Abstract
The tumor suppressor p53 regulates cell cycle arrest and apoptosis by transactivating several genes that are critical for these processes. The transcriptional activity of p53 is often regulated by post-translational modifications and its interactions with various transcriptional coactivators. Here we report a physical interaction between the N-terminal transactivation domain (TAD) of p53 and the C-terminal DNA-binding domain of positive cofactor 4 (PC4CTD). Using NMR spectroscopy, we showed that residues 35–57 (TAD2) interact with PC4. 15N,1H HSQC and fluorescence competition experiments indicated that TAD binds to the DNA-binding site of PC4. Hepta-phosphorylation of the TAD peptide increased its binding affinity. Computer modeling of the p53N-PC4 complex revealed several important interactions that are reminiscent of those in the single-stranded DNA-PC4 complex. The ubiquitous nature of the acidic transactivation domain of p53 in mediating interactions with several transcription cofactors is also manifested as a DNA mimetic.
The tumor suppressor p53 regulates the expression of numerous target genes involved in cell cycle arrest and DNA repair through interaction with various proteins. Interaction of p53 with cellular proteins is essential for its transcriptional activity, stability, or specificity in DNA binding (1). p53 is composed of four structural/functional domains as follows: an N-terminal transactivation domain (TAD),3 a central DNA-binding domain, a tetramerization domain, and a C-terminal regulatory domain (CTD) (2). The p53 N-transactivation domain contains two subdomains TAD1-(1–40) and TAD2-(41–61). Many protein interactions are mediated with one or both of the TAD domains. The interaction between the N-terminal domain of MDM2 and TAD1 is very tight (3, 4). In contrast, the subdomains of p300 bind only weakly to the TAD1 region but with high affinity to a peptide containing the TAD1 and TAD2 region (4). Mutations in either TAD1 (L22Q/W23S = QS1) or TAD2 (W53Q/F54S = QS2) reduce transcriptional activation of p53, whereas the quadruple mutations QS1/QS2 completely abolish transcription (5).
Apart from its interaction with the basal transcription machinery, p53 interacts with other transcriptional cofactors such as JMY, Zac1, CBP/p300, and HMGB-1 (6–10). Recently, p53 was found also to interact with positive cofactor 4 (PC4), and this interaction activates it for specific DNA binding, enhancing its transcriptional activity (10, 11). Post-translational modifications of PC4 affect the DNA binding function of p53. Acetylation of PC4 enhances p53 DNA binding, and phosphorylation of PC4 abolishes this activity (11).
PC4 was initially identified as a transcriptional coactivator that mediates activator-dependent transcription of class II genes through interactions with the basal transcription machinery (12). It plays a vital role in several processes such as replication, transcription, DNA repair, and cell growth (12, 13). PC4 is composed of two domains with distinct functional properties, the N-terminal half of PC4-(1–60) and the DNA-binding C-terminal half of PC4 (PC4CTD-(61–126)). The human PC4CTD binds tightly to melted double-stranded DNA and ssDNA (14). Besides binding to DNA, PC4CTD is essential for interaction with several transcriptional activation domains, such as AP-2α and VP16 (15, 16). The binding of the acidic domain of VP16 to PC4CTD has been investigated in detail, and a structural model of the complex has been recently proposed (16, 17).
The acidic domains of VP16 and p53 share some degree of sequence similarity and are known to interact with common partners such as TATA-binding protein, the CREB-binding protein, the general transcription factor IIB (TFIIB), TATA-binding protein-associated factor TAFII31, and the p62/Tfb1 (human and yeast) subunit of the general transcription factor IIH (TFIIH) (18). Both VP16 and p53 acidic domains are intrinsically disordered under physiological conditions, but an α-helix is induced upon binding to target proteins.
Here we showed that the acidic transactivation domain of p53 interacts with the C-terminal domain of PC4 (PC4CTD). Using a combination of biophysical techniques, we identified the relevant binding sites in both p53 and PC4. To gain insight into the molecular basis of p53-PC4 interaction, we have modeled the p53TAD2-PC4CTD complex using a data-driven protein docking approach. Our model showed a good agreement with the results from mutational analysis and revealed that the p53 peptide acts as ssDNA mimic. We also discuss the general role of acidic transactivation domains as ssDNA mimics.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
cDNA of PC4 full-length and PC4CTD-(61–127) was cloned into a modified PET 24A vector encoding an N-terminal His6 tag, lipoyl domain, and a TeV cleavage site. Both PC4 and PC4CTD constructs were overexpressed in Escherichia coli strain BL21 and purified by Ni2+-affinity column followed by TeV cleavage. Subsequent purification by SP-Sepharose and gel filtration on Superdex 75 yielded a purity of >99%. Full-length p53 and TAD-(1–93) were expressed and purified as described (19).
NMR Experiments
TAD-(1–93) was previously assigned (4). 15N,1H HSQC spectra of free or bound TAD and PC4CTD were acquired on a Bruker (Karlsruhe DRX) 600-MHz spectrometer equipped with a triple-resonance single-axis gradient probe, at 293 K. Resonance assignments for PC4CTD were taken from the published data (20).
Fluorescence Anisotropy Experiments
TAD was labeled with Alexa Fluor 546 at a cysteine introduced at position 91. Fluorescence anisotropy titrations were carried out at 293 K as described (21). The titration buffer was 20 mm HEPES, 150 mm NaCl, 1 mm dithiothreitol, pH 7.2.
Isothermal Titration Calorimetry
ITC measurements were performed as described (19). Injection steps were 10 μl (first injection, 3 μl) with 250-s spacing. Data evaluation was done using the MicroCal OriginTM program.
Peptide Synthesis and Purification
Peptides were synthesized and purified as described (4). Phosphorylated serines were used during synthesis. All peptides were C-terminally labeled with Lys-methoxycoumarin.
Modeling
Structures of two distinct fragments of the p53 N-transactivation domain, comprising residues 33–56 and residues 45–58, have been solved in a complex with replication protein A and Tfb1, respectively (18, 22). In both complexes, the bound p53 transactivation domain forms an amphipathic α-helix. Residues 48–56 of both p53TAD fragments have a virtually identical conformation and superpose with a root mean square deviation of 0.6 Å. This region exhibits the most significant chemical shift changes upon binding to PC4CTD and therefore was used for modeling. Docking was performed using the crystal structure of PC4CTD (23). For less ambiguity, only one of the DNA-binding interfaces of the dimeric PC4CTD was used.
Initial docking, constrained by surface complementarity and electrostatics, was performed using FTDOCK (24). Each possible complex was scored by using the RPSCORE program and an empirical pair potential matrix derived from nonhomologous interfaces observed in the Protein Data Bank. The high scoring docking models were analyzed. For refined docking the HADDOCK program was employed (25). The ambiguous interaction restraints were chosen on the basis of chemical shift perturbation for p53TAD. Residues that constitute the DNA-binding site of PC4 were selected as restraints. These restraints were used as input for the HADDOCK program. Default parameters were used. 1000 independent rigid-body minimizations were performed leading to 1000 docked complexes. The 200 lowest intermolecular energy solutions were the subject of rigid-body simulated annealing followed by semi-flexible simulated annealing. The obtained structures were further refined in explicit solvent employing simulated annealing in Cartesian space. The resulting 200 complexes were clustered and scored according to their HADDOCK scores. The best scoring cluster contained 25 structures with a root mean square deviation from the lowest energy structure of 0.9 ± 0.6 Å.
RESULTS
p53TAD2 Interacts with Positive Cofactor PC4
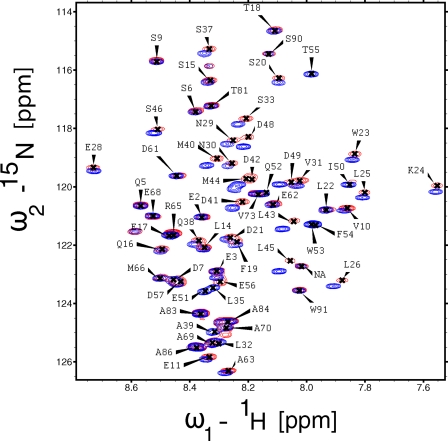
To determine the binding sites on the p53 transactivation domain, we recorded 15N,1H HSQC spectra of p53TAD-(1–93) upon addition of PC4 (Fig. 1). Chemical shift changes were observed mainly for residues 35–57 of the TAD2 domain. In particular, residues 40–45 showed large chemical shift changes, whereas resonances corresponding to residues 50–55 disappeared in the complex. Similar chemical shift changes are observed when p53TAD binds to the TAZ2 domain of p300, human mitochondrial single-stranded DNA-binding protein, and replication protein A, suggesting that these residues can mediate interactions with different targets (4, 26, 27). A few residues (Leu-22 and Trp-23) in the TAD1 domain also showed large chemical shift perturbations. To identify which region of PC4 interacts with p53TAD, we recorded 15N,1H HSQC spectra of TAD with full-length PC4 (PC4fl) and PC4 C-terminal domain (60–127 PC4CTD). The spectra of PC4fl-TAD and PC4CTD-TAD complex overlapped very well indicating that the C-terminal domain of PC4 interacts with p53TAD.
FIGURE 1.
Binding of TAD to PC4 analyzed by NMR spectroscopy. 15N,1H HSQC spectrum of TAD-(1–93) alone (blue) and with the 15N,1H HSQC spectrum of TAD-PC4CTD complex superimposed (red). The complex consists of 15N-labeled TAD and unlabeled PC4CTD.
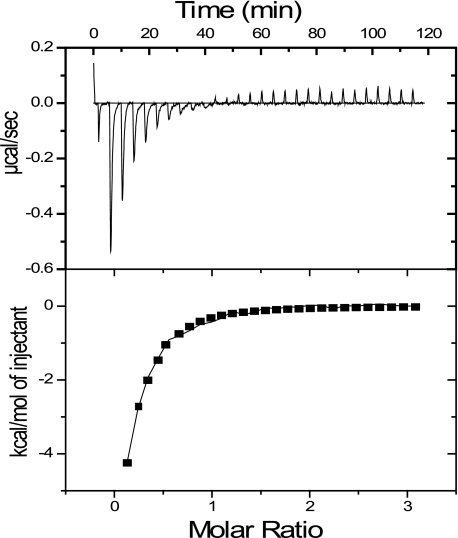
ITC experiments showed that full-length p53 binds PC4 with Kd = 6 μm (Fig. 2). Previous studies, using GST pulldown assays, report that the DNA-binding and C-terminal domains of p53 are involved in interaction with PC4CTD (10). ITC and NMR experiments showed no specific interactions with these domains.
FIGURE 2.
Typical ITC measurements showing the raw data (upper panels) and the fit after integration (lower panels). PC4fl was titrated into the cell containing 20 μm p53fl at 20 °C in a buffer containing 20 mm HEPES, pH 7.2, 150 mm NaCl, 1 mm dithiothreitol.
Binding of p53TAD and ssDNA to PC4 Is Mutually Exclusive
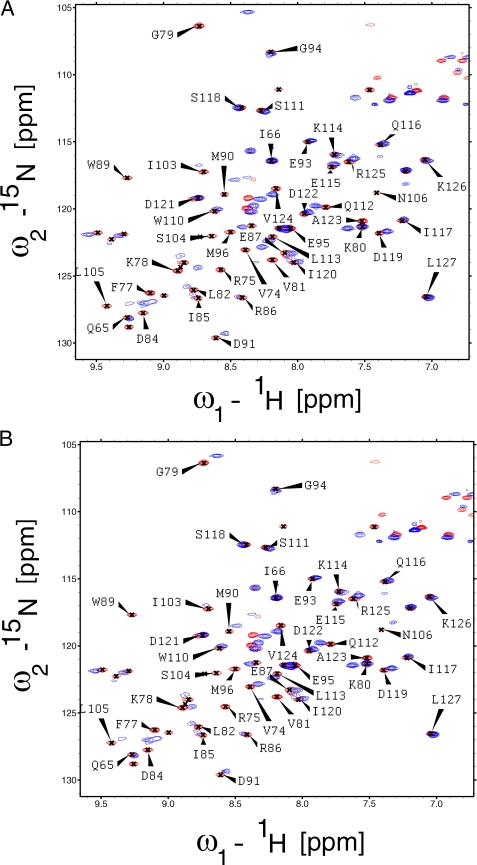
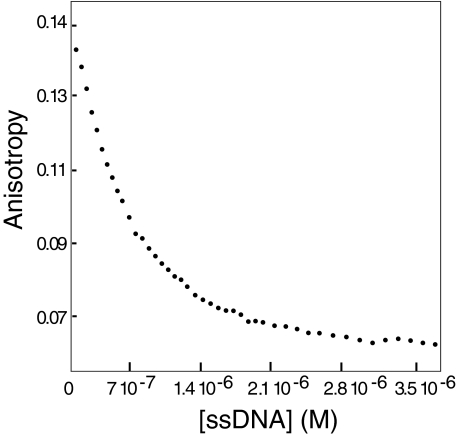
Having identified PC4CTD as the interacting domain, we tested whether the p53TAD and DNA compete for a common binding site on PC4. We recorded 15N,1H HSQC spectra of PC4CTD in the presence and absence of ssDNA (Fig. 3A). Resonance assignments of the PC4CTD and PC4CTD-DNA complex were reported previously (20), and most of the resonances in our spectra could be assigned unambiguously with the exception of a few peaks. Gly-79 and Asn-105 had significant changes in chemical shift upon ssDNA binding consistent with the previous observation. We also recorded 15N,1H HSQC spectra of PC4CTD in the presence of TAD (Fig. 3B). Most of the resonances that showed significant shifts in the PC4CTD-DNA complex were also shifted in the PC4CTD-p53N complex. Gly-79, Asn-105, and Leu-106 showed significant changes in chemical shifts. Resonances for Phe-64, Val-72, and Ser-73 could not be assigned unambiguously in the free PC4CTD. Two of these residues exhibited significant chemical shift change upon binding to TAD and were also perturbed in the PC4CTD-DNA complex. These results indicate that TAD and ssDNA may share the same binding interface on PC4CTD. To confirm this observation, we conducted competition anisotropy titration. The binding constant for PC4 to ssDNA was found to be 5 nm under experimental conditions. ssDNA was titrated into the TAD-PC4CTD complex, with p53N labeled with Alexa Fluor 546 at a cysteine introduced at position 91. As the ssDNA binds PC4CTD, labeled TAD is released into solution (Fig. 4). This experiment clearly showed that ssDNA and TAD compete for the same binding site on PC4CTD.
FIGURE 3.
Binding of PC4CTD to ssDNA and TAD analyzed by NMR spectroscopy. A, 15N,1H HSQC spectrum of PC4CTD alone (red) and with 15N,1H HSQC spectrum of ssDNA-PC4CTD complex superimposed (blue). The complex consists of ssDNA and 15N-labeled PC4CTD. B, 15N,1H HSQC spectrum of PC4CTD alone (red) with 15N,1H HSQC spectrum of TAD-PC4CTD complex superimposed (blue). The complex consists of unlabeled p53N and 15N-labeled PC4CTD.
FIGURE 4.
Fluorescence anisotropy competition assay. Unlabeled ssDNA was titrated into a solution containing Alexa Fluor 546-labeled TAD and PC4CTD.
Phosphorylation of p53N Slightly Increased the Affinity for PC4
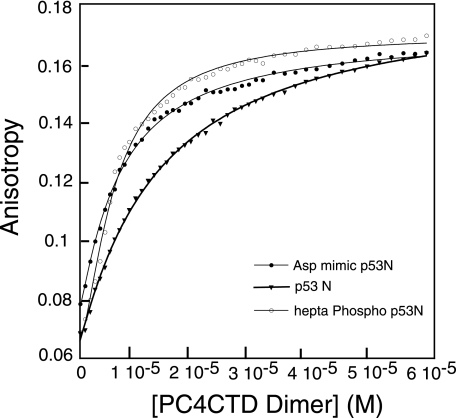
Upon stress, p53 is phosphorylated at multiple sites, and these phosphorylations are important for maintaining its cellular stability or for mediating interactions with other target proteins. Mutations of the phosphorylation sites have deleterious effects on the function of p53 (28). We recently showed that hepta-phosphorylated TAD-(10–57) had enhanced affinity for the CH3 domain of p300, while simultaneously decreasing MDM2 binding (29). To test the effect of phosphorylation on the binding of p53TAD to PC4, we analyzed the binding of peptides singly phosphorylated at Ser-33, Thr-55, Ser-37, or Ser-46. All the peptides had dissociation constants of 5.8–6.9 μm. We also synthesized a peptide comprising residues 10–57 phosphorylated at Ser-15, Thr-18, Ser-20, Ser-33, Ser-37, Ser-46, and Thr-55. All these sites are phosphorylated in the active form of p53. This hepta-phosphorylated peptide (hPp53N) bound with a dissociation constant of 3 μm as compared with 8 μm for wild type (Fig. 5). This increase in affinity by about a factor of 3 is probably because of the nonspecific electrostatic interactions that are primarily dictated by the negative charges, and hence an increase in the total net charge leads to tighter binding. An aspartic acid mutant (hepta-mutant) of the peptide was synthesized, and it had a Kd of 5 μm, further supporting the fact that these interactions are electrostatically driven.
FIGURE 5.
Fluorescence binding isotherms. PC4CTD was titrated into wild-type peptide, hepta-phosphorylated peptide, and with an aspartic acid mutant peptide (residues 10–57). All the peptides were C-terminally labeled with Lys-methoxycoumarin. The experiments were carried out at 20 °C in 20 mm HEPES, 150 mm NaCl, 1 mm dithiothreitol, pH 7.2.
Modeling the p53TAD-PC4CTD Complex
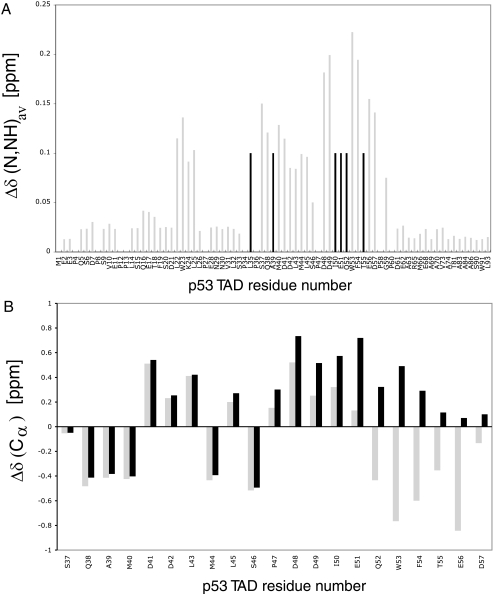
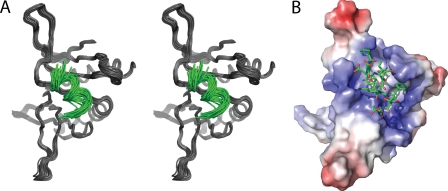
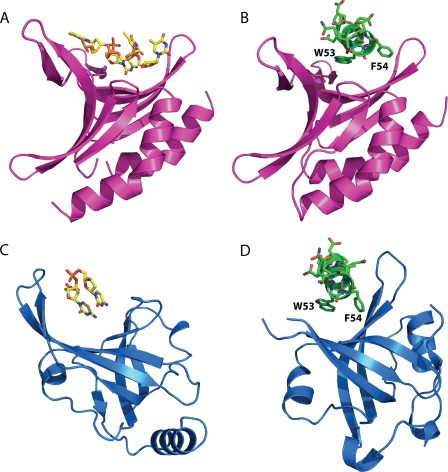
Because of experimental limitations such as line broadening and disappearance of peaks, we were unable to solve the solution structure of the p53N-PC4 complex. Nevertheless, the chemical shift mapping data were sufficient as constraints in molecular docking. NMR titration experiments showed that the amide 1H and 15N shift toward lower parts/million values, whereas positive resonance shifts were observed for 13Cα indicating formation of an α-helix in otherwise intrinsically unstructured p53TAD2 (Fig. 6) (30). Thus, p53TAD2 adopts mainly α-helical conformation when bound to PC4CTD, which is in agreement with the observed conformation of this region in complex with replication protein A (RPA) and Tfb1 (18, 22). The most significant chemical shift perturbations upon p53TAD-PC4CTD complex formation involved residues 48–56 of p53TAD2 indicating these residues as key determinants for binding (Fig. 6). This region has identical conformations in both the p53TAD-RPA and the p53TAD-Tfb1 complexes and therefore was used in our docking experiments. Two different docking approaches were applied. Preliminary docking was performed using FTDOCK without using constraints (24). Most of the high scoring complexes contained the p53 peptide docked to the DNA-binding site of PC4 suggesting that there is sufficient geometric and electrostatic complementarity between them. A refined model of the p53TAD-PC4 complex was obtained using the HADDOCK program (25). Ambiguous interaction restraints for the p53TAD were defined on the basis of the chemical shift perturbations. In addition, the residues that constitute the DNA-binding interface of PC4 were used as restraints. The ensemble of the 25 energetically most favorable structures yielded 0.9 ± 0.6 Å root mean square deviation. The model of the complex is shown in Fig. 7. The peptide is positioned along the concave β surface of PC4. The electrostatic surface of the PC4 DNA-binding site provides a suitable complementary environment for the acidic amino acids of p53TAD. Asp-49 and Glu-51 of p53 may participate in a salt bridge with Arg-100 and Lys-78, respectively. Lys-101 of PC4 interacts with the backbone carbonyl group of Glu-56. Residues Phe-54 and Ile-50 of the TAD make hydrophobic contact with the highly conserved Phe-77 of PC4. Phe-77 is involved in a stacking interaction with ssDNA, and mutation of this residue abolishes the DNA binding (31). Notably, the mode of interaction between the p53 peptide and PC4 in the modeled complex is similar to the ssDNA binding to PC4 (23). Phe-54 and Ile-50 are reminiscent of the DNA bases, whereas the backbone and side-chain carbonyl groups of p53TAD2 mimic the phosphate backbone of DNA (Fig. 8). In addition, Trp-53 is located near Arg-86. In the PC4-ssDNA complex, the latter binds simultaneously two water molecules and thereby positions them for hydrogen bond donation to the ssDNA phosphate group. In our modeled complex, this residue may form an essential cation-π interaction with Trp-53. Cation-π interactions are frequently found in protein-protein interfaces, and they contribute to the stability of the complex as much as any conventional interaction such as salt bridges or hydrogen bonds (32).
FIGURE 6.
Weighted average chemical shift changes of p53TAD-(1–93) and secondary 13Cαchemical shift changes of p53TAD-(37–57). A, bars showing weighted average chemical shift changes Δδ (N,HN)av, calculated from the chemical shifts of amide 1H and 15N between the free and bound states for p53TAD amide resonances by binding to PC4CTD (1:0.4 molar ratio of labeled TAD and PC4CTD was used). Resonances that disappeared in the complex are shown as black bars and are assigned an arbitrary value of 0.10 ppm. In a 1:0.6 (molar ratio) complex, we observed significant broadening and disappearance of many peaks due to exchange between free and the complex p53TAD-PC4CTD. In a 1:1 complex, most of the residues that showed significant chemical shift changes have disappeared. B, 13Cα shifts calculated by subtracting standard random coil values from the experimental 13Cα chemical shifts for free p53TAD-(1–93) and p53TAD-PC4CTD complex are shown in gray and black bars, respectively (50). A positive Δδ value of 13Cα for p53TAD-(47–55) is an indication of α-helical preference. Here 1:0.3 molar ratio of labeled p53TAD and unlabeled PC4CTD was used. Disappearance of peaks was observed at higher molar ratios. Residues 36–57 are shown for clarity. Similar effects are reported in the literature when KIX, a regulatory domain of p300, and human replication protein A (hRPA70) binds p53TAD (27, 51).
FIGURE 7.
Docking model of PC4-p53TAD2 complex. A, superimposition of the 25 lowest energy models representing the docked complex of PC4CTD (black) and TAD2 peptide (green). B, surface and charge complementarity revealed by the model, PC4CTD shown on the surface and TAD peptide as sticks.
FIGURE 8.
ssDNA and p53TAD2 peptide binding by RPA and PC4. A, structure of PC4 bound to ssDNA (23). B, docking model of PC4-p53TAD2 complex. C, structure of RPA bound to ssDNA (52). D, structure of RPA bound to p53TAD2 peptide (22).
Mutational Studies Show That Hydrophobic and Negatively Charged Residues Are Important for the Interaction
We tested our docking model by examining the effect of mutations on the binding. Based on the interactions observed in the modeled p53-PC4 complex, we synthesized two mutant peptides I50A/W53A/F54A p53N-(35–57) and D49K/E51K p53N-(35–57). Both peptides showed very weak binding of 42 and 22 μm, respectively. QS2 (W53Q/F54S) mutations have reduced transcriptional activation of apoptosis and cell cycle arresting gene (5). We tested the binding of QS2 mutant peptide W53Q/F54S p53N-(10–57) to PC4 and found it had a 2-fold decrease in affinity. In addition to the residues identified in the modeled complex, several other hydrophobic and acidic residues had some changes in chemical shift upon PC4 binding.
To investigate the role of these residues and their contribution to the binding, we made peptides bearing the following mutations: L43A/L45A/I50A/W53A/F54A p53N-(35–57) and D41K/D42K/D48K/D49K/E51K p53N-(35–57). Both of them had weak binding, with the latter showing no quantifiable binding. Table 1 lists the binding constants of the mutant peptides tested for binding to PC4CTD. Although mutation of single hydrophobic residues to alanine had a minor effect on the binding constant (KD 14–16 μm) (data not shown), there was a larger change upon multiple mutations (KD 45 μm) indicating the additive nature of hydrophobic residues in forming a stable complex. p53TAD is intrinsically unstructured in its free state (33), and hence mutations introduced had no effect on its secondary structure as evidenced by circular dichroism measurements (data not shown).
TABLE 1.
Dissociation constants obtained for the p53TAD peptides with PC4CTD
| Peptides | Kd |
|---|---|
| μm | |
| p53N-(10–57)-wild type | 8.2 ± 2 |
| hp53N-(10–57)-heptaphosphorylated | 3 ± 1 |
| hDp53N-(10–57)-aspartic acid mimic | 5 ± 1 |
| P53N-(35–57)-wild type | 12 ± 2 |
| Q52L p53N-(35–57) | 16 ± 1 |
| D49K/E51K p53N-(35–57) | 22 ± 3 |
| QS2 W53Q/F54S p53N-(35–57) | 20 ± 3 |
| I50A/W53A/F54A p53N-(35–57) | 42.5 ± 2 |
| L43A/L45A/I50A/W53A/F54A p53N-(35–57) | 45 ± 3 |
| D41K/D42K/D48K/D49K/E51K p53N-(35–57) | Not quantifiable |
We also investigated the role of residues Phe-77, Lys-78, and Arg-86 of PC4 that are likely involved in interactions with TAD. We constructed a quadruple mutant (F77A/K78G/K80G/R86A), in which Phe-77 and Arg-86 were replaced with Ala and Lys-78 and Lys-80 were mutated to Gly. The fluorescence anisotropy experiments showed that the quadruple mutant does not bind TAD. In addition, no binding to ssDNA was observed, which is in agreement with earlier work where F77A/K78G/K80G mutations of PC4 abolished DNA binding (31).
DISCUSSION
We investigated the interaction between tumor suppressor p53 and positive cofactor 4 and showed that TAD2 of p53 mediates interaction with the C-terminal DNA-binding domain of PC4. We found that binding of TAD2 and ssDNA to PC4 were mutually exclusive, suggesting that p53 and ssDNA share a common binding site. Modeling of the p53N-PC4 complex revealed a wide range of stabilizing interactions such as hydrophobic, cation-π, and salt bridges. The binding of the p53 peptide to PC4 in the docking model revealed similarity to the ssDNA binding to PC4.
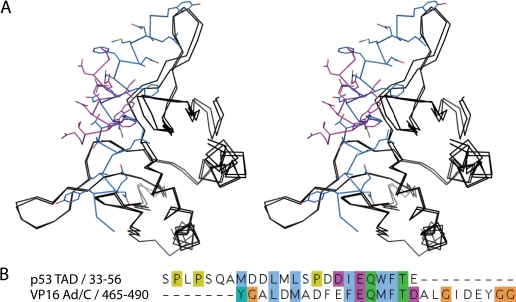
Our docking model was validated with the experimental results of mutational analysis. Interestingly, the p53TAD2 binds PC4 in a similar orientation as VP16ad in the VP16-PC4 model complex (Fig. 9A) (16). Sequence alignment of the activation domains of p53 and VP16 shows considerable degree of similarity (Fig. 9B). Residues of p53 that contribute to the interaction with PC4 are nearly invariant in VP16 and occupy similar spatial positions in both p53/PC4 and VP16/PC4 models.
FIGURE 9.
Superposition of the p53TAD2-PC4 and VP16-PC4 complexes. A, PC4CTD, p53TAD2, and VP16 are shown in black, magenta, and blue, respectively. B, pairwise sequence alignment of p53TAD-(33–56) and VP16Ad/C-(465–490).
Functional Implications of the p53-PC4 Interaction
Tumor suppressor p53 regulates the transcription of numerous response elements, which, in turn, modulates the cell cycle or apoptotic pathways during genotoxic stress (1). PC4 facilitates activator-dependent transcription by RNA polymerase II through mediating interactions with the basal transcriptional machinery (12). PC4 can stimulate the sequence-specific DNA binding of p53 and the C-terminal deletion mutant p53Δ30 (10). PC4 also stimulates the recruitment of p53 and p53Δ30 to p53-response elements such as Bax and p21 in vivo through its interaction with p53 and enhances the apoptosis (11). Using GST pulldown experiments, Banerjee et al. (10) showed that the C terminus of p53 interacts with PC4, which does not clearly explain the observed in vivo results (in the case of p53Δ30). But a false-positive rate of 61% and false-negative rate of 38% are reported for the GST pulldown approach (34). On the other hand, NMR is the gold standard for looking at weak and strong protein-protein interactions in solution at the atomic level (35, 36). However, the in vivo observations reported are reliable and are important and relevant for our present findings. Hence, based on our results and the published results of Banerjee et al. (10), we speculate a possible mechanism in which PC4 can recruit p53 to the response element site by interacting with its TAD. Once the site is reached, PC4 releases p53TAD and binds to DNA. PC4 is a DNA-bending protein and as such it probably provides a bent DNA conformation of the p53 cognate site and thereby enhances the ability of p53 to bind specifically to DNA. Under normal conditions, p53 levels in the cell are low, but upon stress, the levels of p53 rise, and cofactors like PC4, which are highly abundant, may enhance the p53-induced transcription by facilitating the transcriptional complex formation, and this would in turn lead to the increased level of p53-responsive target elements. Alternatively, p53 can exert an antagonizing role in inhibiting PC4-mediated repression. PC4 is shown to repress transcription by binding to melted DNA, and this effect is alleviated in the presence of TFIIH (31). Hence, the functional role of PC4-p53 interaction may be context-dependent and needs further investigation.
General Role of Activation Domains in Target Binding
Acidic activators have been proposed to interact with target proteins through electrostatic interactions by their intrinsically unstructured regions (37). Secondary structure formation upon target binding is observed in these activation domains (38). The isolated TAD is intrinsically disordered, a property that is common in many other activation domains (33). An α-helix is induced in the TAD upon binding to various target proteins like MDM2, TAZ2, RPA, and PC4 (22, 39). Similar secondary structure formation is observed in the VP16-bound complex (16, 40). Initially, it was suggested that acidic domains mediate interactions with the target proteins through ionic interactions (41). However, further studies showed that hydrophobic amino acids are critical for protein-protein interactions between activators and general transcriptional machinery. Mutations of these residues have a deleterious effect on transcriptional activation (42–44). Based on these findings, a target-induced folding model has been proposed, where the electrostatic interactions determine the kinetic stability of the complex formation, whereas the hydrophobic interactions establish the thermodynamic binding affinity of the interaction (37, 38). Our mutational analyses suggest that the p53TAD2-PC4 interaction is a composite of hydrophobic and electrostatic interactions. They probably work synergistically in target protein recognition and binding. The hydrophobic interactions may play an important role in determining the stability of complex formation, whereas electrostatic interactions facilitate the formation of complex. Phosphorylation increases the net negative charge, which can explain its positive effect on the p53/PC4 binding.
Biological Significance of DNA Mimicry of p53 Transactivation Domain
The molecular basis of protein mimics of DNA has been extensively reviewed over the past years (45). The effective DNA mimic usually represents a surface that is complementary in shape and similar in chemical composition (charge) to DNA. Although most of the cases described in the literature concern protein mimics of double-stranded DNA, only few examples of single-stranded DNA mimicry are known. Among those is the crystal structure of the p53TAD-RPA complex that provides direct evidence for ssDNA mimicry by the p53 transactivation domain (22). Recently, we showed that p53TAD interacts with mitochondrial ssDNA (mtSSB)-binding protein, and this interaction enhances the 3′–5′-exonuclease activity of p53 and DNA repair in mitochondria during oxidative stress. We also demonstrated that the p53TAD binds to the DNA binding surface of mtSSB and perturbs the interaction with ssDNA (26). In this study we showed that p53TAD binds to PC4 and its binding site resides in the DNA-binding interface. Taken together, the p53TAD competes with ssDNA for binding to three structurally distinct DNA-binding domains (PC4, mtSSB, and RPA). Although RPA and mtSSB share a common OB fold, PC4CTD forms an intertwined dimer with a globally different structure. The only common feature between these protein domains is the DNA-binding interface, which in all cases is a β-sheet with a characteristic curvature. Despite the structural differences between these p53TAD interaction partners, the p53 peptide binds to them in a very similar way. In both p53TAD/RPA structure and our p53TAD/PC4 model, p53TAD2 is positioned along the concave β-sheet interface with the hydrophobic residues pointing toward the interface and acidic residues to the surface (Fig. 7). Thus the shape and charge of the binding interface rather than the fold of the target protein determine the selective p53TAD2 binding. p53TAD2 could potentially interact with other structurally distinct DNA-binding domains that share these features. DNA mimicry could be a general functional feature of this region of p53.
p53 contains a DNA-binding domain that binds to double-stranded DNA (46). Because the p53 DNA-binding interface is very different from that of PC4, mtSSB, and RPA, we tested whether p53TAD binds to this interface. Our experiments indicated that the p53TAD binds with a weak affinity of >100 μm to the core domain (data not shown). Competition experiments revealed that p53TAD did bind to the DNA-binding surface of p53. But the interaction is completely electrostatic because at higher ionic strengths no binding was observed.
The DNA mimicry role of p53TAD can play an important role in the transcriptional machinery. PC4 was identified as a DNA-binding protein that enhances the activator-dependent transcription of class II genes in vitro by stimulating diverse activators like VP16, thyroid hormone receptor, octamer transcription factor 1, and BRCA-1 by facilitating assembly of the preinitiation complex between the activators and the general transcriptional machinery (12, 47–49). PC4 may be involved in the recruitment of active phosphorylated p53 to the basal transcriptional machinery through its association with the transactivation domain of p53. Apart from the regulation of basal transcription function and DNA repair, other functional DNA mimicry roles of p53TAD need to be investigated. Moreover, many acidic activation domains may share similar tendencies in modulating their functions at the cellular level through the DNA mimicry role.
Acknowledgments
We thank Drs. C. M. Johnson, T. S. Wong, and A. Joerger for invaluable help and useful discussions.
Footnotes
- TAD
- transactivation domain
- ssDNA
- single-stranded DNA
- CTD
- C-terminal regulatory domain
- ITC
- isothermal titration calorimetry
- GST
- glutathione S-transferase
- RPA
- replication protein A
- msSSB
- mitochondrial ssDNA (mtSSB)-binding protein.
REFERENCES
- 1.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2.Joerger A. C., Fersht A. R. (2008) Annu. Rev. Biochem. 77, 557–582 [DOI] [PubMed] [Google Scholar]
- 3.Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 4.Teufel D. P., Freund S. M., Bycroft M., Fersht A. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7009–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venot C., Maratrat M., Sierra V., Conseiller E., Debussche L. (1999) Oncogene 18, 2405–2410 [DOI] [PubMed] [Google Scholar]
- 6.Avantaggiati M. L., Ogryzko V., Gardner K., Giordano A., Levine A. S., Kelly K. (1997) Cell 89, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 7.Huang S. M., Schönthal A. H., Stallcup M. R. (2001) Oncogene 20, 2134–2143 [DOI] [PubMed] [Google Scholar]
- 8.Shikama N., Lee C. W., France S., Delavaine L., Lyon J., Krstic-Demonacos M., La Thangue N. B. (1999) Mol. Cell 4, 365–376 [DOI] [PubMed] [Google Scholar]
- 9.Jayaraman L., Moorthy N. C., Murthy K. G., Manley J. L., Bustin M., Prives C. (1998) Genes Dev. 12, 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee S., Kumar B. R., Kundu T. K. (2004) Mol. Cell. Biol. 24, 2052–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batta K., Kundu T. K. (2007) Mol. Cell. Biol. 27, 7603–7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge H., Roeder R. G. (1994) Cell 78, 513–523 [DOI] [PubMed] [Google Scholar]
- 13.Pan Z. Q., Ge H., Amin A. A., Hurwitz J. (1996) J. Biol. Chem. 271, 22111–22116 [DOI] [PubMed] [Google Scholar]
- 14.Werten S., Langen F. W., van Schaik R., Timmers H. T., Meisterernst M., van der Vliet P. C. (1998) J. Mol. Biol. 276, 367–377 [DOI] [PubMed] [Google Scholar]
- 15.Zhong L., Wang Y., Kannan P., Tainsky M. A. (2003) Gene 320, 155–164 [DOI] [PubMed] [Google Scholar]
- 16.Jonker H. R., Wechselberger R. W., Boelens R., Folkers G. E., Kaptein R. (2005) Biochemistry 44, 827–839 [DOI] [PubMed] [Google Scholar]
- 17.Jonker H. R., Wechselberger R. W., Boelens R., Kaptein R., Folkers G. E. (2006) Biochemistry 45, 5067–5081 [DOI] [PubMed] [Google Scholar]
- 18.Langlois C., Mas C., Di Lello P., Jenkins L. M., Legault P., Omichinski J. G. (2008) J. Am. Chem. Soc. 130, 10596–10604 [DOI] [PubMed] [Google Scholar]
- 19.Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werten S., Wechselberger R., Boelens R., van der Vliet P. C., Kaptein R. (1999) J. Biol. Chem. 274, 3693–3699 [DOI] [PubMed] [Google Scholar]
- 21.Rajagopalan S., Jaulent A. M., Wells M., Veprintsev D. B., Fersht A. R. (2008) Nucleic Acids Res. 36, 5983–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochkareva E., Kaustov L., Ayed A., Yi G. S., Lu Y., Pineda-Lucena A., Liao J. C., Okorokov A. L., Milner J., Arrowsmith C. H., Bochkarev A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werten S., Moras D. (2006) Nat. Struct. Mol. Biol. 13, 181–182 [DOI] [PubMed] [Google Scholar]
- 24.Gabb H. A., Jackson R. M., Sternberg M. J. (1997) J. Mol. Biol. 272, 106–120 [DOI] [PubMed] [Google Scholar]
- 25.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 26.Wong T. S., Rajagopalan S., Townsley F. M., Freund S. M., Petrovich M., Loakes D., Fersht A. R. (2009) Nucleic Acids Res. 37, 568–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vise P. D., Baral B., Latos A. J., Daughdrill G. W. (2005) Nucleic Acids Res. 33, 2061–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 29.Teufel D. P., Bycroft M., Fersht A. R. (2009) Oncogene 28, 2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart D. S., Sykes B. D. (1994) J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 31.Werten S., Stelzer G., Goppelt A., Langen F. M., Gros P., Timmers H. T., van der Vliet P. C., Meisterernst M. (1998) EMBO J. 17, 5103–5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallivan J. P., Dougherty D. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9459–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., Fersht A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comuzzi B., Sadar M. D. (2006) Cellscience 3, 61–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonvin A. M., Boelens R., Kaptein R. (2005) Curr. Opin. Chem. Biol. 9, 501–508 [DOI] [PubMed] [Google Scholar]
- 36.Zuiderweg E. R. (2002) Biochemistry 41, 1–7 [DOI] [PubMed] [Google Scholar]
- 37.Hermann S., Berndt K. D., Wright A. P. (2001) J. Biol. Chem. 276, 40127–40132 [DOI] [PubMed] [Google Scholar]
- 38.Ferreira M. E., Hermann S., Prochasson P., Workman J. L., Berndt K. D., Wright A. P. (2005) J. Biol. Chem. 280, 21779–21784 [DOI] [PubMed] [Google Scholar]
- 39.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 40.Uesugi M., Nyanguile O., Lu H., Levine A. J., Verdine G. L. (1997) Science 277, 1310–1313 [DOI] [PubMed] [Google Scholar]
- 41.Ptashne M., Gann A. A. (1990) Nature 346, 329–331 [DOI] [PubMed] [Google Scholar]
- 42.Sainz M. B., Goff S. A., Chandler V. L. (1997) Mol. Cell. Biol. 17, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regier J. L., Shen F., Triezenberg S. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cress W. D., Triezenberg S. J. (1991) Science 251, 87–90 [DOI] [PubMed] [Google Scholar]
- 45.Dryden D. T., Tock M. R. (2006) Biochem. Soc. Trans. 34, 317–319 [DOI] [PubMed] [Google Scholar]
- 46.Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T. E., Shakked Z. (2006) Mol. Cell 22, 741–753 [DOI] [PubMed] [Google Scholar]
- 47.Luo Y., Ge H., Stevens S., Xiao H., Roeder R. G. (1998) Mol. Cell. Biol. 18, 3803–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haile D. T., Parvin J. D. (1999) J. Biol. Chem. 274, 2113–2117 [DOI] [PubMed] [Google Scholar]
- 49.Malik S., Guermah M., Roeder R. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 51.Lee C. W., Arai M., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2009) Biochemistry, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bochkarev A., Pfuetzner R. A., Edwards A. M., Frappier L. (1997) Nature 385, 176–181 [DOI] [PubMed] [Google Scholar]