Abstract
RDH12 mutations are responsible for early-onset autosomal recessive retinal dystrophy, which results in profound retinal pathology and severe visual handicap in patients. To investigate the function of RDH12 within the network of retinoid dehydrogenases/reductases (RDHs) present in retina, we studied the retinal phenotype of Rdh12-deficient mice. In vivo rates of all-trans-retinal reduction and 11-cis-retinal formation during recovery from bleaching were similar in Rdh12-deficient and wild-type mice matched for an Rpe65 polymorphism that impacts visual cycle efficiency. However, retinal homogenates from Rdh12-deficient mice exhibited markedly decreased capacity to reduce exogenous retinaldehydes in vitro. Furthermore, in vivo levels of the bisretinoid compound diretinoid-pyridinium-ethanolamine (A2E) were increased in Rdh12-deficient mice of various genetic backgrounds. Conversely, in vivo levels of retinoic acid and total retinol were significantly decreased. Rdh12 transcript levels in wild-type mice homozygous for the Rpe65-Leu450 polymorphism were greater than in Rpe65-Met450 mice and increased during postnatal development in wild-type mice and Nrl-deficient mice having an all-cone retina. Rdh12-deficient mice did not exhibit increased retinal degeneration relative to wild-type mice at advanced ages, when bred on the light-sensitive BALB/c background, or when heterozygous for a null allele of superoxide dismutase 2 (Sod2+/−). Our findings suggest that a critical function of RDH12 is the reduction of all-trans-retinal that exceeds the reductive capacity of the photoreceptor outer segments.
RDH12 is a major disease gene for Leber congenital amaurosis, with RDH12 mutations responsible for approximately 2% of cases of childhood-onset severe autosomal recessive retinal dystrophy (1–4). Affected individuals experience poor vision in early life, which progressively declines with age as a result of both rod and cone degeneration (5). Analyses of retinal organization and visual function in patients with mutations in RDH12 or RPE65, another Leber congenital amaurosis gene involved in retinoid metabolism, show distinctly different pathologies associated with defects in each gene that will be important to consider when developing targeted forms of therapy (6).
RDH12 encodes a member of the family of short chain dehydrogenases/reductases that catalyze oxidation and reduction reactions involved in various aspects of metabolism (reviewed in Refs. 7 and 8). In the photoreceptor cells and retinal pigment epithelium (RPE),3 the interconversion of oxidized and reduced retinoids by RDH enzymes is an important feature of the visual cycle, the process responsible for the conversion of vitamin A (all-trans-retinol) to 11-cis-retinal, the chromophore of the visual pigments (reviewed in Ref. 9). On the basis of in vitro assays showing reactivity toward retinaldehyde substrates and expression in photoreceptor cells, RDH12 was initially proposed to function in the reduction of all-trans-retinal released by bleached visual pigments (10). Subsequent in vitro studies showed that RDH12 can also act upon C9 aldehydes resulting from lipid photo-oxidation (11–13), as well as certain steroid substrates (14), suggesting that it contributes to additional pathways important for photoreceptor function. In previous studies, we generated Rdh12-deficient mice and evaluated their retinal phenotype, finding grossly normal retinal histology, visual responses, retinoid content, and assays of oxidative stress (15). We also showed that RDH12 localizes to photoreceptor inner segments in humans and mice, distant from the phototransduction reactions occurring in the outer segment disc membranes. Based on this analysis, we concluded that RDH12 deficiency does not limit chromophore synthesis or visual cycle throughput, in agreement with Maeda et al. (16).
Similarly, earlier attempts to identify an RDH isoform essential for chromophore synthesis failed. These studies included generation and analysis of Rdh5-deficient mice lacking the RDH isoform specific to the RPE (17), Rdh11-deficient mice lacking the RDH isoform most closely related to Rdh12 (18, 19), and Rdh8-deficient mice lacking the RDH isoform that localizes to photoreceptor outer segments (20). In each case, loss-of-function resulted in only minor slowing of dark adaptation without accompanying retinal degeneration, thus raising the possibility of redundancy in RDH function relative to the visual cycle mechanism.
We now report that the retinas of Rdh12-deficient mice exhibit a markedly decreased capacity to reduce retinaldehyde substrates in vitro but do not show decreased synthesis of 11-cis-retinal in vivo or age-related degeneration on various genetic backgrounds. Our data are consistent with a mechanism in which RDH12 contributes to the reduction all-trans-retinal that escapes conversion to all-trans-retinol by enzyme(s) present in the outer segments. Although fundamental differences exist between species, our findings in mice point to an important role of RDH12 in retinoid metabolism occurring in photoreceptor cells, and of likely relevance to disease pathology in the human retina, but which does not limit chromophore synthesis in vivo.
EXPERIMENTAL PROCEDURES
Animals and Antibodies
Transgenic mice of the following genotypes were used for our studies: Rdh12−/− mice homozygous for a Rpe65-Leu450 (L/L) or Rpe65-Met450 (M/M) polymorphism on a mixed Sv129;C57BL/6 (pigmented) background (15); Rdh12−/− mice on the BALB/c (albino) background and homozygous for Rpe65-Leu450 (L/L) that were obtained by breeding (generation F6); Rdh12−/− mice heterozygous for a Sod2 null allele (Sod2+/−) and homozygous for Rpe65-Met450 (M/M) obtained by breeding with mice from The Jackson Laboratory (Bar Harbor, ME; stock No. 002973, strain B6.129S7-Sod2tm1Leb); Rdh11−/− mice homozygous for Rpe65-Leu450 (L/L) (pigmented) characterized by P. Nelson and K. Palczewski (18); and Nrl−/− mice homozygous for Rpe65-Met450 (M/M) that have a model “all-cone” retina (21). Primary antibodies used were: a rabbit anti-Rdh12 polyclonal antibody specific for the mouse protein (against 252SPFFKSTSQGAQ263) (15); a rabbit anti-transducin polyclonal antibody (Genetex); and a rabbit anti-arrestin polyclonal antibody (Affinity Bioreagents).
Analysis of Visual Cycle Retinoids
All-trans-retinal, all-trans-retinol, and 11-cis-retinal levels in mouse eyes were determined using normal-phase high performance liquid chromatography (HPLC) as described previously (3). In brief, dark-adapted mice (∼3 months old) were subjected to bleaching light (5000 lux, 15 min), recovered in the dark for 0–60 min, and euthanized. Under dim red light, whole eyes were homogenized in chloroform:methanol:hydroxylamine, the organic phases were collected, and the solvent evaporated. The extracts were dissolved in hexane and chromatographed on a Supelcosil LC-31 analytical column (25 cm × 4.6 mm, 5 μm) developed with 95% hexane, 5% 1,4-dioxane. Absorbance peaks were identified by comparison with external standards, and molar quantities per eye were calculated by comparison with standard concentrations determined spectrophotometrically using published extinction coefficients (22) and normalizing to total sample volumes. Retinaldehyde content was calculated as the sum of syn- and anti-retinal oximes. Data were analyzed and curves fitted using SigmaPlot (Systat Software Inc.). Standards used were: all-trans-retinol (SigmaAldrich), 11-cis-retinol obtained by NaBH4 reduction of 11-cis-retinal (R. Crouch), and all-trans- and 11-cis-retinal oximes obtained by hydroxylamine reaction of the corresponding aldehydes (23).
Analysis of Lipofuscin Pigment Accumulation
RPE lipofuscin pigments were separated, identified and quantified by HPLC analysis as previously described (24, 25). In brief, posterior segments from 3–6 mouse eyes (from animals ∼3–6 months old) were homogenized in phosphate-buffered saline (PBS) and extracted with chloroform:methanol. After passage through a reverse-phase cartridge (C8 Sep-Pak, Millipore) in 0.1% trifluoroacetic acid in methanol and evaporation of solvent, the extracts were dissolved in 50% methanolic chloroform containing 0.1% trifluoroacetic acid and chromatographed on Atlantis® dC18 (3 μm, 4.6 × 150 mm) and Delta Pak® C4 (5 μm, 3.9 × 150 mm) columns (Waters Corp.) using gradients of water and aceotnitrile with 0.1% of trifluoroacetic acid. Molar quantity per eye was determined from peak areas using calibration curves constructed from known concentrations of synthesized standards and by normalizing to the total sample volume (26–28).
Analysis of Retinoic Acid and Total Retinol
HPLC analysis coupled with tandem mass spectrometry or UV analysis was used to quantitate retinoic acid and total retinol levels, respectively, present in the retinas of dark-adapted mice (∼5 months old) according to methods described previously (29, 30). In brief, retinas were homogenized in 0.9% NaCl, and 4,4-dimethyl-retinoic acid and retinyl acetate in acetonitrile were added as internal standards followed by the addition of ethanolic KOH. The samples were extracted with hexane, and the organic phase, containing nonpolar retinoids, was collected. The aqueous phase was acidified by the addition of HCl, and polar retinoids were extracted in hexane. After evaporation of the organic phases, the extracts were resuspended in acetonitrile. Total retinol content was analyzed by reverse-phase HPLC on a Zorbax SB-C18 column (4.6 × 100 mm, 3.5 μm) developed with gradients of water, acetonitrile, and 0.1% formic acid with quantification by analysis of UV absorbance at 325 nm and comparison to standards. For retinoic acid analysis, extracts were subjected to HPLC on an Ascentis RP-Amide column (Supelco, 2.1 × 150 mm, 3 μm) using gradients of water, acetonitrile, 0.1% of formic acid; tandem mass spectrometry data were acquired with atmospheric pressure chemical ionization operated in positive ion mode. All retinoid standards except 4,4-dimethyl-retinoic acid were purchased from Sigma-Aldrich, with concentrations verified spectrophotometrically using published extinction coefficients (31).
Analysis of Retinal Reductase Activity
Mouse retina homogenates were assayed for retinal reductase activity with exogenous substrates. Light-adapted mice (∼2–5 months old) were euthanized, and dissected retinas from 4–8 animals were pooled and homogenized in 300 μl of 0.25 m sucrose, 25 mm Tris acetate, pH 7, 1 mm dithiothreitol. The homogenates were centrifuged at 1000 × g for 5 min to remove unbroken cells, and then the supernates were sonicated with a microtip probe (30 times for 1 s) on ice. Protein concentrations were determined by a modification of the Lowry procedure (32), and levels of Rdh12 and arrestin (a marker for total photoreceptor proteins) were evaluated by densitometric analysis of Western blots. Assay reactions (200 μl) were prepared that contained 20 μg of retinal protein, 200 μm all-trans-retinal or 11-cis-retinal, and 200 μm NADPH in HEPES buffer (pH 8); reactions were incubated for 0–60 min at 32 °C (a reaction temperature that minimizes thermal isomerization of the retinoid substrates, as well as enzyme inactivation). All-trans-retinol and 11-cis-retinol formation were quantitated using normal-phase HPLC analysis with comparison to standards, as described above, and the data were analyzed using SigmaPlot. Controls for recovery included samples containing known amounts of standard compounds as described previously (3).
Immunohistochemistry
Anesthetized mice (∼3 months old) were perfused with 4% paraformaldehyde in 0.1 m sodium phosphate, pH 7.4, the eyes were enucleated and post-fixed (total fixation time ≤ 15 min) and rinsed in phosphate buffer, and the anterior segments and lenses were removed. The posterior eyecups were transitioned through a sucrose series and flash-frozen in sucrose-OCT. Cryosections (18 μm) were permeabilized with 0.125% Triton X-100 in PBS, treated with biotin blocking reagents (Invitrogen), blocked with 1% bovine serum albumin, 10% normal goat serum, and 0.125% Triton X-100 in PBS for 1 h, and then incubated over night at 4 °C with primary antibody in 1% bovine serum albumin, 1% normal goat serum, and 0.125% Triton X-100 in PBS and washed. Sections were incubated with biotinylated anti-rabbit secondary antibody (1:400; Molecular Probes) for 3 h, washed, and incubated with streptavidin-Alexa Fluor-488 or -555 (Molecular Probes, 1:500). Sections were washed, mounted in ProLong Gold containing 4′,6-diamidino-2-phenylindole (Molecular Probes) to stain nuclei, and imaged using an Olympus FV500 confocal microscope with FluoView FV500 data acquisition software.
Analysis of Histology and Retinal Thickness
Anesthetized mice were perfused, and the eyes were enucleated, post-fixed for at least 1 h in 4% paraformaldehyde or 2% paraformaldehyde plus 2% glutaraldehyde, and embedded in JB-4 plastic. Sections (5 μm) were stained with Lee's stain and imaged on a Nikon Eclipse E800 microscope with Nikon DMX1200 digital camera using the manufacturer's data acquisition software. Measurements of retinal outer nuclear layer thickness on sections parallel to the vertical meridian of the eye were plotted versus distance from the optic nerve head for animals of various ages (33).
Analysis of Transcript Abundance by Quantitative Real-time PCR
Total RNA was isolated from mouse retinas (∼2–3 months old) using the RNeasy mini kit (Qiagen), and quality and quantity were assessed using an Agilent Bioanalyzer 2100. First strand cDNAs were generated using SuperScript II (Invitrogen) and oligo(dT) (Invitrogen). Sequences present in the 3′-untranslated regions of transcripts of interest were amplified using AmpliTaq Gold (Applied Biosystems) and SYBR Green I (Molecular Probes) with a Rotor-Gene3000 thermocycler (Corbett Research) as follows: 95 °C for 10 min, 95 °C for 25 s, 57 °C for 25 s, and 72 °C for 30 s, with steps 2–4 repeated 35 times. Samples in triplicate were each amplified in triplicate and normalized to the values for Hprt transcripts. Primers (sense, antisense) were as follows: Rdh12, 5′-GGCCAATCTGCTCTTCACTC-3′, 5′-ACTTTCCACTCAGGGGCTCTA-3′; Rdh11, 5′-CCACAGCAAACTAGCCAACA-3′, 5′-CCAACTGGCAATCACTGAAA-3; Rdh8, 5′-GATGTGGCCCAGGTCATT-3′, 5′-GACCAAGGTTGAGGAGGTGA-3′; Hprt, 5′-GCAAGCTTGCTGGTGAAAGGA-3′, 5′-CCTGAAGTACTCATTATAGTCAAGGG-3′.
Microarray Analysis of Expression
Total retinal RNA from Rdh12−/− and wild-type mice (∼2 months old) was isolated as described above and converted into biotinylated, fragmented, and cleaned cRNA using Affymetrix reagents. Mouse genome arrays (Affymetrix 430_2.0) were hybridized with fragmented cRNA and processed as described (34). The arrays were optically read with a Hewlett-Packard GeneArray scanner, and CHP/.CEL files were generated using the Affymetrix microarray suite of programs. The quality of the GeneChip hybridizations was excellent according to Affymetrix criteria. Normalization and calculation of signal intensities were performed using the Robust Multi-Chip Average software package (R-Project). Ratios of average signal intensity (log 2) were calculated for probe sets between pairs of genotypes and converted to average -fold change. Statistical validation was performed as described previously (34) by assigning p values and -fold changes of gene responses in a two-step procedure based on the construction of false discovery rate confidence intervals (35, 36). Affymetrix GeneChip data for flow-sorted GFP+ photoreceptor cells from wild-type and Nrl−/− mice at various developmental ages were generated as described previously (37).
RESULTS
Visual Cycle Retinoids in Rdh12-deficient Mouse Eyes
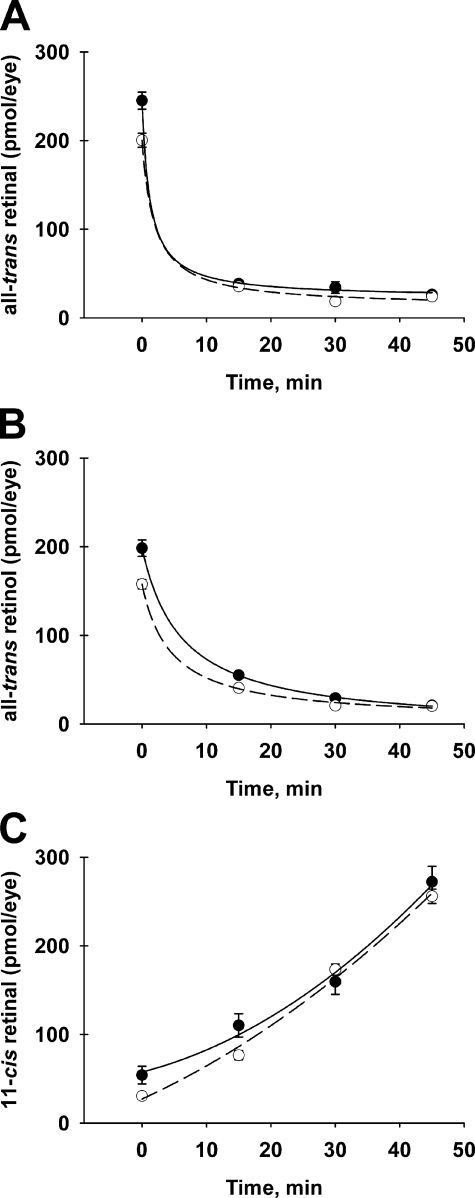
The first step in the recycling aspect of the visual cycle is the conversion of all-trans-retinal, released from bleached visual pigments, to all-trans-retinol, which is returned to the RPE. In studies employing retinoid extraction and HPLC analysis, we showed previously that steady-state levels of visual cycle retinoids were similar in the eyes of Rdh12-deficient and wild-type mice heterozygous for the Rpe65-L450M (L/M) polymorphism that impacts rates of chromophore synthesis (15). We have now used these methods to evaluate retinoid processing during dark adaptation in Rdh12-deficient mice homozygous for the Rpe65-Leu450 (L/L) variant associated with high visual cycle throughput (38). During recovery in the dark after full bleaching, our results show that rates of all-trans-retinal reduction and 11-cis-retinal formation are not significantly different between Rdh12−/− and wild-type animals (Fig. 1).
FIGURE 1.
All-trans-retinal, 11-cis-retinal, and all-trans-retinol levels during recovery from bleaching. Rdh12-deficient and wild-type mice were exposed to bleaching light (5000 lux, 15 min) and recovered in the dark for the times shown. Retinoids extracted from whole eyes in organic solvents were quantitated using normal-phase HPLC analysis. A, all-trans-retinal; B, all-trans-retinol; C, 11-cis-retinal. Experimental values are plotted as mean ± S.E. (n = 3). ○, Rdh12−/− mice; ●, wild-type mice.
RDH Activity in Rdh12-deficient Mouse Retinas
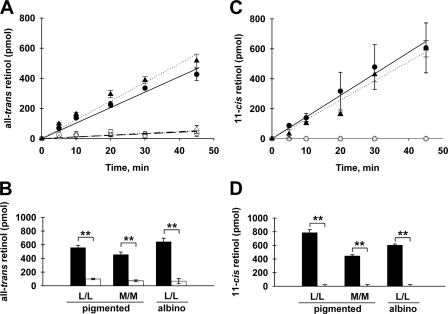
To determine the contribution of Rdh12 to the overall capacity of the mouse retina to reduce retinaldehyde substrates, we performed in vitro assays of retinal homogenates with exogenous all-trans-retinal or 11-cis-retinal as substrate. Retinas from Rdh12−/− and wild-type mice on three different genetic backgrounds were evaluated: Rpe65-L/L or Rpe65-M/M on 129Sv:C57BL/6 (pigmented) and Rpe65-L/L on BALB/c (albino). For wild-type pigmented and albino mice that were Rpe65-L/L, we found that robust retinal reductase activity was present in assays with saturating levels of all-trans-retinal or 11-cis-retinal (Fig. 2, A and C). Initial rates of all-trans-retinol formation were 0.51 and 0.62 pmol min−1 μg protein−1 in the pigmented and albino mice, respectively. Similarly, initial rates of 11-cis-retinol formation were 0.70 and 0.60 pmol min−1 μg protein−1. For Rdh12−/− mice, significantly lower retinal reductase activity was observed with both substrates (Fig. 2, A and C). Initial rates of all-trans-retinol formation were 0.052 and 0.057 pmol min−1 μg protein−1 in pigmented and albino mice, respectively, whereas 11-cis-retinol formation was undetectable. Mice on the Rpe65-M/M background exhibited lower RDH activity than Rpe65-L/L mice; however, the relative decrease in RDH activity seen in Rdh12−/− mice was similar (Fig. 2, B and D).
FIGURE 2.
Effects of Rdh12 deficiency on in vitro retinoid reductase activity. Retinal homogenates were prepared from Rdh12−/− and wild-type mice that were homozygous for Rpe65-Leu450 (pigmented L/L) or Rpe65-Met450 (pigmented M/M) on the 129Sv:C57BL/6 background or for Rpe65-Leu450 (albino L/L) on the BALB/c background. The homogenates were incubated with exogenous retinaldehyde substrates (all-trans-retinal or 11-cis-retinal) and NADPH for various times, and retinol formation was quantitated by HPLC analysis of organic solvent extracts. Formation of all-trans-retinol (A and B) and 11-cis-retinol (C and D) in retinal lysates from Rdh12−/− and wild-type mice is shown. A and C, time course (0–45 min). ○, Rdh12−/− (pigmented L/L); ▵, Rdh12−/− (albino L/L); ●, wild-type (pigmented L/L); ▴, wild-type (albino L/L). B and D, single point assays (60 min), except 11-cis-retinol albino L/L (45 min). Open bars, Rdh12−/− mice; solid bars, wild-type mice. Assays were performed in triplicate and plotted as mean ± S.D. (n > 3). **, p < 0.01.
RDH Activity in Rdh11-deficient Mouse Retina
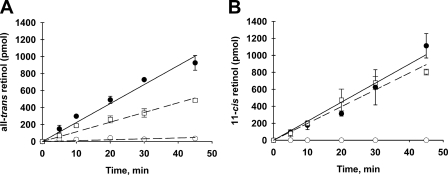
In vitro assays of retina homogenates from Rdh11−/− mice exhibited retinoid reductase activity that was significantly greater than of Rdh12−/− mice on the same genetic background (pigmented Rpe65-L/L) (Fig. 3). At saturating substrate concentrations, initial rates of all-trans-retinol and 11-cis-retinol formation in Rdh11−/− retinal homogenates were 0.57 and 0.99 pmol min−1 μg protein−1, respectively.
FIGURE 3.
Effects of Rdh11 deficiency on in vitro retinoid reductase activity. Retinal homogenates from Rdh11−/− and wild-type mice (pigmented Rpe65-L/L) were assayed for RDH activity with exogenous retinoid substrates and NADPH for the times shown, and retinol formation was quantitated by HPLC analysis of organic solvent extracts and normalized to Rdh12 content. Assays were performed in triplicate and plotted as mean ± S.D. (n = 3). Data from Rdh11−/− mice (□) are shown alongside those from Rdh12−/− mice (○) and wild-type mice (●).
RPE Lipofuscin Pigments in Eyes of Rdh12-deficient Mice
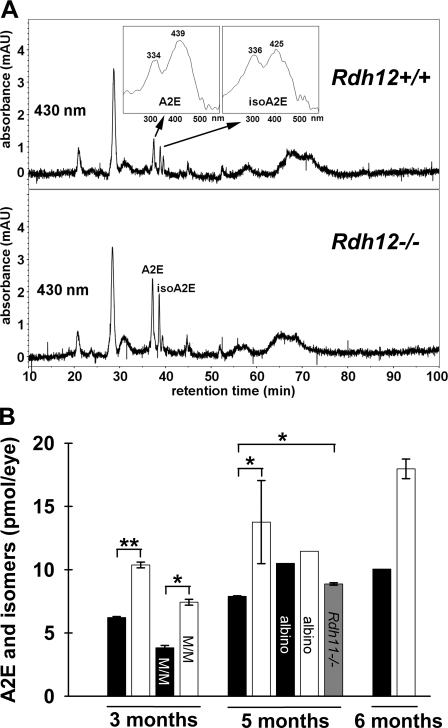
The removal of all-trans-retinal from the photoreceptor outer segments requires its conversion to all-trans-retinol, with inefficient removal resulting in increased accumulation of A2E and other lipofuscin pigments in the RPE (39). To determine the effect of Rdh12 deficiency on lipofuscin accumulation, HPLC analysis of A2E and A2E isomers present in posterior eyecups was performed for pigmented and albino mice on the Rpe65-L/L background (Fig. 4). The data show that A2E levels (the sum of A2E and isomers) increased with age in both Rdh12−/− and wild-type animals at ∼3, 5, and 6 months of age. In addition, Rdh12−/− mice exhibited significantly increased A2E levels relative to wild-type at each age. Levels of A2E were also increased in Rdh12−/− mice on the Rpe65-M/M background (assayed at 5 months old), although levels were less than in Rpe65-L/L animals, as observed previously in studies of wild-type mice (24). In contrast, analysis of eyecups from pigmented Rdh11−/− mice showed only slightly elevated A2E levels. Rdh12−/− mice also exhibited similar increases in levels of atRAL dimer-phosphatidylethanolamine, another aldehyde derivative component of RPE lipofuscin (data not shown).
FIGURE 4.
A2E levels in Rdh12-deficient and wild-type mice. Mouse posterior eyecups of Rdh12-deficient and wild-type mice were extracted using organic solvent, and A2E levels (the sum of A2E and isoA2E) were quantitated using HPLC with comparison to standards. A, representative reverse-phase HPLC chromatograms obtained by monitoring at 430 nm. B, A2E levels in posterior eyecups at the ages indicated. Mice were C57BL/6 and homozygous for Rpe65-Leu450 (pigmented) unless noted, with M/M designating data from mice homozygous for Rpe65-Met450 and albino indicating mice that were BALB/c. □, Rdh12−/−; ■, wild type; ▩, Rdh11−/−. Assays were plotted as mean ± S.E. for n > 3. *, p < 0.05; **, p < 0.01.
Retinoic Acid Content of Rdh12-deficient Mouse Retina
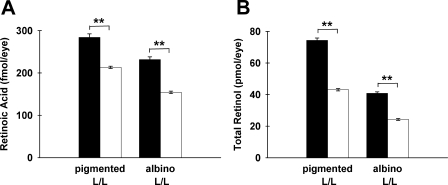
Retinoic acid synthesis involves the oxidation of retinaldehydes in a reaction typically catalyzed by members of the family of aldehyde dehydrogenase enzymes (reviewed in Ref. 41). To determine whether Rdh12 deficiency affects retinoic acid synthesis in the retina, directly or indirectly, we assayed retinal homogenates from dark-adapted mice using HPLC-coupled tandem mass spectrometry. We found that retinoic acid levels were decreased in Rdh12−/− mice relative to the wild type, in both pigmented and albino animals that were Rpe65-L/L (Fig. 5). Downward trends were apparent in the data calculated per retina, and statistical significance (p < 0.01) was reached for data normalized to protein content, suggesting variability in tissue recovery. Similar results were obtained for Rdh12−/− mice that were Rpe65-M/M (data not shown). In addition, significant decreases in total retinol content of retinas from Rdh12−/− mice were detected using HPLC-coupled UV analysis (Fig. 5B). Together, these findings suggest that Rdh12 deficiency in the retina produces a shift in retinoid homeostasis resulting in decreased retinoid content, inconsistent with a previously proposed model suggesting that Rdh12 deficiency increases retinoic acid levels because of increased levels of retinaldehydes (42).
FIGURE 5.
Retinoic acid and total retinol content in Rdh12-deficient and wild-type mice. Isolated retinas from dark-adapted mice were extracted with organic solvent under neutral and acidic conditions to obtain total retinol and retinoic acid-containing extracts, respectively, that were subjected to reverse-phase HPLC. A, retinoic acid was quantitated by coupled tandem mass spectrometry analysis. B, total retinol was quantitated using UV-visible absorbance. Assays were plotted as mean ± S.E. for n = 3. □, Rdh12−/−; ■, wild type. **, p < 0.01.
Rdh12 Expression on Various Genetic Backgrounds and during Development
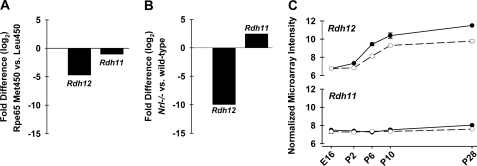
To determine whether Rdh12 expression reflects differences in visual cycle efficiency attributable to the Rpe65-L450M polymorphism, we used quantitative real-time PCR to compare transcript levels in eyes from pigmented wild-type mice that were Rpe65-L/L or Rpe65-M/M. We found that Rdh12 transcripts were ∼4-fold lower in mice with the Rpe65-M/M polymorphism, associated with low levels of 11-cis-retinal synthesis, compared with mice with Rpe65-L/L, which exhibits high visual cycle throughput (Fig. 6A). In Nrl-deficient mice having an all-cone retina, Rdh12 transcript levels were approximately 10-fold lower than in wild-type mice (Rpe65-M/M) (Fig. 6B). Changes of the same magnitude were not seen for Rdh11.
FIGURE 6.
Comparative analysis of Rdh11 and Rdh12 expression in mouse retina. Quantitative real-time PCR was used to amplify RDH transcripts from retina total RNA, and cycle threshold data were normalized to Hprt and plotted as -fold difference. A, transcript levels in wild-type mice on the Rpe65-Met450 background compared with levels in wild-type mice on the Rpe65-Leu450 background (pigmented 110 days old; n = 3). B, RDH transcript levels in Nrl−/− mice compared with levels in wild-type mouse (pigmented Rpe65-M/M, 50 days old; n = 3). C, expression during retinal development in Nrl−/− and wild-type mice. Normalized microarray intensity data for flow-sorted GFP+ photoreceptors isolated and analyzed as described in Ref. 37 is shown for following ages: embryonic day 16 (E16) and postnatal (P) days 2, 6, 10, and 28 (n = 4 for each time point). ○, Nrl−/−; ●, wild-type.
To evaluate Rdh12 expression during development, data from microarray analysis of flow-sorted GFP-positive photoreceptor cells were obtained as described previously (37). The photoreceptor cells were from dissociated retinas of transgenic mice expressing GFP under the control of the Nrl promoter on the wild-type (predominantly rod) or the Nrl−/− (all-cone) background (Rpe65-M/M). Plots of normalized signal intensity showed that Rdh12 transcript levels markedly increased from embryonic day 16 to postnatal day 28 in wild-type mice and to a lesser extent in Nrl−/− mice (Fig. 6C). In contrast, Rdh11 transcript levels remained relatively unchanged in both wild-type and Nrl−/− mice.
Expression of RDH Isoforms in Wild-type and Rdh12-deficient Mice
The absence of retinal pathology in Rdh12-deficient mice suggests, as one possibility, that one or more RDH isoform(s) with overlapping activity compensates for Rdh12 loss-of-function. To compare the expression of RDH isoforms of Rdh12−/− and wild-type mice (Rpe65-L450M) in total retinal RNA, relative transcript abundance was evaluated using Affymetrix microarray profiling. For each of the RDH isoforms reported to be present in photoreceptor cells and represented on gene chips (Rdh11, Rdh13, Rdh14, Dhrs3(retSDR1) (10, 43)), we found no marked differences in expression between the Rdh12−/− and wild-type mice (Table 1). This was also the case for Rdh10 expressed in RPE and Müller cells (44), as well as other known RDH isoforms, including Rdh1, Rdh7, Rdh9, and Rdh16 (data not shown). One exception was Rdh5, for which transcripts were apparently decreased ∼1.7-fold in Rdh12−/− mice. The presence of Rdh5 transcripts in our RNA preparations likely reflects the presence of contaminating RPE cells in our dissected retinal tissue, as Rdh5 is an RPE enzyme involved in the conversion of 11-cis-retinol to 11-cis-retinal (45). A second exception was Rdh8, which localizes to photoreceptor outer segments. Because Rdh8 is not represented by probe sets on the Affymetrix gene chips, Rdh8 expression was evaluated using quantitative real-time PCR analysis, which showed that transcripts were decreased ∼3.1-fold in Rdh12−/− mice. The apparent down-regulation of Rdh5 and Rdh8 suggests that retinoid processing associated with visual cycle function is less robust in the absence of Rdh12. As no known RDH isoforms appear to be up-regulated in Rdh12−/− mice, our findings suggest that the activity provided by the enzyme levels normally present in mouse retina are sufficient to compensate for Rdh12 loss-of-function.
TABLE 1.
Microarray analysis of RDH isoforms expressed in mouse retina
Rdh12−/− and wild-type mice (WT) were heterozygous for Rpe65-L450M. Data shown were calculated from the average intensity values of four replicates. For genes with multiple probes sets, the values shown correspond to those with the highest intensity signals. NA, not applicable.
| Gene I.D. | Probe set | Rdh12−/−vs. WT |
|---|---|---|
| -Fold change | ||
| Rdh12 | 1424256_at | −129.99 |
| Rdh11 | 1418760_at | 1.29 |
| Rdh10 | 1426968_a_at | −1.38 |
| Rdh13 | 1433799_at | 1.15 |
| Rdh14 | 1417438_at | 1.05 |
| Dhrs3 | 1448390_a_at | −1.06 |
| Rdh5 | 1418808_at | −1.68 |
| Rdh8a | NA | −3.15 |
a Values obtained by quantitative real-time PCR analysis.
Rdh12 Localization in Dark- and Light-adapted Mouse Retina
Our previous studies of RDH12 localization showed immunoreactivity in the photoreceptor inner segments and outer nuclear layer in mouse and human retina. To determine whether Rdh12 undergoes light-induced movements similar to the translocation of other photoreceptor proteins (46), we compared the pattern of Rdh12 immunoreactivity in retinas of wild-type albino mice (Rpe65-L/L) that were dark-adapted or exposed to bleaching light (2000 lux for 2 h) (supplemental Fig. 1). Under conditions resulting in redistribution of transducin from photoreceptor outer segments in the dark, to inner segments in the light, Rdh12 immunoreactivity remained confined to the inner segments. Similar results were obtained for wild-type pigmented mice, in which bleaching was less efficient (data not shown).
Effects of Aging and Oxidative stress in Rdh12-deficient mice
To assess potential changes in retinal histology at advanced ages, we performed morphometric analysis of retina/RPE/choroid sections from Rdh12-deficient and wild-type mice reared in normal laboratory lighting. We found no evidence of increased retinal pathology or decreased outer nuclear layer (ONL) thickness in Rdh12−/− mice at 20 months old that were pigmented and Rpe65-L/L or Rpe65-M/M (supplemental Fig. 2, A and B). We also evaluated the retinas of albino Rdh12−/− mice on the BALB/c background, which imparts increased light sensitivity due in part to the presence of the Rpe65-L450 polymorphism (47). Again, no increased retinal pathology or decreased ONL thickness was seen in Rdh12-deficient mice crossed onto the BALB/c background for six generations (supplemental Fig. 2C).
To determine whether increased oxidative stress exacerbates the retinal phenotype of Rdh12-deficient mice, we crossed mice carrying a null allele of the superoxide dismutase 2 gene (Sod2) with Rdh12−/− animals. Although the homozygous null Sod2−/− genotype is lethal, Sod2+/− mice are viable and exhibit increased levels of reactive oxygen species associated with increased oxidative damage to proteins, DNA, and membranes (48, 49). Mice with the Rdh12−/−;Sod2+/− (Rpe65-M/M) genotype were obtained by breeding and were evaluated by morphometric analysis. Retina sections from Rdh12−/−;Sod2+/− mice at 9 months old showed no evidence of increased retinal pathology or decreased ONL thickness relative to age-matched Sod2+/− mice that were wild type for Rdh12 (supplemental Fig. 2D). In addition, the RDH activity of retinal lysates assayed in vitro was equivalent in Rdh12−/−;Sod2+/− and Rdh12−/− animals on the Rpe65-M/M background (data not shown).
DISCUSSION
The present study establishes that Rdh12 loss-of-function markedly impacts the overall capacity of the mouse retina to reduce retinaldehyde substrates, even though in vivo rates of all-trans-retinal reduction during recovery from bleaching are not significantly affected. The decrease in the ability of the Rdh12-deficient retina to reduce all-trans-retinal and profound loss of activity toward 11-cis-retinal suggest that Rdh12 makes a unique contribution to the capacity for retinoid processing in the photoreceptor cells. These findings are consistent with the assumption that defects in retinoid metabolism contribute to the disease phenotype resulting from RDH12 mutations in patients. Together with previous studies, our results suggest that the supply of all-trans-retinol needed for chromophore synthesis is not limited by RDH activity in the photoreceptors (15, 16, 18–20) and that this need may be met by delivery from the circulation and stores of retinyl esters in the RPE. It follows that increased rates of regeneration attributed to interphotoreceptor retinol-binding protein (IRBP) (50) may result from a potential role in reducing photoreceptor levels of all-trans-retinol rather than its delivery to the RPE.
Previous studies of the retinal phenotype of Rdh12-deficient mice did not detect marked changes in retinoid processing or visual cycle function (15, 16). In one study, delayed recovery of the electroretinogram a-wave after bleaching was observed as well as increased levels of 11-cis-retinal, but no decrease in all-trans-retinal reductase activity was detected in vitro (16). Subsequent studies of Rdh12-Rdh8 double knock-out mice showed that Rdh12 loss-of-function significantly decreased the all-trans-retinal reductase activity of Rdh8-deficient mice (51). In contrast, our studies show that retina homogenates from mice deficient in Rdh12 alone have significantly reduced capacity for reduction of both trans- and cis-retinaldehydes on all genetic backgrounds tested, but do not exhibit delayed dark adaptation or increased levels of retinaldehydes in vivo (this work and Ref. 15). In addition, the decrease of retinal reductase activity in Rdh12-deficient mice is greater than that observed for Rdh11-deficient mice. We also found that Rdh12-deficient mice exhibit elevated A2E levels, and that these levels increase with aging, in agreement with earlier studies (16). In addition, wild-type mice with the Rpe65-M/M polymorphism exhibit lower levels of retinal reductase activity than mice with the Rpe65-L/L polymorphism, associated with higher visual cycle activity (38, 47). Thus, a key finding of our study is that Rdh12 activity appears to correlate with visual cycle activity, with loss-of-function resulting in a shift in the equilibrium toward products that exit the visual cycle. As Rdh12 deficiency does not result in elevated levels of total retinaldehydes, such increases are likely to occur focally, transiently, or in both ways. Although differing in mechanistic detail and degree of severity, the resulting accumulation of A2Es may reflect a situation akin to that due to mutations in ABCA4, which delay the removal of all-trans-retinal from the disc membranes (53).
Our studies showed that retinoic acid levels are decreased in the retinas of Rdh12-deficient mice. This finding was unexpected, as a previous study of recombinant RDH12 activity in vitro found increased levels of retinoic acid in transfected cells expressing loss-of-function mutants relative to cells expressing the wild-type protein (42). The previous study suggested that RDH12 reduction of retinaldehydes is needed to protect against the potentially damaging effects of high levels of retinoic acid. However, our data do not support the view that RDH12 acts exclusively as a reductase in vivo to decrease levels of retinoic acid, as in vivo levels of both retinoic acid and total retinol were decreased in Rdh12−/− mice. The decrease in retinoic acid levels resulting from Rdh12 deficiency points to a complex mechanism involved in regulating retinoic acid biosynthesis, suggesting the possibility that RDH12 may play a role in this mechanism in the retina, acting either as a reductase or a dehydrogenase depending on its metabolic environment. Our findings further suggest that some aspects of the disease phenotype associated with human RDH12 mutations may be a consequence of impaired retinoic acid production and/or availability.
The broad substrate specificity of RDH12 has prompted suggestions that this enzyme functions in diverse aspects of retinal cell biology. In vitro assays of recombinant RDH12 activity have shown it to be active in reducing certain lipid photo-oxidation products including the C9-aldehydes nonanal and 4-hydroxynonenal (11), as well as certain steroids including dihydrotestosterone (14), albeit with lower affinity and catalytic efficiency compared with all-trans-retinal. In our initial analysis of the phenotype of Rdh12-deficient mice, we found no increase in oxidative products measured as thiobarbituric acid-reactive substances in the retina (15). We have now studied the retinal phenotype of Rdh12-deficient mice that were heterozygous for a null allele of manganese superoxide dismutase 2 (Sod2+/−). Mice heterozygous for Sod2 loss-of-function were used as model to test for effects of increased oxidative stress, as reduction of this enzyme activity results in increased mitochondrial oxidative damage and susceptibility to apoptotic cell death (54), whereas the Sod2−/− genotype is lethal (48, 49), and severe Sod2 deficiency alone results in profound retinal degeneration (55, 56). Our results show no evidence of increased retinal cell death in Rdh12−/−;Sod2+/− mice, as measurements of ONL thickness, as well as retinoid reductase activity,4 were similar to those of Rdh12−/− mice on the wild-type background. Previous studies also reported increased apoptotic cell death in retinas of Rdh12-deficient mice exposed to high intensity illumination (16). However, we detected no increase in age-related pathology in Rdh12−/− mice, including animals on the albino BALB/c background that were reared in normal laboratory lighting. Thus, our findings do not rule out the possibility that RDH12 plays a role in protecting against oxidative stress and light-damage, but they do not support the view that this is its exclusive function.
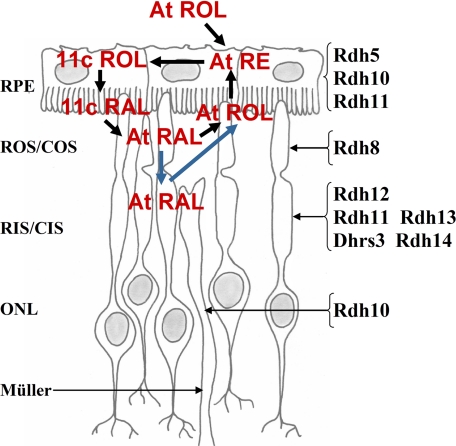
Efforts to identify the RDH enzymes essential for visual cycle function have been confounded by the fact that photoreceptor cells express a number of isoforms which exhibit the necessary specificity in vitro (10, 43, 57) (Fig. 7). Elegant studies of retinoid processing in retina slices and isolated photoreceptor cells have shown that all-trans-retinol formation localizes to the outer segments during recovery from bleaching (40, 58). Of the RDH isoforms known to be expressed in retina, only RDH8 has been localized to the outer segments (51, 57). However, Rdh8-deficient mice exhibit only mild perturbations in retinoid processing (decreased rates of dark adaptation) without accompanying pathology (20). In Rdh12-Rdh8 double knock-out mice, rates of dark adaptation are further slightly decreased, and outer segments are shorter, but significant levels of chromophore synthesis persist (51). These findings are consistent with recent measurements of in vivo rates of all-trans-retinol synthesis suggesting that the rate-limiting step in the production of the 11-cis-retinal chromophore is the release of all-trans-retinal from opsin (50). Thus, the primary role of RDH isoforms present in the photoreceptors may be to reduce potentially toxic levels of retinaldehydes in various cellular compartments. It seems possible that RDH8, which recognizes trans- but not cis-retinaldehydes, is important for the rapid reduction of all-trans-retinal in the vicinity of the rhodopsin signaling complex, whereas the contribution of RDH12, with its broader substrate specificity and localization near the metabolic source of NADPH, becomes more important under conditions of continuous illumination. As RDH12 is restricted to inner segments in both human and mouse retinas (15, 16), and we found no evidence of light-induced translocation, this implies that all-trans-retinal can move between outer and inner segment compartments, perhaps associated with retinoid-binding proteins or with opsin found in the inner segment plasma membrane.
FIGURE 7.
Schematic of RDH isoforms and retinoids present in the outer retina and RPE..
It is not uncommon for mouse models to fail to show the human disease phenotype unless subjected to appropriate stress conditions or placed on a permissive genetic background (52). Accordingly, mice deficient in Rdh12 do not suffer the devastating consequences of RDH12 loss-of-function seen in humans, and they fail to exhibit retinal pathology or limited visual cycle function (15, 16). In this case, differences between human and mouse seem likely to reflect differences in the expression and/or catalytic activity of a second RDH isoform that maintains activity levels above a critical threshold in mouse, but not in human, retinas. If so, RDH11 is a likely candidate for providing this activity, as it is apparently expressed in the photoreceptor inner segment, and no other RDH isoforms appear to be significantly up-regulated in Rdh12-deficient mice. In addition, the genes encoding RDH11 and RDH12 are highly related evolutionarily and are located in head-to-head orientation on the same chromosome in human and mouse. An important challenge for the future will be to generate animal models of the human pathology resulting from RDH12 mutations.
In summary, our analysis of the retinal phenotype of Rdh12-deficient mice supports the view that defects in retinoid metabolism are a contributing factor to disease resulting from RDH12 loss-of-function mutations in patients, providing new insights into the visual cycle mechanism and localization of retinoid processing reactions. Future studies of the relative contributions of the RDH isoforms to photoreceptor function will be needed to fully understand the role of RDH12 in retinal physiology, as well as for the development of strategies for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Austra Liepa, Mitchell Gillett, Lin Jia, and Cameron Strong for technical assistance. Rdh11−/− mice were from P. Nelson (Fred Hutchinson Cancer Research Center) and K. Palczewski (Case Western University). 11-cis-Retinal was from Rosalie Crouch (Medical University of South Carolina), and 4,4-dimethyl-retinoic acid was from Marcia Dawson (Burnham Institute, La Jolla, CA) and Peter Hobbs (SRI International, Menlo Park, CA).
This work was supported, in whole or in part, by National Institutes of Health Grants P30-EY07003 (Michigan Vision Core Grant) and P60-DK-20572 (Michigan Diabetes Research and Training Center). EY11115 (to A. S.), AG13566 (to J. L. N.), and EY12951 (to J. R. S.). This work was also supported by Functional Genomics of the Retina in Health and Disease (EVI-GENORET) Grant LSHG-CT-2005-512036 (to A. G.), the Foundation Fighting Blindness (to D. A. T. and A. S.), Research to Prevent Blindness (to D. A. T. and A. S.), Midwest Eye Banks (to J. D. C.), and the Kaplan Foundation (to J. R. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- RPE
- retinal pigment epithelium
- RDH
- retinol dehydrogenase/reductase
- A2E
- diretinoid-pyridinium-ethanolamine
- HPLC
- high performance liquid chromatography
- GFP
- green fluorescent protein
- L/L
- Leu450 polymorphism
- M/M
- Met450 polymorphism
- PBS
- phosphate-buffered saline
- ONL
- outer nuclear layer.
REFERENCES
- 1.Janecke A. R., Thompson D. A., Utermann G., Becker C., Hübner C. A., Schmid E., McHenry C. L., Nair A. R., Rüschendorf F., Heckenlively J., Wissinger B., Nürnberg P., Gal A. (2004) Nat. Genet. 36, 850–854 [DOI] [PubMed] [Google Scholar]
- 2.Perrault I., Hanein S., Gerber S., Barbet F., Ducroq D., Dollfus H., Hamel C., Dufier J. L., Munnich A., Kaplan J., Rozet J. M. (2004) Am. J. Hum. Genet. 75, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson D. A., Janecke A. R., Lange J., Feathers K. L., Hübner C. A., McHenry C. L., Stockton D. W., Rammesmayer G., Lupski J. R., Antinolo G., Ayuso C., Baiget M., Gouras P., Heckenlively J. R., den Hollander A., Jacobson S. G., Lewis R. A., Sieving P. A., Wissinger B., Yzer S., Zrenner E., Utermann G., Gal A. (2005) Hum. Mol. Genet. 14, 3865–3875 [DOI] [PubMed] [Google Scholar]
- 4.Sun W., Gerth C., Maeda A., Lodowski D. T., Van Der Kraak L., Saperstein D. A., Héon E., Palczewski K. (2007) Vision Res. 47, 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster A., Janecke A. R., Wilke R., Schmid E., Thompson D. A., Utermann G., Wissinger B., Zrenner E., Gal A. (2007) Invest. Ophthalmol. Vis. Sci. 27, 1370–1379 [DOI] [PubMed] [Google Scholar]
- 6.Jacobson S. G., Cideciyan A. V., Aleman T. S., Sumaroka A., Schwartz S. B., Windsor E. A., Roman A. J., Heon E., Stone E. M., Thompson D. A. (2007) Invest. Ophthalmol. Vis. Sci. 48, 332–338 [DOI] [PubMed] [Google Scholar]
- 7.Líden M., Eriksson U. (2006) J. Biol. Chem. 281, 13001–13004 [DOI] [PubMed] [Google Scholar]
- 8.Napoli J. L. (2001) Mol. Cell. Endocrinol. 171, 103–109 [DOI] [PubMed] [Google Scholar]
- 9.Schuster A., Janecke A. R., Wilke R., Schmid E., Thompson D. A., Utermann G., Wissinger B., Zrenner E., Gal A. (2007) Invest. Ophthalmol. Vis. Sci. 48, 1824–1831 [DOI] [PubMed] [Google Scholar]
- 10.Haeseleer F., Jang G. F., Imanishi Y., Driessen C. A., Matsumura M., Nelson P. S., Palczewski K. (2002) J. Biol. Chem. 277, 45537–45546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belyaeva O. V., Korkina O. V., Stetsenko A. V., Kim T., Nelson P. S., Kedishvili N. Y. (2005) Biochemistry 44, 7035–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S. A., Belyaeva O. V., Kedishvili N. Y. (2008) Biochim. Biophys. Acta 1782, 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wicker L. D., Kasus-Jacobi A. (2008) Invest. Ophthalmol. Vis. Sci. 49, ( Abstr. 4397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller B., Adamski J. (2007) J. Steroid Biochem. Mol. Biol. 104, 190–194 [DOI] [PubMed] [Google Scholar]
- 15.Kurth I., Thompson D. A., Rüther K., Feathers K. L., Chrispell J. D., Schroth J., McHenry C. L., Schweizer M., Skosyrski S., Gal A., Hübner C. A. (2007) Mol. Cell. Biol. 27, 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda A., Maeda T., Imanishi Y., Sun W., Jastrzebska B., Hatala D. A., Winkens H. J., Hofmann K. P., Janssen J. J., Baehr W., Driessen C. A., Palczewski K. (2006) J. Biol. Chem. 281, 37697–37704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driessen C. A., Winkens H. J., Hoffmann K., Kuhlmann L. D., Janssen B. P., Van Vugt A. H., Van Hooser J. P., Wieringa B. E., Deutman A. F., Palczewski K., Ruether K., Janssen J. J. (2000) Mol. Cell. Biol. 20, 4275–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim T. S., Maeda A., Maeda T., Heinlein C., Kedishvili N., Palczewski K., Nelson P. S. (2005) J. Biol. Chem. 280, 8694–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasus-Jacobi A., Ou J., Birch D. G., Locke K. G., Shelton J. M., Richardson J. A., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Edwards A. O. (2005) J. Biol. Chem. 280, 20413–20420 [DOI] [PubMed] [Google Scholar]
- 20.Maeda A., Maeda T., Imanishi Y., Kuksa V., Alekseev A., Bronson J. D., Zhang H., Zhu L., Sun W., Saperstein D. A., Rieke F., Baehr W., Palczewski K. (2005) J. Biol. Chem. 280, 18822–18832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001) Nat. Genet. 29, 447–452 [DOI] [PubMed] [Google Scholar]
- 22.Garwin G. G., Saari J. C. (2000) Methods Enzymol. 316, 313–324 [DOI] [PubMed] [Google Scholar]
- 23.Landers G. M. (1990) Methods Enzymol. 189, 70–80 [DOI] [PubMed] [Google Scholar]
- 24.Kim S. R., Fishkin N., Kong J., Nakanishi K., Allikmets R., Sparrow J. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11668–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S. R., Jang Y. P., Jockusch S., Fishkin N. E., Turro N. J., Sparrow J. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19273–19278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai N., Decatur J., Nakanishi K., Eldred G. E. (1996) J. Am. Chem. Soc. 118, 1559–1560 [Google Scholar]
- 27.Parish C. A., Hashimoto M., Nakanishi K., Dillon J., Sparrow J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14609–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fishkin N. E., Sparrow J. R., Allikmets R., Nakanishi K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7091–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kane M. A., Folias A. E., Wang C., Napoli J. L. (2008) Anal Chem. 80, 1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane M. A., Folias A. E., Napoli J. L. (2008) Anal. Biochem. 378, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barua A. B., Furr H. C. (1998) Mol. Biotechnol. 10, 167–182 [DOI] [PubMed] [Google Scholar]
- 32.Peterson G. L. (1977) Anal. Biochem. 83, 346–356 [DOI] [PubMed] [Google Scholar]
- 33.LaVail M. M., Matthes M.T., Yasumura D., Faktorovich E.G., Steinberg R.H. (1997) Histological Method to Assess Photoreceptor Light Damage and Protection by Survival Factors, pp. 369–384, Plenum Press, New York [Google Scholar]
- 34.Yoshida S., Mears A. J., Friedman J. S., Carter T., He S., Oh E., Jing Y., Farjo R., Fleury G., Barlow C., Hero A. O., Swaroop A. (2004) Hum. Mol. Genet. 13, 1487–1503 [DOI] [PubMed] [Google Scholar]
- 35.Reiner A., Yekutieli D., Benjamini Y. (2003) Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y., Yekutieli D. (2005) Genetics 171, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akimoto M., Cheng H., Zhu D., Brzezinski J. A., Khanna R., Filippova E., Oh E. C., Jing Y., Linares J. L., Brooks M., Zareparsi S., Mears A. J., Hero A., Glaser T., Swaroop A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenzel A., Reme C. E., Williams T. P., Hafezi F., Grimm C. (2001) J. Neurosci. 21, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparrow J. R., Boulton M. (2005) Exp. Eye Res. 80, 595–606 [DOI] [PubMed] [Google Scholar]
- 40.Tsina E., Chen C., Koutalos Y., Ala-Laurila P., Tsacopoulos M., Wiggert B., Crouch R. K., Cornwall M. C. (2004) J. Gen. Physiol. 124, 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Napoli J. L. (1999) Biochim. Biophys. Acta 1440, 139–162 [DOI] [PubMed] [Google Scholar]
- 42.Lee S. A., Belyaeva O. V., Popov I. K., Kedishvili N. Y. (2007) J. Biol. Chem. 282, 35621–35628 [DOI] [PubMed] [Google Scholar]
- 43.Haeseleer F., Huang J., Lebioda L., Saari J. C., Palczewski K. (1998) J. Biol. Chem. 273, 21790–21799 [DOI] [PubMed] [Google Scholar]
- 44.Wu B. X., Chen Y., Chen Y., Fan J., Rohrer B., Crouch R. K., Ma J. X. (2002) Invest. Ophthalmol. Vis. Sci. 43, 3365–3372 [PubMed] [Google Scholar]
- 45.Simon A., Hellman U., Wernstedt C., Eriksson U. (1995) J. Biol. Chem. 270, 1107–1112 [PubMed] [Google Scholar]
- 46.Calvert P. D., Strissel K. J., Schiesser W. E., Pugh E. N., Jr., Arshavsky V. Y. (2006) Trends Cell Biol. 16, 560–568 [DOI] [PubMed] [Google Scholar]
- 47.Danciger M., Matthes M. T., Yasamura D., Akhmedov N. B., Rickabaugh T., Gentleman S., Redmond T. M., La Vail M. M., Farber D. B. (2000) Mamm. Genome 11, 422–427 [DOI] [PubMed] [Google Scholar]
- 48.Lebovitz R. M., Zhang H., Vogel H., Cartwright J., Jr., Dionne L., Lu N., Huang S., Matzuk M. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9782–9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Huang T. T., Carlson E. J., Melov S., Ursell P. C., Olson J. L., Noble L. J., Yoshimura M. P., Berger C., Chan P. H., Wallace D. C., Epstein C. J. (1995) Nat. Genet. 11, 376–381 [DOI] [PubMed] [Google Scholar]
- 50.Wu Q., Blakeley L. R., Cornwall M. C., Crouch R. K., Wiggert B. N., Koutalos Y. (2007) Biochemistry 46, 8669–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maeda A., Maeda T., Sun W., Zhang H., Baehr W., Palczewski K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19565–19570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Argmann C. A., Chambon P., Auwerx J. (2005) Cell Metab. 2, 349–360 [DOI] [PubMed] [Google Scholar]
- 53.Weng J., Mata N. L., Azarian S. M., Tzekov R. T., Birch D. G., Travis G. H. (1999) Cell 98, 13–23 [DOI] [PubMed] [Google Scholar]
- 54.Williams M. D., Van Remmen H., Conrad C. C., Huang T. T., Epstein C. J., Richardson A. (1998) J. Biol. Chem. 273, 28510–28515 [DOI] [PubMed] [Google Scholar]
- 55.Sandbach J. M., Coscun P. E., Grossniklaus H. E., Kokoszka J. E., Newman N. J., Wallace D. C. (2001) Invest. Ophthalmol. Vis. Sci. 42, 2173–2178 [PubMed] [Google Scholar]
- 56.Justilien V., Pang J. J., Renganathan K., Zhan X., Crabb J. W., Kim S. R., Sparrow J. R., Hauswirth W. W., Lewin A. S. (2007) Invest. Ophthalmol. Vis. Sci. 48, 4407–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rattner A., Smallwood P. M., Nathans J. (2000) J. Biol. Chem. 275, 11034–11043 [DOI] [PubMed] [Google Scholar]
- 58.Chen C., Tsina E., Cornwall M. C., Crouch R. K., Vijayaraghavan S., Koutalos Y. (2005) Biophys. J. 88, 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.