Abstract
Neuronal communication relies on the fusion of neurotransmitter-containing vesicles with the plasma membrane. The soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) proteins initiate membrane fusion through the formation of the SNARE complex, a process tightly regulated by Sec1/Munc18-1 (SM) proteins. The emerging trend is that SM proteins promote SNARE-mediated membrane fusion by binding to a Syntaxin N-terminal motif. Here we report that mutations in the hydrophobic pocket of Munc18-1 (F115E and E132A), predicted to disrupt the N-terminal Sx1a interaction have a modest effect on binding to Sx1a in its free state, but abolish binding to the SNARE complex. Overexpression of the Munc18-1 mutant in PC12 cells lacking Munc18-1 rescues both neuroexocytosis and the plasma membrane localization of Syntaxin. However, total internal reflection fluorescence microscopy analysis reveals that expression of a Munc18-1 double mutant reduces the rate of vesicle fusion, an effect only detectable at the onset of stimulation. The Munc18-1 hydrophobic pocket is therefore critical for SNARE complex binding. However, mutations abrogating this interaction have a limited impact on Ca2+-dependent exocytosis in PC12 cells.
Following stimulation of neurons, a number of well orchestrated protein/protein (1) and protein/lipid (2) interactions underpin the fusion of secretory vesicles with the presynaptic plasma membrane. In this sequence of interactions, vesicles approach the plasma membrane (tethering and docking), undergo priming and, upon Ca2+ influx, fuse with the plasma membrane, thereby releasing neurotransmitter into the synaptic cleft (1). Vesicular exocytosis relies on the function of soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE)2 proteins as demonstrated by the blockade of neuroexocytosis following SNARE protein cleavage by clostridial neurotoxins (3). One of the key players in SNARE regulation is the cytosolic regulatory protein, Munc18-1 (Munc18a, nsec-1) (4–7). Although the function of SNARE proteins in mediating exocytosis is well established (2, 8), the precise role of Munc18-1 in exocytosis is still a subject of heated debate (6, 7, 9, 10).
Munc18-1 belongs to the Sec1/Munc18 (SM) family of proteins that are involved in mediating membrane trafficking events (11–13). Mutations in these proteins have recently been associated with infantile epileptic encephalopathy (14). Although the function of Munc18-1 and its interaction with SNAREs have been studied for over 10 years, the molecular mechanism of Munc18-1 regulation of membrane fusion is still not clear. Munc18-1 was originally characterized as a negative regulator of exocytosis as it binds to the target membrane SNARE, Syntaxin 1a (Sx1a) (5) in a conformation that sequesters the Sx1a SNARE helix and inhibits SNARE complex formation (7, 15). Other SM proteins have been shown to bind to their cognate syntaxins via an N-terminal motif (16–19), allowing interactions that are associated with a positive role for SM proteins in SNARE-mediated membrane fusion (20). Despite biochemical evidence supporting a negative regulatory role for Munc18-1, there is strong genetic evidence for a critical positive role for Munc18-1 in exocytosis, as demonstrated by a Munc18-1 knock-out mouse that exhibits a complete blockage of neurotransmission (21).
Recently, a short N-terminal peptide from Sx1a was also shown to bind to Munc18-1 via a novel interaction that promotes SNARE-mediated fusion of liposomes in vitro (6). Moreover, the N-terminal truncation of Sx1a only affects the binding of the open conformation of Sx1a to Munc18-1 occurring near the plasma membrane (10). Oddly, the Munc18-1-Sx1a interaction in solution predominantly involves the closed conformation (10), which raises the question of the relative contribution of the Sx1a N terminus to the overall Munc18-1-Sx1a interaction.
To address the functional significance of the N-terminal interaction, we took a reverse strategy and investigated the molecular nature of the binding of Munc18-1 to the Sx1a N terminus and to the SNARE complex. We mutated Munc18-1:Phe115 and Glu132, which are located in an evolutionarily conserved surface pocket of Munc18-1 (17, 22). We found that whereas these mutations have only a mild effect on the binding of Munc18-1 to Sx1a in its free state, they completely abrogate Munc18-1 binding to the SNARE complex. Expression of these mutants in PC12 cells lacking Munc18-1 rescues exocytosis to a similar extent to that obtained with Munc18-1-WT. Total internal reflection fluorescence (TIRF) microscopy of vesicle docking at the cell surface reveals that these mutations support exocytosis. However, the rate of vesicle fusion is markedly reduced at the onset of stimulation, arguing for a role of Munc18-1 in regulating the dynamics of SNARE-mediated vesicle fusion during exocytosis.
EXPERIMENTAL PROCEDURES
Munc18-1-Sx1a Model
Coordinates for the Munc18c-Sx4-(1–29) complex (Protein Data Bank code 2PJX) and the Munc18-1:Sx1a complexes (PDB code 1DN1) were obtained from the Protein Data Bank. Analysis was performed using MacPymol (PyMol, W. L. DeLano). A model for Sx1a peptide binding to domain 1 of Munc18-1 was constructed in the following way: the crystal structure of Munc18-1 was superimposed onto the crystal structure of Munc18c (from the Munc18c-Sx4-(1–29) complex). The peptide binding site of the Munc18c-Sx4-(1–29) complex was compared with the corresponding region in the overlaid Munc18-1 crystal structure. Residues in the Sx4 peptide of the crystal structure of the Munc18c-Sx4-(1–29) complex were mutated in silico to the corresponding Sx1a residues. Sequences were aligned using ClustalW (23). Figures showing model or crystal structures were generated using MacPymol.
Constructs
Expression of recombinant full-length rat GST-Munc18-1 (GST-Munc18-1-WT) in the pGEX-KG vector has been described previously (24). Various point mutants (GST-Munc18-1-F115E, GST-Munc18-1-E132A, and GST-Munc18-1-F115E/E132A) were constructed from the GST-Munc18-1 pGEX-KG vector using a QuikChange site-directed mutagenesis kit (Stratagene) and mutation primers as follows: GST-Munc18-1-F115E, forward 5′-CATGTCCAGATGCCCTGGAAAACGAGCTGGTAAAATC-3′ and reverse, 5′-GATTTTACCAGCTCGTTTTCCAGGGCATCTGGACATG-3′; GST-Munc18-1-E132A, forward 5′-CATCAAGACGCTGACGGCAATCAACATTGCGTTTC-3′ and reverse, 5′-GTAGTTCTGCGACTGCCGTTAGTTGTAACGCAAAG-3′.
A C-terminal truncated GST-Munc18-1 mutant (GST-Munc18-1-dom1, residues 1–132) was constructed by introducing a STOP codon at bp 394 (Gly to Thr) using the QuikChange kit. An N-terminal truncated Munc18-1 mutant (GST-Munc18-1-dom2/3, residues 132–594) was constructed by introducing an EcoRI site at residue 132 by mutagenesis (primers: forward, 5′-CATCAAGACGCTGACGGAATTCAACATTGCGTTTC-3′ and reverse, 5′-GAAACGCAATGTTGAATTCCGTCAGCGTCTTGATG-3′). After digestion with EcoRI, the fragment corresponding to residues 132–594 was ligated into pGEX-KG also digested with EcoRI. pCMV5-Munc18-1-F115E/E132A plasmids containing the silent mutations were constructed using the same strategy using pCMV5-Munc18-1-WT as template. Neuropeptide Y (NPY)-Venus and pXGHF-human growth hormone (hGH) were also used to transfect PC12-KD43 cells as previously described (25).
Recombinant Proteins and SNARE Complex Production
GST-VAMP2 (residues 1–94) and GST-SNAP25 (residues 1–206) were both expressed in pGEX-KG. All GST fusion proteins were expressed in Escherichia coli by autoinduction in pGEX-KG. His6-tagged Sx1a (Sx1a-His) (residues 1–265) was also expressed in pHO4c using the autoinduction method. All GST fusion proteins were purified using similar methods to those previously described (18). The GST tag was cleaved from purified GST-Munc18-1 proteins using bovine thrombin overnight at room temperature in standard buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm β-mercaptoethanol) containing 3 mm CaCl2. GST-SNAP25 was eluted from glutathione beads with standard buffer containing 20 mm glutathione. His6-tagged Sx1a was purified using Co2+-affinity beads (Scientifix) in standard buffer containing 20 mm imidazole, pH 7.4, then eluted with standard buffer containing 500 mm imidazole, pH 7.4.
The SNARE complex was produced by combining Sx1a-His, GST-SNAP25, and VAMP2 in a 1:1.5:2 ratio and incubating overnight at 4 °C in standard buffer containing 0.1% Triton X-100 and 20 mm imidazole, pH 7.4. The complex was isolated on Co2+-affinity beads via the His tag on Sx1a.
Circular Dichroism
Detagged Munc18-1-WT (1.3 mg/ml), Munc18-1-F115E (1.3 mg/ml), Munc18-1-E132A (2.3 mg/ml), and Munc18-1-F115E/E132A (2.3 mg/ml) were analyzed by circular dichroism to whether the mutation caused changes in the protein secondary structure compared with Munc18-1-WT. Five replicates of each spectrum were measured on a J-810 polarimeter (Jasco) in a quartz demountable cuvette with a path length of 0.01 cm (Hellma) using a dwell time of 1 s, a scanning speed of 50 nm/min, and data pitch of 1 nm.
Pulldown Binding Assays
Pulldown assays were performed by first isolating tagged proteins on affinity beads, then incubating with a molar excess of purified protein in solution. For all pulldown assays, the concentration of protein bound to the beads was quantified by densitometry using ImageJ and a bovine serum albumin standard curve. For the pulldown assay with Munc18-1-WT point and truncated mutants, a 3-fold molar excess of detagged Munc18 was incubated with Sx1a-His beads in standard buffer containing 20 mm imidazole, pH 7.4, and 0.1% Triton X-100. Munc18-1-WT or mutant binding to Sx1a was quantified by densitometry using ImageJ and compared for three independent experiments. For SNARE complex pulldown assays, SNARE complex-bound beads were incubated with a 3-fold molar excess of Munc18-1-WT, Munc18-1-F115E, or Munc18-1-F115E/E132A at 4 °C in standard buffer containing 0.1% Triton X-100 and 20 mm imidazole, pH 7.4. After extensive washing, proteins bound to beads were analyzed by SDS-PAGE using standard procedures. Western blot was performed for detection of Munc18-1 using anti-Munc18-1 antibody (rabbit polyclonal, Synaptic Systems).
Isothermal Titration Calorimetry (ITC)
Syntaxin1a-(2–243) was prepared as previously described (26). Munc18-1-WT and Munc18-1-F115E/E132A used for the ITC experiment were prepared as previously described (18). All proteins were further purified by gel filtration chromatography into 20 mm sodium phosphate buffer, pH 7.5, 150 mm NaCl, and 1 mm dithiothreitol (ITC buffer). ITC was carried out at 298 K using a MicroCal iTC200, with 13 × 3.1-μl injections of 25 μm Sx1a-(2–243) (30 μg in 40 μl) into 2.5 μm Munc18-1-WT or Munc18-1-F115E/E132A (32 μg in 200 μl). Integration of the titration curves was performed using ORIGIN to extract thermodynamic parameters, stoichiometry N, equilibrium association constant Ka (=Kd−1) and the binding enthalpy ΔH. The Gibbs free energy of binding ΔG was calculated from the relation ΔG = −RT ln(Ka) and the binding entropy ΔS was deduced from the equation (ΔG = ΔH − TΔS). Experiments were performed with protein concentrations well within the recommended range for the c-value (concentration of protein in cell/Kd ∼ 100). Binding paramaters were calculated as the average of three independent experiments mean ± S.E.
Immunofluorescence Staining and Confocal Microscopy of PC12 Cells
For rescue of Sx1a localization, Munc18-1 knockdown PC12-KD43 cells were transfected with empty pCMV5, pCMV5-Munc18-1-WT, or pCMV5-Munc18-1-F115E/E132A using Lipofectamine 2000 (Invitrogen). After 48 h, the cells were briefly washed, fixed with 4% paraformaldehyde and double-immunolabeled with anti-Munc18-1 (Synaptic Systems) and anti-Syntaxin1a (HPC-1, Sigma) antibodies followed by Alexa 488 and Alexa 546 secondary antibodies (Invitrogen), respectively. For NPY-Venus and hGH colocalization experiments, PC12-KD43 cells were co-transfected with plasmids pCMV-NPY-Venus and pXGH5-hGH, expressing Venus-tagged NPY and hGH, respectively. 48 h after transfection, the cells were washed, fixed (4% paraformaldehyde), and immunolabeled with anti-hGH antibody, followed by Alexa 546 secondary antibody. Cells were visualized and images acquired using a Zeiss LSM 510 Meta confocal microscope as previously described (27).
Human Placental Alkaline Phosphatase (hPLAP) Secretion Assay from Co-transfected PC12 Cells
Munc18-1 knockdown PC12 cells (KD43) at 70–80% confluence in 10-cm dishes were co-transfected with 2 μg of pCMV-NPY-hPLAP (to express NPY fused with soluble human placental alkaline phosphatase) and 15 μg of empty pCMV5 (for control) or pCMV-Munc18-1 (silent mutations) (for rescue of the wild type or the indicated mutant) using electroporation (25). After 48 h, the cells were harvested and re-plated in 24-well plates. 6 or 7 days after electroporation, the plated cells were washed once with physiological saline solution (PSS) containing 145 mm NaCl, 5.6 mm KCl, 2.2 mm CaCl2, 0.5 mm MgCl2, 5.6 mm glucose, and 15 mm HEPES, pH 7.4, and NPY-hPLAP secretion was stimulated with 200 μl of PSS or high K+-PSS (containing 81 mm NaCl and 70 mm KCl). Secretion was terminated after a 20-min incubation at 37 °C by chilling to 0 °C, and samples were centrifuged at 4 °C for 3 min. Supernatants were removed, and the pellets were solubilized in 200 μl of PSS containing 0.1% Triton X-100. The amounts of NPY-hPLAP secreted into the medium and retained in the cells were measured by the Phospha-Light Reporter Gene Assay System (Applied Biosystems). Samples treated at 65 °C for 30 min to inactivate non-placental alkaline phosphatases and an aliquot (10 μl) was assayed for placental alkaline phosphatase activity with the kit. The total volume of the assay was 120 μl. After 5 to 10 min, chemiluminescence was quantified by a FB12 luminometer (Berthold Detection Systems).
hGH Release from Transfected PC12-KD43 or pSuper Cells
PC12-KD43 cells were plated on a 6-well tissue culture plate (Techno Plastic Plate). pXGH5 vector encoding hGH (1.6 μg/well) either alone or together with mammalian expression vectors encoding proteins as indicated in the figure (0.8 μg/well) were co-transfected in PC12-KD43 cells with Lipofectamine LTX (Invitrogen) in Opti-MEM I (Invitrogen) according to the manufacturer's instructions. 72 h after transfection, PC12-KD43 cells were briefly washed with PSS and either stimulated with PSS or high K+ PSS buffer (containing 81 mm NaCl and 70 mm KCl) for 20 min at 37 °C. Secretion was terminated by chilling to 0 °C and supernatants were used for the hGH assay as previously described (28). For totals, cells were solubilized on ice in 1% Triton X-100 in PSS buffer containing protease inhibitors (Calbiochem). The amount of secretion from co-transfected cells was determined as the percentage of unstimulated hGH released using an hGH ELISA kit (Roche Diagnostics) as previously described (29). Plotted data are representative of three independent experiments, each carried out in triplicate.
Evanescent Wave Imaging
TIRF imaging was done using an Olympus IX81 inverted microscope with a ×60 TIRF objective and 1.45 N.A. objective illuminated with a 10-milliwatt argon laser (Melles Griot). Images were captured using the Hamamatsu ORCA1-ER camera controlled by Metamorph 5.0 software (Universal Imaging). Time-lapse images were acquired every 5 s for 5 min at room temperature. PC12-KD43 cells were co-transfected with GFP-tagged human growth hormone (hGH-GFP) and either pCMV5-Munc18-1-WT or pCMV5-Munc18-1-F115E/E132A using Lipofectamine 2000 (Invitrogen). The cells were then plated on poly-l-lysine-coated glass bottom microwell dish (35 mm) (MatTek) and allowed to express for 48 h. Cells were then washed in Buffer A (145 mm NaCl, 5 mm KCl, 1.2 mm Na2HPO4, 20 mm HEPES-NaOH, pH 7.4) and imaged in Buffer A supplemented with Ca2+ (2 mm). Cells were stimulated using 100 μm nicotine 30 s following the onset of acquisition.
TIRF Image Analysis
Following TIRF image acquisition, hGH-GFP-labeled granules entering the evanescence field were analyzed by particle tracking using Slidebook software (Version 4.2.0.13, Intelligent Innovation Imaging). The path lifetime of single vesicle tracks was followed immediately after application of nicotine until the disappearance of hGH-GFP-labeled vesicles. The mean square displacement (MSD) of tracked vesicle motion was calculated from the Slidebook software and plotted against increment time interval. MSD plots can be fitted to different equations describing vesicle motions in three different categories (30).
The simple diffusion motion is described by the following linear equation,
where C is an offset constant related to the accuracy of the tracking algorithm.
From the second-order polynomial fit information about the velocity v and diffusion coefficient D was obtained by fitting MSD to,
The third case, restricted diffusion, is described by the following equation in terms of the radius of the cage R, diffusion coefficient, and two constants (a1 = 0.9, a2 = 0.85).
When analyzing the MSD of hGH-GFP positive granules for both Munc18-1-WT and Munc18-1-F115E/E132A, the best fit was obtained with Equation 2. The velocity was therefore extracted from Equation 2.
Image Processing and Data Analysis
Background correction and contrast adjustment of raw images were done either using Adobe Photoshop CS2 (version 9) or SlideBook (version 4.2.0.13; Intelligent Imaging Innovations). Vesicle tracking and analysis were performed using Slidebook and the results were expressed as mean ± S.E.
Statistical Analysis
Data analysis was carried out using unpaired Student's t tests. Experiments were performed at least 3 times. Values are expressed as mean ± S.E., and data are considered significant at: *, p < 0.05; **, p < 0.01.
RESULTS AND DISCUSSION
Defining the Munc18-1 Hydrophobic Pocket Interaction with the Syntaxin1a N Terminus
Phe115 and Glu132 are key Munc18-1 residues predicted to mediate the interaction between the N-terminal peptide of Sx1a and a predominantly hydrophobic pocket on the surface of Munc18-1 (Fig. 1). Initially, we used a model of the Munc18-1-Sx1a N-terminal interaction and compared interactions with the crystal structure of the closely related SM protein, Munc18c (Fig. 1B, top panel). Our Munc18-1-Sx1a model (Fig. 1B, middle panel) was supported by the newly refined model for the crystal structure of the Munc18-1-Sx1a complex recently reported by Burkhardt and colleagues (22) (Fig. 1B, bottom panel). The key interaction is formed between Munc18c/Phe119 and Sx4/Leu8 (17, 18), which is conserved in the Munc18-1-Sx1a interaction (Fig. 1, B and C) as well as other SM:Syntaxin cognate partners exhibiting the N-terminal binding mode (17). Furthermore, a conserved DRT motif of Sx1a (D3, R4, T5) has been shown to contribute to interactions with Munc18-1 via the hydrophobic pocket (17, 22, 31). Based on the structural analysis, Glu132 can form an electrostatic interaction with Arg4 in this motif. We aimed to investigate the impact of two key Munc18-1 residues, Glu132 and Phe115, on binding to Sx1a. We constructed two point mutants of Munc18-1 at residue Phe115 (Munc18-1-F115E) and at residue Glu132 (Munc18-1-E132A), as well as a double mutant (Munc18-1-F115E/E132A), to disrupt the binding pocket and examine their contribution to the interactions with Sx1a alone and with the SNARE complex.
FIGURE 1.
The N-terminal interaction of Munc18-1 and Sx1a. A, the Munc18-1-Sx1a complex represented as ribbon (left) and surface (right) models with the two key binding sites indicated. These involve the Sx1a N-terminal peptide (residues 2–9), and the combined Sx1a Habc and H3 domains (residues 26–248). B, key residues required for forming the SM protein/N-peptide interaction in the Munc18c:Sx4-(1–29) crystal structure (top, PDB code 2PJX) (16) are present in the Munc18-1:Sx1a model (middle) and the newly refined Munc18-1:Sx1a structure (bottom, PDB code 3C98)(20). Conserved hydrophobic residues defining the pocket are shown (blue). C, sequences of Syntaxin1a (Homo sapiens, Rattus norvegicus, and Mus musculus), Syntaxin4 (H. sapiens, R. norvegicus, and M. musculus), and Sed5p (S. cerevisiae) N termini were aligned. Conserved residues are highlighted (orange).
Mutations in the Munc18-1 Hydrophobic Pocket Alter Its Binding to Syntaxin1a
We first compared the interaction of Sx1a with our four Munc18-1 constructs: Munc18-1-WT, Munc18-1-F115E, Munc18-1-E132A, and Munc18-1-F115E/E132A. In addition, two other truncated Munc18-1 constructs were tested for Sx1a binding, Munc18-1-dom1, which contains only domain 1 of Munc18-1 (residues 1–132), and Munc18-1-dom2/3, consisting of domains 2 and 3 only (residues 132–594) (Fig. S1). His-tagged Sx1a was tethered to Co2+-affinity beads and incubated with de-tagged Munc18-1-WT and the three mutants, then analyzed by SDS-PAGE (Fig. 2, A and B). In this strategy, the GST tag was removed from the N terminus of the Munc18-1 proteins and the Sx1a was tethered to the beads via its C-terminal His tag.
FIGURE 2.
Munc18-1 mutants have reduced binding to Syntaxin1a. A, Munc18-1 representation showing the locations of the F115E, E132A, and F115E/E132A mutations. B, Munc18-1 (WT and mutant) binding to Sx1a-His tethered to Co2+-affinity beads was analyzed by Coomassie-stained SDS-PAGE. Munc18-1 mutant binding to Sx1a was reduced compared with Munc18-1-WT. C, Munc18-1 (WT and mutant) binding to Sx1a tethered to Co2+-affinity beads was quantified and compared (n = 3). D, overlaid circular dichroism spectra measured for purified Munc18-1-WT (♦), Munc18-1-F115E (■), Munc18-1-E132A (▲), and Munc18-1-F115E/E132A (♦) proteins.
As expected, Munc18-1-WT bound to Sx1a-His. However, when compared with Munc18-1-WT, the Munc18-1 mutants showed a significant reduction in Sx1a binding (Fig. 2B) or in the case of the truncated mutants, no detectable binding to Sx1a at all (Fig. S1). It should be noted that the point mutants still displayed a relatively high affinity as discerned by the strong intensity bands detected in Coomassie-stained gels. This is not surprising considering the independent peptide binding surface represents only ∼25% of the total buried surface area when full-length Sx1a is bound to Munc18-1 (790 Å2 of a total of 4940 Å2) (22). Neither of the truncated Munc18-1 proteins, dom1 and dom2/3, bound to Sx1a-His indicating that the intact full-length Munc18-1 protein is required (Fig. S1). The reduced Sx1a binding observed for Munc18-1-F115E, Munc18-1-E132A, and double mutant Munc18-1-F115E/E132A was quantified and compared with WT Sx1a binding (n = 3). The Phe115 mutation resulted in ∼40% reduction in binding to Sx1a compared with Munc18-1-WT (Fig. 2C), whereas the Glu132 mutation resulted in ∼30% decrease in binding to Sx1a (Fig. 2C). The double mutation resulted in a marked ∼65% reduction in binding to Sx1a compared with Munc18-1-WT (Fig. 2C). Circular dichroism analysis of the Munc18-1 point mutants revealed no detectable secondary structure differences compared with the Munc18-1-WT, eliminating the possibility that reduced binding could be caused by a non-functional, unfolded protein (Fig. 2D).
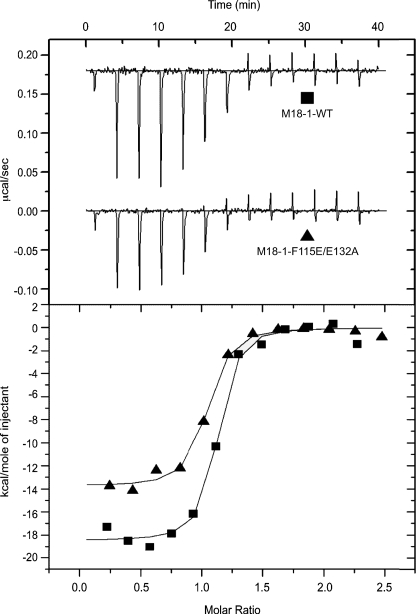
To further characterize these interactions and determine whether the reduction in binding to Sx1a was a result of affinity changes, ITC experiments were conducted. Sx1a-(2–243) was titrated into either Munc18-1-WT or the Munc18-1-F115E/E132A double mutant. Sx1a-(2–243) exothermically binds to Munc18-1-WT with a dissociation constant (Kd) of 27.3 ± 7.5 nm (Fig. 3, Table 1). This is higher than the Kd value of 1.4 nm reported by Burkhardt et al. (22) but is in good agreement with the 7.5–20.5 nm values obtained using ITC by Deak et al. (26), and comparable with 10–20 nm values obtained by other methods (31, 32). Interestingly, despite the reduced association detected in pulldowns, the dissociation constant of the Sx1a binding affinity to Munc18-1-F115E/E132A was not significantly changed (25.77 ± 4.76 nm) when compared with that of Munc18-1-WT (Fig. 3, Table 1). In contrast, the binding enthalpy (ΔH) was significantly altered by the mutations, with −19.98 ± 0.88 kcal/mol obtained for the Munc18-1-WT and −14.17 ± 0.17 kcal/mol for Munc18-1-F115E/E132A (Fig. 3, Table 1). This result is consistent with that of Burkhardt et al. (22) who also observed a reduction of ∼50% in the enthalpy of binding when the Sx1a N-terminal peptide was removed. Thermodynamically, the binding of Munc18-1-F115E/E132A is characterized by a more favorable entropic contribution (ΔTΔS = −5.3 kcal/mol). A possible explanation for the decreased enthalpic and increased entropic contributions to binding is that the overall binding surface area is reduced when the Sx1a peptide is unable to bind, but there is a smaller entropic penalty incurred by the folding of the unstructured Sx1a N-terminal region upon association. Our results suggest that the Munc18-1 mutant has altered binding kinetics to Sx1, based on the reduced Sx1a binding observed in pulldowns, which unlike ITC is not an equilibrium measurement. Surface plasmon resonance experiments may be useful to probe these binding kinetics further. Taken together our results indicate that mutation of the Munc18-1 hydrophobic pocket, predicted to mediate binding to the Sx1a N-terminal peptide, affects the thermodynamics and kinetic properties of association, but surprisingly does not cause a major change in the equilibrium binding affinity.
FIGURE 3.
The N-terminal interaction of Syntaxin1a with Munc18-1 measured by ITC. Raw data (top panel) and integrated normalized ITC data (bottom panel) for binding of Sx1a-(2–243) to Munc18-1 WT (closed squares) and Munc18-1-F115E/E132A (closed triangles). Experiments were performed by injecting 3.1-μl aliquots of 25 μm Sx1a-(2–243) into 2.5 μm Munc18-1 proteins at 298 K.
TABLE 1.
Thermodynamic parameters for the binding of Sx1a-(2–243) to Munc18-1 determined by ITC
| Proteins interaction | ΔH | TΔS | ΔG | Kd | n |
|---|---|---|---|---|---|
| kcal mol−1 | nm | ||||
| Munc18-1-WT-Sx1a-(2–243) | −19.98 ± 0.88 | −9.59 ± 1.04 | −10.39 ± 0.20 | 27.27 ± 7.51 | 1.05 ± 0.01 |
| Munc18-1-F115E/E132A-Sx1a-(2–243) | −14.17 ± 0.17a | −3.79 ± 0.28a | −10.38 ± 0.11 | 25.77 ± 4.76 | 0.96 ± 0.01 |
a Values were expressed as mean ± S.E. (n = 3, p < 0.01). See ″Experimental Procedures″ for sample concentrations used. All experiments were performed at 298 K.
Mutations in the Munc18-1 Hydrophobic Pocket Abrogate Its Binding to the SNARE Complex
In view of the recent discovery that Munc18-1 binds to the SNARE complex, we next investigated the role of the Munc18-1 hydrophobic pocket in mediating this interaction (6, 24). As previously demonstrated (6), we found that Munc18-1-WT can bind to the ternary SNARE complex (consisting of Sx1a, SNAP25, and VAMP2) (Fig. 4, A and B). Importantly, a single mutation in the Munc18-1 hydrophobic pocket, Munc18-1-F115E, abrogates Munc18-1 interaction with the assembled SNARE complex (Fig. 4A). As expected, the double mutant Munc18-1-F115E/E132A was also unable to bind the assembled SNARE complex (Fig. 4, B and C). The Munc18-1 hydrophobic pocket is therefore critical to enable binding to the SNARE complex, an interaction that has been shown to promote membrane fusion (6). This is in agreement with recent reports demonstrating that the SNARE complexes produced using either Sx1a containing the complementary Leu8 mutation (6) or an N-terminal truncation of Sx1a (33) do not interact with Munc18-1.
FIGURE 4.
Munc18-1-WT but not Munc18-1-F115E or Munc18-1-F115E/E132A binds to the SNARE complex. A, assembled SNARE complex (comprising Sx1a-His, GST-SNAP25, and VAMP2) was isolated on Co2+ affinity beads, incubated with Munc18-1-WT or Munc18-1-F115E for 2 h at 4 °C. Following extensive washing, beads were analyzed for bound proteins using Coomassie-stained SDS-PAGE. Munc18-1-WT and Munc18-1-F115E binding to the assembled SNARE complex was confirmed by analyzing both boiled (95 °C for 5 min, denatured complex) and not boiled (SDS-resistant complex) samples. B, Western blot of the SNARE complex-mediated pulldown of Munc18-1-WT and Munc18-1-F115E/E132A probed with anti-Munc18-1 antibody. C, the amount Munc18-1-WT and Munc18-1-F115E/E132A binding to the SNARE complex on beads was averaged and expressed as percentage of Munc18-1-WT binding (mean values ± S.E., n = 4).
There are three key points of contact between Sx1a and Munc18-1, involving the N-terminal peptide, Habc domain, and SNARE H3 helix of Sx1a (22) (Fig. 1). As the H3 helix will be unable to participate in Munc18-1 binding when complexed with cognate SNAREs, it is expected that the relative contributions of the Habc domain and N-terminal peptide will be significantly increased when associated with SNARE complexes. In support of this, recent work by Deak and colleagues (26) examined specific mutations in the Munc18-1 protein contacting the Sx1a Habc domain, and found that they are important for SNARE complex association but do not significantly affect binding to free Sx1a. These results almost perfectly mirror our own experiments analyzing the importance of the Sx1a N-terminal peptide interface, and support a model whereby Munc18-1 has at least two productive binding modes, a high affinity mode utilizing all three Sx1a conserved domains, and a lower affinity mode utilizing only Sx1a N-terminal peptide and Habc domains that is important for Munc18-1 association with SNARE complexes. Evidence also suggests that VAMP2, within the SNARE complex, may interact directly with Munc18-1 and that these interactions are involved in exerting a positive regulatory role on SNARE-mediated fusion (6, 33, 34). In summary our results demonstrate that the hydrophobic pocket of Munc18-1 is essential for interaction with the assembled SNARE complex and contributes to a lesser extent to the free Sx1a binding.
Expression of the Munc18-1 Double Mutant Lacking the Interaction with Sx1a N Terminus Rescues the Plasma Membrane Localization of Syntaxin1a in PC12-KD43 Cells
Having demonstrated that mutations in key residues of the Munc18-1 hydrophobic pocket impaired SNARE complex binding (and to a lesser extent association with free Sx1a), we investigated the functional significance of this interaction. Converging lines of evidence have recently shown that Munc18-1 facilitates Sx1a targeting to the plasma membrane (10, 25, 35). Moreover, a switch in the Munc18-1-Sx1a interaction has been found to occur at the level of the plasma membrane (10). In the vicinity of the plasma membrane, Munc18-1 binding mainly occurs through the N terminus of Sx1a, thereby favoring the subsequent interaction of Sx1a with SNAP-25 (10). This raises the question as to whether the N-terminal interaction is necessary and/or sufficient for the actual delivery of Sx1 to the plasma membrane. We therefore examined whether the Munc18-1 double mutant lacking the ability to bind to the N terminus of Sx1a was capable of delivering Sx1a to the plasma membrane. The knockdown of Munc18-1 in PC12-KD43 cells greatly affects the delivery of Sx1a to the plasma membrane (Fig. 5) as previously reported (25). In these cells, expression of Munc18-1-WT efficiently restores plasma membrane localization (Fig. 5) as shown previously (25). Surprisingly, expression of Munc18-1-F115E/E132A was also capable of delivering Sx1a to the plasma membrane (Fig. 5, Fig. S2). This result does not support a major role for the N-terminal interaction in the actual delivery of Sx1a to the plasma membrane.
FIGURE 5.
Munc18-1 restores membrane transport and localization of Syntaxin1a in Munc18-1 knockdown PC12-KD43 cells. Representative confocal images of PC12-KD43 cells transfected with pCMV5 empty vector (top panel), pCMV5-Munc18-1-WT (middle panel), or pCMV5-Munc18-1-F115E/E132A (bottom panels). Cells were fixed 48 h after transfection and immunostained for Munc18-1 (green) and Sx1a (red). Scale bars, 10 μm.
Expression of SNARE Binding-deficient Munc18-1 Mutants Rescues Exocytosis in PC12 Cells Lacking Munc18-1
We next examined whether Munc18-1-WT and mutants were capable of restoring secretion when expressed in Munc18-1 knockdown (KD43) PC12 cells. These cells were co-transfected with a reporter plasmid allowing the expression of NPY fused with a soluble domain (residues 18–506) of hPLAP, which can be secreted in a Ca2+-dependent manner and quantified by measurements of alkaline phosphatase activity of hPLAP as previously described (25). Expression of Munc18-1-WT rescued the secretion of NPY-hPLAP by 71.5 ± 8.1% (Fig. 6, A–C). Munc18-1-WT overexpression is also known to rescue exocytosis in Munc18-1 knock-out chromaffin cells (36, 37). Expression of Munc18-1-F115E produced a significant rescue of exocytosis, albeit slightly reduced when compared with the rescue generated by Munc18-1-WT (Fig. 6B). Importantly, expression of Munc18-1-E132A and Munc18-1-F115E/E132A was also able to fully rescue exocytosis (Fig. 6A), with the functional rescue not significantly different from that of Munc18-1-WT (Fig. 6, B and C). The level of secretion in control PC12 cells (Psuper) is indicated in Fig. 6D.
FIGURE 6.
NPY secretion defects in Munc18-1 knockdown PC12-KD43 cells are rescued upon reintroduction of Munc18-1 (WT and the indicated mutants) by transfection. A, immunoblot analysis of transfected Munc18-1 proteins in KD43 cells. 30 mg of total homogenates from KD43 cells electroporated with 20 μg of the empty plasmid (pCMV5 as control) or the plasmid that expresses Munc18-1 proteins were analyzed by SDS-PAGE and immunoblotting using anti-Munc18-1 and anti-VCP/p97 antibodies. VCP/p97, a ubiquitous membrane trafficking protein (42), was used as a loading control. B, secretion of transfected NPY-hPLAP from the knockdown (KD43) cells that were co-transfected with the control plasmid or the Munc18-1-WT, Munc18-1-F115E, or Munc18-1-E132A expression plasmids (n = 16). C, secretion of NPY-hPLAP from the KD43 cells that were transfected with the control plasmid or the Munc18-1-WT or Munc18-1-F115E/E132A expression plasmids (n = 15). D, average secretion of transfected NPY-hPLAP from 5 different clones of control PC12 cells (pSuper) that were co-transfected with the control plasmid. Data were expressed as mean ± S.E. and considered significant at * p < 0.05. ns, not significant.
We next checked whether this result could be reproduced using the stimulated release of another hormone. PC12-KD43 cells were first transfected with NPY-Venus and hGH. Cells were processed for hGH immunoreactivity and imaged by confocal microscopy. As expected, NPY-Venus staining overlapped with hGH immunoreactivity (Fig. 7A), demonstrating that both overexpressed hormones were packaged in the same secretory vesicles. PC12-KD34 cells were next co-transfected with hGH and either Munc18-1-WT or Munc18-1-F115E/E132A. Both Munc18-1-WT and Munc18-1-F115E/E132A were capable of rescuing depolarization-induced hGH release (Fig. 7B). Furthermore, no significant differences were found in the WT and mutant rescuing effect (Fig. 7B), thereby confirming our original finding using NPY release.
FIGURE 7.
hGH secretion defects in Munc18-1 knockdown PC12-KD43 cells are rescued upon reintroduction of Munc18-1 (WT and double mutant) by transfection. A, Munc18-1 knockdown PC12 KD43 cells were co-transfected with NPY-Venus and hGH and immunostained with anti-hGH (red) and visualized by confocal microscopy. NPY-Venus (yellow) co-localizes with hGH in secretory vesicles. Scale bar, 10 μm. B, Munc18-1 knockdown PC12-KD43 cells transiently transfected with hGH and control plasmid (pCMV5), Munc18-1-WT, or Munc18-1-F115E/E132A were incubated for 20 min in the absence or presence of high K+ to stimulate exocytosis. hGH release is expressed as the percentage of hGH released in unstimulated conditions. Data are given as the mean ± S.E. (n = 3) and are representative of results obtained in three independent experiments. n.s., not significant (Student's t test; n = 3).
Although the release of NPY and hGH was rescued to a large extent in PC12-KD43 cells upon expression of Munc18-1-F115E/E132A, our experiment did not allow deeper examination of the actual process of exocytosis. To investigate this further, evanescent wave imaging was conducted. PC12-KD43 cells were co-transfected with hGH-GFP and either Munc18-1-WT or Munc18-1-F115E/E132A. Both Munc18-1-WT or Munc18-1-F115E/E132A were able to restore hGH-GFP exocytosis in these cells (Fig. 8, A and B). Noticeably, during a short window of time following nicotine stimulation, the rate of fusion of the hGH-GFP vesicle in cells expressing Munc18-1-F115E/E132A was significantly slower than in cells expressing Munc18-1-WT (Fig. 8, B and C). However, this effect was transient and no significant difference was detected at longer time points. Immediately after stimulation, the hGH-GFP-positive vesicle path lifetime (residence time) of Munc18-1-F115E/E132A-expressing cells was increased as compared with that of Munc18-1-WT-expressing cells (Fig. 8D). Although the difference path lifetime was not significant, the velocity of hGH-GFP-positive vesicles was significantly reduced in Munc18-1-F115E/E132A-expressing cells (Fig. 8E).
FIGURE 8.
Munc18-1 rescues hGH-GFP exocytosis in Munc18-1 knockdown PC12-KD43 cells. A, footprint of Munc18-1 knockdown cells expressing hGH-GFP and co-transfected with Munc18-1-WT or Munc18-1-F115E/E132A as indicated. The arrows indicate the vesicles highlighted in B. B, time series of single vesicles. The vesicle in cells transfected with Munc18-1-WT fused ∼40 s following nicotine stimulation (100 μm). Fusion of the vesicle from cells expressing Munc18-1-F115E/E132A was significantly delayed. C, time course of hGH-GFP positive vesicle fusion following nicotine stimulation at time 0 s. Vesicle fusion in cells expressing Munc18-1-WT (n = 11 cells) was significantly more rapid at the onset of stimulation compared with vesicles in cells expressing Munc18-1-F115E/E132A (n = 11 cells). D, path lifetime; and E, velocity of hGH-GFP positive vesicles in cells expressing Munc18-1-WT (n = 11 from 5 cells) and Munc18-1-F115E/E132A (n = 37 from 8 cells) following nicotine stimulation. Data are normalized to the Munc18-1-WT value, expressed as mean ± S.E. and significant at *, p < 0.05 (unpaired Student's t test).
The effect of Munc18-1-F115E/E132A on the rate of vesicle fusion and their path lifetime and velocity was only transient and the overall impact of these changes to the release of hGH and NPY was barely detectable. However, it is likely that such changes in the kinetic behavior of large dense core vesicles could produce more dramatic effects in phasic neurotransmitter release from neurons. Indeed, efficient neurotransmitter release relies on the synchronous release of many small synaptic vesicles. This process is highly dependent on the timing of vesicular fusion and is greatly affected by botulinum neurotoxin-induced VAMP2 cleavage (38) or through genetic alteration of Synaptotagmin 1 (39). Furthermore, two recent studies have found that mutations of Munc18-1 that also alter SNARE complex binding do not rescue phasic neurotransmitter release in Munc18-1 knock-out mice and Unc18-null Caenorhabditis elegans (26, 40). However, in these cortical neurons isolated from the Munc18-1 KO mice, the effect was attributed to a reduction in the readily releasable pool of synaptic vesicles. Bearing in mind the effect of Munc18-1 on the fusion pore (41), an alternative possibility is that the Munc18-1 mutations affecting SNARE complex binding could affect the duration of opening of the fusion pore. Clearly more work is required to investigate the nature of these differences.
Despite the newly proposed theory that the Sx1a N-terminal interaction with Munc18-1 is critical for SNARE complex-induced fusion of exocytic vesicles (6, 31), our results indicate that this does not contribute to Sx1a delivery to the plasma membrane and has only a limited impact on Ca2+-dependent neuroexocytosis in PC12 cells. It will be interesting to determine whether genetically ablating this interaction has more functional consequences in an animal model.
Supplementary Material
Acknowledgments
We thank Dirk Fasshauer and Josep Rizo for their kind gifts of the Sx1a and Munc18-1 constructs, Jenny Martin for critical reading of an earlier version of the manuscript, Nathan Cowieson for assistance with initial CD data measurements, Lotten Ragnarsson for help in designing some of the Munc18-1 constructs, and Jianyuan Sun for helpful discussions on the manuscript. We also express our gratitude to Alpha Yap, Hayley Cox, and Luke Hammond for access and help with the TIRF microscopy.
This work was supported by a grant from the Australian National Health and Medical Research Council (to F. A. M.), a Linkage Infrastructure, Equipment and Facilities grant from the Australian Research Council (to F. A. M.), and Canadian Institute of Health Research Grant MOP-57825 (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- SNARE
- soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor
- SM
- Sec1/Munc18
- TIRF
- total internal reflection fluorescence
- MSD
- mean square displacement
- NPY
- neuropeptide Y
- hPLAP
- human placental alkaline phosphatase
- Sx1a
- Syntaxin 1a
- WT
- wild type
- GST
- glutathione S-transferase
- hGH
- human growth hormone
- PSS
- physiological saline solution
- PDB
- Protein Data Bank
- GFP
- green fluorescent protein.
REFERENCES
- 1.Sudhof T. C. (2004) Annu. Rev. Neurosci. 27, 509–547 [DOI] [PubMed] [Google Scholar]
- 2.Söllner T. H. (2003) Mol. Membr. Biol. 20, 209–220 [DOI] [PubMed] [Google Scholar]
- 3.Schiavo G., Benfenati F., Poulain B., Rossetto O., Polverino de Laureto P., DasGupta B. R., Montecucco C. (1992) Nature 359, 832–835 [DOI] [PubMed] [Google Scholar]
- 4.Garcia E. P., Gatti E., Butler M., Burton J., De Camilli P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2003–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hata Y., Slaughter C. A., Südhof T. C. (1993) Nature 366, 347–351 [DOI] [PubMed] [Google Scholar]
- 6.Shen J., Tareste D. C., Paumet F., Rothman J. E., Melia T. J. (2007) Cell 128, 183–195 [DOI] [PubMed] [Google Scholar]
- 7.Yang B., Steegmaier M., Gonzalez L. C., Jr., Scheller R. H. (2000) J. Cell Biol. 148, 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahn R., Lang T., Südhof T. C. (2003) Cell 112, 519–533 [DOI] [PubMed] [Google Scholar]
- 9.Peng R., Gallwitz D. (2004) EMBO J. 23, 3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickman C., Medine C. N., Bergmann A., Duncan R. R. (2007) J. Biol. Chem. 282, 12097–12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallwitz D., Jahn R. (2003) Trends Biochem. Sci. 28, 113–116 [DOI] [PubMed] [Google Scholar]
- 12.Toonen R. F., Verhage M. (2003) Trends Cell Biol. 13, 177–186 [DOI] [PubMed] [Google Scholar]
- 13.Latham C. F., Meunier F. A. (2007) Int. J. Biochem. Cell Biol. 39, 1576–1581 [DOI] [PubMed] [Google Scholar]
- 14.Saitsu H., Kato M., Mizuguchi T., Hamada K., Osaka H., Tohyama J., Uruno K., Kumada S., Nishiyama K., Nishimura A., Okada I., Yoshimura Y., Hirai S., Kumada T., Hayasaka K., Fukuda A., Ogata K., Matsumoto N. (2008) Nat. Genet. 40, 782–788 [DOI] [PubMed] [Google Scholar]
- 15.Misura K. M., Scheller R. H., Weis W. I. (2000) Nature 404, 355–362 [DOI] [PubMed] [Google Scholar]
- 16.Bracher A., Weissenhorn W. (2002) EMBO J. 21, 6114–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S. H., Latham C. F., Gee C. L., James D. E., Martin J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8773–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latham C. F., Lopez J. A., Hu S. H., Gee C. L., Westbury E., Blair D. H., Armishaw C. J., Alewood P. F., Bryant N. J., James D. E., Martin J. L. (2006) Traffic 7, 1408–1419 [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi T., Dulubova I., Min S. W., Chen X., Rizo J., Südhof T. C. (2002) Dev. Cell 2, 295–305 [DOI] [PubMed] [Google Scholar]
- 20.Peng R., Gallwitz D. (2002) J. Cell Biol. 157, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhage M., Maia A. S., Plomp J. J., Brussaard A. B., Heeroma J. H., Vermeer H., Toonen R. F., Hammer R. E., van den Berg T. K., Missler M., Geuze H. J., Südhof T. C. (2000) Science 287, 864–869 [DOI] [PubMed] [Google Scholar]
- 22.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. (2008) EMBO J. 27, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latham C. F., Osborne S. L., Cryle M. J., Meunier F. A. (2007) J. Neurochem. 100, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 25.Arunachalam L., Han L., Tassew N. G., He Y., Wang L., Xie L., Fujita Y., Kwan E., Davletov B., Monnier P. P., Gaisano H. Y., Sugita S. (2008) Mol. Biol. Cell 19, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deák F., Xu Y., Chang W. P., Dulubova I., Khvotchev M., Liu X., Südhof T. C., Rizo J. (2009) J. Cell Biol. 184, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen P. J., Osborne S. L., Morrow I. C., Parton R. G., Domin J., Meunier F. A. (2008) Mol. Biol. Cell 19, 5593–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne S. L., Wen P. J., Boucheron C., Nguyen H. N., Hayakawa M., Kaizawa H., Parker P. J., Vitale N., Meunier F. A. (2008) J. Biol. Chem. 283, 2804–2813 [DOI] [PubMed] [Google Scholar]
- 29.Meunier F. A., Osborne S. L., Hammond G. R., Cooke F. T., Parker P. J., Domin J., Schiavo G. (2005) Mol. Biol. Cell 16, 4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steyer J. A., Almers W. (1999) Biophys. J. 76, 2262–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khvotchev M., Dulubova I., Sun J., Dai H., Rizo J., Südhof T. C. (2007) J. Neurosci. 27, 12147–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pevsner J., Hsu S. C., Braun J. E., Calakos N., Ting A. E., Bennett M. K., Scheller R. H. (1994) Neuron 13, 353–361 [DOI] [PubMed] [Google Scholar]
- 33.Dulubova I., Khvotchev M., Liu S., Huryeva I., Südhof T. C., Rizo J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zilly F. E., Sørensen J. B., Jahn R., Lang T. (2006) PLoS Biol. 4, e330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam A. D., Tryoen-Toth P., Tsai B., Vitale N., Stuenkel E. L. (2008) Mol. Biol. Cell 19, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulyás-Kovács A., de Wit H., Milosevic I., Kochubey O., Toonen R., Klingauf J., Verhage M., Sørensen J. B. (2007) J. Neurosci. 27, 8676–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voets T., Toonen R. F., Brian E. C., de Wit H., Moser T., Rettig J., Südhof T. C., Neher E., Verhage M. (2001) Neuron 31, 581–591 [DOI] [PubMed] [Google Scholar]
- 38.Meunier F. A., Schiavo G., Molgó J. (2002) J. Physiol. Paris 96, 105–113 [DOI] [PubMed] [Google Scholar]
- 39.Geppert M., Goda Y., Hammer R. E., Li C., Rosahl T. W., Stevens C. F., Südhof T. C. (1994) Cell 79, 717–727 [DOI] [PubMed] [Google Scholar]
- 40.Johnson J. R., Ferdek P., Lian L. Y., Barclay J. W., Burgoyne R. D., Morgan A. (2009) Biochem. J. 418, 73–80 [DOI] [PubMed] [Google Scholar]
- 41.Fisher R. J., Pevsner J., Burgoyne R. D. (2001) Science 291, 875–878 [DOI] [PubMed] [Google Scholar]
- 42.Peters J. M., Walsh M. J., Franke W. W. (1990) EMBO J. 9, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.