Abstract
The small GTPases RalA and RalB are multifunctional proteins regulating a variety of cellular processes. Like other GTPases, the activity of Ral is regulated by the opposing effects of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Although several RalGEFs have been identified and characterized, the molecular identity of RalGAP remains unknown. Here, we report the first molecular identification of RalGAPs, which we have named RalGAP1 and RalGAP2. They are large heterodimeric complexes, each consisting of a catalytic α1 or α2 subunit and a common β subunit. These RalGAP complexes share structural and catalytic similarities with the tuberous sclerosis tumor suppressor complex, which acts as a GAP for Rheb. In vitro GTPase assays revealed that recombinant RalGAP1 accelerates the GTP hydrolysis rate of RalA by 280,000-fold. Heterodimerization was required for this GAP activity. In PC12 cells, knockdown of the β subunit led to sustained Ral activation upon epidermal growth factor stimulation, indicating that the RalGAPs identified here are critical for efficient termination of Ral activation induced by extracellular stimuli. Our identification of RalGAPs will enable further understanding of Ral signaling in many biological and pathological processes.
Members of the Ras superfamily of small GTPases act as molecular switches by cycling between an inactive GDP-bound conformation and an active GTP-bound conformation. In their GTP-bound form, small GTPases interact with their effector proteins to induce downstream signaling events. The GDP/GTP cycling is strictly regulated by two classes of regulatory proteins termed guanine nucleotide exchange factors (GEFs)7 and GTPase-activating proteins (GAPs) (1). GEFs activate GTPases by inducing the release of bound GDP for subsequent GTP binding. GAPs inactivate GTPases by accelerating the slow intrinsic rate of GTP hydrolysis.
The Ras-like small GTPases, RalA and RalB, regulate a large variety of cellular processes including transcription, translation, cytoskeletal organization, membrane trafficking, cytokinesis, cell migration, cell proliferation, and cell survival (for review, see Ref. 2–4). RalA and RalB are highly similar to each other (81% amino acid identity) and share the same effectors, such as the exocyst complex (5–7) and RalBP1/RLIP76 (8, 9). Despite this similarity, functional differences between the two isoforms have been described; RalA is indispensable for the anchorage-independent proliferation of tumor cells (10–13), whereas RalB counters apoptotic signaling to support survival of tumor cells (10, 14). To date, six RalGEF proteins have been identified and characterized. They are structurally classified into two major groups. Members of the RalGDS family, which includes RalGDS (15), Rgl (16), Rlf (17), and Rgl3 (18), possess a Ras binding domain and are directly activated by binding to Ras-GTP (19, 20). Another class of RalGEFs consists of RalGPS1/RalGEF2 (21, 22) and RalGPS2 (21, 23). This novel type of RalGEF lacks the Ras binding domain but, instead, has a pleckstrin homology domain.
The finding that Ras can activate most RalGEFs in addition to Raf and phosphoinositide 3-kinases suggested that Ral GTPases mediate some functions of oncogenic Ras. Recent studies using human cells have revealed an essential contribution of this Ras-RalGEFs-Ral signaling pathway to Ras-induced tumorigenic transformation (24, 25). Although these studies highlight the importance of appropriate regulation of Ral activity in normal cells, the negative regulators for Ral GTPases, namely RalGAPs, have not yet been identified. A report by Feig and co-workers (26) demonstrated the existence of Ral-specific GAP activity in the cytosolic fraction of brain and testis. However, the molecule(s) responsible for this GAP activity still remains elusive.
In this study we report the molecular identification of RalGAPs, which we have named RalGAP1 and RalGAP2. They are large heterodimeric complexes, each composed of a catalytic α1 or α2 subunit and a common β subunit. Interestingly, the RalGAP complexes have structural and catalytic similarities to the tuberous sclerosis tumor suppressor complex.
EXPERIMENTAL PROCEDURES
Purification of the p240-p170 Complex from Porcine Brain Cytosol
All purification procedures were performed at 4 °C unless otherwise noted. Eight porcine brains were homogenized in 1.6 liters of buffer A (50 mm HEPES/KOH, pH 7.4, 78 mm KCl, 4 mm MgCl2, 2 mm EGTA, 0.2 mm CaCl2, 1 mm dithiothreitol) containing protease inhibitors (protease inhibitor mixture for mammalian cells; Sigma) using a Waring blender followed by a glass-Teflon homogenizer. The homogenate was centrifuged at 2800 × g for 30 min to remove coarse particulates. The supernatant was dialyzed overnight against 5 liters of buffer A. The dialyzed material was further centrifuged at 120,000 × g for 1 h to obtain clarified cytosol (∼5 mg of protein ml−1, 1.2 liters). The cytosol was supplemented with the non-hydrolyzable GTP analogue guanosine 5′-(β,γ-imido)triphosphate (GppNHp) (10 μm; Sigma) and used for RalAQ72L affinity chromatography as described below.
For preparation of the affinity column, glutathione S-transferase (GST)-RalAQ72L (residues 9–183, 160 mg) was immobilized on 8 ml of glutathione beads (Glutathione-Sepharose 4B; GE Healthcare). RalAQ72L-bound nucleotides were exchanged for GppNHp by incubating the beads in buffer A containing 10 mm EDTA and 1 mm GppNHp at room temperature for 90 min. The GppNHp-bound form of RalAQ72L was stabilized by adding MgCl2 to a final concentration of 14 mm. These affinity beads were packed into the PolyPrep columns (Bio-Rad), and porcine brain cytosol prepared as above was loaded onto these columns. After washing the columns with 20 column volumes of buffer A, bound proteins were eluted with 2 column volumes of buffer B (50 mm HEPES, KOH, pH 7.4, 78 mm KCl, 10 mm EDTA, 1 mm dithiothreitol, 5 mm GDP). The eluate (16 ml) was passed through a glutathione beads column (0.4 ml) to remove GST-RalAQ72L that leaked from the affinity column during the elution step and then concentrated to ∼0.6-ml using a Centriprep YM-100 (Millipore). The concentrated sample was applied to a Superose 6 10/300 GL gel filtration column (GE Healthcare) equilibrated with buffer A, and fractions of 0.6 ml were collected. Fractions were analyzed by SDS-PAGE and Coomassie Blue staining. Protein bands of p240 and p170 were excised from the gel and subjected to mass spectrometry analyses as described previously (27, 28).
Purification of the p220-p170 Complex from Rat Lung Cytosol
We performed essentially the same purification scheme used for the p240-p170 complex purification except that purification was performed on a 1/10 scale with freshly isolated lungs of thirty male Wister rats. The affinity column eluate was fractionated on a Superose 6 PC3.2/30 column (SMART chromatography system; GE Healthcare), and fractions containing p170 (fractions 17–22) were pooled and used for GTPase assays. As a comparison, we treated rat brains in the same fashion.
Plasmid Constructs and Recombinant Proteins
We amplified full-length p240 cDNA by PCR from human brain Marathon-Ready cDNA (BD Biosciences) and cloned it into the XhoI site of pBluescript II SK(+) (Stratagene). We sequenced six independent clones and found that all of them contained a 141-bp insertion after bp 2266 of the deposited cDNA sequence for Homo sapiens GTPase-activating Rap/Ran GAP domain-like 1 (NM_014990). This insertion encodes a 47-amino acid sequence without changing the reading frame. The N1950K mutation was introduced by PCR-based site-directed mutagenesis. The p170 cDNA (KIAA1219) was obtained from Kazusa DNA Research Institute (Chiba, Japan). These cDNAs were subcloned into pFastBac HT to generate recombinant baculoviruses (Bac-to-Bac Baculovirus Expression System; Invitrogen). The cDNAs for RalA, RalB, H-Ras, Rap1A, Rheb, RhoA (27), Rab27A, Ran, Sec5 Ral binding domain (residues 1–120) (28), and Raf1 Ras binding domain (residues 51–131) were amplified by PCR from human brain or bone marrow Marathon-Ready cDNA and cloned into pGEX-2T (GE Healthcare). The truncated version of RalA (residues 9–183) (29) with a Q72L mutation was generated by PCR and subcloned into pGEX-2T. All constructs were verified by DNA sequencing.
His6-tagged p240 and p170 were expressed in Sf9 insect cells and affinity-purified on nickel beads (nickel-nitrilotriacetic acid-agarose; Qiagen). Purified proteins were concentrated and further purified on a Superose 6 PC3.2/30 gel filtration column equilibrated with buffer A. The peak fractions were stored at 4 °C and used for GTPase assays within 3 days. GST fusion proteins were expressed in Escherichia coli strain BL21 (DE3) transformed with the corresponding pGEX plasmids and affinity-purified on glutathione beads. Purified proteins were dialyzed against buffer A, snap-frozen in liquid nitrogen, and stored at −80 °C until use. Protein concentrations were determined by the Bradford assay (Bio-Rad) or by densitometry of Coomassie Blue-stained gels using bovine serum albumin as a standard in both cases.
GTPase Assays
We assessed GAP activity using a nitrocellulose filter binding assay or thin layer chromatography. For the filter binding assay, GST fusion GTPases were loaded with [γ-32P]GTP in buffer A containing 10 mm EDTA and 10 μm [γ-32P]GTP (∼3000 cpm pmol−1; PerkinElmer Life Sciences) at 30 °C for 15 min. Loading reactions were stopped by chilling on ice and adding MgCl2 to a final concentration of 14 mm. GTPase assays were performed at 30 °C in 20-μl mixtures containing 20 pmol of labeled GTPases and test materials. Reactions were stopped by diluting the 20-μl mixtures into 0.8 ml of ice-cold buffer A and filtering on 0.45-μm nitrocellulose filters (Whatman). Filters were washed twice with 4 ml of ice-cold buffer A, dried, and counted in a liquid scintillation counter. For the thin layer chromatography analysis, GTPase assays were performed with GST fusion GTPases preloaded with [α-32P]GTP (∼6000 cpm pmol−1; PerkinElmer). Nucleotides trapped on the filters were eluted at 85 °C for 3 min with buffer A containing 0.1% SDS, 10 mm EDTA, 1 mm GTP, and 1 mm GDP. Released nucleotides were spotted on polyethyleneimine cellulose plates (Merck) and chromatographed with 1 m LiCl and 1 m formic acid. Plates were dried and processed for autoradiography.
Antibodies
Rabbit anti-p240 polyclonal antibodies were raised against a recombinant fragment of p240 (residues 803–1080) and the following synthetic peptides: MFSKKPHGDVKKSTQKVLD (residues 1–19) and SMSDQEKPEEPPTSNEC (residues 1630–1646). We used the anti-recombinant p240 antibody for Western blot analysis and the anti-peptide antibodies for immunoprecipitation assays. Rabbit anti-p170 antibody was raised against the peptide KRLESDSYSPPHVRRKQKI (residues 1442–1460). Rabbit anti-p220 antibody was raised against the peptide AQEDADKLGLSETDSKE (residues 451–467). Other antibodies used were anti-RalA, -Ras, -Sec6, -Sec8, (BD Biosciences), anti-tubulin α (Sigma), anti-phospho-extracellular signal-regulated kinase (ERK) 1/2 (Santa Cruz Biotechnology), anti-Sec5 (28), and horseradish peroxidase-labeled anti-rabbit or -mouse IgG (GE Healthcare).
Immunoprecipitation and Immunodepletion Assays
For immunoprecipitation assays, porcine brain cytosol (Fig. 1E), rat testis cytosol (Fig. 3B), or rat lung cytosol (Fig. 3E) were incubated at 4 °C for 3 h with the indicated antibodies prebound to protein A beads (Protein A-agarose; Roche Applied Science). Where indicated, immunoprecipitates were prepared in the presence of 50 μg of antigen peptides. Beads were washed 4 times with buffer A containing 0.1% (w/v) Triton X-100, and proteins bound to the beads were analyzed by Western blotting. In Fig. 3E, washed beads were directly incubated with radiolabeled GTPases. For immunodepletion assays, 10 μl of packed protein A beads were incubated at 4 °C for 1 h with the indicated antisera (20 μl each) and then washed 5 times with buffer A. Aliquots of fraction 18 (Fig. 1F) or the pooled lung fraction (Fig. 3C) were incubated with the IgG-coated beads at 4 °C for 3 h. Beads were removed by brief centrifugation, and the supernatants were used for GTPase assays.
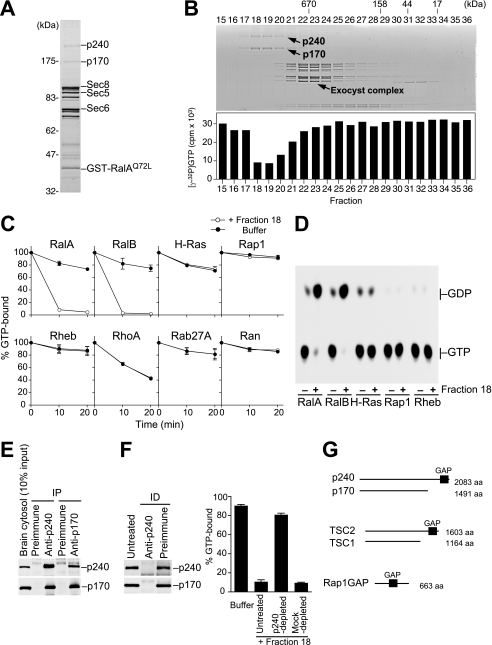
FIGURE 1.
The p240-p170 complex purified from porcine brain has RalGAP activity. A, RalAQ72L affinity column eluate was analyzed by SDS-PAGE and Coomassie Blue staining. Components of the exocyst complex (Sec8, Sec5, and Sec6) and GST-RalAQ72L that had leaked from the column are indicated, as identified by Western blotting (not shown). B, upper panel, the mixture of RalAQ72L-binding proteins was separated on a Superose 6 gel filtration column, and the resulting fractions were analyzed by SDS-PAGE and Coomassie blue staining. Lower panel, each fraction was tested for RalGAP activity by a filter binding assay using [γ-32P]GTP-loaded RalA as described under “Experimental Procedures.” C, various Ras family GTPases preloaded with [γ-32P]GTP were incubated at 30 °C for the indicated periods with (open circles) or without (closed circles) fraction 18, and the [γ-32P]GTP remaining bound to GTPases was measured by the filter binding assay (mean ± S.E., n = 2). D, Ras subfamily GTPases preloaded with [α-32P]GTP were incubated at 30 °C for 10 min with (+) or without (−) fraction 18, and bound nucleotides were analyzed by thin layer chromatography and autoradiography. E, co-immunoprecipitation of p240 and p170 from porcine brain cytosol. IP, immunoprecipitation. F, immunodepletion of p240 from fraction 18 results in a concomitant depletion of p170 (left panel) and abolishes the GAP activity for RalA (right graph) (mean ± S.E., n = 3). ID, immunodepletion. G, schematic comparison of the domain structure of the p240-p170 complex, the TSC2-TSC1 complex, and Rap1GAP. aa, amino acids.
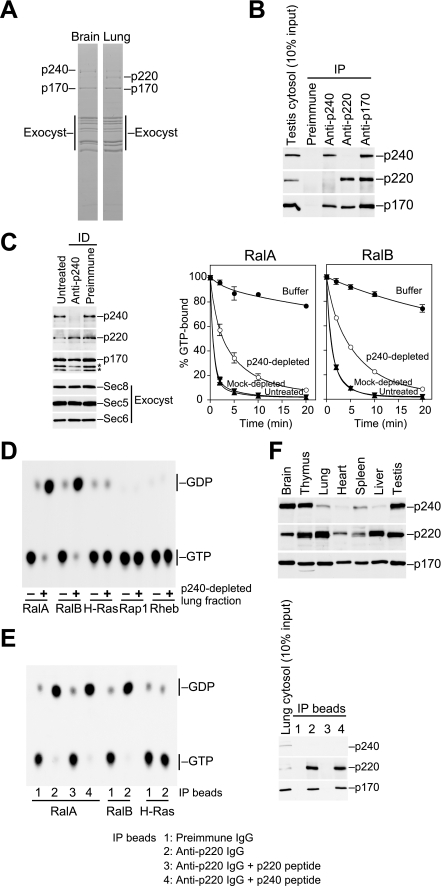
FIGURE 3.
The p220-p170 complex; a second RalGAP identified from rat lung. A, comparison of RalAQ72L-binding proteins from rat brain and lung cytosol. RalAQ72L affinity column eluates were fractionated by gel filtration, and fractions containing p170 (fractions 17–22) were pooled and analyzed by SDS-PAGE and Coomassie Blue staining. p240, p220, p170, and the components of the exocyst complex are indicated, as identified by Western blotting and mass spectrometry. B, co-immunoprecipitation of p220 and p170 from rat testis cytosol. Note that p220 is not co-purified with p240. IP, immunoprecipitation. C, the pooled lung fraction depleted of p240 retains RalGAP activity. Left panel, the pooled lung fraction was treated with anti-p240 IgG- or preimmune IgG-coated beads, and the resulting supernatants were analyzed for the specified proteins. Asterisks denote degradation products of p170. Right graphs, RalGAP activity of the immunodepleted fractions was measured by the filter-binding assay (mean ± S.E., n = 2). ID, immunodepletion. D, autoradiograph showing that the p240-depleted lung fraction has Ral-specific GAP activity. Ras subfamily GTPases preloaded with [α-32P]GTP were incubated at 30 °C for 10 min with (+) or without (−) the p240-depleted lung fraction. E, immunoprecipitates prepared from rat lung cytosol with preimmune IgG (IP beads 1), anti-p220 IgG (IP beads 2), or anti-p220 IgG absorbed with p220 antigen peptide (IP beads 3) or p240 antigen peptide (IP beads 4) were used in GTPase assays with the indicated GTPases preloaded with [α-32P]GTP. After 10 min of incubation at 30 °C, bound radiolabeled nucleotides were analyzed by thin layer chromatography and autoradiography. The right panel shows Western blot analysis of the immunoprecipitates with the indicated antibodies. F, Western blot analysis of p240, p220, and p170 in rat tissues.
Tissue Distribution Analysis
Various organs from male Wister rats were homogenized in buffer A containing protease inhibitors. The homogenates were centrifuged at 2,000 × g for 10 min, and the supernatants were further centrifuged at 200,000 × g for 10 min. Aliquots of the resulting supernatants (100 μg total protein) were analyzed by Western blotting with the indicated antibodies.
Knockdown and Pulldown Assays
We used two distinct short interfering RNAs (siRNAs) targeting rat/mouse p170 (StealthTM RNAi; Invitrogen): 5′-GAAACCUGGGAAGUCUUACUGUUGU-3′ (p170 siRNA#1) and 5′-GAUUGAUGUUGUGGUUCCUUACUUU-3′ (p170 siRNA#2). Their scrambled sequences were used as controls. PC12 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 10% horse serum in 6-cm culture dishes. Cells at ∼40% confluency were transfected with the individual siRNAs (50 nm) with RNAiMAX transfection regent (Invitrogen). Seventy-two hours after transfection cells were rinsed once with phosphate-buffered saline and lysed in 0.8 ml of ice-cold buffer C (50 mm HEPES/KOH, pH 7.4, 100 mm NaCl, 4 mm MgCl2, 2 mm EGTA, 1 mm dithiothreitol, 1% (w/v) Triton X-100, 10 mm β-glycerophosphate, 10 mm NaF, 1 mm Na3VO4) containing protease inhibitors. Lysates were rotated at 4 °C for 5 min and then centrifuged at 200,000 × g for 5 min. The supernatants were incubated at 4 °C for 30 min with glutathione beads precoated with 20 μg of GST-Sec5 Ral binding domain. After washing the beads three times with buffer C, bead-associated RalA was analyzed by Western blotting. Part of the supernatants was taken before the incubation and used for Western blot analysis to assess knockdown efficiencies. NIH3T3 cells were lysed at 48 h after siRNA transfection and processed as described above. For epidermal growth factor (EGF) stimulation assays, we used the p170 siRNA#1 and siRNA#2 in combination at 25 nm each. PC12 cells treated with the siRNAs were serum-starved for 8 h and then stimulated with EGF (20 ng ml−1; R&D Systems). After stimulation, cells were harvested in buffer C at the indicated time points, and the GTP-bound forms of RalA and Ras were pulled down using glutathione beads precoated with both GST-Sec5 Ral binding domain and GST-Raf1 Ras binding domain (20 μg each).
RESULTS AND DISCUSSION
Affinity Purification of RalGAP Complex from Porcine Brain Cytosol
To purify RalGAP, we applied an affinity chromatography approach using RalAQ72L as bait. In general, Ras family GTPases with a glutamine to leucine substitution in the switch II region (Q61L in Ras) are resistant to GAP-stimulated GTP hydrolysis, whereas these mutants can still bind to their cognate GAPs (1). RasQ61L, although unable to hydrolyze GTP efficiently, has higher affinities than wild type for RasGAPs, p120GAP, and NF1/neurofibromin (22, 23). Therefore, we reasoned that the analogous mutant RalAQ72L would be ideal bait for RalGAP. We prepared an affinity column immobilized with GST-RalAQ72L preloaded with GppNHp, a non-hydrolyzable GTP analogue, and searched for binding proteins in porcine brain cytosol.
We observed 14 proteins eluting from the column with EDTA and GDP (Fig. 1A). Among them, we identified components of the exocyst complex, a known effector of Ral GTPases (5–7), thus confirming the specificity of the affinity purification. We then separated the mixture of RalAQ72L-binding proteins by gel filtration chromatography and tested the fractions for RalGAP activity. Activity was monitored by incubating each fraction with [γ-32P]GTP-loaded RalA and measuring the decrease in radioactivity that remained bound to RalA. We detected a marked activity in fractions with an apparent molecular mass of >1000 kDa (Fig. 1B, fractions 18–20). Interestingly, proteins of 240 kDa (p240) and 170 kDa (p170) both co-fractionated with this activity (Fig. 1B, upper panel). As clearly seen in Fig. 1C, the activity in fraction 18 was highly specific for RalA and RalB among the Ras family GTPases tested. Moreover, the activity was indeed due to acceleration of GTP hydrolysis, but not to GTP dissociation, because [α-32P]GTP bound to RalA or RalB was specifically hydrolyzed to GDP after incubation with fraction 18, as analyzed by thin layer chromatography (Fig. 1D). These results indicated that fraction 18, which contained only p240 and p170, had GAP activity toward Ral GTPases.
Molecular Identification of p240 and p170
Mass spectrometry analysis revealed p240 to be the porcine orthologue of human GTPase-activating Rap/RanGAP domain-like 1 (GARNL1, also known as tuberin-like protein 1 or KIAA0884), the gene for which was identified as a candidate for 14q13-linked neurological disorders (32). p170 was identified to be the porcine orthologue of human KIAA1219, an evolutionarily conserved protein of unknown function. We generated polyclonal antibodies against the two proteins and confirmed these results (data not shown).
Because p240 and p170 co-migrated in the gel filtration (Fig. 1B), we tested whether the two proteins formed a complex. As shown in Fig. 1E, p240 and p170 were co-immunoprecipitated from porcine brain crude cytosol with either the anti-p240 or anti-p170 antibodies. Immunodepletion of p240 from fraction 18 with anti-p240 IgG-coated beads resulted in a concomitant depletion of p170 (Fig. 1F, left) and abolished the activity in the fraction (Fig. 1F, right). Thus, p240 and p170 form a stoichiometric complex that acts as RalGAP.
The C-terminal ∼200-amino acid region of p240 shows a high degree of sequence identity to the catalytic domain of TSC2/tuberin (31% amino acid identity) and to a lesser extent that of Rap1GAP (24% identity) (supplemental Fig. 1A). Apart from this region, p240 has no other discernible domains. TSC2 is a tumor suppressor protein that acts as a GAP toward Rheb (33–37), a Ras subfamily GTPase that promotes cell growth through activation of the mTOR pathway. TSC2 functions in a complex with a non-catalytic subunit TSC1/hamartin. Loss of either TSC2 or TSC1 leads to constitutive activation of Rheb and causes the benign tumor syndrome tuberous sclerosis (38). Notably, the p240-p170 complex has a similar domain organization as the TSC2-TSC1 complex (Fig. 1G), although there is no sequence similarity between p170 and TSC1.
Biochemical and Catalytic Properties of the Recombinant p240-p170 Complex
Next, we characterized the biochemical and catalytic properties of the complex using His6-tagged recombinant forms of human p240 and p170. We could purify the proteins at a nearly stoichiometric ratio from Sf9 insect cells co-infected with baculoviruses encoding each protein. Gel filtration analysis showed that the purified proteins eluted together at >1000 kDa (Fig. 2A, top), which is similar to the elution properties of the brain native complex (Fig. 1B). In contrast, p240 alone or p170 alone, which were purified from separately infected cells, eluted around 450 or 700 kDa, respectively (Fig. 2A, middle and bottom). This observation again indicates that the two proteins interact directly to form a stable complex. The recombinant complex exhibited potent GAP activity toward RalA and RalB (Fig. 2B). Kinetic analysis revealed that the complex acts on RalA with Km and kcat values of 12.1 μm and 46.8 s−1, respectively (Fig. 2C). Because the intrinsic GTP hydrolysis rate of RalA is calculated to be 0.010 min−1, the complex accelerates the GTP hydrolysis rate of RalA by 280,000-fold. These kinetic values are within an order of magnitude of those reported for other GAPs (30, 31).
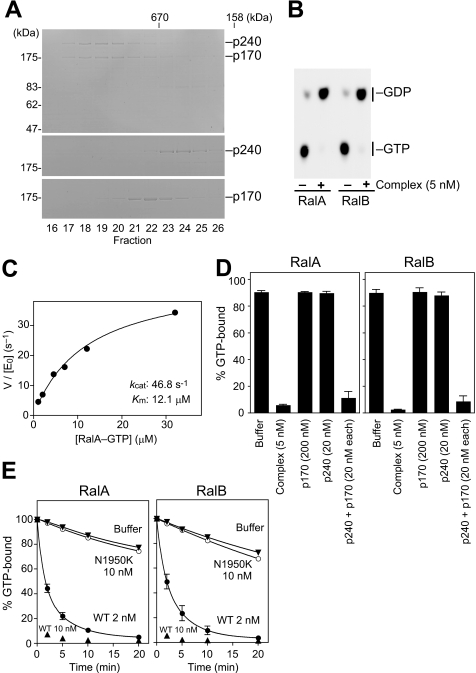
FIGURE 2.
Biochemical and catalytic properties of the recombinant p240-p170 complex. A, chromatographic comparison of the recombinant p240-p170 complex (top), p240 alone (middle), and p170 alone (bottom) on Superose 6 gel filtration chromatography. Gels were Coomassie Blue-stained. B, autoradiograph showing that the recombinant complex stimulates Ral GTPase activity. [α-32P]GTP-loaded RalA or RalB were incubated at 30 °C for 10 min with (+) or without (−) the recombinant complex (5 nm). C, kinetic analysis of the recombinant p240-p170 complex. Initial rates of GTP hydrolysis on RalA were determined at 30 °C at increasing concentrations of GTP-bound RalA by the filter binding assay. The recombinant p240-p170 complex was used at 1 nm ([E0]). GTPase reaction rates were fitted to the Michaelis-Menten equation to give Km (12.1 μm) and kcat (46.8 s−1) values of the reaction. D, p240 requires p170 for GAP activity. [γ-32P]GTP-loaded RalA or RalB were incubated at 30 °C for 10 min with the recombinant complex, p240 alone, p170 alone, or a mixture of separately purified p240 and p170 at the indicated concentrations (mean ± S.E., n = 3). E, the N1950K mutation abrogates p240 GAP activity. [γ-32P]GTP-loaded RalA or RalB were incubated at 30 °C for the indicated periods with the wild-type complex (WT; 2 nm or 10 nm) or the mutant complex (N1950K; 10 nm) (mean ± S.E., n = 3).
We next tested whether dimerization with p170 is required for p240 GAP activity. As shown in Fig. 2D, p240 alone had no detectable GAP activity. However, the addition of equimolar p170 to the reaction dramatically enhanced the activity to an extent comparable with that achieved by the complex (Fig. 2D). Because p170 alone had no effect on the GTPase activity even at the high concentration (200 nm; Fig. 2D), this result indicates that p240 requires p170 for its GAP activity.
Most GAPs, including RasGAPs and RhoGAPs, supply an arginine residue, called the “arginine finger,” into the active site of the GTPase to directly catalyze GTP hydrolysis (1). RapGAPs and TSC2 differ from these GAPs in that they do not possess a catalytic arginine but instead use an asparagine residue, termed the “asparagine thumb,” for catalysis (39–41). Sequence comparison suggested that p240 also employs the asparagine thumb machinery (supplemental Fig. 1A). To test this we mutated the corresponding asparagine of p240 to lysine, which mimics a disease-causing mutation in TSC2 (42), and asked whether this mutant p240N1950K retained RalGAP activity. Although the mutation did not affect the ability of p240 to interact with p170 (data not shown), the p240N1950K-p170 complex showed no appreciable activity for RalA or RalB (Fig. 2E). Thus, RalGAP shares the same catalytic mechanism as other asparagine thumb-type GAPs.
Identification of a Second RalGAP Complex from Rat Lung
Tissue distribution analysis revealed that, whereas p170 was abundantly expressed in all rat tissues examined, expression levels of p240 were highly variable between tissues and did not correlate with the p170 expression profile. Specifically, we detected a considerably low level of p240 in the lung (see below). This observation led us to search this tissue for another p170 binding partner that would functionally substitute for p240. To this end, we performed RalAQ72L affinity chromatography with rat lung and brain cytosol. The affinity column eluates were fractionated by gel filtration using the SMART micropurification system, and fractions containing p170 were pooled and analyzed (Fig. 3A). We detected similar amounts of p170 and the components of the exocyst complex in both brain and lung fractions (Fig. 3A). However, the band intensity of p240 was much weaker in the lung fraction compared with that in the brain fraction (Fig. 3A). Instead, a protein of 220 kDa (p220) was detected (Fig. 3A). Mass spectrometry analysis identified p220 as the rat orthologue of human C20orf74 (also known as AS250 (43) or KIAA1272). The overall structure of p220 is very similar to p240 (54% amino acid identity); in particular, the C-terminal ∼200-amino acid region of p220 shows striking homology to the catalytic region of p240 (85% identity; supplemental Fig. 2A). This structural similarity and the fact that p220 was purified as a RalAQ72L-binding protein strongly suggested that p220 also acts as RalGAP, likely as a complex with p170. Using an anti-p220 antibody, we confirmed that p220 immunoprecipitated with p170 from rat testis cytosol (Fig. 3B). Importantly, p220 was not co-purified with p240 by either the anti-p220 or p240 antibodies (Fig. 3B). This result indicates that the p220-p170 complex is physically distinct from the p240-p170 complex.
To determine whether the p220-p170 complex possesses RalGAP activity, we immunodepleted p240 (and p170 complexed to p240) from the pooled lung fraction with the antibody specific to p240 and then examined the activity remaining in this immunodepleted fraction. The p240-depleted fraction, which contained only the p220-p170 complex and the exocyst complex (Fig. 3C, left), still exhibited substantial GAP activity, albeit less efficiently than mock-depleted or untreated fractions (Fig. 3C, right). This residual activity was highly specific for RalA and RalB (Fig. 3D). Because the exocyst complex had no effect on Ral GTPase activity (Fig. 1B), these results suggest that the p220-p170 complex also has RalGAP activity. To more directly examine the activity of the p220-p170 complex, we tested the activity of the complex immunoprecipitated with the p220-specific peptide antibody. The p220 immunoprecipitates prepared from rat lung cytosol, which also contained p170 but were devoid of p240 (Fig. 3E, right), exhibited Ral-specific GAP activity (Fig. 3E, left). When immunoprecipitates were prepared in the presence of excess p220 antigen peptide, the activity of the immunoprecipitates disappeared completely (Fig. 3E). This result indicates that the p220-p170 complex actually has specific GAP activity for Ral GTPases. Consistent with its role as a second RalGAP catalytic subunit, p220 is abundantly expressed in tissues where p240 expression levels are relatively low, such as the lung and liver (Fig. 3F). Thus, there exist the two structurally similar catalytic subunits, p240 and p220, with complementary tissue expression patterns.
p170 Knockdown Leads to Sustained Activation of RalA in EGF-stimulated PC12 Cells
Finally, we performed RNA interference experiments to test whether the two complexes function as RalGAPs in intact cells. Two distinct siRNAs targeting p170, but not scrambled controls, suppressed the levels of p170 expression in PC12 cells (Fig. 4A). Suppression of p170 expression also led to the reduction in p240 and p220 levels (Fig. 4A), suggesting that p170 contributes to stability of its binding partners. In p170 knockdown cells, RalA existed in a relatively activated state with an ∼3-fold increase in the GTP-bound form (Fig. 4A). We obtained similar results with NIH3T3 cells, which predominantly expressed p220 (Fig. 4A). These results indicate that the complexes play a substantial role in maintaining RalA in an inactive state.
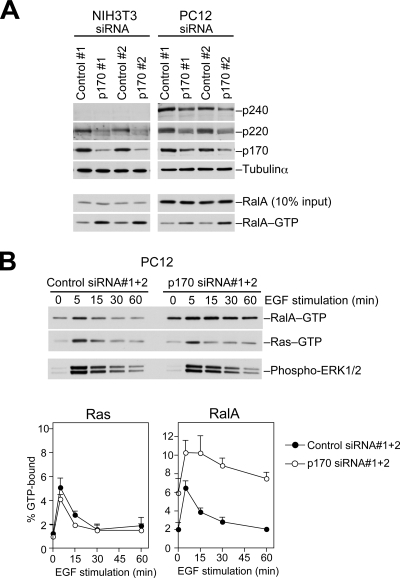
FIGURE 4.
p170 knockdown leads to sustained activation of RalA. A, PC12 or NIH3T3 cells transfected with the indicated siRNAs were lysed, and the levels of the GTP-bound form of RalA (RalA-GTP) were determined by a pulldown assay using Sec5 Ral binding domain as described under “Experimental Procedures.” Lysates were also analyzed for the levels of the specified proteins. B, PC12 cells transfected with the indicated siRNAs were serum-starved and then stimulated with EGF (20 ng ml−1). At the indicated time points, cells were lysed, and the amounts of RalA-GTP and Ras-GTP in the same lysate were determined. Lysates were also probed for phosphorylated ERK1/2. The lower graphs show quantification of Ras-GTP and RalA-GTP levels (means ± S.E., n = 3).
We then examined the effects of reducing p170 on the activation/inactivation kinetics of RalA after stimulation with EGF. Many growth factors, including EGF, have been shown to activate RalA through the Ras-RalGEF pathway (44). In control PC12 cells, we observed a transient activation of both Ras and RalA with peak activation at 5 min (Fig. 4B). By contrast, in p170 knockdown cells the level of GTP-bound RalA further increased from baseline and remained elevated over 15 min followed by a gradual decline, whereas Ras was rapidly inactivated as in control cells (Fig. 4B). Phosphorylation of ERKs, an indicator of the Ras-Raf pathway, was not affected in p170 knockdown cells (Fig. 4B). These results demonstrate that p170 knockdown specifically impairs RalGAP activity in the cells and that the complexes are critical for efficient termination of Ral signaling after growth factor stimulation.
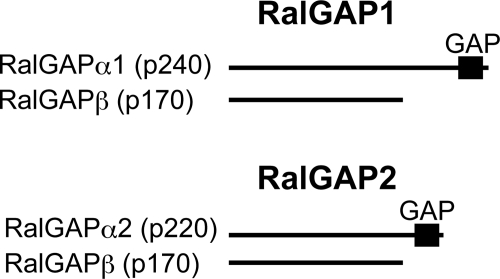
On the basis of these findings, we define the RalGAP complexes as RalGAP1, composed of the catalytic subunit RalGAPα1 (p240) and the common subunit RalGAPβ (p170), and RalGAP2, composed of RalGAPα2 (p220) and RalGAPβ (p170) (Fig. 5). RalGAPα1 and -α2 along with their relatives TSC2 and RapGAPs define a distinct family of GAPs that use a catalytic asparagine (supplemental Fig. 1B). Both catalytic and common subunits are highly conserved from fly to mammals, which suggests that the heterodimeric structure of RalGAP is strictly conserved across species (supplemental Fig. 2). Heterodimer dissociation may be involved in the regulation of GAP activity, as shown for the TSC2-TSC1 complex (45–47).
FIGURE 5.
The RalGAP complexes. Schematic representation of RalGAP1 and RalGAP2. RalGAP1 is composed of the catalytic subunit RalGAPα1 (p240) and the common subunit RalGAPβ (p170). RalGAP2 is composed of the catalytic subunit RalGAPα2 (p220) and the common subunit RalGAPβ (p170).
Increasing evidence indicates that Ral GTPases play crucial roles in tumor cell growth (10–13), survival (10, 14), and metastasis (12, 48). A recent in vivo study showing that mice deficient for RalGDS, the founding member of the Ras-regulated RalGEFs, are resistant to Ras-mediated skin carcinogenesis (49) further supports the role for Ral activation in Ras-induced tumorigenesis. Our identification of RalGAPs now allows investigation into the biological roles of negative regulation of Ral signaling. Considering the aberrant activation of Ral GTPases in many human cancer tissues (12, 50), it is possible that RalGAP function is impaired in human cancers.
Supplementary Material
Acknowledgments
We are grateful to the KAZUSA DNA Research Institute for providing the KIAA1219 clone. We thank T. Matsubara for technical assistance.
This work was supported by Research Grants 15081206 and 20013201 (to H. H.) and 07J08426 (to R. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Takeda Science Foundation (to H. H.) This work was also supported in part by Global COE Program “Center for Frontier Medicine” from the Ministry of Education, Culture, Sports, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The nucleotide sequence reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession numbers AB511280.
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- GST
- glutathione S-transferase
- GppNHp
- guanosine 5′-(β,γ-imido)triphosphate
- EGF
- epidermal growth factor
- ERK
- extracellular signal-regulated kinase
- siRNA
- short interfering RNA.
REFERENCES
- 1.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 2.Feig L. A. (2003) Trends Cell Biol. 13, 419–425 [DOI] [PubMed] [Google Scholar]
- 3.Camonis J. H., White M. A. (2005) Trends Cell Biol. 15, 327–332 [DOI] [PubMed] [Google Scholar]
- 4.Bodemann B. O., White M. A. (2008) Nat. Rev. Cancer 8, 133–140 [DOI] [PubMed] [Google Scholar]
- 5.Brymora A., Valova V. A., Larsen M. R., Roufogalis B. D., Robinson P. J. (2001) J. Biol. Chem. 276, 29792–29797 [DOI] [PubMed] [Google Scholar]
- 6.Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. (2002) Nat. Cell Biol. 4, 66–72 [DOI] [PubMed] [Google Scholar]
- 7.Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., Ohta Y. (2002) Nat. Cell Biol. 4, 73–78 [DOI] [PubMed] [Google Scholar]
- 8.Cantor S. B., Urano T., Feig L. A. (1995) Mol. Cell. Biol. 15, 4578–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jullien-Flores V., Dorseuil O., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J. H. (1995) J. Biol. Chem. 270, 22473–22477 [DOI] [PubMed] [Google Scholar]
- 10.Chien Y., White M. A. (2003) EMBO Rep. 4, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. (2005) Cancer cell 7, 533–545 [DOI] [PubMed] [Google Scholar]
- 12.Lim K. H., O'Hayer K., Adam S. J., Kendall S. D., Campbell P. M., Der C. J., Counter C. M. (2006) Curr. Biol. 16, 2385–2394 [DOI] [PubMed] [Google Scholar]
- 13.Sablina A. A., Chen W., Arroyo J. D., Corral L., Hector M., Bulmer S. E., DeCaprio J. A., Hahn W. C. (2007) Cell 129, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien Y., Kim S., Bumeister R., Loo Y. M., Kwon S. W., Johnson C. L., Balakireva M. G., Romeo Y., Kopelovich L., Gale M., Jr., Yeaman C., Camonis J. H., Zhao Y., White M. A. (2006) Cell 127, 157–170 [DOI] [PubMed] [Google Scholar]
- 15.Albright C. F., Giddings B. W., Liu J., Vito M., Weinberg R. A. (1993) EMBO J. 12, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi A., Demo S. D., Ye Z. H., Chen Y. W., Williams L. T. (1994) Mol. Cell. Biol. 14, 7483–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolthuis R. M., Bauer B., van 't Veer L. J., de Vries-Smits A. M., Cool R. H., Spaargaren M., Wittinghofer A., Burgering B. M., Bos J. L. (1996) Oncogene 13, 353–362 [PubMed] [Google Scholar]
- 18.Shao H., Andres D. A. (2000) J. Biol. Chem. 275, 26914–26924 [DOI] [PubMed] [Google Scholar]
- 19.Urano T., Emkey R., Feig L. A. (1996) EMBO J. 15, 810–816 [PMC free article] [PubMed] [Google Scholar]
- 20.Wolthuis R. M., de Ruiter N. D., Cool R. H., Bos J. L. (1997) EMBO J. 16, 6748–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebhun J. F., Chen H., Quilliam L. A. (2000) J. Biol. Chem. 275, 13406–13410 [DOI] [PubMed] [Google Scholar]
- 22.de Bruyn K. M., de Rooij J., Wolthuis R. M., Rehmann H., Wesenbeek J., Cool R. H., Wittinghofer A. H., Bos J. L. (2000) J. Biol. Chem. 275, 29761–29766 [DOI] [PubMed] [Google Scholar]
- 23.Ceriani M., Scandiuzzi C., Amigoni L., Tisi R., Berruti G., Martegani E. (2007) Exp. Cell Res. 313, 2293–2307 [DOI] [PubMed] [Google Scholar]
- 24.Hamad N. M., Elconin J. H., Karnoub A. E., Bai W., Rich J. N., Abraham R. T., Der C. J., Counter C. M. (2002) Genes Dev. 16, 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangarajan A., Hong S. J., Gifford A., Weinberg R. A. (2004) Cancer Cell 6, 171–183 [DOI] [PubMed] [Google Scholar]
- 26.Emkey R., Freedman S., Feig L. A. (1991) J. Biol. Chem. 266, 9703–9706 [PubMed] [Google Scholar]
- 27.Higashi T., Ikeda T., Shirakawa R., Kondo H., Kawato M., Horiguchi M., Okuda T., Okawa K., Fukai S., Nureki O., Kita T., Horiuchi H. (2008) J. Biol. Chem. 283, 8746–8755 [DOI] [PubMed] [Google Scholar]
- 28.Kawato M., Shirakawa R., Kondo H., Higashi T., Ikeda T., Okawa K., Fukai S., Nureki O., Kita T., Horiuchi H. (2008) J. Biol. Chem. 283, 166–174 [DOI] [PubMed] [Google Scholar]
- 29.Fukai S., Matern H. T., Jagath J. R., Scheller R. H., Brunger A. T. (2003) EMBO J. 22, 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gideon P., John J., Frech M., Lautwein A., Clark R., Scheffler J. E., Wittinghofer A. (1992) Mol. Cell. Biol. 12, 2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brownbridge G. G., Lowe P. N., Moore K. J., Skinner R. H., Webb M. R. (1993) J. Biol. Chem. 268, 10914–10919 [PubMed] [Google Scholar]
- 32.Schwarzbraun T., Vincent J. B., Schumacher A., Geschwind D. H., Oliveira J., Windpassinger C., Ofner L., Ledinegg M. K., Kroisel P. M., Wagner K., Petek E. (2004) Genomics 84, 577–586 [DOI] [PubMed] [Google Scholar]
- 33.Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A. (2003) J. Biol. Chem. 278, 32493–32496 [DOI] [PubMed] [Google Scholar]
- 34.Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. (2003) Mol. Cell 11, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 35.Inoki K., Li Y., Xu T., Guan K. L. (2003) Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. (2003) Nat. Cell Biol. 5, 578–581 [DOI] [PubMed] [Google Scholar]
- 38.Crino P. B., Nathanson K. L., Henske E. P. (2006) N. Engl. J. Med. 355, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 39.Daumke O., Weyand M., Chakrabarti P. P., Vetter I. R., Wittinghofer A. (2004) Nature 429, 197–201 [DOI] [PubMed] [Google Scholar]
- 40.Scrima A., Thomas C., Deaconescu D., Wittinghofer A. (2008) EMBO J. 27, 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Inoki K., Guan K. L. (2004) Mol. Cell. Biol. 24, 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maheshwar M. M., Cheadle J. P., Jones A. C., Myring J., Fryer A. E., Harris P. C., Sampson J. R. (1997) Hum. Mol. Genet. 6, 1991–1996 [DOI] [PubMed] [Google Scholar]
- 43.Gridley S., Chavez J. A., Lane W. S., Lienhard G. E. (2006) Cell. Signal. 18, 1626–1632 [DOI] [PubMed] [Google Scholar]
- 44.Wolthuis R. M., Zwartkruis F., Moen T. C., Bos J. L. (1998) Curr. Biol. 8, 471–474 [DOI] [PubMed] [Google Scholar]
- 45.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 46.Potter C. J., Pedraza L. G., Xu T. (2002) Nat. Cell Biol. 4, 658–665 [DOI] [PubMed] [Google Scholar]
- 47.Cai S. L., Tee A. R., Short J. D., Bergeron J. M., Kim J., Shen J., Guo R., Johnson C. L., Kiguchi K., Walker C. L. (2006) J. Cell Biol. 173, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin J., Pollock C., Tracy K., Chock M., Martin P., Oberst M., Kelly K. (2007) Mol. Cell. Biol. 27, 7538–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.González-García A., Pritchard C. A., Paterson H. F., Mavria G., Stamp G., Marshall C. J. (2005) Cancer Cell 7, 219–226 [DOI] [PubMed] [Google Scholar]
- 50.Smith S. C., Oxford G., Baras A. S., Owens C., Havaleshko D., Brautigan D. L., Safo M. K., Theodorescu D. (2007) Clin. Cancer Res. 13, 3803–3813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.