using Bcl-x minigenes (36, 41) transfected in HeLa cells identified an 82-nt region (B1AU, Fig. 2A), starting 10 nt upstream of the 5′ splice site of Bcl-xS that regulates Bcl-x splicing. When this region is deleted from the X2.13 minigene carrying the two 5′ splice sites of Bcl-x, the Bcl-xL/Bcl-xS splicing ratio is increased by 10-fold, as judged by RT-PCR analysis (Fig. 2B, compare lane 2 with lane 1). Thus, B1AU behaves as an enhancer for the Bcl-xS 5′ splice site. Further dissection of B1AU was performed by dividing the element into three parts. Deletion of the upstream B1AG portion (32 nt) also decreased the relative usage of the Bcl-xS site (Fig. 2B, compare lane 4 to lane 3). Removing the central portion of B1AU (B1d; 25 nt) had a similar effect (Fig. 2B, lane 5). The deletion of both B1d and B1AG to produce ΔB1AC increased splicing to the Bcl-xL 5′ splice site more than the impact of the two individual deletions (Fig. 2B, lane 6), indicating the presence of at least two functionally distinct enhancer elements. Furthermore, the impact of the B1AC deletion was even stronger than that of removing B1AU, suggesting that the downstream portion of B1AU (B1u; 25 nt) might possess silencer activity. Indeed, removing only B1u enhanced the use of the Bcl-xS splice site, consistent with the presence of a splicing silencer in this region (Fig. 2B, lane 7). Deleting both B1d and B1u (B1, Fig. 2B, lane 8) also decreased the effect relative to the deletion of B1d alone (lane 8). Thus, B1AU contains two antagonistic regions composed of at least three elements as follows: B1AG and B1d acting as enhancers, and B1u behaving as a repressor of Bcl-xS usage. The activities of the enhancer (B1AC and B1d) and silencer (B1u) elements were similarly detected when the analysis was carried out in human 293 and PC-3 cells (supplemental Fig. 1).
FIGURE 2.
Deletion mutagenesis of the B1 region leads to the discovery of three novel splicing regulating elements. A, locations of the deletions are indicated by the bars, and their sizes are shown in nucleotides. B, radiolabeled RT-PCR assays on total RNA extracted from HeLa cells transfected with the different deletion mutations. After separation on acrylamide gels, the bands were analyzed using a PhosphorImager. The corresponding Bcl-xL and Bcl-xS bands are shown, as well as the Bcl-xL/Bcl-xS ratio displayed above the lane numbers. The results presented are representative of many independent experiments.
Mutational Analysis of B1AG, B1d, and B1u
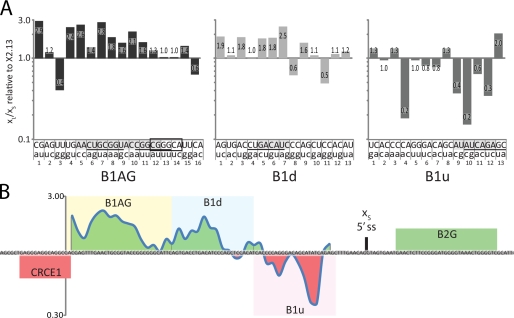
To identify nucleotides important for the activity of the different elements, a collection of 42 mutants was constructed, each one containing a dinucleotide mutation in B1AG, B1d, or B1u (Fig. 3). Each mutant was transfected in HeLa cells, and the xL/xS splicing ratio of plasmid-derived transcripts was determined by RT-PCR. The analysis was performed at least three times for most mutations (supplemental Fig. 2). The results of one experiment are expressed relative to the ratio obtained with the wild-type X2.13 construct (Fig. 3A). Because most of the mutations in B1AG reduced splicing to the Bcl-xS site (Fig. 3A, left panel), we may conclude that B1AG is an enhancer. Disruptions anywhere in the contiguous stretch of the central 16 nt (AACUGCGGUACCGGCG) shifted splicing toward Bcl-xL. In silico analysis of these nucleotides using ESEfinder (46, 47) identified putative ASF/SF2-binding sites (underlined in Fig. 3), and most of the mutations that reduce the strength of the sites also compromised usage of Bcl-xS. Intriguingly, mutating the nucleotides recently described as important for the binding of RBM25 (44) (boxed in Fig. 3) had no effect.
FIGURE 3.
Point mutations in the three elements affect Bcl-x splicing in HeLa cells. A, mutations for every two nucleotides are represented in lowercase below the wild-type sequence. Shown in the graph are Bcl-xL/Bcl-xS ratios for each mutation, relative to the Bcl-xL/Bcl-xS ratio of the minigene X2.13 which is represented by the horizontal line at 1.0. The nucleotides deemed important are highlighted in gray, and the putative binding sites for ASF/SF2 (B1AG and B1d) and SRp40 (B1u) are underlined. The RBM25-binding site recently discovered is boxed. B, topological map representing the average of the effect of the mutational analysis. The value for each mutation was calculated as an average of it and both adjoining mutations. For example, the value for mutation B1d.7 is an average of the values in A for the mutations B1d.6, B1d.7, and B1d.8.
A similar mutational analysis of B1d yielded a more nuanced conclusion (Fig. 3A, middle panel). The impact of mutating the first 14 nucleotides of B1d was consistent with the existence of a splicing enhancer targeting Bcl-xS, and a contiguous stretch of six nucleotides (GACAUC) that may represent a binding site for ASF/SF2 (underlined) appeared important for this activity. In contrast, the mutations that had the most impact in the remaining downstream portion of B1d improved Bcl-xS usage, a result that is more consistent with the presence of a silencer element. If a silencer element exists in B1d, it is not the dominant activity because deleting or transplanting B1d (see Fig. 4) indicates that its global activity is that of an enhancer.
FIGURE 4.
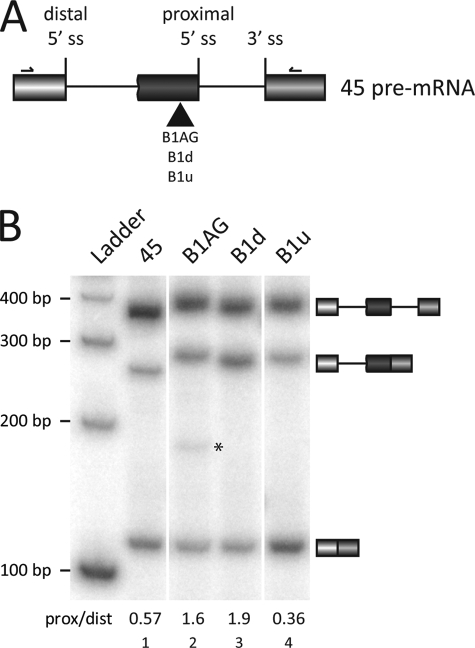
I nsertion of the enhancer elements in the minigene 45 replicates their activity. A, each element was inserted upstream of the proximal 5′ splice site (ss) of the minigene 45 as indicated. The primers used for subsequent RT-PCR analysis are shown. B, transcripts derived from minigene 45 were incubated in HeLa nuclear extracts in in vitro splicing conditions. RT-PCR analysis was done on total RNA. The splicing products are indicated as well as the proximal/distal (prox/dist) ratios. A new product presumably produced by a cryptic splice site is indicated with an asterisk.
Finally, dinucleotide mutations in B1u generally increased the use of Bcl-xS, consistent with this region being a silencer (Fig. 3A, right panel). The most important sequence was a stretch of eight nucleotides (AUAUCAGA) that represent putative binding sites for SRp40. However, mutations that did not affect the putative SRp40-binding site had a strong effect on Bcl-x splicing (Fig. 3A, right panel, mutation 9). Moreover, knocking down SRp40 in a variety of cell lines did not significantly affect Bcl-x splicing (data not shown).
Several mutants were also tested in 293 cells with an impact similar to what was observed in HeLa cells (supplemental Fig. 2). Globally, the results of the mutational analysis match very well the results of the deletions. Because splicing regulatory factors like SR and hnRNP proteins often have degenerate binding sites, not all mutations may alter in the same way the activity of a control element. Moreover, some mutations may create an element that imposes new splicing control. To facilitate the graphical representation of our results and to minimize the above caveats, we assigned to each mutation a value that is an average of the impact of the mutation and the two adjoining mutations, thus more accurately displaying the contribution of existing elements (Fig. 3B). The most important feature of this map is a transition from enhancer to silencer activities occurring in B1d. Although B1d globally behaves as an enhancer because of its 5′ portion, its 3′ portion appears to represent the extremity of a silencer that extends into B1u.
Enhancer Elements Can Function When Placed in a Different Context
Mutations may modify alternative splicing by changing the secondary structure near a splice site or because they compromise the interaction with a trans-acting regulatory factor. To assess the intrinsic modulatory activity of the elements on alternative splicing, we inserted each one into a previously characterized reporter gene (45) containing the 5′ splice sites of exon 7 and 7B of hnRNP A1, their surrounding sequences, and the 3′ splice site of the adenovirus major late exon L2 (Fig. 4A). The resulting minigenes were transcribed in vitro, and the resulting pre-mRNAs were incubated in HeLa extracts for 2 h. RT-PCR analysis was conducted to assess the splicing behavior of the inserted element. Placing B1AG upstream of the proximal 5′ splice site improved its use (Fig. 4B, compare lane 2 with lane 1). When the other enhancer element (B1d) was inserted at the same position, proximal 5′ splice site usage was also increased (Fig. 4B, lane 3). Finally, the silencer element B1u had very little impact on splicing (Fig. 4B, lane 4). These results were confirmed by testing the various transcripts in several batches of nuclear extracts (not shown). Our results therefore suggest that the enhancer activities of B1AG and B1d are mediated by trans-acting factor(s). The activity of B1u does not appear transplantable (Fig. 4B, lane 4), suggesting that B1u may promote splicing repression by preventing the activity of the upstream enhancers.
hnRNP K Binds to the B1 Element
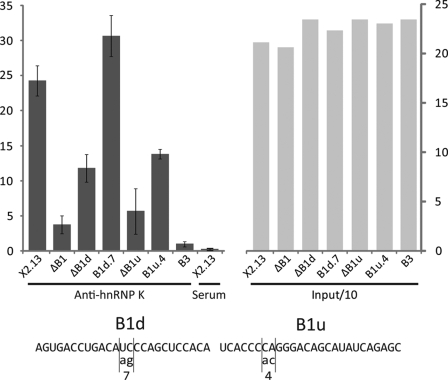
To isolate factors that mediate the activity of these control elements, we carried out affinity chromatography with RNA portions covalently linked to agarose beads using HeLa nuclear extracts. The bound material was eluted with increasing amounts of KCl. The content was fractionated on acrylamide gels and revealed by silver staining; bands were cut, and proteins were analyzed by nanoliquid chromatography coupled on line with tandem mass spectrometry (supplemental Fig. 3). The strongest hit obtained came with B1d for which a 60-kDa protein that eluted at 250 mm KCl was identified as hnRNP K. Binding of hnRNP K may be explained by the presence of UCCCAG and UCCACAU, which are sequences very similar to the high affinity RNA-binding site for hnRNP K (UC3–4(AU)/(UA)) identified through SELEX (48). Our previous analysis in Fig. 3A showed that mutating the two central cytidines of either sequence stimulated splicing to the 5′ splice site of Bcl-xS, consistent with the view that hnRNP K might repress the production of Bcl-xS.
To further analyze the interaction of hnRNP K with the Bcl-x pre-mRNA, we carried out an RNA immunoprecipitation assay using our model Bcl-x pre-mRNA (X2.13) and versions carrying various deletions. Radiolabeled transcripts were incubated in HeLa nuclear extracts under in vitro splicing conditions. hnRNP K antibody (a kind gift from G. Dreyfuss) conjugated to protein A-Sepharose was then added. After several washes, the labeled RNA was recovered and quantitated on the gel. This assay revealed that the binding of hnRNP K to X2.13 strongly decreased when B1, B1d, or B1u were deleted (Fig. 5). This interaction of hnRNP K with B1u may be explained by the presence of a contiguous stretch of four cytidines, a known recognition site for hnRNP K (49). Consistent with this possibility, the mutation B1u.4, which changes the last cytidine of that stretch and severely compromises the activity of the silencer, diminished the recovery of the RNA by anti-hnRNP K immunoprecipitation. Mutating the abutting CC into AA did not compromise recovery with the anti-hnRNP K antibody nor did it affect Bcl-x splicing (Fig. 3A). Notably, mutation B1d.7, which has an effect opposite that of deleting B1u (Fig. 2B and Fig. 3A), slightly but significantly increased recovery of the RNA by anti-hnRNP K immunoprecipitation. This mutation may enhance hnRNP K binding directly or indirectly by disrupting a binding site for an enhancer protein that competes with hnRNP K for binding in the region where the transition between enhancer and silencer activities occurs. We could not confirm direct binding of hnRNP K because our recombinant hnRNP K protein produced in bacteria lacked any type of binding activity.
FIGURE 5.
RNA co-immunoprecipitation of a portion of the Bcl-x pre-mRNA using hnRNP K antibodies is dependent on the B1 region. The transcripts were incubated in HeLa nuclear extracts in splicing conditions and then on beads coupled with hnRNP K antibodies (Anti-hnRNP K) or preimmune serum (Serum). After five washes, the RNA was precipitated, migrated on an acrylamide gel, and quantified on a PhosphorImager. The left panel is a graph of the values of radioactivity measured. The right panel shows the relative levels of a tenth of the input RNA. The sequence of the point mutations that were tested is indicated in the bottom panel.
Knockdown of hnRNP K Affects Bcl-x Splicing in a B1-dependent Manner
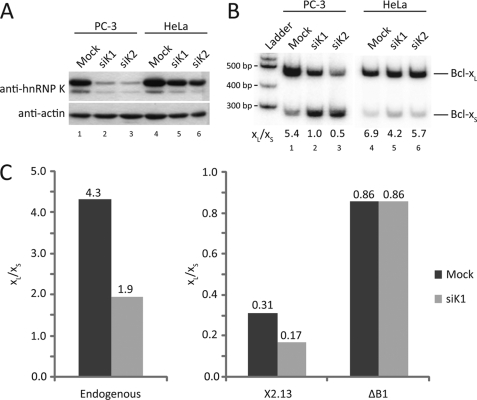
A few studies have documented a role for hnRNP K in splicing control (50–52). The related Nova proteins contain KH domains found in prototypical hnRNP K, and they regulate brain-specific alternative splicing events (52–54). To clarify the contribution of hnRNP K to Bcl-x splicing, we knocked down hnRNP K expression by RNA interference using two different nonoverlapping siRNAs (siK1 and siK2) in PC-3 and HeLa cells. The knockdown was successful in PC-3 cells but was less efficient in HeLa cells (Fig. 6A). Assessing the impact of the knockdowns on the endogenous Bcl-x splicing profiles in PC-3 cells revealed an increase in the use of the Bcl-xS 5′ splice site (Fig. 6B, lanes 1–3), consistent with the notion that hnRNP K represses the splicing of Bcl-xS. The effect was less dramatic in HeLa cells (Fig. 6B, lanes 4–6), as expected from a more limited depletion. We observed increased Bcl-xS isoform formation upon siRNA treatment in several other cell lines (data not shown). To confirm that the modulation of splicing by hnRNP K requires the B1 element, we looked at the impact of the hnRNP K depletion on the splicing of transcripts produced from minigenes X2.13 containing or lacking the B1 element (Fig. 6C). Only the wild-type X2.13 minigene experienced a decrease in the Bcl-xL/Bcl-xS ratio when hnRNP K was depleted in PC-3 cells. Thus, our results indicate that the activity of hnRNP K is mediated through B1, most likely through the C-rich elements important for hnRNP K binding in B1d and B1u.
FIGURE 6.
K nockdown of hnRNP K affects Bcl-x splicing in a B1-dependent way. A, knockdown of hnRNP K was done in PC-3 and HeLa cells using two different siRNAs (siK1 and siK2) or mock-transfected (Mock). Actin was used as a loading control. Proteins loaded on an acrylamide gel were revealed using antibodies against hnRNP K and actin. B, RT-PCR analysis of endogenous Bcl-x mRNAs using the RNAi-treated PC-3 and HeLa cells. The Bcl-xL/Bcl-xS ratios are indicated above the lane numbers. C, siK1 RNAi-treated PC-3 cells were mock-transfected or transfected with the wild-type minigene or its ΔB1 version. Twenty four h later, total RNA was extracted and analyzed using radiolabeled RT-PCR with primers for endogenous Bcl-x (for mock-transfected cells) or primers specific for the minigenes. The graphs represent the Bcl-xL/Bcl-xS ratios for each sample.
DISCUSSION
Given the functional importance of the major Bcl-x splice isoforms in apoptosis and their antagonistic roles, it is not surprising that the splicing decisions that occur on the Bcl-x pre-mRNA are regulated by a variety of elements and factors (35–41, 44, 55). Many of these elements may help link splicing regulation with specific pathways that monitor the integrity of various cellular components and compartments as well as the availability of the nutrients and growth conditions.
In this study, we document the activity of an 82-nt region (B1AU) located immediately upstream of the 5′ splice site of Bcl-xS. The 5′ portion of this region displays enhancer activity because deleting or mutating specific nucleotides decreases the use of the Bcl-xS site. Because portions of this region can function in a heterologous context, the activity of the enhancer likely requires trans-acting factor(s). The region contains three putative high affinity binding sites for ASF/SF2. However, a role for ASF/SF2 in the activity of these elements is unlikely because overexpressing recombinant ASF/SF2 in a variety of cell lines increases the production of Bcl-xL rather than that of Bcl-xS (40, 42, 56, 57). Recently, RBM25 was identified as binding to a six-nucleotide element (CGGGCA) in B1AG (44). Although the activity of RBM25 in HeLa cells is consistent with the enhancer activity of B1AG, our mutational analysis did not reveal a role for these sequences both in 293 and HeLa cells (Fig. 3 and data not shown). Perhaps mutating only two nucleotides at a time was not enough to destroy the activity of this sequence element. The SR protein SC35 has also been implicated as mediating the increase in Bcl-xS upon treatment of various cell lines with doxycycline or cyclophosphamide (43). However, these drugs and the knockdown of SC35 did not affect Bcl-x splicing in HeLa and PC-3 cells (data not shown). Further work will be required to identify the factors that are responsible for the enhancing activity of B1AG and B1d.
We also mapped a silencer occupying the 3′ half of the B1AU element. RNA affinity chromatography using a subregion identified hnRNP K, a protein that was previously reported to stimulate inclusion of the β-tropomyosin exon 6A (50) but acted as a repressor in the glucose-6-phosphate dehydrogenase pre-mRNA (51). hnRNP K can also bind to the Nova1 pre-mRNA and can decrease the inclusion of alternative exon 4 (52). Recently, it was shown that hnRNP K can play a prominent role in alternative splicing control because nearly half of 56 alternative splicing events in apoptotic genes were affected upon hnRNP K depletion, either enhancing or suppressing exon inclusion (58). This broad role in splicing regulation may come from its ability to interact with several splicing-related proteins, including other hnRNP proteins (49, 59). As with other hnRNP proteins, hnRNP K has also been implicated in other steps of gene expression, including transcription and translation (49). For example, hnRNP K activates transcription of c-myc and serves as a cofactor for p53-dependent transcriptional activation following DNA damage (60, 61), but it can also repress transcription of thymidine kinase and CD43 (62, 63). hnRNP K also represses translation of p21 and c-src during neuronal and erythroid differentiation, respectively (64, 65).
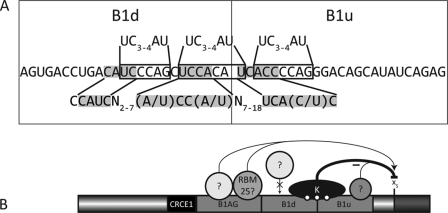
In the case of Bcl-x, hnRNP K binds to portions of B1 containing C-rich elements that are similar to the high affinity sites identified by SELEX (48). These sites are located in the silencer portion of B1AU. Depleting hnRNP K by RNAi provokes a B1-dependent increase in the production of Bcl-xS. Although we cannot completely eliminate off-target effects of the RNAi approach, our results are consistent with a role for hnRNP K in repressing the use of Bcl-xS. Our mutational analysis indicates that the silencer element occupies the 3′ end of B1d and all of B1u. Individually mutating the two central cytidines of the two putative high affinity binding sites for hnRNP K in B1d abrogated the repression, consistent with a direct role for hnRNP K in the activity of this portion of B1d. Thus, B1d likely represents a pivotal region because it is bound by both positive and negative control factors. Another putative binding site for hnRNP K is present in B1u (ACCCCA) (Fig. 7A). Mutating the last two nucleotides strongly diminished the repression of Bcl-xS, as well as decreased binding of hnRNP K. Another high affinity binding site that was previously identified using yeast three-hybrid screens is CCAUCN2–7(A/U)CC(A/U)N7–18UCA(C/U)C (66, 67). B1d and B1u contain ACAUCCCAGCUCCACA|UCACCCCA (the vertical line indicates the division between both elements). Thus, three putative hnRNP K-binding sites exist within a stretch of 25 nucleotides, and cooperative interactions may stabilize the binding of hnRNP K and antagonize the binding of positive factors in the central portion of B1d. The fact that B1u displays little activity when transplanted in a different pre-mRNA is consistent with a model in which the role of the silencer would be to antagonize the binding or activity of flanking activators. The silencer element is likely to be more complex than the binding of hnRNP K alone because the 3′ end of B1u is the most active silencer region and yet it lacks putative hnRNP K-binding sites. A simple working model for the antagonizing activities associated with B1AU is presented in Fig. 7B. The enhancer activity would be mediated by factor(s) interacting with the 5′ half, perhaps RBM25. The central portion contains sequences bound by hnRNP K, but this region may overlap with binding sites for the enhancing factors. Two putative hnRNP K-binding sites already exist in B1d, and a third one is likely present in B1u. As indicated in the Introduction, overlapping binding sites for factors with different activities are often used to control splicing decisions in other pre-mRNAs (33, 34, 68, 69).
FIGURE 7.
A, putative binding sites for hnRNP K in the B1d and B1u elements. The three sites resembling the high affinity binding sites are boxed and compared with those identified by SELEX (48), shown above the RNA sequence for the B1 element. Below is the binding site defined using yeast three-hybrid screens, compared with the shaded binding site found on the Bcl-x pre-mRNA. B, hnRNP K binding to B1 may compete with the binding of an unknown enhancer protein, thus inhibiting splicing to the Bcl-xS site. Deletion of the enhancer element B1AG augments the use of the Bcl-xL splice site. When the other enhancer element, B1d, is removed, the inhibition by hnRNP K on the Bcl-xS 5′ splice site would be slightly diminished, perhaps by the stronger binding of a protein to B1AG or reduced binding affinity of hnRNP K to the pre-mRNA. Removal of the silencer B1u would abrogate hnRNP K binding leading to a strong increase in the production of the Bcl-xS isoform, as does a knockdown of hnRNP K by RNA interference.
hnRNP K can interact with Sam68 in vitro and in vivo (70, 71), a protein known to regulate Bcl-x splicing (40). However, although we have shown that hnRNP K represses the production of Bcl-xS, Sam68 stimulates it. Although it is unclear where Sam68 binds, hnRNP K may neutralize Sam68 by interacting with it when these proteins are in close proximity. However, performing a knockdown of Sam68 had no effect in our conditions and cell lines (data not shown).
The ability of hnRNP K to repress the production of the pro-apoptotic Bcl-xS isoform would confer to hnRNP K an anti-apoptotic function that may help cancer cells escape death signals. As shown recently, the knockdown of hnRNP K can influence the alternative splicing of several apoptotic genes (58), including favoring the production of the pro-apoptotic splice form of MCL1.3 MCL1 is another BCL2 family member, and its pro-apoptotic variant is consistently underproduced in breast cancer tissues (72). An anti-cell death function for hnRNP K is also suggested by the increase in the nuclear concentration of hnRNP K that occurs in proliferating cells as well as in tumors (73) and the higher level of hnRNP K observed in a variety of cancers compared with the corresponding healthy tissues (74–80). Overexpression of hnRNP K also led to the transformation of Rat1A cells (81). Because DNA damage transiently increases the production of hnRNP K in many human cancer cell lines (60), this response may be used to further repress the production of apoptotic isoforms, hence creating conditions to implement DNA repair without triggering apoptosis. Through its role in splicing, hnRNP K may therefore help coordinate the DNA damage response with apoptotic regulation.
Supplementary Material
Acknowledgments
We thank Gideon Dreyfuss for the hnRNP K antibodies, Shu-Ching Huang for the RBM25 antibodies, and Catherine Desrosiers for Taq polymerase. We also thank Uli Froehlich and the Laboratory of Functional Genomics of the Université de Sherbrooke for providing reagents. We are grateful to Laetitia Michelle and Lulzim Shkreta for discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
B. Chabot, unpublished results.
- snRNP
- small nuclear ribonucleoprotein
- hnRNP
- heterogeneous nuclear ribonucleoprotein particles
- RT
- reverse transcription
- siRNA
- small interfering RNA
- siK
- siRNA targeting hnRNP K
- PTB
- polypyrimidine tract-binding protein
- nt
- nucleotide
- RNAi
- RNA interference.
REFERENCES
- 1.Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 2.Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G. S., Cooper T. A. (2007) Nat. Rev. Genet. 8, 749–761 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Blanco M. A., Baraniak A. P., Lasda E. L. (2004) Nat. Biotechnol. 22, 535–546 [DOI] [PubMed] [Google Scholar]
- 5.Baralle D., Baralle M. (2005) J. Med. Genet. 42, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venables J. P. (2006) BioEssays 28, 378–386 [DOI] [PubMed] [Google Scholar]
- 7.Shkreta L., Revil T., Bell B., Venables V. P., Prinos P., Abou Elela S., Chabot B. (2008) Mol. Cancer Ther. 7, 1398–1409 [DOI] [PubMed] [Google Scholar]
- 8.Baehrecke E. H. (2002) Nat. Rev. Mol. Cell Biol. 3, 779–787 [DOI] [PubMed] [Google Scholar]
- 9.Hipfner D. R., Cohen S. M. (2004) Nat. Rev. Mol. Cell Biol. 5, 805–815 [DOI] [PubMed] [Google Scholar]
- 10.Green D. R., Evan G. I. (2002) Cancer Cell 1, 19–30 [DOI] [PubMed] [Google Scholar]
- 11.Fulda S., Debatin K. M. (2006) Oncogene 25, 4798–4811 [DOI] [PubMed] [Google Scholar]
- 12.Viktorsson K., Lewensohn R., Zhivotovsky B. (2005) Adv. Cancer Res. 94, 143–196 [DOI] [PubMed] [Google Scholar]
- 13.Tsuruo T., Naito M., Tomida A., Fujita N., Mashima T., Sakamoto H., Haga N. (2003) Cancer Sci. 94, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwerk C., Schulze-Osthoff K. (2005) Mol. Cell 19, 1–13 [DOI] [PubMed] [Google Scholar]
- 15.Akgul C., Moulding D. A., Edwards S. W. (2004) Cell. Mol. Life Sci. 61, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 17.Lebedeva I., Rando R., Ojwang J., Cossum P., Stein C. A. (2000) Cancer Res. 60, 6052–6060 [PubMed] [Google Scholar]
- 18.Watanabe J., Kushihata F., Honda K., Mominoki K., Matsuda S., Kobayashi N. (2002) Int. J. Oncol. 21, 515–519 [PubMed] [Google Scholar]
- 19.España L., Fernández Y., Rubio N., Torregrosa A., Blanco J., Sierra A. (2004) Breast Cancer Res. Treat. 87, 33–44 [DOI] [PubMed] [Google Scholar]
- 20.Linden M., Kirchhof N., Carlson C., Van Ness B. (2004) Blood 103, 2779–2786 [DOI] [PubMed] [Google Scholar]
- 21.Castilla C., Congregado B., Chinchón D., Torrubia F. J., Japón M. A., Sáez C. (2006) Endocrinology 147, 4960–4967 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z. B., Zhang Y., Liu Y. Q., Guo Y., Xu H., Dong B., Cui Y. F. (2006) Cell Biol. Int. 30, 15–20 [DOI] [PubMed] [Google Scholar]
- 23.Sumantran V. N., Ealovega M. W., Nuñez G., Clarke M. F., Wicha M. S. (1995) Cancer Res. 55, 2507–2510 [PubMed] [Google Scholar]
- 24.Hossini A. M., Eberle J., Fecker L. F., Orfanos C. E., Geilen C. C. (2003) FEBS Lett. 553, 250–256 [DOI] [PubMed] [Google Scholar]
- 25.Mercatante D. R., Mohler J. L., Kole R. (2002) J. Biol. Chem. 277, 49374–49382 [DOI] [PubMed] [Google Scholar]
- 26.Mercatante D. R., Bortner C. D., Cidlowski J. A., Kole R. (2001) J. Biol. Chem. 276, 16411–16417 [DOI] [PubMed] [Google Scholar]
- 27.Wilusz J. E., Devanney S. C., Caputi M. (2005) Nucleic Acids Res. 33, 6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Contreras R., Fisette J. F., Nasim F. U., Madden R., Cordeau M., Chabot B. (2006) PLoS Biol. 4, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir-Ahmady B., Boutz P. L., Markovtsov V., Phillips M. L., Black D. L. (2005) RNA 11, 699–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Förch P., Merendino L., Martínez C., Valcárcel J. (2001) RNA 7, 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla S., Del Gatto-Konczak F., Breathnach R., Fisher S. A. (2005) RNA 11, 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlet-B N., Logan P., Singh G., Cooper T. A. (2002) Mol. Cell 9, 649–658 [DOI] [PubMed] [Google Scholar]
- 33.Paradis C., Cloutier P., Shkreta L., Toutant J., Klarskov K., Chabot B. (2007) RNA 13, 1287–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahler A. M., Damgaard C. K., Kjems J., Caputi M. (2004) J. Biol. Chem. 279, 10077–10084 [DOI] [PubMed] [Google Scholar]
- 35.Li C. Y., Chu J. Y., Yu J. K., Huang X. Q., Liu X. J., Shi L., Che Y. C., Xie J. Y. (2004) Cell Res. 14, 473–479 [DOI] [PubMed] [Google Scholar]
- 36.Revil T., Toutant J., Shkreta L., Garneau D., Cloutier P., Chabot B. (2007) Mol. Cell. Biol. 27, 8431–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalfant C. E., Rathman K., Pinkerman R. L., Wood R. E., Obeid L. M., Ogretmen B., Hannun Y. A. (2002) J. Biol. Chem. 277, 12587–12595 [DOI] [PubMed] [Google Scholar]
- 38.Massiello A., Salas A., Pinkerman R. L., Roddy P., Roesser J. R., Chalfant C. E. (2004) J. Biol. Chem. 279, 15799–15804 [DOI] [PubMed] [Google Scholar]
- 39.Massiello A., Roesser J. R., Chalfant C. E. (2006) FASEB J. 20, 1680–1682 [DOI] [PubMed] [Google Scholar]
- 40.Paronetto M. P., Achsel T., Massiello A., Chalfant C. E., Sette C. (2007) J. Cell Biol. 176, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garneau D., Revil T., Fisette J. F., Chabot B. (2005) J. Biol. Chem. 280, 22641–22650 [DOI] [PubMed] [Google Scholar]
- 42.Cloutier P., Toutant J., Shkreta L., Goekjian S., Revil T., Chabot B. (2008) J. Biol. Chem. 283, 21315–21324 [DOI] [PubMed] [Google Scholar]
- 43.Merdzhanova G., Edmond V., De Seranno S., Van den Broeck A., Corcos L., Brambilla C., Brambilla E., Gazzeri S., Eymin B. (2008) Cell Death Differ. 15, 1815–1823 [DOI] [PubMed] [Google Scholar]
- 44.Zhou A., Ou A. C., Cho A., Benz E. J., Jr., Huang S. C. (2008) Mol. Cell. Biol. 28, 5924–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasim F. U., Hutchison S., Cordeau M., Chabot B. (2002) RNA 8, 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith P. J., Zhang C., Wang J., Chew S. L., Zhang M. Q., Krainer A. R. (2006) Hum. Mol. Genet 15, 2490–2508 [DOI] [PubMed] [Google Scholar]
- 48.Thisted T., Lyakhov D. L., Liebhaber S. A. (2001) J. Biol. Chem. 276, 17484–17496 [DOI] [PubMed] [Google Scholar]
- 49.Bomsztyk K., Denisenko O., Ostrowski J. (2004) BioEssays 26, 629–638 [DOI] [PubMed] [Google Scholar]
- 50.Expert-Bezançon A., Le Caer J. P., Marie J. (2002) J. Biol. Chem. 277, 16614–16623 [DOI] [PubMed] [Google Scholar]
- 51.Griffith B. N., Walsh C. M., Szeszel-Fedorowicz W., Timperman A. T., Salati L. M. (2006) Biochim. Biophys. Acta 1759, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B. J., Darnell R. B. (2006) Nature 444, 580–586 [DOI] [PubMed] [Google Scholar]
- 53.Ule J., Darnell R. B. (2007) Adv. Exp. Med. Biol. 623, 148–160 [DOI] [PubMed] [Google Scholar]
- 54.Licatalosi D. D., Mele A., Fak J. J., Ule J., Kayikci M., Chi S. W., Clark T. A., Schweitzer A. C., Blume J. E., Wang X., Darnell J. C., Darnell R. B. (2008) Nature 456, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boon-Unge K., Yu Q., Zou T., Zhou A., Govitrapong P., Zhou J. (2007) Chem. Biol. 14, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massiello A., Chalfant C. E. (2006) J. Lipid Res. 47, 892–897 [DOI] [PubMed] [Google Scholar]
- 57.Li X., Wang J., Manley J. L. (2005) Genes Dev. 19, 2705–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venables J. P., Koh C. S., Froehlich U., Lapointe E., Couture S., Inkel L., Bramard A., Paquet E. R., Watier V., Durand M., Lucier J. F., Gervais-Bird J., Tremblay K., Prinos P., Klinck R., Elela S. A., Chabot B. (2008) Mol. Cell. Biol. 28, 6033–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mikula M., Dzwonek A., Karczmarski J., Rubel T., Dadlez M., Wyrwicz L. S., Bomsztyk K., Ostrowski J. (2006) Proteomics 6, 2395–2406 [DOI] [PubMed] [Google Scholar]
- 60.Moumen A., Masterson P., O'Connor M. J., Jackson S. P. (2005) Cell 123, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 61.Lee M. H., Mori S., Raychaudhuri P. (1996) J. Biol. Chem. 271, 3420–3427 [DOI] [PubMed] [Google Scholar]
- 62.Da Silva N., Bharti A., Shelley C. S. (2002) Blood 100, 3536–3544 [DOI] [PubMed] [Google Scholar]
- 63.Hsieh T. Y., Matsumoto M., Chou H. C., Schneider R., Hwang S. B., Lee A. S., Lai M. M. (1998) J. Biol. Chem. 273, 17651–17659 [DOI] [PubMed] [Google Scholar]
- 64.Yano M., Okano H. J., Okano H. (2005) J. Biol. Chem. 280, 12690–12699 [DOI] [PubMed] [Google Scholar]
- 65.Naarmann I. S., Harnisch C., Flach N., Kremmer E., Kühn H., Ostareck D. H., Ostareck-Lederer A. (2008) J. Biol. Chem. 283, 18461–18472 [DOI] [PubMed] [Google Scholar]
- 66.Klimek-Tomczak K., Wyrwicz L. S., Jain S., Bomsztyk K., Ostrowski J. (2004) J. Mol. Biol. 342, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 67.Paziewska A., Wyrwicz L. S., Bujnicki J. M., Bomsztyk K., Ostrowski J. (2004) FEBS Lett. 577, 134–140 [DOI] [PubMed] [Google Scholar]
- 68.Expert-Bezançon A., Sureau A., Durosay P., Salesse R., Groeneveld H., Lecaer J. P., Marie J. (2004) J. Biol. Chem. 279, 38249–38259 [DOI] [PubMed] [Google Scholar]
- 69.Swanson A. K., Stoltzfus C. M. (1998) J. Biol. Chem. 273, 34551–34557 [DOI] [PubMed] [Google Scholar]
- 70.Yang J. P., Reddy T. R., Truong K. T., Suhasini M., Wong-Staal F. (2002) Oncogene 21, 7187–7194 [DOI] [PubMed] [Google Scholar]
- 71.Gorla L., Cantù M., Miccichè F., Patelli C., Mondellini P., Pierotti M. A., Bongarzone I. (2006) Cell. Signal. 18, 2272–2282 [DOI] [PubMed] [Google Scholar]
- 72.Venables J. P., Klinck R., Bramard A., Inkel L., Dufresne-Martin G., Koh C., Gervais-Bird J., Lapointe E., Froehlich U., Durand M., Gendron D., Brosseau J. P., Thibault P., Lucier J. F., Tremblay K., Prinos P., Wellinger R. J., Chabot B., Rancourt C., Elela S. A. (2008) Cancer Res. 68, 9525–9531 [DOI] [PubMed] [Google Scholar]
- 73.Ostrowski J., Bomsztyk K. (2003) Br. J. Cancer 89, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandal M., Vadlamudi R., Nguyen D., Wang R. A., Costa L., Bagheri-Yarmand R., Mendelsohn J., Kumar R. (2001) J. Biol. Chem. 276, 9699–9704 [DOI] [PubMed] [Google Scholar]
- 75.Carpenter B., McKay M., Dundas S. R., Lawrie L. C., Telfer C., Murray G. I. (2006) Br. J. Cancer 95, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roychoudhury P., Chaudhuri K. (2007) Br. J. Cancer 97, 574–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dejgaard K., Leffers H., Rasmussen H. H., Madsen P., Kruse T. A., Gesser B., Nielsen H., Celis J. E. (1994) J. Mol. Biol. 236, 33–48 [DOI] [PubMed] [Google Scholar]
- 78.Li C., Hong Y., Tan Y. X., Zhou H., Ai J. H., Li S. J., Zhang L., Xia Q. C., Wu J. R., Wang H. Y., Zeng R. (2004) Mol. Cell. Proteomics 3, 399–409 [DOI] [PubMed] [Google Scholar]
- 79.Hatakeyama H., Kondo T., Fujii K., Nakanishi Y., Kato H., Fukuda S., Hirohashi S. (2006) Proteomics 6, 6300–6316 [DOI] [PubMed] [Google Scholar]
- 80.Pino I., Pío R., Toledo G., Zabalegui N., Vicent S., Rey N., Lozano M. D., Torre W., García-Foncillas J., Montuenga L. M. (2003) Lung Cancer 41, 131–143 [DOI] [PubMed] [Google Scholar]
- 81.Lynch M., Chen L., Ravitz M. J., Mehtani S., Korenblat K., Pazin M. J., Schmidt E. V. (2005) Mol. Cell. Biol. 25, 6436–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.