Abstract
Aberrant activity of the phosphatidylinositol 3-kinase (PI3K) pathway supports growth of many tumors including those of breast, lung, and prostate. Resistance of breast cancer cells to targeted chemotherapies including tyrosine kinase inhibitors (TKI) has been linked to persistent PI3K activity, which may in part be due to increased membrane expression of epidermal growth factor (EGF) receptors (HER2 and HER3). Recently we found that proteins of the RGS (regulator of G protein signaling) family suppress PI3K activity downstream of the receptor by sequestering its p85α subunit from signaling complexes. Because a substantial percentage of breast tumors have RGS16 mutations and reduced RGS16 protein expression, we investigated the link between regulation of PI3K activity by RGS16 and breast cancer cell growth. RGS16 overexpression in MCF7 breast cancer cells inhibited EGF-induced proliferation and Akt phosphorylation, whereas shRNA-mediated extinction of RGS16 augmented cell growth and resistance to TKI treatment. Exposure to TKI also reduced RGS16 expression in MCF7 and BT474 cell lines. RGS16 bound the amino-terminal SH2 and inter-SH2 domains of p85α and inhibited its interaction with the EGF receptor-associated adapter protein Gab1. These results suggest that the loss of RGS16 in some breast tumors enhances PI3K signaling elicited by growth factors and thereby promotes proliferation and TKI evasion downstream of HER activation.
The role of the PI3K3 pathway in cell proliferation and survival, adhesion, metabolism, migration, drug resistance, and cytoskeletal rearrangement is well documented (1–3). Mutations in PI3K and dysregulation of the PI3K pathway have been implicated in many human cancers including lymphoma, multiple myeloma, and melanoma (4–8). Because the PI3K signal is a gatekeeper for tumor growth, an understanding of its regulation is critical for the therapeutic intervention of cancer.
PI3K, which catalyzes the production of phosphatidylinositol 3,4,5-trisphosphate from phosphatidylinositol 3,4-bisphosphate (9, 10), is activated by extracellular receptor tyrosine kinases including the EGF receptor (EGFR or HER) family, platelet-derived growth factor receptor, and the insulin growth factor receptor. HER stimulation activates Class IA PI3Ks consisting of dimers of p85α or β and either p110α, β, and δ catalytic subunits (11). Tyrosine phosphorylation of the adapter protein Grb2-associated binder 1 (Gab1) recruits p85 to the EGFR complex through a Src homology 2 (SH2) domain in p85 (12), which co-localizes the catalytic p110 subunit and membrane phospholipid substrates at the plasma membrane. Phosphatidylinositol 3,4,5-trisphosphate generated by PI3K activity recruits phosphoinositide-dependent kinase 1 through its pleckstrin homology domain, which in turn phosphorylates the mitogenic and antiapoptotic kinase Akt. Substrates of Akt include mTOR, BAD, IKK, FOXO, p27, MDM2, and GSK3β, all of which are signaling molecules with vital functions in cell cycle regulation and growth (3). Overexpression of Akt has been shown in several tumors such as ovarian and breast carcinoma and may lead directly to transformation of malignant melanoma (5).
Proteins of the RGS (regulator of G protein signaling) family mediate cellular desensitization to G protein-coupled receptor stimulation. RGS proteins act as GTPase-accelerating proteins to reduce the life span of activated (GTP-bound) Gα subunits of the G protein-coupled receptor signal-transducing heterotrimeric G protein (13). The R4 subfamily of RGSs (RGS1, 2, 4, 5, 8, 13, 16, 18, and 21) are the smallest members of the family, containing few residues outside of the ∼120-amino acid RGS domain that mediates binding to Gα proteins and GTPase-accelerating protein activity. We found recently that several R4 RGS proteins interacted with the phosphorylated p85α subunit of PI3K (14). In mast cells, RGS13 inhibited PI3K activation induced by high affinity IgE receptor (FcϵRI) cross-linking by antigen. FcϵRI stimulates PI3K by recruiting its catalytic p110δ subunit through p85 binding to a multi-protein complex that includes Gab2 and Grb2 at the plasma membrane (15). PI3K has an essential function in allergic responses (16). As a result of increased PI3K activation, mice deficient in RGS13 had more IgE-mediated mast cell degranulation and anaphylaxis (14).
RGS16, an R4 RGS protein homologous to RGS13, was identified originally as a p53 target gene in breast and colon cancer cells (17, 18). Recent analysis of 222 primary breast cancers found a high rate (50%) of genomic instability at the RGS16 locus (19). Because RGS16 associates with both EGFR (20) and p85α (14), we investigated how it affected the growth and survival of breast cancer cells. We found that RGS16 directly bound the amino-terminal SH2 and inter-SH2 domains of phosphorylated p85α, which mediate p110 and adapter binding and membrane localization (21). RGS16 overexpression in MCF7 breast cancer cells suppressed proliferation and EGF-induced Akt phosphorylation, whereas extinction of RGS16 expression increased cell growth and resistance to TKI treatment. Thus, through regulation of PI3K activity, RGS16 may limit proliferation of mammary cells and render cancer cells more susceptible to TKIs or other therapeutic compounds.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, Lentiviral Transduction, and Gene Transfection
HEK293T cells were cultured in complete Dulbecco's modified Eagle's medium (Invitrogen) (containing 10% (v/v) fetal bovine serum, 1% penicillin and streptomycin at 37 °C and 5% CO2). MCF7 and BT474 cells were cultured in complete RPMI (Invitrogen). MCF7 cells expressing either scrambled control or RGS16-specific shRNAs were constructed and maintained in complete RPMI supplemented with neomycin (0.4 mg/ml) as described elsewhere (18). In brief, the plasmid pSUPER (from Reuven Agami), which contains the human H1 RNA polymerase promoter, was used as the backbone for insertion of short sequence-specific shRNAs. Small interfering RNA sequences were: human RGS16, CGCTTCCTGAAGTCGCCTG (RGS16 shRNA1); or scrambled, GCGCGCTTTGTAGGATTCG (control shRNA1). Lentiviral constructs expressing either scrambled shRNA (catalog number SCH002; control shRNA2) and RGS16-specific shRNA (catalog number TRCN0000014292; small interfering RNA sequence TTCCTGAAGTCGCCTGCTT; RGS16 shRNA #2) were purchased from Sigma. RGS16 wild type and a mutant containing amino acids (aa) 91–202 were generated by PCR and cloned into pLenti6.3-V5 (Invitrogen). The target gene or shRNA constructs were transfected into HEK293T cells together with a lentiviral packaging mix (Invitrogen) using Lipofectamine 2000. After 72 h virus was harvested in the supernatant and concentrated 3-fold by centrifugation. Concentrated virus was added directly to MCF7 cells, and gene expression and function was assayed 48 h later. The plasmid pCEFL-hmycEGFP2-PI3Kp85α was the gift of J. Silvio Gutkind (NIDCR, National Institutes of Health). Plasmids encoding p85α truncation mutants were generated by PCR using full-length p85 as a template and subcloned into pCEFLhmycEGFP2. The plasmid pCDA3.1-V5/His-RGS16 was described previously (18).
Recombinant Protein Pulldowns, Immunoprecipitation, and Immunoblotting
MCF7 cells were stimulated with EGF for 15 min. prior to lysis in buffer containing 50 mm Tris pH 7.5, 150 mm NaCl, 5 mm MgS04, 1 mm dithiothreitol, 1 mm Na3VO4, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture tablet (Roche Applied Science). The lysates were incubated with either 2.5 μg of GST or GST-RGS16 (18) coupled to glutathione-Sepharose beads (Amersham Biosciences). After three washes with lysis buffer, pulldowns were resolved by SDS-PAGE and immunoblotted as indicated. We did GST binding assays using recombinant p85α (250 ng; Jena Bioscience) and GST or GST-RGS16 (2.5 μg). p85α was preincubated in buffer containing 60 mm HEPES, pH 7.5, 3 mm MgCl2, 3 mm MnSO4, 3 μm Na3VO4, 1.2 mm dithiothreitol, 50 μm ATP, 1 mm phenylmethylsulfonyl fluoride with or without 25 ng of recombinant Lyn (Upstate Biotechnology) for 30 min. at 30 °C prior to 10-fold dilution in buffer containing 50 mm HEPES, pH 8, 300 mm NaCl, 2 mm MgCl2, 10% glycerol, 0.1% Triton X-100 and purification with glutathione-Sepharose (GE Biosciences). Binding of His-p85α (200 ng; Jena Biosciences) to GST-Gab1 (200 ng; Axxora LLC) in the presence or absence of His-RGS16 expressed in bacteria and purified by nickel affinity chromatography (18) was determined after phosphorylation of both p85α and Gab1 and precipitation with glutathione beads as above. The interaction of GST-RGS16 to phosphorylated His-p85α/p110α (200 ng, Jena Bioscience) was assessed by incubation with nickel-nitriloacetic acid-agarose beads (Qiagen) in buffer containing 20 mm HEPES, pH 8, 150 mm NaCl, 6 mm MgCl2, 10% glycerol, 0.04% Triton X-100, and 10 mm imidazole. For co-immunoprecipitations, the cells were processed in buffer containing 50 mm Tris, pH 7.5, 250 mm NaCl, 5 mm MgCl2, 5% glycerol, 1% Triton X-100, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor tablets followed by immunoprecipitation with Myc antibody coupled to protein G-Sepharose (Fast Flow 4B; Amersham Biosciences). All of the blots are representative of two to four independent experiments.
Antibodies
Rabbit polyclonal antibodies against Akt, phosphorylated Akt (Thr308), PI3K-p85α, Gab1, PTEN, HER2, and mouse anti-Myc antibody were purchased from Cell Signaling Technology. β-Actin antibody was from Sigma, and anti-phosphotyrosine (4G10) was from Upstate Biotechnology. GST antibody was purchased Santa Cruz, GFP antibodies were from University of Alberta, Edmonton, Canada, and V5 antibody was from Invitrogen. Affinity-purified rabbit polyclonal anti-RGS16 antibody was described previously (22). Secondary antibodies were from Thermo Scientific.
EdU Incorporation
Click-iT EdU Imaging kit (Invitrogen) was used for S phase entry measurements. The cells were maintained in culture chambers and serum-starved for 24 h prior to stimulation with EGF (50 ng/ml; Sigma) or FBS (10% in RPMI) for 16 h. EdU was added into cell culture medium at a final concentration of 10 μm for 2 h before harvest. Cell fixation, permeabilization, and EdU detection were performed according to the manufacturer's instructions. 4′,6-Diamidino-2-phenylindole staining was used to identify nuclei.
Cell Viability Assays
These assays were performed as described (23). Briefly, the cells were cultured in eight separate wells of a 96-well plate. The cell viability was measured using a luminescence assay kit (CellTiter-Glo, Promega), and the results were read on a plate luminometer (Fluoroskan Ascent FL, Thermo Labsystems). MCF7 cells were transduced with lentiviruses expressing scrambled or RGS16-specific shRNAs or viruses encoding RGS16-V5 (wild type), RGS16-V5 (91–202), or β-galactosidase-V5 in 6-well plates. 24 h post-transduction the cells were split and seeded into eight separate wells of a 96-well plate for each condition. After incubation for another 24 h, the cells were serum-starved for an additional 24 h before stimulation with EGF (50 ng/ml) or FBS (10%) in RPMI for the indicated times. The EGF receptor tyrosine kinase inhibitor PD168393 was purchased from EMD Biosciences.
Statistical Analysis
Student's t test was used for analysis of two groups and analysis of variance for multiple groups. The data are expressed in the form of the means ± S.E. Probability values (P) of less than 0.05 were considered significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001. The Image J software program (National Institutes of Health) was used for densitometric analysis.
RESULTS
RGS16 Knockdown Enhances Breast Cancer Cell Growth and Cell Cycle Progression
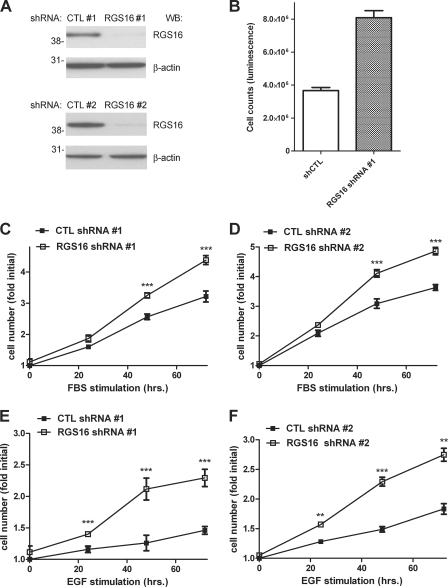
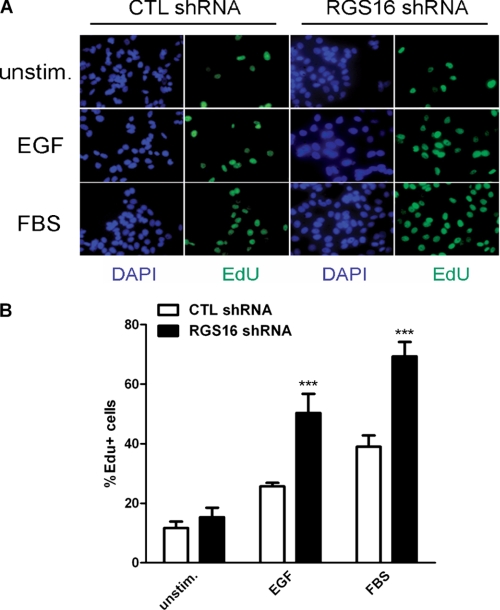
To investigate how RGS16 affects proliferation of breast cancer cells, we compared the growth of MCF7 cells stably expressing scrambled or RGS16-specific short shRNA (18). The RGS16-specific shRNA (RGS16 shRNA1) reduced RGS16 protein expression more than 75% compared with a control shRNA (control shRNA1) (Fig. 1A, top two panels). Initial examination of control and RGS16-depleted cells revealed an obvious difference in growth between the two populations. After 72 h of incubation with serum-containing medium, the number of RGS16-specific shRNA-expressing cells was double that of control cells (Fig. 1B). Further analysis showed that serum-starved MCF7 cells with reduced expression of RGS16 proliferated significantly more than control cells did over time upon exposure to medium containing either FBS (Fig. 1C) or EGF (Fig. 1E). To evaluate the specificity of this RGS16-targeted shRNA, we treated MCF7 cells with lentiviruses encoding either scrambled or RGS16-specific shRNAs distinct from those used to generated the stable cell line described above. RGS16 knockdown in MCF7 cells infected with the RGS16-targeted shRNA virus (RGS16 shRNA2) was similar to that observed in the stable cell line (Fig. 1A, bottom two panels). These MCF7 cells depleted of RGS16 also grew significantly faster after exposure to medium containing either FBS (Fig. 1D) or EGF (Fig. 1F). Finally, to test whether extinction of RGS16 expression affected cell cycle progression in MCF7 cells, we measured incorporation of EdU, which represents cells in the S phase of the cell cycle. Nearly twice as many serum-starved MCF7 cells depleted of RGS16 incorporated EdU than control cells did after exposure to either EGF or FBS (Fig. 2). Collectively, these results suggest that RGS16 negatively regulates growth of MCF7 cells induced by specific factors including EGF.
FIGURE 1.
Enhanced growth of RGS16-depleted MCF7 cells. A, RGS16 (top panels) and β-actin (bottom panels) quantities in lysates of MCF7 cells expressing either of two scrambled control (CTL) or RGS16-specific shRNAs determined by immunoblotting. B, viability of MCF7 cells stably expressing a scrambled control shRNA (CTL) or RGS16 shRNAs cultured in 96-well plates (5000 cells/well) for 48 h. The data are from a single experiment representative of three similar experiments. C and D, viability of MCF7 cells expressing plasmid (designated 1) or lentiviral (designated 2) control (CTL) or RGS16-specific shRNAs after serum starvation followed by treatment with FBS (10% v/v in culture medium) for 24–72 h (mean ± S.E. of the fold increase in initial cell number, set as 1, in three independent experiments). E and F, proliferation of MCF7 cells expressing control or RGS16-targeted shRNAs after EGF (50 ng/ml) stimulation for 24–72 h. WB, Western blot.
FIGURE 2.
Stable RGS16 knockdown increases the number of S phase MCF7 cells in response to EGF. A, cells in S phase determined by EdU (green) uptake under serum-starved conditions (top row) or after stimulation with EGF (middle row) or FBS (bottom row). 4′,6-Diamidino-2-phenylindole (DAPI) (blue) staining identifies nuclei. B, percentage of EdU-positive cells determined by counting at least 500 cells/condition in three independent experiments (means ± S.E.). CTL, control.
RGS16 Regulates EGF-induced PI3K Activation and Modulates Resistance to an EGFR Tyrosine Kinase Inhibitor
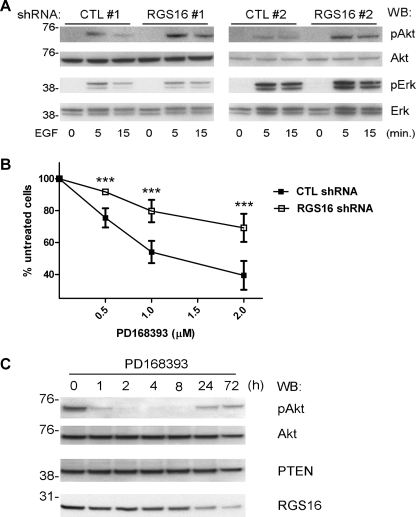
Receptors for EGF include the HER family members HER1 (EGFR) and HER2–4. Dysregulated expression and/or activation of HER proteins, particularly HER2 and HER3, have been found frequently in breast cancer cells. Approximately 20–30% of primary human breast cancers overexpress HER2, which presages aggressive tumor growth and poor response to therapy (24). Because RGS16 knockdown did not affect HER2 expression in MCF7 cells (supplemental Fig. S1), we hypothesized that it may control cell growth by regulating the downstream signaling pathway activated by EGF rather than receptor expression or phosphorylation. Given the importance of PI3K activity in breast cancer cell growth (25), we examined PI3K activation in MCF7 cells stimulated with EGF by measuring Akt phosphorylation. Cells expressing either of the RGS16-specific shRNAs had significantly more Akt phosphorylation after EGF stimulation than cells expressing control shRNAs (Fig. 3A). In contrast, ERK phosphorylation in response to EGF stimulation was not affected by RGS16 knockdown. These results suggest that the increased growth of MCF7 cells expressing RGS16 shRNA is due to increased PI3K activity and that RGS16 regulates EGF-evoked MCF7 cell growth primarily by affecting the PI3K-Akt signaling route.
FIGURE 3.
EGF-evoked PI3K activation in RGS16-depleted MCF7 cells and response to an EGFR-targeted TKI. A, immunoblot analysis of phosphorylated Akt (pAkt, top row), total Akt (second row from top), phosphorylated ERK (pERK, third row from top), and total ERK (bottom row) in MCF7 cells expressing either of two control (CTL) or RGS16 shRNAs serum-starved for 24 h and treated with EGF (50 ng/ml) for indicated times. B, viability of MCF7 cells expressing control or RGS16 shRNAs pulsed with the indicated concentrations of PD168393 for 6 h followed by incubation in complete medium for an additional 72 h (means ± S.E. of the percentage of untreated cell number in 3 independent experiments). C, immunoblot analysis of pAkt (top row), Akt (second row from top), PTEN (third row from top), or RGS16 (bottom row) expression in cell extracts of BT474 cells incubated with PD168393 (500 nm) for indicated times. WB, Western blot.
Enhanced and/or resurgent PI3K-dependent Akt phosphorylation has been linked to breast cancer resistance to several antitumor therapies such as doxorubicin (26), endocrine deprivation (8), and TKIs (25). However, the molecular mechanisms by which PI3K-Akt signaling evades these treatments remain incompletely defined. To determine whether the loss of RGS16 affects susceptibility of breast cancer cells to targeted chemotherapy, we treated cells with an EGFR-targeted TKI (PD168393) and measured viability. MCF7 cells expressing RGS16-specific shRNA were significantly more resistant to the antiproliferative effect of this compound than cells expressing the control shRNA (Fig. 3B).
Because these findings indicated a relationship between quantities of RGS16 and resistance of MCF7 cells to PD168393, we explored the relationship between RGS16 expression and PI3K activity in wild type MCF7 cells by examining Akt phosphorylation after TKI treatment. However, because we could not detect Akt phosphorylation in MCF7 cells without EGF stimulation (Fig. 3A), we incubated BT474 breast cancer cells, which also express RGS16, with PD168393. Consistent with prior studies, continuous exposure of BT474 cells to this compound led to a transient decrease in Akt phosphorylation (Fig. 3C), which suggested an antitumor effect of the TKI (25). After 24 h, however, Akt phosphorylation rebounded, which correlated with a diminution in RGS16 amounts (Fig. 3C). Amounts of PTEN, a phosphatidylinositol phosphatase that is mutated in a wide array of tumors (27) and negatively regulates PI3K activity, did not vary substantially during this time period. These results demonstrate a clear link between PI3K activity and the level of RGS16 expression in TKI-treated breast cancer cells and suggest another possible mechanism by which such cells escape the antiproliferative effect of the TKI.
RGS16 Binds PI3K Directly through Its p85α Subunit
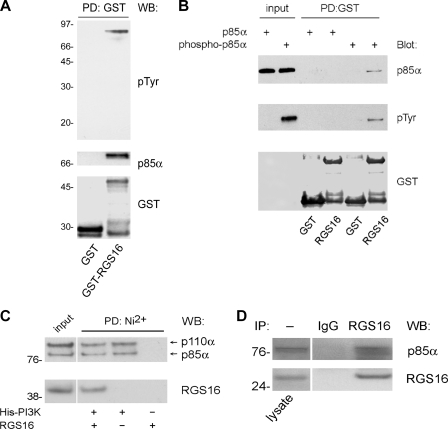
To determine the mechanism(s) whereby RGS16 regulates PI3K activity in breast cancer cells, we performed immunoprecipitation assays in HEK293T cells using an overexpression strategy. In addition to RGS13, several R4 RGS proteins including RGS1, 4, 5, and 16 co-immunoprecipitated the PI3K p85α subunit (supplemental Fig. S2). To determine whether RGS16 interacted with PI3K in breast cancer cells, we incubated the lysates of EGF-stimulated MCF7 cells with recombinant GST or GST-RGS16 immobilized on glutathione beads. Using an anti-phosphotyrosine antibody, we detected a single protein (∼85 kDa) that co-purified with GST-RGS16 but not GST (Fig. 4A). By immunoblot analysis, we identified this protein as p85α. A pulldown assay showed that recombinant, His-p85α directly bound recombinant GST-RGS16 but not GST expressed in bacteria and precipitated with glutathione-Sepharose (Fig. 4B). Similar results were obtained in reciprocal experiments in which bacterially expressed GST-RGS16 fusion protein co-precipitated with His-PI3K bound to nickel beads (Fig. 4C). Although the Src family kinases such as Lyn phosphorylate tyrosine residues on p85, its phosphorylation is not required for p110 or adapter interactions (28, 29). In this case, however, p85α tyrosine phosphorylation was necessary for RGS16 binding (Fig. 4B). In contrast, p110 was not required for the direct interaction of RGS16 with p85α (Fig. 4B), nor did RGS16 affect binding of p85 to p110 (Fig. 4C). These results suggest that RGS16, p85, and p110 can co-exist in a complex. Finally, co-immunoprecipitation experiments showed that endogenous RGS16 interacted with p85α in MCF7 cells under steady-state conditions (FBS) (Fig. 4D).
FIGURE 4.
RGS16 interacts with the p85α component of PI3K. A, MCF7 cell lysates treated with EGF (50 ng/ml) incubated with either GST or GST-RGS16 bound to glutathione-Sepharose beads. Pulldowns were analyzed by immunoblotting as indicated. B, recombinant p85α incubated with or without recombinant Lyn kinase followed by incubation with either GST or GST-RGS16 coupled to glutathione-Sepharose. Binding reactions were washed extensively and immunoblotted as indicated. 20% of the input protein was run as a blotting control. C, purified recombinant His-p85α/p110α PI3K was phosphorylated by Lyn kinase as in B, incubated with recombinant GST-RGS16, and precipitated with nickel-nitriloacetic acid beads. Co-purification of RGS16 with immobilized PI3K was assessed by immunoblotting. No binding of GST-RGS16 to nickel-nitriloacetic acid beads was observed in the absence of PI3K (Far right lane). 10% of the input protein was run as an immunoblot control. D, association of RGS16 and p85α in MCF7 cells, assessed by immunoprecipitation (IP) with RGS16 antibody and immunoblotting as indicated. WB, Western blot; PD, pulldown.
Interaction between the RGS Domain of RGS16 and the Amino-terminal SH2 and Inter-SH2 Domains of p85α Affect p85α Binding to Gab1
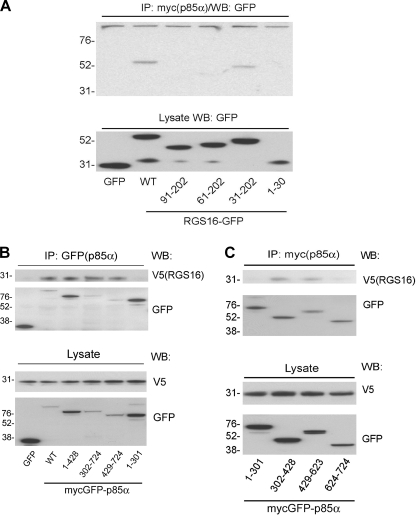
To further define the molecular association of RGS16 and PI3K, we determined the regions in RGS16 and p85α, mediating their interaction using deletion mutants (supplemental Figs. S3 and S4). Co-immunoprecipitations of lysates from HEK293T cells transiently transfected with full-length p85α, and wild type RGS16 (or its mutants; supplemental Fig. S3) showed that a mutant containing aa 31–202 interacted with p85α, whereas mutants comprised of aa 61–202 or 1–30 did not. This experiment suggests that p85α binding required aa 31–60 of RGS16 (Fig. 5A). To identify the RGS16-binding site on p85α, we constructed p85α deletion mutants lacking one or more domains (supplemental Fig. S4). p85α is a modular protein containing an amino-terminal SH3 domain (aa 1–77), a BCR/RacGAP homology domain (aa 78–301), and Src homology 2 (nSH2 and cSH2) domains (aa 302–428 and 624–724) separated by the p110 binding inter-SH2 (iSH2) domain (aa 429–623) (30). Co-immunoprecipitation of full-length RGS16 by p85α mutants containing deletions of the carboxyl terminus demonstrated that aa 302–724 of p85α were required for RGS16 binding because a mutant containing only residues 1–301 did not co-precipitate RGS16 (Fig. 5B). Further, p85α mutants containing only nSH2 (aa 302–428) or iSH2 domains (aa 429–623) co-immunoprecipitated RGS16 (Fig. 5C). These results indicate that either the nSH2 or iSH2 domains are sufficient for RGS16 binding.
FIGURE 5.
RGS16 binds the niSH2 domain of p85α. A, immunoassay of HEK293T cells transfected with plasmids encoding GFP-tagged RGS16 truncation mutants (numbers at the bottom indicate aa included] together with Myc-GFP-p85α. The lysates were immunoprecipitated (IP) with Myc antibody followed by immunoblotting with GFP antibody (top panel). Protein expression in total cell lysates blotted with GFP antibody (bottom panel). B and C, immunoassay of HEK293T cells transfected with full-length Myc-GFP-tagged p85α or truncation mutants (numbers at the bottom indicate aa included) together with V5-tagged RGS16. Anti-GFP immunoprecipitations (top panels) or total cell lysates (bottom panels) analyzed by immunoblot analysis with V5 and GFP antibodies. WB, Western blot.
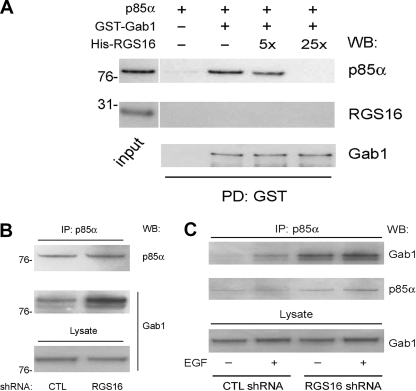
The nSH2 domain of p85α is involved in PI3K-adapter interactions. EGFR activation induces Gab1 tyrosine phosphorylation, which promotes engagement of the nSH2 domain of p85α by Gab1 (1, 12). We determined whether RGS16 binding to this region affected the association of phosphorylated p85α with phospho-Gab1 in competition experiments. We first incubated recombinant GST-Gab1 and p85α with Lyn kinase to phosphorylate both proteins. Phosphorylated Gab1 and p85α were then mixed together in the presence or absence of RGS16, and GST-Gab1 was precipitated with glutathione beads. As shown in Fig. 6A, a 5–25-fold molar excess of RGS16 progressively inhibited p85α co-precipitation with GST-Gab1. Importantly, recombinant RGS16 was not pulled down by GST-Gab1 in the presence or absence of p85α (Fig. 6A, middle panel), indicating that RGS16 does not bind Gab1.
FIGURE 6.
RGS16 inhibits the interaction between p85α and Gab1. A, recombinant GST-Gab1 expressed in bacteria and His-p85α were phosphorylated by recombinant Lyn kinase as in Fig. 4. Phosphorylated proteins were precipitated with glutathione beads in the presence or absence of His-RGS16 in 5–25 molar excess of p85α. B, lysates of MCF7 cells expressing either control or RGS16 shRNAs incubated in serum-containing medium immunoprecipitated (IP) with p85α antibody followed by immunoblotting with Gab1 or p85α antibodies (top panel). Protein expression in total lysates evaluated by immunoblotting (bottom panel). C, lysates of MCF7 cells expressing either control or RGS16 shRNA treated or not with EGF (50 ng/ml for 5 min.) immunoprecipitated with p85α antibody followed by immunoblotting with Gab1 or p85α antibodies. WB, Western blot; PD, pulldown; CTL, control.
These results suggested that RGS16 prevents p85α binding to its cognate adapter Gab1. To evaluate the effect of RGS16 binding on p85α protein-protein interactions in vivo, we analyzed association of p85α and Gab1 in RGS16-depleted MCF7 cells. Although RGS16 knockdown in MCF7 cells did not alter expression of either p85α or Gab1 (Fig. 6B), more Gab1 was associated with p85α in the presence of FBS (Fig. 6B) or in response to EGF treatment (Fig. 6C). These results suggest that the loss of RGS16 in these cells enhanced PI3K activity as a result of increased Gab1-p85α complex formation.
RGS16 Overexpression Inhibits Breast Cancer Cell Growth
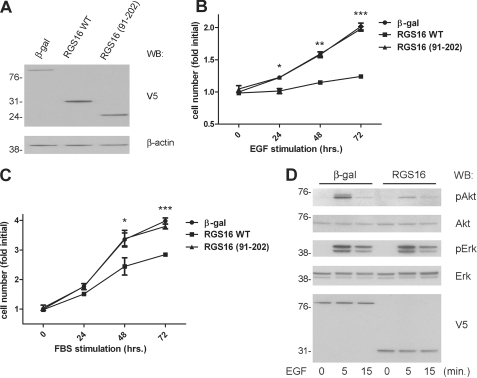
We used lentiviruses to express V5 epitope-tagged RGS16 (wild type or a mutant containing aa 91–202, which lacks the p85α-binding site) in MCF7 cells and detected these proteins in cell lysates by immunoblotting with anti-V5 antibody (Fig. 7A). RGS16 overexpression significantly inhibited proliferation of serum-starved MCF7 cells in response to EGF (Fig. 7B) or FBS (Fig. 7C) compared with cells expressing a control protein (β-galactosidase). In contrast, expression of RGS16 (91–102) had no effect on EGF- or FBS-induced proliferation (Fig. 7, B and C). This finding indicates that RGS16 interaction with p85α is critical for its regulation of MCF7 cell growth. To determine how RGS16 overexpression affected EGF-induced PI3K activity, we assessed Akt phosphorylation in MCF7 cells after EGF stimulation. Cells expressing RGS16 had reduced Akt phosphorylation upon exposure to EGF compared with cells expressing β-galactosidase (Fig. 7D). On average, Akt phosphorylation (as determined by densitometry) was inhibited 57 ± 7% in such cells compared with control. By contrast, EGF-evoked ERK phosphorylation was not affected by the expression of RGS16 in these cells. These findings support the hypothesis that RGS16 impairs EGF-induced growth of breast cancer cells specifically by binding p85α and suppressing activation of the PI3K-Akt signaling pathway.
FIGURE 7.
RGS16 suppresses EGF-induced MCF7 cell growth and PI3K activation. A, expression of V5-tagged proteins from cells infected with corresponding lentiviruses assessed by immunoblotting with anti-V5 antibody (top panel). Immunoblotting with β-actin antibody was used to assess protein loading (bottom panel). B, viability of serum-starved MCF7 cells infected with lentiviruses encoding β-galactosidase-V5, RGS16-V5 wild type, or RGS16-V5 (aa 91–202) plated in 96-well plates (5000 cells/well) and restimulated with EGF (50 ng/ml) for the indicated times. C, viability of serum-starved MCF7 cells expressing the indicated proteins in response to FBS (10% in RPMI) treatment for the indicated times. D, immunoblot analysis of Akt phosphorylation (pAkt, top panel), total Akt (second panel from top), phospho-ERK (third panel from top), or total ERK (bottom panel) in serum-starved MCF7 cells infected with lentiviruses encoding either RGS16 wild type or control protein (β-galactosidase (β-gal)) and stimulated or not with EGF (50 ng/ml) for the indicated times. Expression of the V5-tagged proteins is shown in the bottom panel. WB, Western blot.
DISCUSSION
Although a major predictor of breast cancer growth is hormone receptor (estrogen and progesterone receptor) status, ∼30% of estrogen receptor- and progesterone receptor-positive breast tumors are resistant to endocrine therapy (8). The EGF signaling pathway and specifically PI3K activation induced by HER family members have emerged as important determinants of breast cancer cell growth. Mutations in genes for signaling molecules downstream of EGF such as p110 (PIKC3A), PTEN, and Akt are also common in breast tumors (31). Loss of PTEN activity has been associated with resistance to anti-EGFR monoclonal antibody (trastuzumab) therapy (27). Thus, dysregulated activation of the EGFR-PI3K signaling axis may promote breast cancer progression and resistance to therapy.
Our studies suggest that RGS16 could act as a tumor suppressor by inhibiting PI3K-dependent mammary epithelial cell growth. RGS16 was identified recently as a potential breast cancer susceptibility gene. A high rate of allelic imbalance (50%) was found at the RGS16 locus (1q25.3) in a series of clinical breast tumors (19). Detailed mapping of chromosomal breakpoints revealed microdeletions in the putative RGS16 promoter region, and in 10% of these tumors the RGS16 promoter was methylated. A substantial portion (∼67%) of tumors with such mutations had reduced RGS16 protein expression (30–60% of nonmalignant epithelial breast tissue amounts) (19). We showed here that RGS16 depletion in breast cancer cells enhances EGF-evoked PI3K activity and growth. An inverse relationship exists between RGS16 quantities and PI3K activity in MCF7 and BT474 cells. Together, these findings suggest that abnormal RGS16 expression or function contributes to breast carcinogenesis.
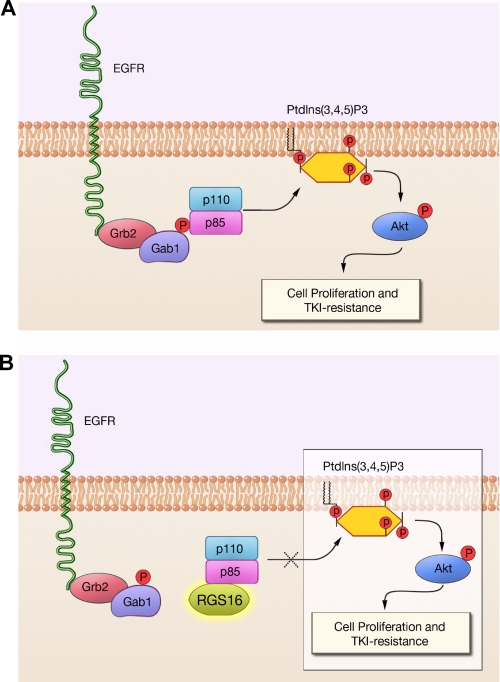
Our mutational and co-immunoprecipitation analyses indicate that RGS16 inhibits PI3K activation in MCF7 cells by constraining protein-protein interactions induced by growth factors. p85α exists in two pools: free and complexed with p110 catalytic subunits (30). p85 contacts the adapter-binding domain and C2 domains of p110 through its nSH2 domain and the coiled-coil iSH2 domain (together referred to as niSH2), and this binding inhibits p110 catalytic activity (21, 32). Upon stimulation of cellular receptors, nSH2 binds phosphotyrosine residues on the receptor itself (as for platelet-derived growth factor receptor) or on adapter proteins associated with the receptor (such as Gab1 for EGFR) (Fig. 8A) (12). Because RGS16 contacts the PI3K complex through the niSH2 domain of p85α, such binding may prevent p85α from engaging phosphotyrosines on receptors or adapters (Fig. 8B).
FIGURE 8.
A model for the regulation of EGF-induced PI3K activity by RGS16. A, in the absence of RGS16, EGF stimulation elicits phosphorylation of the adapter protein Gab1, which recruits the nSH2 domain of p85α to the EGFR-Gab1-Grb2 complex. Such targeting promotes conversion of membrane phosphatidylinositol 3,4-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) by the catalytic p110 subunit, which facilitates phosphorylation of Akt and activation of downstream mitogenic pathways. B, RGS16 binds the phosphorylated p85α subunit of PI3K, preventing binding to Gab1 and co-localization with the EGFR-Gab1-Grb2 complex, thereby inhibiting PI3K activation.
In mast cells, RGS13 inhibited p85α binding to Gab2, the adapter phosphorylated upon activation of FcϵRI (14). Although collectively these findings suggest that RGS proteins regulate PI3K activity by inhibiting signaling complex formation, they do not exclude other processes. For example, a basic stretch in the iSH2 domain of p85α is thought to contact membrane phospholipids directly (21), and RGS16 binding to niSH2 could interfere with membrane localization of PI3K. Although p110 and RGS both bind iSH2, our preliminary studies have not shown that RGS proteins inhibit p85-p110 interactions (Ref. 14 and Fig. 4C). Thus, RGS16 may bind a surface on p85α distinct from that which contacts p110 or may occupy a unique region within iSH2.
The temporal and spatial regulation of the p85-RGS interaction itself requires further study. Most R4 RGS proteins such as RGS13 and RGS16 are localized in the cytoplasm under steady-state conditions but may be recruited to the membrane or other organelles (33–35). Preliminary evidence suggests that in mast cells, RGS13 is rapidly and transiently localized at the plasma membrane after IgE-antigen stimulation of FcϵRI on mast cells.4 Thus, cellular localization patterns of RGS proteins may vary based on the distinct binding partners they acquire during the course of cellular stimulation, and future studies will address the timing and stability of RGS16-p85α complex formation in resting and activated cells.
In summary, using two different cell types with distinct receptors mediating PI3K activation (Ref. 14 and the current study), we have described a general mechanism of PI3K regulation induced by RGS protein binding to the regulatory p85α subunit, which inhibits its recruitment to signaling complexes. Further studies such as co-crystallization of the RGS-PI3K complex should provide mechanistic details as well as provide a basis for the design of new compounds that could mimic RGS regulation of PI3K. Given the importance of PI3K for many physiological processes, such drugs may be useful in a variety of disorders involving dysregulated metabolism, cell proliferation, and immunity, among others.
Supplementary Material
Acknowledgments
We thank John A. Olson and James Koh (Duke University School of Medicine) for several breast cancer cell lines.
This work was supported, in whole or in part, by Intramural Research funding from NIAID, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PI3K
- phosphatidylinositol 3-kinase
- TKI
- tyrosine kinase inhibitor(s)
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- Gab1
- Grb2-associated binder 1
- SH
- Src homology
- shRNA
- small hairpin RNA
- aa
- amino acid(s)
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- FBS
- fetal bovine serum
- EdU
- 5-ethynyl-2′-deoxyuridine
- iSH
- inter-SH
- ERK
- extracellular signal-regulated kinase
- HER
- human epidermal growth factor receptor
- RGS
- regulator of G protein signaling.
REFERENCES
- 1.Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2.Chang F., Lee J. T., Navolanic P. M., Steelman L. S., Shelton J. G., Blalock W. L., Franklin R. A., McCubrey J. A. (2003) Leukemia 17, 590–603 [DOI] [PubMed] [Google Scholar]
- 3.Harvey R. D., Lonial S. (2007) Future Oncol. 3, 639–647 [DOI] [PubMed] [Google Scholar]
- 4.Bahlis N. J., King A. M., Kolonias D., Carlson L. M., Liu H. Y., Hussein M. A., Terebelo H. R., Byrne G. E., Jr., Levine B. L., Boise L. H., Lee K. P. (2007) Blood 109, 5002–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindarajan B., Sligh J. E., Vincent B. J., Li M., Canter J. A., Nickoloff B. J., Rodenburg R. J., Smeitink J. A., Oberley L., Zhang Y., Slingerland J., Arnold R. S., Lambeth J. D., Cohen C., Hilenski L., Griendling K., Martínez-Diez M., Cuezva J. M., Arbiser J. L. (2007) J. Clin. Invest. 117, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opel D., Poremba C., Simon T., Debatin K. M., Fulda S. (2007) Cancer Res. 67, 735–745 [DOI] [PubMed] [Google Scholar]
- 7.Tazzari P. L., Cappellini A., Ricci F., Evangelisti C., Papa V., Grafone T., Martinelli G., Conte R., Cocco L., McCubrey J. A., Martelli A. M. (2007) Leukemia 21, 427–438 [DOI] [PubMed] [Google Scholar]
- 8.Tokunaga E., Kimura Y., Mashino K., Oki E., Kataoka A., Ohno S., Morita M., Kakeji Y., Baba H., Maehara Y. (2006) Breast Cancer 13, 137–144 [DOI] [PubMed] [Google Scholar]
- 9.Fruman D. A., Meyers R. E., Cantley L. C. (1998) Annu. Rev. Biochem. 67, 481–507 [DOI] [PubMed] [Google Scholar]
- 10.Datta S. R., Brunet A., Greenberg M. E. (1999) Genes Dev. 13, 2905–2927 [DOI] [PubMed] [Google Scholar]
- 11.Scaltriti M., Baselga J. (2006) Clin. Cancer Res. 12, 5268–5272 [DOI] [PubMed] [Google Scholar]
- 12.Mattoon D. R., Lamothe B., Lax I., Schlessinger J. (2004) BMC Biol. 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willars G. B. (2006) Semin Cell Dev. Biol. 17, 363–376 [DOI] [PubMed] [Google Scholar]
- 14.Bansal G., Xie Z., Rao S., Nocka K. H., Druey K. M. (2008) Nat. Immunol. 9, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu H., Saito K., Klaman L. D., Shen J., Fleming T., Wang Y., Pratt J. C., Lin G., Lim B., Kinet J. P., Neel B. G. (2001) Nature 412, 186–190 [DOI] [PubMed] [Google Scholar]
- 16.Gilfillan A. M., Tkaczyk C. (2006) Nat. Rev. Immunol. 6, 218–230 [DOI] [PubMed] [Google Scholar]
- 17.Buckbinder L., Velasco-Miguel S., Chen Y., Xu N., Talbott R., Gelbert L., Gao J., Seizinger B. R., Gutkind J. S., Kley N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7868–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson E. N., Seasholtz T. M., Waheed A. A., Kreutz B., Suzuki N., Kozasa T., Jones T. L., Brown J. H., Druey K. M. (2003) Nat. Cell Biol. 5, 1095–1103 [DOI] [PubMed] [Google Scholar]
- 19.Wiechec E., Overgaard J., Hansen L. L. (2008) Genes Chromosomes Cancer 47, 766–780 [DOI] [PubMed] [Google Scholar]
- 20.Derrien A., Druey K. M. (2001) J. Biol. Chem. 276, 48532–48538 [DOI] [PubMed] [Google Scholar]
- 21.Huang C. H., Mandelker D., Schmidt-Kittler O., Samuels Y., Velculescu V. E., Kinzler K. W., Vogelstein B., Gabelli S. B., Amzel L. M. (2007) Science 318, 1744–1748 [DOI] [PubMed] [Google Scholar]
- 22.Estes J. D., Thacker T. C., Hampton D. L., Kell S. A., Keele B. F., Palenske E. A., Druey K. M., Burton G. F. (2004) J. Immunol. 173, 6169–6178 [DOI] [PubMed] [Google Scholar]
- 23.Liang G., Yang J., Wang Z., Li Q., Tang Y., Chen X. Z. (2008) Hum. Mol. Genet. 17, 3254–3262 [DOI] [PubMed] [Google Scholar]
- 24.Ross J. S., Fletcher J. A. (1998) Stem. Cells 16, 413–428 [DOI] [PubMed] [Google Scholar]
- 25.Sergina N. V., Rausch M., Wang D., Blair J., Hann B., Shokat K. M., Moasser M. M. (2007) Nature 445, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Lu Y., Liang K., Liu B., Fan Z. (2005) Breast Cancer Res. 7, R589–R597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata Y., Lan K. H., Zhou X., Tan M., Esteva F. J., Sahin A. A., Klos K. S., Li P., Monia B. P., Nguyen N. T., Hortobagyi G. N., Hung M. C., Yu D. (2004) Cancer Cell 6, 117–127 [DOI] [PubMed] [Google Scholar]
- 28.Cuevas B. D., Lu Y., Mao M., Zhang J., LaPushin R., Siminovitch K., Mills G. B. (2001) J. Biol. Chem. 276, 27455–27461 [DOI] [PubMed] [Google Scholar]
- 29.Yamanashi Y., Fukui Y., Wongsasant B., Kinoshita Y., Ichimori Y., Toyoshima K., Yamamoto T. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L. C. (2005) Trends Biochem. Sci. 30, 194–204 [DOI] [PubMed] [Google Scholar]
- 31.Stemke-Hale K., Gonzalez-Angulo A. M., Lluch A., Neve R. M., Kuo W. L., Davies M., Carey M., Hu Z., Guan Y., Sahin A., Symmans W. F., Pusztai L., Nolden L. K., Horlings H., Berns K., Hung M. C., van de Vijver M. J., Valero V., Gray J. W., Bernards R., Mills G. B., Hennessy B. T. (2008) Cancer Res. 68, 6084–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C. H., Mandelker D., Gabelli S. B., Amzel L. M. (2008) Cell Cycle 7, 1151–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark M. A., Sethi P. R., Lambert N. A. (2007) FEBS Lett. 581, 764–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A. A., Lemberg K. E., Chidiac P. (2003) Mol. Pharmacol. 64, 587–593 [DOI] [PubMed] [Google Scholar]
- 35.Hiol A., Davey P. C., Osterhout J. L., Waheed A. A., Fischer E. R., Chen C. K., Milligan G., Druey K. M., Jones T. L. (2003) J. Biol. Chem. 278, 19301–19308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.