Abstract
Huntington disease (HD) is a neurodegenerative disorder caused by an expansion of a polyglutamine (polyQ) domain in the N-terminal region of huntingtin (htt). PolyQ expansion above 35–40 results in disease associated with htt aggregation into inclusion bodies. It has been hypothesized that expanded polyQ domains adopt multiple potentially toxic conformations that belong to different aggregation pathways. Here, we used atomic force microscopy to analyze the effect of a panel of anti-htt antibodies (MW1–MW5, MW7, MW8, and 3B5H10) on aggregate formation and the stability of a mutant htt-exon1 fragment. Two antibodies, MW7 (polyproline-specific) and 3B5H10 (polyQ-specific), completely inhibited fibril formation and disaggregated preformed fibrils, whereas other polyQ-specific antibodies had widely varying effects on aggregation. These results suggest that expanded polyQ domains adopt multiple conformations in solution that can be readily distinguished by monoclonal antibodies, which has important implications for understanding the structural basis for polyQ toxicity and the development of intrabody-based therapeutics for HD.
Huntington disease (HD)5 is a fatal neurodegenerative disorder that is caused by an expansion of a polyglutamine (polyQ) domain in the protein huntingtin (htt), which leads to its aggregation into fibrils (1). HD is part of a growing group of diseases that are classified as “conformational diseases,” which include Alzheimer disease (AD), Parkinson disease (PD), the prion encephalopathies, and many more (2–4). The length of polyQ expansion in HD is tightly correlated with disease onset, and a critical threshold of 35–40 glutamine residues is required for disease manifestation (5). Biochemical and electron microscopic studies with htt fragments demonstrated that expanded polyQ repeats (>39) form detergent-insoluble aggregates that share characteristics with amyloid fibrils (6–8), and the formation of amyloid-like fibrils by polyQ was confirmed by studies with synthetic polyQ peptides (9). Collectively, these studies demonstrated a correlation between polyQ length and the kinetics of aggregation. This phenomenon has been recapitulated in cell-culture models that express htt fragments (10–12). Although it is clear that proteins with expanded polyQ repeats assemble into fibrils in vitro, recent studies have reported that htt fragments can also assemble into spherical and annular oligomeric structures (13–16) similar to those formed by Aβ and α-synuclein, which are implicated in AD and PD, respectively.
While the major hallmark of HD is the formation of intranuclear and cytoplasmic inclusion bodies of aggregated htt (17), the role of these structures in the etiology of HD remains controversial. For instance, the onset of symptoms in a transgenic mouse model of HD follows the appearance of inclusion bodies (18), while other studies indicate that inclusion body formation may protect against toxicity by sequestering diffuse, soluble forms of htt (10, 19, 20). Based on the direct correlation between polyQ length, htt aggregation propensity, and toxicity (6), it has been hypothesized that the aggregation of htt may mediate neurodegeneration in HD. However, there is no clear consensus on the aggregate form(s) that underlie toxicity, and there likely exist bioactive, oligomeric aggregates undetectable by traditional biochemical and electron microscopic approaches whose formation precedes disease symptoms. Although identification of the one or more toxic species of htt that trigger neurodegeneration in HD remains elusive, such species might exist in a diffuse, mobile fraction rather than in inclusion bodies (19). A thioredoxin-polyQ fusion protein was recently reported to exhibit toxicity in a meta-stable, β-sheet-rich, monomeric conformation (21), suggesting that polyQ can adopt multiple monomeric conformations, only some of which may be toxic. Consistent with such a scenario, molecular dynamic simulations and fluorescence correlation spectroscopy experiments with synthetic polyQ peptides indicate that polyQ domains can adopt a heterogeneous collection of collapsed conformations that are in equilibrium before aggregation (22–25).
Although biochemical, biophysical, and computational approaches have yielded insight into the structures formed by polyQ in vitro, whether such structures form in vivo remains largely unknown. Indeed, determining the conformational state of any misfolded/aggregated protein in situ and/or in vivo remains a major technical challenge.
Toward this goal, antibodies have been explored as a potentially powerful tool for detecting specific conformations or multimeric states of aggregated proteins in situ. Antibodies specific for amyloid fibrils often do not react with natively folded globular proteins from which they are derived, suggesting that such antibodies recognize a conformational epitope (26, 27). Several antibodies display conformation-dependent interactions with amyloids, aggregation intermediates, or natively folded precursor proteins. For example, there are antibodies specific for paired helical filaments of Tau (28–31), of aggregated forms of Aβ ranging from dimers to fibrils (32–34), and of native (35) or disease-related (36) forms of the prion protein. Antibodies have also been developed that are specific for common structural motifs associated with amyloid diseases, such as oligomers (37) and fibrils (38), independent of the peptide sequence of the amyloid forming protein from which they are derived, suggesting the potential for a common mechanism of aggregation and toxicity for these diseases.
With regard to htt, several antibodies (MW1, MW2, MW3, MW4, MW5, IC2, and IF8), which are specific for polyQ repeats, stain Western blots of htt with expanded polyQ repeats much more strongly than htt with normal polyQ length (39, 40), suggesting that these antibodies may recognize abnormal polyQ conformations. Furthermore, these polyQ-specific antibodies have distinct staining patterns in immunohistochemical studies of brain tissue sections (39). In one study, the affinity and stoichiometry of MW1 binding to htt increased with polyQ length, suggesting a “linear lattice” model for polyQ (41). This model is supported by a crystal structure of polyQ bound to MW1, which showed that polyQ can adopt an extended, coil-like structure (42). However, an independent structural study showed that the anti-polyQ antibody 3B5H10 binds to a compact β-sheet-like structure of polyQ in a monomeric htt fragment.6 These results clearly indicate that polyQ domains can fold into at least two unique, stable, monomeric conformations and suggest that the “linear lattice” model is not generally applicable to all polyQ structures.
Not only are antibodies useful for understanding what polyQ structures exist in situ, especially in the diffuse htt fraction of neurons, but antibodies and/or intrabodies may also have potential as therapeutic agents. For example, several studies showed that intrabodies reduce htt toxicity in cellular models (44–49). Moreover, one intrabody (C4) slows htt aggregation and prolongs lifespan in a Drosophila model of HD (50, 51), while another (mEM48) ameliorates neurological symptoms in a mouse model of HD (48).
Three of the antibodies examined in this study (MW1, MW2, and MW7) modulate htt-induced cell death when co-transfected as single-chain variable region fragment antibodies (scFvs) in 293 cells with htt exon 1 containing an expanded polyQ domain (46). In these studies MW1 and MW2, which bind to the polyQ repeat in htt, increased htt-induced toxicity and aggregation (46). Conversely, MW7, which binds to the polyproline (polyP) regions adjacent to the polyQ repeat in htt, decreased its aggregation and toxicity (46). Interestingly, MW7 has also been shown to increase the turnover of mutant htt in cultured cells and reduce its toxicity in corticostriatal brain slice explants (49).
Given the difficulty in understanding which specie(s) of htt exist and mediate pathogenesis in the putative toxic diffuse fraction of neurons, we sought to rigorously characterize the conformational specificity of a panel of anti-htt antibodies, the best in situ probes currently available for distinguishing specie(s) of htt. We reasoned that if htt can adopt multiple conformations that mediate different aggregation pathways, then anti-htt antibodies should differentially alter htt aggregation pathways by stabilizing or sequestering the specific conformers or aggregates they recognize. We therefore examined the effects of various antibodies on mutant htt fragment fibril formation and stability by atomic force microscopy (AFM). Our results are consistent with the hypothesis that monoclonal antibodies recognize distinct conformational epitopes formed by polyQ in a mutant htt fragment.
EXPERIMENTAL PROCEDURES
Protein Purification
GST-HD53Q fusion proteins were purified as described (52). Cleavage of the GST moiety by PreScission Protease (Amersham Biosciences) initiates aggregation. Fresh, unfrozen GST-HD53Q was used for each experiment. GST-HD53Q was centrifuged at 20,000 × g for 30 min at 4 °C to remove any preexisting aggregates before the addition of the PreScission protease. MW series of antibodies were obtained as described previously (39). 3B5H10 was purified as described before (53).
Western Blot Analysis
For Western blotting analysis, purified GST-HD53Q proteins were incubated at 37 °C with shaking at 1400 rpm. Solutions were sampled at 0, 5, and 20 h after the addition of PreScission Protease. Proteins and aggregates were separated by SDS-PAGE and then transferred onto Protran BA85 nitrocellulose membranes (pore-size = 0.45 μm, Whatman) by standard Western transfer techniques. The membranes were incubated for 1 h at 37 °C with MW1, MW2, MW3, MW4, MW5, MW7, MW8, or 3B5H10 at a dilution of 1:1000. The membranes were then incubated with horseradish peroxidase-conjugated rabbit anti-mouse IgG or IgM (Jackson ImmunoResearch) at a 1:5000 dilution for 1 h at room temperature. The horseradish peroxidase was detected using an ECL Advance Western blotting Detection System (Amersham Biosciences), and the membranes were exposed to x-ray films.
Neuronal Culture, Transfection, and Immunocytochemistry
Primary cultures of rat striatal neurons were prepared from embryos (embryonic days 16–18) and transfected with plasmids (6–7 days in vitro) as described (10). Neurons were co-transfected with pGW1-Httex1-Q46 or 97-GFP in a 1:1 molar ratio, using a total of 1–4 μg of DNA in each well of a 24-well plate. After transfection, neurons were maintained in serum-free medium. All immunocytochemistry was performed as described (54). Cy3-conjugated secondary antibodies targeted to the appropriate primary antibody were acquired from Jackson Immunolabs.
Atomic Force Microscopy
For experiments on monomeric preparations, GST-HD53Q was incubated at 20 μm alone or with anti-htt antibodies (MW1, MW2, MW3, MW4, MW5, MW7, MW8, or 3B5H10) in a 1:1 ratio of protein to antigen binding sites in buffer A (50 mm Tris-HCl, pH 7, 150 mm NaCl, 1 mm dithiothreitol). PreScission protease (4 units/100 μg of fusion protein) was added at time zero to initiate GST cleavage and aggregation. Samples were incubated at 37 °C with shaking at 1400 rpm for the duration of the experiment. At time 1, 5, 8, and 24 h after cleavage of the GST, a sample (5 μl) of each incubation solution was deposited onto freshly cleaved mica (Ted Pella Inc., Redding, CA) and allowed to sit for 1 min. The substrate was washed with 200 μl of ultrapure water and dried under a gentle steam of air. For experiments on preformed fibrils, 40 μm solutions of HD53Q were incubated alone for 5–6 h after the removal of the GST tag to allow the formation of fibrils. Buffer or anti-htt antibodies (MW1, MW2, MW3, MW4, MW5, MW7, MW8, or 3B5H10) were added so that the final concentration of HD53Q was 20 μm, and the ratio of HD53Q to anti-htt antigen binding sites was 1:1. These solutions were sampled immediately and 3 h after the addition of the buffer or anti-htt antibody. Dose dependence studies of fibril disaggregation by MW7 and 3B5H10 were performed similarly, except that the ratio of HD53Q to antibody binding site varied (10:1, 5:1, and 1:1) and solutions were sampled at 0, 1, and 3 h after the addition of the antibodies.
Each sample was imaged ex situ using an MFP3D scanning probe microscope (Asylum Research, Santa Barbara, CA). Images were taken with silicon cantilevers with nominal spring constants of 40 newtons (N)/m and resonance frequency of ∼300 kHz. Typical imaging parameters were: drive amplitude 150–500 kHz with set points of 0.7–0.8 V, scan frequencies of 2–4 Hz, image resolution 512 by 512 points, and scan size of 5 μm. All experiments were performed in triplicate.
For in situ AFM experiments tracking individual fibrils, solutions containing preformed fibrils of HD53Q were allowed to rest on mica until several fibrils were present on the surface. Then, the substrate was washed with buffer A to remove proteins remaining in solution. The deposited fibrils were either imaged in clean buffer as a control or in the presence of anti-htt antibodies (2.5 μm final concentration). Images were taken with V-shaped oxide-sharpened silicon nitride cantilevers with a nominal spring constants of 0.5 N/m. Scan rates were set at 1–2 Hz with cantilever drive frequencies ranging from ∼8–12 kHz.
Statistics
All error bars in quantification of ex situ AFM experiments (number of fibrils or oligomers per μm2) represent the standard error of at least three independent experiments and were compared using a t test. All error bars in quantification of in situ AFM experiments (change in fibril length) represent the standard error measured from at least eight individual fibrils and were compared using a t test. Aggregate populations based on height were compared using a Spearman's rank correlation performed with GraphPad Prism.
RESULTS
Anti-htt Antibodies Recognize a Variety of SDS-stable Oligomeric Species of HD53Q
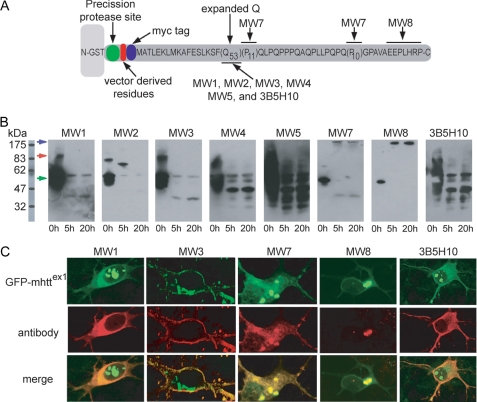
All experiments in this study, with the exception of the immunocytochemistry studies described later, were performed with a mutant htt fragment that expresses exon 1 with 53Q (HD53Q). HD53Q was purified from Escherichia coli as a soluble fusion with glutathione S-transferase (GST) (Fig. 1) (52). After purification, GST-HD53Q appeared non-aggregated as determined by AFM analysis and size-exclusion chromatography (data not shown). The HD53Q fragment contains epitopes specifically recognized by the panel of eight independent monoclonal anti-htt antibodies (Fig. 1A) used in this study. MW1, MW2, MW3, MW4, MW5, and 3B5H10 are specific for the polyQ domain. MW7 is specific for the polyP domains. MW8 is specific for the last seven residues of the C terminus of htt exon 1.
FIGURE 1.
Anti-htt antibodies recognize a variety species of HD53Q in vitro and in situ. A, a schematic representation of the GST-htt exon 1 fusion protein with 53Q (HD53Q) shows a PreScission protease site located between GST and the htt fragment (not drawn to scale) and the locations of epitopes for the antibodies that were used in this study. B, Western blots of HD53Q after incubation with protease for varying times, probed with antibodies as labeled. The location of bands representing intact GST-HD53Q fusion protein at ∼53 kDa is indicated by a green arrow. A band that migrated at an apparent molecular mass of a dimer of the fusion protein (∼106 kDa) is indicated by a red arrow. A blue arrow indicated the location of the wells of the gel where larger HD53Q aggregates are observed. C, primary cultures of rat striatal neurons expressing a GFP-labeled mutant htt-exon1 fragment with 97Q were analyzed by immunocytochemistry with antibodies as labeled.
Cleavage of a unique peptide sequence between the GST moiety and HD53Q with a site-specific protease (PreScission protease) released the HD53Q fragment, initiating aggregation in a time-dependent manner as reported (7, 15). Western blots of HD53Q were used to monitor cleavage 0, 5, and 20 h after the addition of the protease (Fig. 1B). Before proteolytic cleavage (t = 0 h), most antibodies specific for the polyQ domain detected a prominent band of intact htt-GST fusion protein that migrated at an apparent molecular mass of ∼53 kDa, and a less intense band that migrated at an apparent molecular mass of a dimer of the fusion protein (∼106 kDa). At later time points, MW1 and MW3 recognized the intact fusion protein and a band that migrated at a lower apparent molecular mass that may represent monomeric HD53Q (∼40 kDa). MW2 did not recognize this ∼40-kDa species after proteolytic cleavage but did react with a larger, potentially dimeric species (∼80 kDa) at later time points. MW4, MW5, and 3B5H10 recognized a ∼40-kDa species and a variety of SDS-stable bands of HD53Q, some of which may be fragments of HD53Q. Only antibodies that were not specific for the polyQ domain (MW7 and MW8) recognized large aggregated forms of HD53Q that remained in the wells of the gel, indicating that the polyQ epitopes recognized by these anti-polyQ antibodies are not accessible or absent in large aggregates. Of the two antibodies that bound the large aggregated form, only MW7 stained the ∼40-kDa species of HD53Q. These results indicate quite remarkably that six independent anti-polyQ antibodies (MW1–5 and 3B5H10) detect a variety of stable polyQ epitopes formed by HD53Q, even after apparent htt denaturation in SDS. Two antibodies against regions outside the polyQ stretch of htt exon1 (MW7 and -8) appear to expand the repertoire of recognizable htt species further.
Anti-htt Antibodies Recognize a Variety of htt Species in Neurons in Situ
To determine if these anti-htt antibodies could distinguish different htt epitopes in neurons, we applied immunocytochemistry to an established neuronal model (19) in which primary striatal neurons are transiently transfected with a mutant htt exon1 fragment fused to enhanced green fluorescent protein (GFP) (Fig. 1C). We compared the GFP signal, which exhibited fluorescence in a diffuse cytoplasmic localization and in inclusion bodies, to that detected by specific antibodies. Consistent with the results with Western blots, only MW7 and MW8 labeled large htt inclusion bodies based on co-localization with the GFP signal from htt. MW7 also stained diffuse htt. PolyQ-specific antibodies did not stain inclusion bodies; rather, they recognized a diffuse population of htt proteins. All of these results were consistent with Western blots from Fig. 1B. This diffuse population might contain a heterogeneous mix of monomeric conformers and soluble, oligomeric aggregates. The Western blot and immunocytochemistry studies suggest that these antibodies recognized different conformers or oligomeric forms of HD53Q.
Anti-htt Antibodies Modulate htt Aggregation Differentially
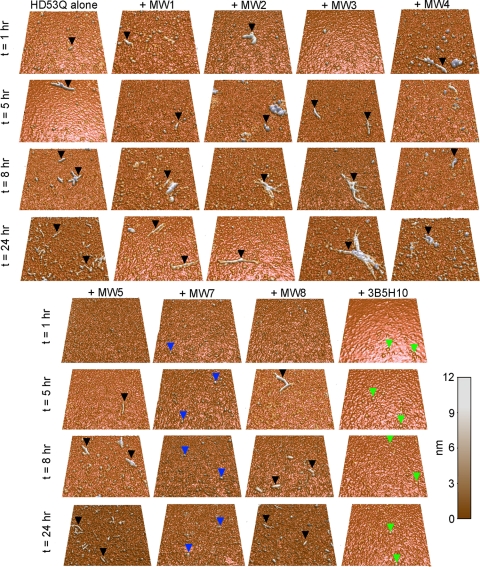
We next used AFM to analyze the effects of anti-htt antibodies on HD53Q aggregation. Co-incubation experiments were performed with monomeric preparations of HD53Q and each antibody. Representative AFM images of aliquots removed from solutions of HD53Q in the presence and absence of anti-htt antibodies after 1, 5, 8, and 24 h of incubation are shown in Fig. 2. The concentration of HD53Q in all solutions was 20 μm, and the ratio of antigen binding sites on the antibody to HD53Q was 1:1. In an effort to quantify the effect the anti-htt antibodies on fibril formation, AFM images from all incubations were analyzed by counting the number of fibrils per μm2 (Fig. 3A). For this analysis, the number of fibrils in the AFM images for a given sample was divided by the total area covered by the AFM images. Fibrils were defined as objects with a height larger than 5 nm and a length-to-width (aspect) ratio >3.
FIGURE 2.
Anti-htt antibodies modulate htt aggregation differentially. Representative 2 μm × 2 μm AFM images of 20 μm HD53Q incubated in the absence or presence of antibodies as labeled for 1, 5, 8, and 24 h after cleavage of the GST moiety. The ratio of antigen binding sites to HD53Q was 1:1. For HD53Q alone and with MW1-MW5 or MW8, fibrillar structures (black arrows) appeared after 1–5 h of incubation. The number of fibrils increased at 8 and 24 h. However, it appeared that there were more fibrils for HD53Q alone. For incubations of HD53Q with MW7 or 3B5H10, no fibrillar structures appeared throughout the 24-h experiment. In incubations with MW7, globular aggregates (blue arrows) around ∼3.5 nm tall were the dominant species observed at all time points. For incubation with 3B5H10, smaller globular species (green arrows) ∼2.5 nm tall were present at all time points. Shown are representative AFM images. Quantification of the number of fibrils per μm2 in these experiments is shown in Fig. 3.
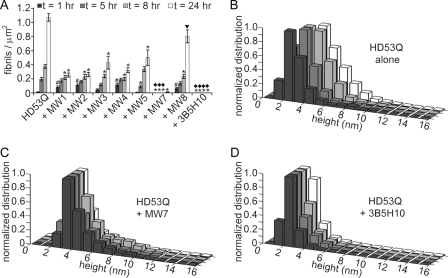
FIGURE 3.
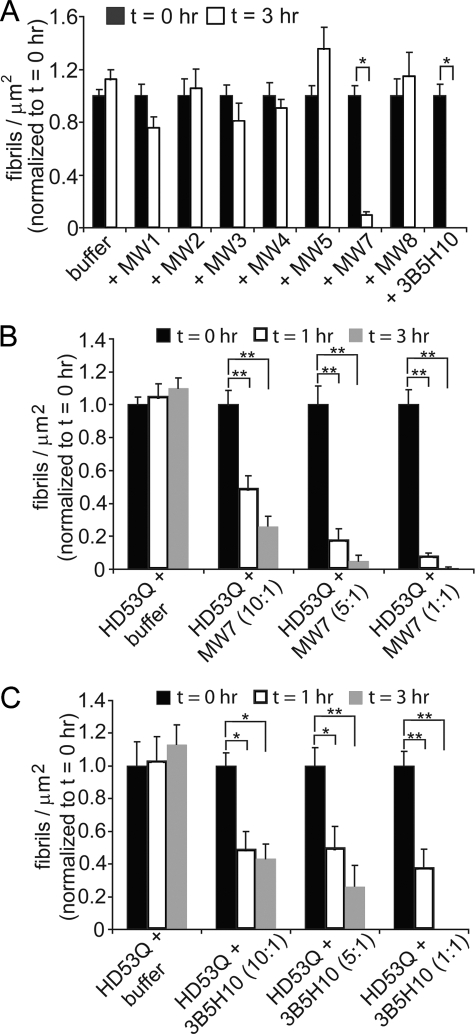
Quantification over time of HD53Q aggregates in the absence and presence of anti-htt antibodies. A, the number of fibrils/μm2 was calculated from AFM images of HD53Q incubated in the absence and presence of anti-htt antibodies analyzed at 1, 5, 8, and 24 h of incubation. Compared with control experiments of HD53Q alone, all of the antibodies significantly reduced the number of fibrils formed at later time points. However, there was a significant increase in the number of fibrils formed after 1 h for incubations with MW1, MW2, MW4, and MW8. MW7 and 3B5H10 completely inhibited the formation of fibrils over the time course of the experiments. #, a significant increase (p < 0.05) in the number of fibrils/μm2 in comparison to HD53Q alone at the same time point (Student's t test). * and ▼ denote significant decreases (* = p < 0.01, ▼ = p < 0.05) in the number of fibrils/μm2 in comparison to HD53Q alone at the same time point (Student's t test). ♦ indicates that no fibrils were observed. The experiment was replicated six times, and the error bars represent standard error. B–D, height histograms for globular structures observed in HD53Q alone (B) and with MW7 (C) or 3B5H10 (D) as a function of time. Whereas the height of HD53Q oligomers gradually increased over time, both MW7 and 3B5H10 stabilized distinct globular structures that likely represent complexes of HD53Q and antibody. The legend applies to all panels in the figures.
The AFM images of HD53Q incubated alone displayed fibril growth and an increase in fibril abundance per unit area over the 24-h time course of the experiment (Figs. 2 and 3A). At 1 h after removal of GST, only a small number of fibrils were present, and these increased in number and grew from several hundred nanometers to ∼1 μm in length at later time points. The fibrils were ∼6–8 nm tall and 12 nm wide (measured at half height). Fibril formation in solutions of HD53Q co-incubated with MW1, MW2, or MW4 altered aggregation similarly (Figs. 2 and 3A). After 1 h of incubation, the number of fibrils/μm2 significantly increased in the presence of these antibodies. Despite this early increase in the number of fibrils, MW1, MW2, and MW4 all had significantly fewer fibrils than the controls at later time points. Co-incubation of HD53Q with MW8 also resulted in an initial increase in the number of fibrils formed, with a significant reduction compared with controls at later time points. However, MW8 appeared to be the least effective antibody in reducing fibril formation after 24 h. At early time points, the number of fibrils formed in the presence of MW3 and MW5 did not significantly differ from controls (Figs. 2 and 3A). By 24 h of co-incubation, however, both MW3 and MW5 had significantly inhibited HD53Q fibril formation. These results suggest that MW1–5 may recognize one or more conformers of mutant htt that are required for efficient fibril formation.
Unlike all other antibodies tested, MW7 and 3B5H10 completely prevented fibril formation of HD53Q over the entire time course of the experiment (Figs. 2 and 3A). Instead of fibrils, compact globular structures were observed in co-incubations of HD53Q with MW7 or 3B5H10. The height of individual globular structures was analyzed at all time points for HD53Q with or without MW7 or 3B5H10 (Fig. 3, B–D). Height was chosen for analysis because it is the most accurately measured dimension in AFM, and it does not contain artifacts due to the finite shape and size of the probe tip. Fibrillar structures were not included in the analysis with HD53Q alone. In incubations of HD53Q alone, globular oligomers gradually increased in height as a function of time (Figs. 2 and 3B). The oligomers observed at 1 and 24 h represented distinct populations of HD53Q aggregates, because the height distributions were no longer similar based on a Spearman's rank correlation coefficient (p = 0.37). MW7 and 3B5H10 appeared to stabilize distinct globular structures, which likely are complexes of antibody and HD53Q, with globular structures observed for co-incubations of HD53Q with MW7 being slightly larger than those observed with 3B5H10 (compare Fig. 3C with 3D). Whereas the mean height of HD53Q oligomers observed in controls at 24 h was 5.3 ± 1.65 nm, globular species observed from co-incubations of HD53Q with MW7 and 3B5H10 were 4.4 ± 1.76 nm and 2.6 ± 0.74 nm tall, respectively. The height distributions under each condition did not change over time based on Spearman's rank correlation coefficient (p < 0.001). That is, the size of globular species formed upon co-incubation of HD53Q with MW7 was the same at all time points, as was true for co-incubations of HD53Q with 3B5H10. This indicated that globular species observed in these co-incubations were different from those formed in incubations of HD53Q alone. Overall, the quantitative AFM analyses demonstrate that antibodies specific for the polyQ domain modulate HD53Q aggregation differentially and that antibodies with specificity for other domains of htt can also alter this process.
We next performed biochemical experiments to confirm the AFM results, in which antibodies were added to monomeric preparations of GST-HD53Q before initiating aggregation with protease. 20 μm HD53Q solutions were sampled after 8 h for Western blot analysis of aggregate formation by staining with MW8 (supplemental Fig. S1). Before addition of protease (t = 0 h), no aggregated HD53Q was detected. Aggregated HD53Q was detected in the wells for HD53Q alone after 8 h of incubation; however, there appeared to be fewer aggregates detected for HD53Q incubated with MW1–MW5 and MW8. For co-incubations of HD53Q with MW7 and 3B5H10, no aggregates were detected in the well, confirming the complete inhibition of aggregate formation by these antibodies.
Anti-htt Antibodies MW7 and 3B5H10 Disassemble htt Aggregates
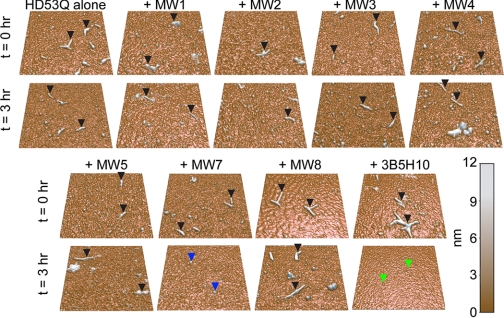
To test the effects of different antibodies on pre-aggregated HD53Q, GST was first removed from HD53Q by proteolytic cleavage, and then HD53Q was incubated for 6–8 h prior to addition of anti-htt antibodies. The preincubation resulted in a large population of HD53Q fibrils (time point 0 h in Fig. 4). After the initial incubation time, aliquots were deposited on mica, dried, and imaged. Approximately 10–20 fibrils were observed per 5 μm2 by ex situ AFM. These pre-aggregated HD53Q solutions were divided into several aliquots to which buffer (for control) or antibodies were added to a final antigen binding site to HD53Q ratio 1:1, with a final HD53Q concentration of 20 μm. Immediately after buffer or antibody were added, the HD53Q solutions were re-sampled and imaged to verify that fibrils were still present to obtain a time point 0-h measurement (Fig. 4). The solutions were then incubated for an additional 3 h, sampled, and imaged (Fig. 4). Preformed fibrils that were treated with buffer, MW1, MW2, MW3, MW4, MW5, or MW8 appeared to be stable, as the number of fibrils per μm2 was unchanged between 0 and 3 h (Figs. 4 and 5A). Importantly, the two anti-htt antibodies that prevented fibril formation (MW7 and 3B5H10) also significantly reduced the number of preformed fibrils. At the 1:1 ratio of antigen binding sites to HD53Q, MW7 and 3B5H10 completely disaggregated preformed fibrils.
FIGURE 4.
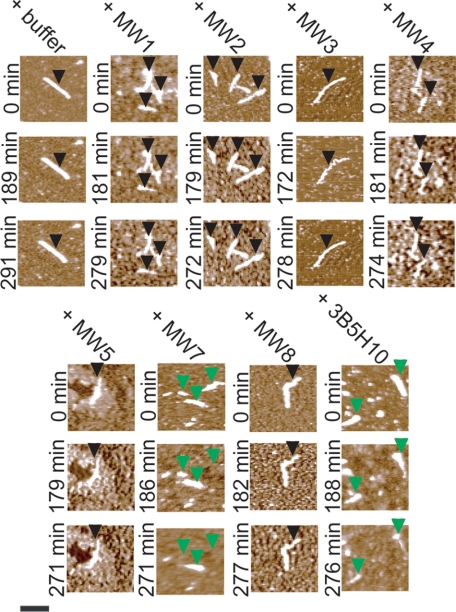
Ex situ AFM analysis indicates that the anti-htt antibodies MW7 and 3B5H10 disassemble htt aggregates. Samples of HD53Q were incubated for 6–8 h after removal of the GST moiety to form a large population of fibrils. Then, buffer (as control), MW1-MW5, MW7, MW8, or 3B5H10 was added. The ratio of antigen binding sites to HD53Q was 1:1. The solutions were sampled directly after the addition of buffer/antibodies (t = 0 h) and deposited on mica for AFM imaging. Fibrils (black arrows) were present in all samples at this time. The solutions were incubated for 3 h after the addition of buffer or antibodies and re-sampled. Fibrils (black arrows) were still present in samples that had been treated with buffer, MW1-MW5 or MW8. However, fibrils were no longer detected in samples treated with MW7 or 3B5H10. Treatment with MW7 resulted in a large population of globular species (blue arrows) that varied greatly in size with the majority of species ranging in height from 4 to 8 nm. Treatment with 3B5H10 resulted in globular species (green arrows) that were only ∼2.5 nm tall. Shown are representative 2 μm × 2 μm AFM images. Quantification of the number of fibrils per μm2 in these experiments is shown in Fig. 5.
FIGURE 5.
Quantification of the number of fibrils/μm2 for pre-aggregated HD53Q treated with buffer or anti-htt antibodies. A, the number of fibrils/μm2 was calculated from AFM images of incubations of fibrillar preparations of HD53Q taken immediately after (t = 0 h) and 3 h after the addition of buffer, MW1-MW5, MW7, MW8, or 3B5H10. The ratio of antigen binding sites to HD53Q was 1:1. For comparison, all bars are normalized to the number of fibrils/μm2 at t = 0 h for that sample. With the addition of buffer (control), MW1-MW5, or MW8, there was no change in the number of fibrils present after 3 h. With MW7 and 3B5H10, the number of fibrils was significantly reduced, indicating that these antibodies were able to disassemble preformed fibrils. *, p < 0.001 (Student's t test). Error bars represent standard error. B and C, the dose dependence of fibril disaggregation was studied by quantitative analysis of AFM images of fibrillar preps of HD53Q taken immediately after (t = 0 h), 1 h, and 3 h after the addition of B, MW7 or C, 3B5H10. The ratio of antigen binding sites to HD53Q was 10:1, 5:1, and 1:1. For comparison, all bars are normalized to the number of fibrils/μm2 at t = 0 h for that sample. The disaggregation of fibrils by MW7 (B) and 3B5H10 (C) appeared to be dose-dependent. *, p < 0.01; **, p < 0.001 (Student's t test).
We next evaluated the dose dependence of HD53Q fibril disaggregation by MW7 and 3B5H10 (Fig. 5, B and C). Preformed fibrils of HD53Q were treated with MW7 or 3B5H10 at an antigen binding site to HD53Q ratio of 1:10, 1:5, and 1:1. Controls consisting of preformed HD53Q fibrils treated with buffer were also prepared. The final concentration of HD53Q was 20 μm in all experiments. These solutions were sampled at 0, 1, and 3 h after the addition of buffer, MW7, or 3B5H10 and imaged with AFM. Preformed fibrils present on mica were significantly reduced at all ratios of antibody:htt, with a clear antibody dose dependence for the disaggregation.
Tracking the Fates of Individual HD53Q Fibrils Exposed to Anti-htt Antibodies in Situ
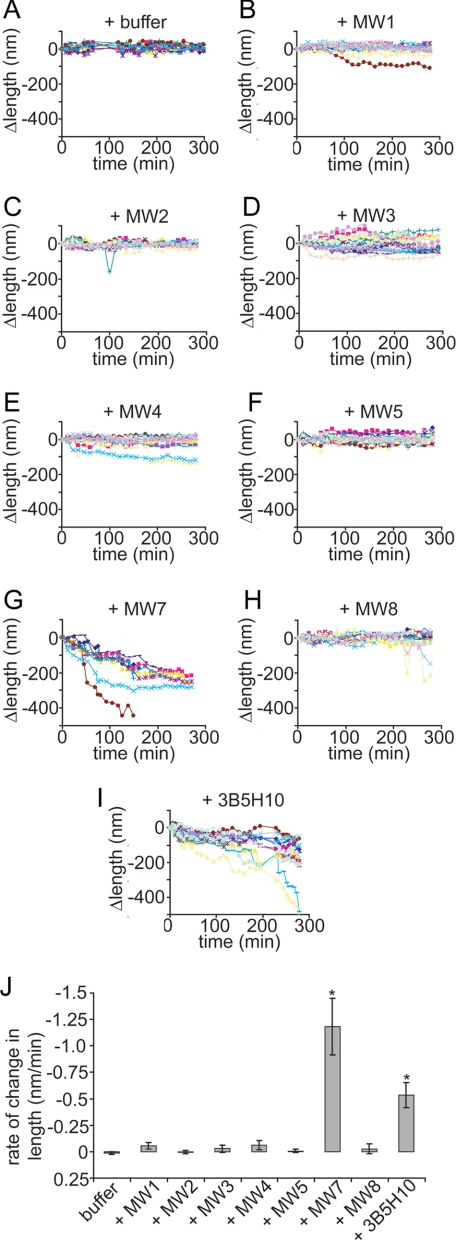
To further explore the stability of preformed fibrils of HD53Q, we took advantage of the ability of AFM in solution to track morphological changes of individual fibrils as a function of time (Fig. 6 and supplemental movies S1 and S2). Preformed HD53Q fibrils were deposited on mica and imaged continuously. Buffer (control) or anti-htt antibodies were injected directly into the fluid cell of the AFM. This allowed for the tracking of the fate of individual fibrils exposed to different anti-htt antibodies. Fibrils that were treated with buffer remained stable with no apparent change in length for over 300 min, verifying that the continual scanning of the AFM probe tip was not sufficient to invoke mechanical disruption of fibril integrity (supplemental movie S1). Similarly, the majority of fibrils treated with MW1, MW2, MW3, MW4, MW5, or MW8 did not exhibit large morphological changes for up to 300 min during continuous imaging (data not shown). Consistent with the co-incubation experiments described above, fibrils exposed to MW7 and 3B5H10 gradually shortened in length (supplemental movie S2). In the case of MW7, some fibrils completely disappeared from the surface. We then quantified the change in length of individual fibrils as a function of time (Fig. 7, A–I) by subtracting the length at time 0 from the length of the fibril at any given time. While the length of fibrils did not vary as a function of time for HD53Q treated with buffer or MW1-MW5 or MW8 (Fig. 7, A–F and H), all fibrils treated with MW7 or 3B5H10 displayed a negative change in length. The average rate of change in fibril length was calculated based on measurements on individual fibrils under all conditions (Fig. 7J). Fibrils exposed to MW7 or 3B5H10 exhibited significant rates of decreasing contour length compared with control fibrils, with MW7 disaggregating fibrils at a faster rate than 3B5H10. The other antibodies did not differ significantly from the buffer control. These results indicate that some, but not all, anti-htt antibodies can disassemble fibrils in solution.
FIGURE 6.
Monitoring disassembly of single htt aggregates incubated with MW7 or 3B5H10 by in situ AFM. Samples of HD53Q were incubated for 6–8 h after removal of the GST moiety to form a large population of fibrils. These fibrils were deposited on mica and imaged using in situ AFM, which allows for the tracking of the fate of individual fibrils as a function of time. These fibrils were imaged in the absence or presence of anti-htt antibodies. Fibrils appeared to be stable after treatment with buffer, MW1-MW5, or MW8 (location of stable fibrils indicated by black arrows). However, treatment with MW7 or 3B5H10 caused fibrils to disaggregate and/or shorten in length (location of disaggregating fibrils indicated by green arrows). Scale bar represents 500 nm and is applicable to all images. See also supplemental movies S1 and S2.
FIGURE 7.
Quantification of change in length and rate of change of fibrils treated with anti-htt antibodies. A–I, the change in length (Δlength) of individual fibrils imaged in the absence and presence of anti-htt antibodies was tracked as a function of time as measured by in situ AFM. Fibril length appeared stable with the addition of buffer (A), MW1-MW5 (B–F), or MW8 (H). The length of individual fibrils steadily decreased after treatment with MW7 (G) or 3B5H10 (I). J, the average rate of change of fibril length for fibrils treated with buffer (as control), MW1-MW5, MW7, MW8, or 3B5H10 was calculated, showing that only MW7 and 3B5H10 caused a significant change in fibril length (*, p < 0.01 with a Student's t test).
MW7 and 3B5H10 Disassemble Fibrils by Forming Different Complexes with htt
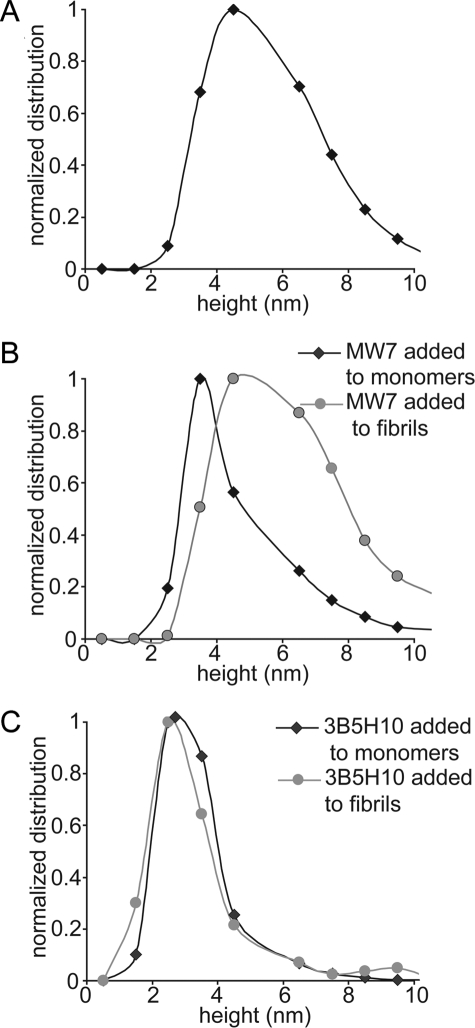
Because MW7 and 3B5H10 both prevented fibril formation and destabilized preformed fibrils, we next compared the height of the globular complexes formed by htt with the antibodies when the antibodies were added to monomeric or fibrillar HD53Q (Fig. 8). Globular species formed after incubation of HD53Q in the absence of antibodies were predominately 4–5 nm tall with a large number of oligomers taller than 6 nm (Fig. 8A). In contrast, globular species observed from co-incubations of MW7 or 3B5H10 with monomeric HD53Q were only 3–4 and 2–3 nm tall, respectively (Fig. 8, B and C). Interestingly, when MW7 was added to preformed fibrillar HD53Q and allowed to completely disaggregate the fibrils (3 h after addition MW7), the resulting oligomeric species were much larger than those observed following incubation of this antibody with monomeric HD53Q (Fig. 8B). These globular structures were predominately 5–6 nm tall with a large number of globular structures taller than 6 nm. Based on a Spearman's rank correlation coefficient, this difference in size was statistically significant, demonstrating that the final size of the complex formed between MW7 and HD53Q can vary, depending upon the initial aggregation state of HD53Q. This result may indicate that MW7 can recognize both monomeric and aggregated forms of htt, consistent with the immunocytochemical experiments and Western blot analysis (Fig. 1). Surprisingly, the globular structures observed from the complete disaggregation (3 h after the addition of antibody) of preformed HD53Q fibrils by 3B5H10 were precisely the same size as those formed when 3B5H10 was added to monomeric HD53Q, based on Spearman's rank correlation coefficient. This indicates that, in contrast to MW7, 3B5H10, which has been previously shown to bind a monomer of htt,6 forms the same complex with HD53Q regardless of its initial aggregation state (Fig. 8C). This suggests that 3B5H10 is incapable of recognizing oligomeric species of htt. Because MW7 apparently recognizes both aggregated and diffuse forms of htt, MW7 may be physically disrupting fibril structure by stabilizing a population of oligomeric structures. However, as 3B5H10 only recognizes soluble, non-aggregated forms of htt, it may be tightly binding and sequestering a monomeric conformation of htt that is in direct equilibrium with fibril ends.
FIGURE 8.
Size analysis of aggregate observed with MW7 or 3B5H10 added to monomeric or fibrillar HD53Q. A, HD53Q oligomers (HD53Q incubated alone) after 5 h of incubation were predominantly 4–5 nm in height with several as tall as 6–8 nm. B, when MW7 was incubated (added at t = 0 h) with monomeric HD53Q (black diamonds), the height of globular aggregates formed after 5 h of co-incubation were predominantly 3–4 nm tall, although there was a large portion of taller globular aggregates (shoulder on the right of the histogram). In contrast, when MW7 was incubated with pre-aggregated fibrillar HD53Q (gray circles), globular aggregates (conditions where fibrils disaggregated) observed when imaged 3 h after addition of MW7 were much taller (4–5 nm) in comparison to those formed by adding MW7 to monomeric HD53Q, with a larger portion of aggregates being 5–10 nm tall. C, when 3B5H10 was incubated (added at t = 0 h) with monomeric HD53Q (black diamonds), the majority of globular aggregates observed after 5 h co-incubation were 2–3 nm in height. Similarly, when 3B5H10 was incubated with pre-aggregated fibrillar HD53Q (gray circles), globular species (conditions where fibrils disaggregated) observed 3 h after the addition of 3B5H10 again were predominantly 2–3 nm tall.
DISCUSSION
Expanded polyQ repeats in htt have been postulated to adopt multiple conformations, but it is unclear which conformations may exist in neurons and are pathogenic. To study the existence and effects of different htt conformations in neurons, appropriate conformational probes must be first be established and characterized. The ability of antibodies to be used in situ makes them attractive tools to measure htt conformations in neurons and to ultimately determine their functional significance in HD pathogenesis. We therefore set out to characterize the range of htt conformations that can be detected by a panel of anti-htt antibodies, including many that are specific for expanded polyQ repeats. Because various htt conformations have been linked to different aggregation pathways in vitro (15), we reasoned that different anti-htt antibodies may have disparate effects on aggregation if the antibodies are recognizing different htt conformational epitopes.
In this study we showed that a panel of antibodies (MW1–MW5 and 3B5H10) that are all specific for polyQ sequences detected different aggregated species of HD53Q in Western blots and in cultured neurons. These antibodies also had widely varying effects on HD53Q aggregation, and some even disassembled preformed htt fibrils. MW1, MW2, and MW4 initially increased fibril formation before suppressing it at later time points. MW3, MW5, and MW8 slowed fibril formation. MW7 (polyP-specific) and 3B5H10 (polyQ-specific) completely prevented the formation of fibrillar structures. These two antibodies also destabilized preformed fibrils despite being specific for different regions of htt. These results are consistent with the hypothesis that expanded polyQ repeats can adopt multiple conformation-specific epitopes that can be easily discriminated by the immune system.
While compared with controls at later time points, all of the polyQ-specific antibodies at least partially inhibited the formation of fibrils. MW1, MW2, and MW4 appeared to initially boost fibril formation. This initial increase in aggregation is consistent with previous reports that MW1 and MW2 enhanced aggregation, which was associated with increased htt-induced toxicity, when they were expressed as scFvs in a cellular model of HD (46). Among the polyQ-specific antibodies we tested, 3B5H10 appears to recognize a unique polyQ conformation, because it was the only polyQ-specific antibody to completely prevent fibril formation and destabilize preformed fibrils. Recent structural studies lend further support to the notion that polyQ repeats can exist in stable conformations of different structure. For example, a crystal structure of a polyQ peptide bound to MW1 showed that polyQ can adopt an extended, coil-like structure (42). However, an independent structural study showed that 3B5H10 binds to a compact β-sheet-like structure of polyQ.6 We speculate that MW1 binding to a range of conformations on single-stranded polyQ may initially catalyze the collapse of polyQ into aggregation-prone structures, accounting for the early increase in fibril formation for HD53Q incubated with MW1 compared with HD53Q incubated in buffer. However, as aggregation starts, the accumulation of MW1 antibody on each HD53Q molecule may eventually sterically hinder further aggregation, accounting for the late attenuation in fibril formation for HD53Q incubated with MW1 compared with HD53Q incubated in buffer. In contrast, 3B5H10's binding to a compact, double-stranded structure of polyQ may fully bury the edges of the polyQ conformation that seeds aggregation, accounting for 3B5H10's ability to completely block aggregation. Therefore, our results indicate unequivocally that polyQ domains can sample at least two unique monomeric conformations, but the polyQ domains are likely to adopt other stable or meta-stable structures as well. For example, fluorescence correlation spectroscopy experiments and molecular dynamics simulations (23) indicate that polyQ peptides can form a heterogeneous population of collapsed structures in aqueous solution. In the absence of antibodies, htt appears to be able to sample multiple conformations; however, a collapsed conformation appears to be the dominant species as detected by small-angle x-ray scattering.6
The antibodies MW7 (anti-polyP) and 3B5H10 (anti-polyQ) both destabilized polyQ fibrils. However, the mechanisms appear to be different, based on size analysis of the aggregate/complex after disaggregation. Although MW7 and 3B5H10 are specific for different regions of htt, there are other notable differences between the two antibodies. MW7 is an IgM while 3B5H10 is an IgG. MW7 recognizes aggregated and diffused forms of htt by Western blot and immunocytochemistry, whereas 3B5H10 does not recognize aggregates of htt. MW7 can block fibril formation from monomeric HD53Q by binding to a specific conformer, resulting in a stable complex with a narrow size distribution. However, MW7 can also bind aggregates and may physically bind to fibrils, disrupting their stability, and resulting in a different population of oligomeric complexes with a broader size distribution. Although we did not observe any direct binding of MW7 to htt fibrils by in situ AFM, this possibility cannot be ruled out because the ∼8-min interval between images may not be fast enough to capture such an event. Although 3B5H10 also formed a stable complex with monomeric HD53Q, it did not appear to bind to large htt aggregates observed by Western blot, biochemical, and immunocytochemical methods, suggesting that 3B5H10 disaggregates fibrils by sequestering monomeric HD53Q and shifting the equilibrium toward soluble forms of HD53Q.7 This notion is supported by the finding that 3B5H10 forms stable complexes of the same size regardless of whether it was added to monomeric or fibrillar HD53Q. Our AFM data also suggest that 3B5H10 is unable to bind oligomeric species of htt, consistent with 3B5H10's demonstrated conformational specificity for a compact, double-stranded conformation of monomeric htt.6
Of the polyQ-specific antibodies used in this study, only MW1 and 3B5H10 are IgG-type antibodies; the rest are IgM. We attempted to control for this difference by calculating the ratio of HD53Q to antibody in all experiments based on antigen binding sites on the respective antibody. Antibody type did not appear to have a clear impact on htt aggregation. For instance, co-incubation of MW1 (IgG) or MW2 (IgM) with monomeric HD53Q resulted in very similar aggregation profiles; whereas, 3B5H10 (IgG) prevented fibril formation. In regards to fibril destabilization, antibody type did not appear to play a role, because 3B5H10 reduced the number of fibrils even at a ratio of five peptides per antigen binding site, which is analogous to the dilution factor used to control for IgM type antibodies. 3B5H10 was still effective in destabilizing fibrils even at a dilution of 10:1, yet none of the polyQ-specific IgM type antibodies destabilized fibrils. The other IgG-type polyQ-specific antibody, MW1, did not disaggregate fibrils even at a ratio of 1:1. Therefore, the effects of these antibodies on htt aggregation and aggregate stability cannot be simply correlated to their antibody type. This notion is further supported by the observations that MW7 (polyP-specific), which is an IgM type antibody, was able to completely prevent fibril formation and destabilize preformed fibrils.
The ability of MW7 to prevent fibril formation and destabilize preformed HD53Q fibrils provides additional support for the importance of the polyP domains in htt aggregation. More broadly, it also indicates the critical importance of flanking sequences on polyQ structure and aggregation. Studies on synthetic peptides revealed that the addition of a 10-residue polyP sequence to the C terminus of a polyQ peptide altered both aggregation kinetics and conformational properties of the polyQ tract (56). Flanking polyP sequences can also inhibit the formation of β-sheet structure in polyQ peptides by inducing a PPII-like helix structure, extending the length of the polyQ domain necessary to induce fibril formation (57). Flanking sequences in htt exon1 of various polyQ domain lengths modulate toxicity in yeast models, not only in cis, but also in trans during aggregation (58, 59). Interestingly, the proline-rich regions of htt exon1 reduced polyQ-related toxicity in these studies (58, 59).
Protein interactions with the polyP sequence in htt may have a major influence on the conformation of the adjacent polyQ domain. Other studies have demonstrated that the polyP domain of htt interacts with vesicle trafficking proteins (i.e. HIP1, SH3GL3, and dynamin), which may lead to sequestration of these proteins in inclusion bodies (61). By analogy, MW7 binding to the polyP domains of HD53Q may stabilize a conformation of the polyP domains that can, in turn, prevent the necessary conformational changes in the polyQ domain that lead to fibril formation. Such findings underscore the critical importance of protein context in polyQ aggregation and aggregate stability. There are currently nine diseases related to polyQ expansions in proteins that are broadly expressed, and the nature of the proteins that contain the polyQ domain and their associated pathologies differ substantially. That is, each mutant polyQ protein causes a distinct neurodegenerative disease that is associated with a different population of affected neurons. It is likely that the protein context of the expanded polyQ domains associated with each disease, and concomitant protein interactions that vary due to protein context, could help explain, at least in part, the striking cell specificity that is observed in each disease.
Because MW7 and MW8 displayed similar behavior in recognizing aggregated forms of htt by Western blot analysis and immunocytochemical studies of a HD neuronal model, it is interesting that MW7 was much more effective in preventing htt aggregation from monomers. This provides further evidence that the polyP region plays an important role in htt aggregation compared with the specific motif recognized my MW8. Further, it appears that binding of an antibody to aggregated forms of htt is not sufficient to disrupt aggregate stability as MW8, which recognized aggregated forms of htt, was not able to disaggregate preformed fibrils.
The AFM studies presented here are consistent with previous reports that MW7 suppresses aggregation and toxicity when it is expressed as a scFv in cellular (46, 49) and Drosophila (60) models of HD. Co-transfection of MW7 with mutant htt exon 1 in corticostriatal rat brain slices increased the number of healthy medium spinal neurons (49). Interestingly, expression of the MW7 scFV promotes turnover of htt in cellular models of HD (49). These results indicate that the ability of MW7 to prevent htt aggregation and destabilize htt fibrils, observed in this study, may play a pivotal role in the ability of MW7 to reduce cellular toxicity in a variety of HD models.
An important finding in the present study is that htt aggregation can be reversed by antibodies. There is a great deal of interest in the use of antibodies and intrabodies as potential therapeutic agents to treat HD and other polyQ disorders (44–47, 50). Our observations point to the potential for preventing aggregation and also destabilizing pre-existing aggregated forms of htt. By promoting formation of soluble forms of htt, antibodies and intrabodies may increase htt turnover, as was shown with a htt fusion protein system in HEK293 cells cotransfected with a scFv of the antibody MW7 (49). This observation is consistent with MW7-promoting soluble forms of HD53Q when added to monomeric or fibrillar forms of the protein, as demonstrated here. Such a notion is supported by mouse models, which demonstrate that continuous expression of mutant htt is required to maintain inclusion integrity and disease symptoms (62). However, without clear knowledge of what constitutes a toxic species or conformation, altering the aggregation process could also conceivably lead to detrimental effects. For example, if a particular antibody recognizes a non-toxic htt conformer, in principle it might actually shift the equilibrium of aggregate species in a manner that would increase the concentration of a toxic conformer(s). Although we show here that the equilibrium of htt aggregation can be altered in vitro by antibodies, other exogenous factors, including molecular chaperones, may possess similar activities (15, 43, 55). Because our panel of anti-polyQ antibodies displayed dramatically different properties, we speculate that polyQ repeats can display a wide variety of conformation-specific epitopes in vivo and that polyQ misfolding and aggregation within the context of the htt protein may be a far more complex process than previously imagined. Thus, additional analyses of which polyQ structures anti-htt antibodies recognize, whether or not they can be used to track the fate of specific conformers and/or oligomeric species of htt in vulnerable neuronal populations in situ, and the evaluation of their effects in vivo on disease progression in animal models of HD are clearly warranted.
Supplementary Material
Acknowledgments
We acknowledge Carl Johnson for insightful discussions and Gary Howard for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS047237 and R01NS054753 (to P. J. M.), P01AG022074 (to S. F.), R01NS039074 (to S. F.), and R01NS045091 and R01NS055298 (to P. H. P.). This work was also supported by the Hereditary Disease Foundation and the Cure Huntington's Disease Initiative.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Movies S1 and S2.
C. Peters-Libeu, E. Rutenber, J. Miller, Y. Newhouse, P. Krishnan, K. Cheung, E. Brooks, K. Widjaja, T. Tran, D. Hatters, S. Mitra, M. Arrasate, L. Mosquera, D. Taylor, K. Weisgraber, and S. Finkbeiner, submitted for publication.
M. Arrasate, J. Miller, E. Brooks, C. Peters-Libeu, J. Legleiter, D. Hatters, J. Curtis, K. Cheung, P. Krishnana, S. Mitra, K. Widjaja, B. Shaby, Y. Newhouse, G. Lotz, V. Thulasiramin, F. Sandou, P. J. Muchowski, M. Segal, K. Weisgraber, and S. Finkbeiner, submitted for publication.
- HD
- Huntington disease
- polyQ
- polyglutamine
- htt
- huntington
- PD
- Parkinson disease
- polyP
- polyproline
- AFM
- atomic force microscopy
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein.
Addendum
Consistent with the data we present here, a recent study showed that a mutant htt fragment can misfold into distinct amyloid conformations, and, depending on whether or not the polyQ domain was exposed or buried in a β-sheet, the amyloids can be either toxic or nontoxic, respectively (Nekooki-Machida et al. (63)).
REFERENCES
- 1.Vonsattel J. P., Myers R. H., Stevens T. J., Ferrante R. J., Bird E. D., Richardson E. P., Jr. (1985) J. Neuropathol. Exp. Neurol. 44, 559–577 [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum J. N. (2003) Trends Biochem. Sci. 28, 585–592 [DOI] [PubMed] [Google Scholar]
- 3.Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 4.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, S10–S17 [DOI] [PubMed] [Google Scholar]
- 5.Tobin A. J., Signer E. R. (2000) Trends Cell Biol. 10, 531–536 [DOI] [PubMed] [Google Scholar]
- 6.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 7.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díaz-Hernandez M., Moreno-Herrero F., Gómez-Ramos P., Morán M., Ferrer I., Baró A. M., Avila J., Hernández F., Lucas J. J. (2004) J. Neurosci. 24, 9361–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Berthelier V., Hamilton J. B., O'Nuallain B., Wetzel R. (2002) Biochemistry 41, 7391–7399 [DOI] [PubMed] [Google Scholar]
- 10.Saudou F., Finkbeiner S., Devys D., Greenberg M. E. (1998) Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 11.Lunkes A., Mandel J. L. (1998) Hum. Mol. Genet. 7, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 12.Hackam A. S., Singaraja R., Wellington C. L., Metzler M., McCutcheon K., Zhang T., Kalchman M., Hayden M. R. (1998) J. Cell Biol. 141, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka M., Morishima I., Akagi T., Hashikawa T., Nukina N. (2001) J. Biol. Chem. 276, 45470–45475 [DOI] [PubMed] [Google Scholar]
- 14.Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 15.Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 16.Dahlgren P. R., Karymov M. A., Bankston J., Holden T., Thumfort P., Ingram V. M., Lyubchenko Y. L. (2005) Nanomedicine 1, 52–57 [DOI] [PubMed] [Google Scholar]
- 17.Zoghbi H. Y., Orr H. T. (2000) Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 18.Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 19.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 20.Muchowski P. J., Ning K., D'Souza-Schorey C., Fields S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagai Y., Inui T., Popiel H. A., Fujikake N., Hasegawa K., Urade Y., Goto Y., Naiki H., Toda T. (2007) Nat. Struct. Mol. Biol. 14, 332–340 [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Vitalis A., Wyczalkowski M. A., Pappu R. V. (2006) Proteins 63, 297–311 [DOI] [PubMed] [Google Scholar]
- 23.Crick S. L., Jayaraman M., Frieden C., Wetzel R., Pappu R. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16764–16769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitalis A., Wang X., Pappu R. V. (2007) Biophys. J. 93, 1923–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitalis A., Wang X., Pappu R. V. (2008) J. Mol. Biol. 384, 279–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linke R. P., Zucker-Franklin D., Franklin E. D. (1973) J. Immunol. 111, 10–23 [PubMed] [Google Scholar]
- 27.Franklin E. C., Zucker-Franklin D. (1972) Proc. Soc. Exp. Biol. Med. 140, 565–568 [DOI] [PubMed] [Google Scholar]
- 28.Carmel G., Mager E. M., Binder L. I., Kuret J. (1996) J. Biol. Chem. 271, 32789–32795 [DOI] [PubMed] [Google Scholar]
- 29.Wolozin B., Pruchnicki A., Dickson D. W., Davies P. (1986) Science 232, 648–650 [DOI] [PubMed] [Google Scholar]
- 30.Jicha G. A., Lane E., Vincent I., Otvos L., Jr., Hoffmann R., Davies P. (1997) J. Neurochem. 69, 2087–2095 [DOI] [PubMed] [Google Scholar]
- 31.Ghoshal N., García-Sierra F., Fu Y., Beckett L. A., Mufson E. J., Kuret J., Berry R. W., Binder L. T. (2001) J. Neurochem. 77, 1372–1385 [DOI] [PubMed] [Google Scholar]
- 32.Yang A. J., Knauer M., Burdick D. A., Glabe C. (1995) J. Biol. Chem. 270, 14786–14792 [DOI] [PubMed] [Google Scholar]
- 33.Soreghan B., Pike C., Kayed R., Tian W., Milton S., Cotman C., Glabe C. G. (2002) Neuromolecular Med. 1, 81–94 [DOI] [PubMed] [Google Scholar]
- 34.Lambert M. P., Viola K. L., Chromy B. A., Chang L., Morgan T. E., Yu J., Venton D. L., Krafft G. A., Finch C. E., Klein W. L. (2001) J. Neurochem. 79, 595–605 [DOI] [PubMed] [Google Scholar]
- 35.Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paramithiotis E., Pinard M., Lawton T., LaBoissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., Francoeur G. P., Papadopoulos M., Haghighat A., Spatz S. J., Head M., Will R., Ironside J., O'Rourke K., Tonelli Q., Ledebur H. C., Chakrabartty A., Cashman N. R. (2003) Nat. Med. 9, 893–899 [DOI] [PubMed] [Google Scholar]
- 37.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 38.O'Nuallain B., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko J., Ou S., Patterson P. H. (2001) Brain Res. Bull 56, 319–329 [DOI] [PubMed] [Google Scholar]
- 40.Trottier Y., Lutz Y., Stevanin G., Imbert G., Devys D., Cancel G., Saudou F., Weber C., David G., Tora L., et al. ( 1995) Nature 378, 403–406 [DOI] [PubMed] [Google Scholar]
- 41.Bennett M. J., Huey-Tubman K. E., Herr A. B., West A. P., Jr., Ross S. A., Bjorkman P. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11634–11639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li P., Huey-Tubman K. E., Gao T., Li X., West A. P., Jr., Bennett M. J., Bjorkman P. J. (2007) Nat. Struct. Mol. Biol. 14, 381–387 [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Smith D. L., Meriin A. B., Engemann S., Russel D. E., Roark M., Washington S. L., Maxwell M. M., Marsh J. L., Thompson L. M., Wanker E. E., Young A. B., Housman D. E., Bates G. P., Sherman M. Y., Kazantsev A. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colby D.W., Chu Y., Cassady J. P., Duennwald M., Zazulak H., Webster J. M., Messer A., Lindquist S., Ingram V. M., Wittrup K. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17616–17621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colby D. W., Garg P., Holden T., Chao G., Webster J. M., Messer A., Ingram V. M., Wittrup K. D. (2004) J. Mol. Biol. 342, 901–912 [DOI] [PubMed] [Google Scholar]
- 46.Khoshnan A., Ko J., Patterson P. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecerf J. M., Shirley T. L., Zhu Q., Kazantsev A., Amersdorfer P., Housman D. E., Messer A., Huston J. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4764–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C. E., Zhou H., McGuire J. R., Cerullo V., Lee B., Li S. H., Li X. J. (2008) Cell Biol. 181, 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Southwell A. L., Khoshnan A., Dunn D. E., Bugg C. W., Lo D. C., Patterson P. M. (2008) J. Neurosci. 28, 9013–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfgang W. J., Miller T. W., Webster J. M., Huston J. S., Thompson L. M., Marsh J. L., Messer A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11563–11568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLear J. A., Lebrecht D., Messer A., Wolfgang W. J. (2008) FASEB J. 22, 2003–2011 [DOI] [PubMed] [Google Scholar]
- 52.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters-Libeu C., Newhouse Y., Krishnan P., Cheung K., Brooks E., Weisgraber K., Finkbeiner S. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 1065–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks E., Arrasate M., Cheung K., Finkbeiner S. M. (2004) Methods Mol. Biol. 277, 103–128 [DOI] [PubMed] [Google Scholar]
- 55.Ehrnhoefer D. E., Duennwald M., Markovic P., Wacker J. L., Engemann S., Roark M., Legleiter J., Marsh J. L., Thompson L. M., Lindquist S., Muchowski P. J., Wanker E. E. (2006) Hum. Mol. Genet. 15, 2743–2751 [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharyya A., Thakur A. K., Chellgren V. M., Thiagarajan G., Williams A. D., Chellgren B. W., Creamer T. P., Wetzel R. (2006) J. Mol. Biol. 355, 524–535 [DOI] [PubMed] [Google Scholar]
- 57.Darnell G., Orgel J. P., Pahl R., Meredith S. C. (2007) J. Mol. Biol. 374, 688–704 [DOI] [PubMed] [Google Scholar]
- 58.Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11051–11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson G. R., Sang T., Khoshnan A., Ko J., Patterson P. H. (2004) Soc. Neurosce. Abstr. 30: 938.5 [Google Scholar]
- 61.Qin Z. H., Wang Y., Sapp E., Cuiffo B., Wanker E., Hayden M. R., Kegel K. B., Aronin N., DiFiglia M. (2004) J. Neurosci. 24, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto A., Lucas J. J., Hen R. (2000) Cell 101, 57–66 [DOI] [PubMed] [Google Scholar]
- 63.Nekooki-Machida Y., Kurosawa M., Nukina N., Ito K., Oda T., Tanaka M. (2009) Proc. Nat. Acad. Sci. U.S.A. 106, 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.