Abstract
Early onset generalized dystonia (DYT1) is an autosomal dominant neurological disorder caused by deletion of a single glutamate residue (torsinA ΔE) in the C-terminal region of the AAA+ (ATPases associated with a variety of cellular activities) protein torsinA. The pathogenic mechanism by which torsinA ΔE mutation leads to dystonia remains unknown. Here we report the identification and characterization of a 628-amino acid novel protein, printor, that interacts with torsinA. Printor co-distributes with torsinA in multiple brain regions and co-localizes with torsinA in the endoplasmic reticulum. Interestingly, printor selectively binds to the ATP-free form but not to the ATP-bound form of torsinA, supporting a role for printor as a cofactor rather than a substrate of torsinA. The interaction of printor with torsinA is completely abolished by the dystonia-associated torsinA ΔE mutation. Our findings suggest that printor is a new component of the DYT1 pathogenic pathway and provide a potential molecular target for therapeutic intervention in dystonia.
Early onset generalized torsion dystonia (DYT1) is the most common and severe form of hereditary dystonia, a movement disorder characterized by involuntary movements and sustained muscle spasms (1). This autosomal dominant disease has childhood onset and its dystonic symptoms are thought to result from neuronal dysfunction rather than neurodegeneration (2, 3). Most DYT1 cases are caused by deletion of a single glutamate residue at positions 302 or 303 (torsinA ΔE) of the 332-amino acid protein torsinA (4). In addition, a different torsinA mutation that deletes amino acids Phe323–Tyr328 (torsinA Δ323–328) was identified in a single family with dystonia (5), although the pathogenic significance of this torsinA mutation is unclear because these patients contain a concomitant mutation in another dystonia-related protein, ϵ-sarcoglycan (6). Recently, genetic association studies have implicated polymorphisms in the torsinA gene as a genetic risk factor in the development of adult-onset idiopathic dystonia (7, 8).
TorsinA contains an N-terminal endoplasmic reticulum (ER)3 signal sequence and a 20-amino acid hydrophobic region followed by a conserved AAA+ (ATPases associated with a variety of cellular activities) domain (9, 10). Because members of the AAA+ family are known to facilitate conformational changes in target proteins (11, 12), it has been proposed that torsinA may function as a molecular chaperone (13, 14). TorsinA is widely expressed in brain and multiple other tissues (15) and is primarily associated with the ER and nuclear envelope (NE) compartments in cells (16–20). TorsinA is believed to mainly reside in the lumen of the ER and NE (17–19) and has been shown to bind lamina-associated polypeptide 1 (LAP1) (21), lumenal domain-like LAP1 (LULL1) (21), and nesprins (22). In addition, recent evidence indicates that a significant pool of torsinA exhibits a topology in which the AAA+ domain faces the cytoplasm (20). In support of this topology, torsinA is found in the cytoplasm, neuronal processes, and synaptic terminals (2, 3, 15, 23–26) and has been shown to bind cytosolic proteins snapin (27) and kinesin light chain 1 (20). TorsinA has been proposed to play a role in several cellular processes, including dopaminergic neurotransmission (28–31), NE organization and dynamics (17, 22, 32), and protein trafficking (27, 33). However, the precise biological function of torsinA and its regulation remain unknown.
To gain insights into torsinA function, we performed yeast two-hybrid screens to search for torsinA-interacting proteins in the brain. We report here the isolation and characterization of a novel protein named printor (protein interactor of torsinA) that interacts selectively with wild-type (WT) torsinA but not the dystonia-associated torsinA ΔE mutant. Our data suggest that printor may serve as a cofactor of torsinA and provide a new molecular target for understanding and treating dystonia.
EXPERIMENTAL PROCEDURES
Expression Constructs and Antibodies
Full-length human printor cDNA (KIAA1384, GenBankTM accession number AB037805) was obtained from Kazusa DNA Institute, Japan. Conventional molecular biological techniques (34) were used to subclone the printor cDNA into mammalian vectors expressing N-terminal HA, Myc, or FLAG tags for transfection into cells. DNA fragments encoding torsinA WT, WTΔ40, ΔE, Δ323–328, K108A, and E171Q were subcloned into mammalian vectors expressing C-terminal HA, Myc, or FLAG tags. A rabbit polyclonal anti-printor antibody was raised against a synthetic peptide encoding amino acids 1–18 of human printor and affinity purified using the immunogen peptide coupled to a Pierce column as we described previously (35–37). Other antibodies used in this study are as follows: anti-torsinA (16); anti-EEA1 and anti-TIM23 (BD transduction); anti-LAMP2 (H4B4; DSHB, University of Iowa); anti-FLAG (M2; Sigma); anti-calnexin and anti-KDEL (Stressgen); anti-β-actin (Sigma); mouse monoclonal anti-HA (12CA5); and anti-Myc (9E10) (35). Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used for immunoblotting. Fluorescein isothiocyanate (FITC)-, Texas Red-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used for immunofluorescence microscopy.
Yeast Two-hybrid Screen
The bait plasmids were constructed by subcloning the full-length human torsinA (WT) or N-terminal-truncated torsinA (WTΔ40) into the vector pPC97 (35, 37, 38). For yeast two-hybrid screens, the yeast strain CG-1945 (Clontech) was sequentially transformed with the WT or WTΔ40 bait plasmid and a rat hippocampal/cortical two-hybrid cDNA library (35, 36, 39). Positive clones were selected on 3-aminotriazole-containing medium lacking leucine, tryptophan, and histidine, and confirmed by filter assay for β-galactosidase activity. Prey plasmids from positive clones were rescued and re-transformed into fresh yeast cells with the torsinA bait(s) to confirm the interaction.
Immunohistochemistry
Mouse whole brains were fixed in 4% paraformaldehyde and cut coronally into 1- to 2-mm slices. Brain slices were dehydrated in a graded series of alcohols and xylenes, infiltrated with paraffin using an automated tissue processor (Shandon Hypercenter XP), and then embedded in paraffin blocks. Eight-micrometer sections were cut using a Shandon AS325 microtome and subjected to immunohistochemical analysis as described (40). Briefly, sections were deparaffinized, blocked with normal serum, and then incubated with purified anti-printor antibody and biotinylated secondary antibody, followed by detection using an avidin-biotin-peroxidase complex method (ABC Elite Kit; Vector Laboratories, Burlingame, CA). The chromagen used for color development was 3,3′-diaminobenzidine, and sections were counterstained with hematoxylin. Omission of the primary antibody was used as a negative control.
Cell Transfections and Co-immunoprecipitation
Transfections of HeLa and SH-SY5Y cells were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cell lysates were prepared from transfected cells and immunoprecipitations were carried out as described previously (16, 35, 41–43) using the indicated antibodies, followed by the recovery of immunocomplexes with protein G-Sepharose beads (Upstate). For co-immunoprecipitations of endogenous proteins, the Seize Primary Mammalian Immunoprecipitation Kit (Pierce) was used according to the manufacturer's instructions. Anti-printor antibody or pre-immune serum was conjugated to the Amino-Link column (Pierce) and used to purify endogenous protein complexes from mouse cerebellum extracts prepared as described (44). After eluting from column or beads, immunocomplexes were analyzed by SDS-PAGE and immunoblotted with appropriate antibodies and horseradish peroxidase-conjugated secondary antibodies. Results were visualized using enhanced chemiluminescence (ECL).
Subcellular Fractionation
Subcellular fractionation was performed as previously described (36, 44, 45). SH-SY5Y cells were collected by centrifugation and the pellet was homogenized in 1 ml of homogenization buffer (250 mm sucrose, 10 mm HEPES/KOH, pH 7.4, 10 mm KCl, 0.1 mm EGTA, 0.1 mm EDTA) containing protease inhibitors and dithiothreitol. After a 10-min centrifugation at 1,000 × g to remove unbroken cells and nuclear material, the supernatant was subjected to a 30-min centrifugation at 100,000 × g to separate into the cytosolic (supernatant) and membrane (pellet) fractions. Aliquots representing an equal percentage of each fraction were analyzed by SDS-PAGE and immunoblotting. Protein bands on the immunoblots were quantified using NIH Scion Image Software.
Opti-Prep Gradient Fractionation
Opti-Prep gradient fractionation was performed as previously described (46). SH-SY5Y cells were homogenized in 1 ml of fractionation buffer (10 mm HEPES/KOH, pH 7.4, 1 mm EDTA) containing 250 mm sucrose. After a 10-min centrifugation at 1,000 × g, the supernatant was layered on a 10–30% Opti-Prep (Nycomed) gradient formed in the fractionation buffer containing 42 mm sucrose, and centrifuged at 4 °C for 5 h at 100,000 × g. Following centrifugation, the gradient was harvested into 250-μl fractions using an Auto Densi-Flow gradient harvester (Labconco). Equal volumes of each fraction were analyzed by SDS-PAGE and by sequential immunoblotting for torsinA, printor, and the ER marker calnexin. Protein bands on the immunoblots were quantified using NIH Scion Image Software.
Immunofluorescence Confocal Microscopy
Cells were fixed in 4% paraformaldehyde, stained with appropriate primary and secondary antibodies, and processed for indirect immunofluorescence microscopy as described previously (16, 35, 41–43). Analysis and acquisition were performed using a Zeiss LSM 510 confocal laser-scanning microscope. Images were imported in TIFF format using LSM-510 software (Carl Zeiss MicroImaging, Inc.) and processed using Adobe Photoshop CS version 8.0 (Adobe Systems, Inc.) to adjust the contrast and brightness.
Quantitative Analysis of the NE/ER Distribution and Protein Co-localization
Quantification of the NE/ER distribution of printor, torsinA, or KDEL was performed on unprocessed images of cells double labeled for printor and KDEL or torsinA and KDEL, by using the MetaMorph Imaging System Software (Molecular Devices) as previously described (16). Quantification of torsinA-printor co-localization was performed on unprocessed images of cells double labeled for printor and torsinA by using the MetaMorph Imaging System Software as previously described (47). For each torsinA genotype (torsinA WT, ΔE, Δ323–328, K108A, or E171Q), 30–45 cells were randomly selected for analysis. Analysis was carried out in a blinded manner by an investigator without any knowledge of torsinA genotype of the cells and the result was confirmed by a second, independent investigator. Experiments were repeated at least three times, and the data were subjected to statistical analysis by unpaired Student's t test.
RESULTS
Identification of Printor, A Novel TorsinA-interacting Protein
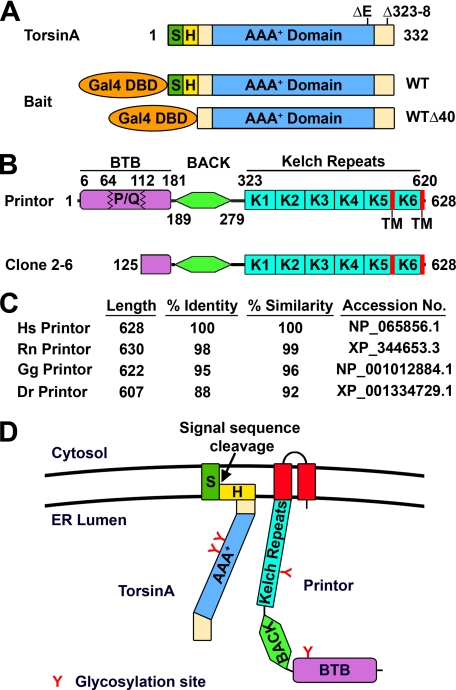
To identify proteins that interact with torsinA in the brain, we first screened a rat hippocampal/cortical cDNA library by yeast two-hybrid selection using the full-length torsinA WT as bait. This screen did not result in isolation of any positive clones, which could be due to the possibility that the purported signal sequence and hydrophobic domain of torsinA interfered with the ability of the bait to translocate into the nucleus for yeast two-hybrid interaction. To overcome this problem, we generated a bait construct encoding an N-terminal-truncated torsinA (torsinA WTΔ40) lacking these hydrophobic regions (Fig. 1A) and used it to screen the cDNA library. Of 15 million yeast transformants screened, we isolated three positive clones, all of which encoded part of a novel protein that we named printor (Fig. 1B) because it is a protein interactor of torsinA. Data base searches revealed that the printor is the rat homologue of human KIAA1384 (also known as Kelch-like protein 14 (KLHL14), GenBankTM accession number NP_065856), an uncharacterized protein discovered by the Kazusa DNA Research Institute human cDNA sequencing project (48).
FIGURE 1.
Isolation of printor as a torsinA-interacting protein from a yeast two-hybrid screen. A, domain structure of torsinA. S, ER signal sequence; H, hydrophobic region. The locations of dystonia-associated torsinA mutations are indicated on the domain structure. Baits used for the yeast two-hybrid screens are indicated below the domain structure. Gal4 DBD, Gal4 DNA-binding domain. B, domain structure of printor (top) and the torsinA-interacting clone isolated from the yeast two-hybrid screen (bottom). BTB, broad complex, tramtrack, and bric-a-brac domain; P/Q, proline/glutamine-rich region; BACK, BTB and C-terminal Kelch domain; K, kelch repeat; TM, transmembrane domain. C, the amino acid sequence homology between human printor and its orthologues. Hs, Homo sapiens; Rn, Rattus norvegicus; Gg, Gallus gallus; Dr, Danio rerio. D, model of membrane topology of torsinA and printor at the ER. Potential glycosylation sites for both torsinA (Asn143 and Asn158) and printor (Asn146 and Asn433) are indicated.
Printor is 628 amino acids in length, with a calculated molecular mass of 70.7 kDa and a theoretical isoelectric point (pI) of 6.68. Sequence analysis revealed that printor contains no signal sequence. Two potential transmembrane domains (residues 562–582 and 609–626) were identified at the C terminus of printor (Fig. 1B) by the TMpred program, which predicted a topology model with an N-terminal ER lumenal orientation (Fig. 1D). In support of this model, two putative N-linked glycosylation sites (Asn146 and Asn433) were identified by the NetNGlyc 1.0 Server in the region of printor that is N-terminal to the first transmembrane domain (Fig. 1D). Printor also contains an N-terminal region with homology to the BTB (broad complex, tramtrack, and bric a brac) domain (also known as POZ (poxvirus and zinc finger) domain), a protein-protein interaction domain that can mediate dimerization (49) or binding to cullin3, a component of a multisubunit E3 ubiquitin-protein ligase complex (50). It is unclear whether this region is a functional BTB domain because it is interrupted by the insertion of a 48-amino acid proline/glutamine (P/Q)-rich sequence (Fig. 1B), which is not found in any other BTB-containing protein. The BTB homology region of printor is followed by a BACK (BTB and C-terminal Kelch) domain of unknown function (51) and six kelch repeats (Fig. 1B). The kelch repeat is a motif of ∼50 amino acids originally identified in the Drosophila ovarian ring canal protein Kelch (52). Kelch repeats form a β-propeller structure with multiple protein-protein interaction sites (53) and are best characterized as an actin-interacting domain (52, 54), although a number of kelch-containing proteins have no association with actin (53).
Database searches revealed the presence of printor orthologues as uncharacterized or predicted proteins from genome projects in a number of organisms, including human, dog, cow, rat, mouse, chicken, and zebrafish. Printor appears to be highly conserved among euteleostomi, or bony vertebrates, but is not found in Caenorhabditis elegans or Drosophila. The amino acid sequence of printor is highly conserved, with the human printor sequence sharing 99% overall amino acid identity with rat printor, 95% overall amino acid identity with chicken printor, and 88% overall amino acid identity with zebrafish printor (Fig. 1C). Moreover, the identified two transmembrane domains, two glycosylation sites, BTB domain, BACK domain, and Kelch repeats are highly conserved among printor orthologues across different species, indicating that printor is an evolutionarily conserved protein among vertebrates.
Printor Co-distributes with TorsinA in Brain as Well as Other Tissues
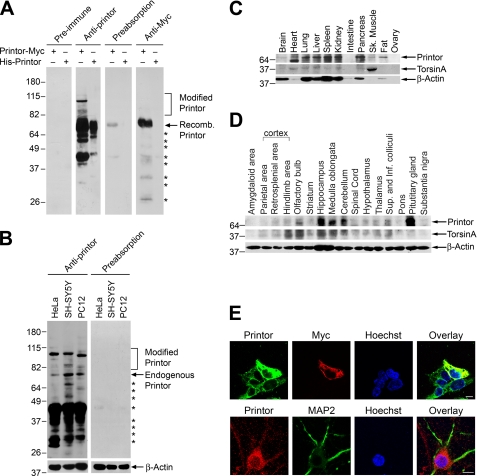
To characterize printor protein and its interaction with torsinA, we generated a rabbit anti-printor antibody against an N-terminal printor peptide (amino acids 1–18), which has 100% amino acid identity across species. Immunoblot analysis showed that the anti-printor antibody, but not the pre-immune serum, specifically recognized ∼70 kDa recombinant His-tagged printor proteins purified from Escherichia coli as well as Myc-tagged printor protein expressed in transfected SH-SY5Y cells, which was also detected by anti-Myc antibody (Fig. 2A). In addition, the anti-printor antibody also recognized the 70-kDa endogenous printor protein in human non-neuronal HeLa cells, human dopaminergic SH-SY5Y cells, and rat dopaminergic PC12 cells (Fig. 2B), demonstrating that the antibody was capable of recognizing endogenous printor in different cell types and across species. We also observed a number of printor-immunoreactive bands with lower and higher molecular weights (Fig. 2, A and B). Preabsorption with recombinant His-tagged printor protein virtually abolished the immunoreactivity of the anti-printor antibody to both recombinant (Fig. 2A) and endogenous (Fig. 2B) printor proteins, further confirming the specificity of our anti-printor antibody. The preabsorption result indicates that the lower and higher molecular weight bands are specific (Fig. 2A). The lower bands probably represent printor degradation products, whereas the upper bands may represent post-translationally modified forms of printor (e.g. glycosylated or ubiquitinated printor).
FIGURE 2.
Printor co-distributes with torsinA in multiple tissues and brain regions. A, lysates from transfected SH-SY5Y cells expressing Myc-tagged printor or purified His-tagged printor protein were analyzed by immunoblotting with preimmune serum, anti-printor antibody, anti-printor antibody preabsorbed with recombinant printor protein, or anti-Myc antibody. Recomb., recombinant. B, equal amounts of lysates (50 μg) from the indicated cells were analyzed by immunoblotting using anti-printor, preabsorbed anti-printor, and anti-β-actin antibodies. The asterisks indicate bands that probably represent printor degradation products. C, equal amounts of homogenates (100 μg) from the indicated rat tissues were analyzed by immunoblotting using anti-printor, anti-torsinA, and anti-β-actin antibodies. Sk., skeletal. D, equal amounts of homogenates (100 μg) from the indicated rat brain regions were analyzed by immunoblotting using anti-printor, anti-torsinA, and anti-β-actin antibodies. Sup., superior; Inf., inferior. E, SH-SY5Y cells overexpressing Myc-tagged printor (top) were immunostained with primary antibodies against printor and the Myc tag, followed by detection with secondary antibodies conjugated to Texas Red (Myc, red) or FITC (printor, green). Primary cortical neurons (bottom) were immunostained with primary antibodies against printor and MAP2, followed by detection with secondary antibodies conjugated to Texas Red (printor, red) or FITC (MAP2, green). Hoechst stain was used to visualize the nucleus. Scale bar, 10 μm. All data are representative of at least three independent experiments.
To characterize the tissue distribution of printor, Western blot analysis of multiple rat tissue samples was performed using the anti-printor antibody. The result showed that printor is expressed in many tissues, including brain, heart, lung, liver, spleen, kidney, and pancreas (Fig. 2C), which agrees with the mRNA expression profile found in the HUGE (Human Unidentified Gene Encoded) database. The tissue distribution pattern of printor is similar to that of torsinA (Fig. 2C). Western blot analysis of protein extracts from various rat brain regions revealed that printor is widely expressed throughout the brain with high protein expression levels in the hippocampus, medulla oblongata, and cerebellum, similar to the expression pattern of torsinA (Fig. 2D).
To determine whether our anti-printor antibody could be used for immunocytochemistry, we performed double label immunofluorescence confocal microscopic analysis of the intracellular distribution of endogenous and recombinant printor protein in SH-SY5Y cells. We observed immunoreactivity to anti-printor antibody in untransfected SH-SY5Y cells and printor immunoreactivity was significantly increased by expression of Myc-tagged printor (Fig. 2E, top). Furthermore, the staining pattern detected by anti-printor antibody in transfected SH-SY5Y cells expressing Myc-tagged printor showed substantial overlap with staining by anti-Myc antibody (Fig. 2E, top). The immunoreactivity to the anti-printor antibody was completely eliminated by preabsorption with recombinant His-tagged printor protein (data not shown). Moreover, no immunoreactivity was observed when preimmune serum was used (data not shown). Together, these results indicate that anti-printor antibody is able to specifically recognize printor protein by immunostaining. Next, we performed double label immunofluorescence confocal microscopic analysis of mouse primary cortical neurons using anti-printor antibody and an antibody to MAP2, a neuron-specific microtubule-associated protein (Fig. 2E, bottom). As expected, the morphology of primary cortical neurons looks very different from that of SH-SY5Y cells (Fig. 2E, top), with multiple MAP2-positive neurites extending from the cell body (Fig. 2E, bottom). We found that printor is localized in the cell body as well as in the MAP2-containing neurites (Fig. 2E, bottom).
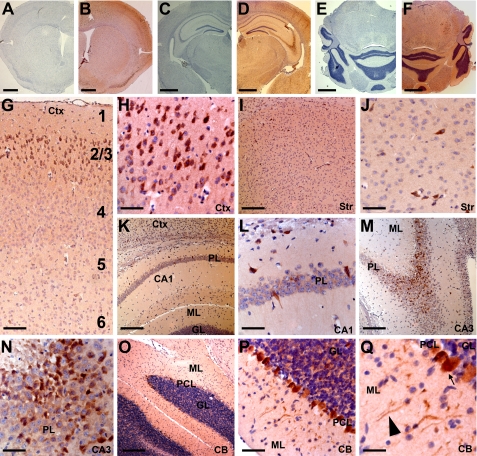
We then used our anti-printor antibody to determine printor protein distribution in mouse brain by immunohistochemistry. Printor immunoreactivity was observed in many neurons throughout the brain (Fig. 3, B, D, and F). The specificity of printor immunoreactivity was confirmed in controls where anti-printor antibody was omitted (Fig. 3, A, C, and E). In the cerebral cortex, printor staining appeared limited to layers 2–3 (Fig. 3G), where neuronal cell bodies and processes were labeled (Fig. 3H). Very little printor immunoreactivity was observed in other cortical layers. Printor was also observed in select neurons of the striatum (Fig. 3, I and J). Consistent with the results of Western blot analysis (Fig. 2D), intense printor immunostaining was observed in the hippocampus (Fig. 3, K–N) and cerebellum (Fig. 3, O–Q). Pyramidal cell neurons of the CA1 (Fig. 3, K and L) and CA3 (Fig. 3, M and N) contained printor immunoreactivity in their cell bodies, although staining appeared stronger in the CA3 region compared with the CA1 region. Purkinje cells of the cerebellum displayed strong immunoreactivity in their cell bodies and processes (Fig. 3, P and Q). Granular cells of the cerebellum also displayed printor immunoreactivity, but to a lesser degree than Purkinje cells (Fig. 3, P and Q). Printor immunoreactivity was also observed in the medial septum, ventral pallidum, thalamus, hypothalamus, amygdala, inferior colliculi, locus caeruleus, peripyramidal nucleus, raphe nucleus, reticular formation, spinal trigeminal nucleus, and vestibular nuclei (data not shown). Very few glial cells were labeled and no immunoreactivity was observed in the corpus callosum (data not shown). Together, our results indicate that printor is primarily expressed in neurons but not glia.
FIGURE 3.
Immunohistochemical analysis of printor protein distribution in mouse brain. Coronal sections through striatum (A and B), hippocampus (C and D), and cerebellum (E and F) were immunostained with either anti-printor antibody (B, D, and F) or no primary antibody (A, C, and E) and counterstained with hematoxylin. G and H, printor immunostaining in the cortex (Ctx). Numbers indicate cortical layers. Neurons in the layers 2–3 were strongly stained. I and J, printor immunostaining in the striatum (Str). K and L, printor immunostaining in the hippocampal CA1 region. Printor immunoreactivity was seen in the pyramidal layer (PL) but not in the molecular (ML) or granular (GL) layers. M and N, printor immunostaining in the hippocampal CA3 region. Printor immunoreactivity was found in the PL but not in the ML. O–Q, printor immunostaining in the cerebellum (CB). Purkinje cell layer (PCL) neurons display intense immunostaining compared with the granular layer (GL). Purkinje cell projections in the molecular layer were also labeled. Arrow indicates cell body and the arrowhead indicates neuronal projection. Scale bar is 1.25 mm in A-F; 125 μm in G; 250 μm in I, K, M, and O; 60 μm in H, J, L, N, and P; 30 μm in Q.
Printor Associates with TorsinA in Cells and in the Brain
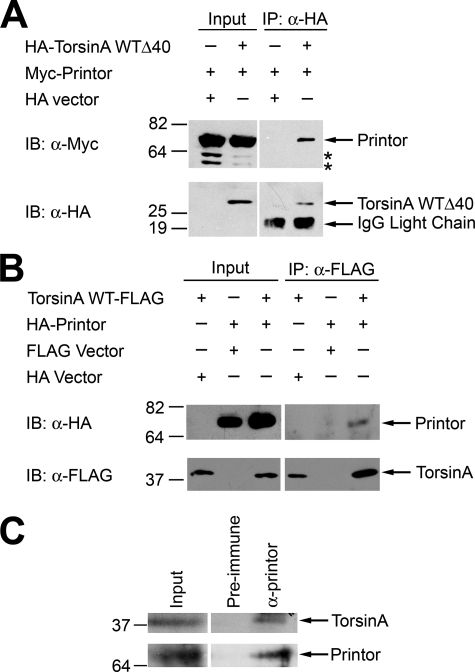
To verify that the interaction between torsinA and printor detected in the yeast two-hybrid screen occurs in vivo, co-immunoprecipitation assays were performed. Because the interaction was identified using torsinA WTΔ40 bait, HeLa cells transfected with Myc-tagged printor and either HA-tagged torsinA WTΔ40 or HA vector were subjected to immunoprecipitation using anti-HA antibody as previously described (39). The result showed that printor co-immunoprecipitated with HA-tagged torsinA WTΔ40, but not with HA alone (Fig. 4A), indicating a specific interaction. We next examined the interaction between full-length torsinA WT and printor and found that torsinA WT, but not the vector control, was able to co-immunoprecipitate printor protein (Fig. 4B), confirming a specific association between torsinA and printor in transfected cells. We then performed additional co-immunoprecipitation experiments to examine the association of endogenous printor and torsinA in mouse cerebellum homogenates (Fig. 4C). Anti-printor antibody, but not the preimmune serum, was able to co-immunoprecipitate printor and torsinA from the homogenates, indicating the existence of an endogenous torsinA-printor complex.
FIGURE 4.
Printor and torsinA interact in vivo. A, co-immunoprecipitation of printor with torsinA WTΔ40. Lysates from HeLa cells expressing either HA-tagged torsinA WTΔ40 or HA vector and Myc-tagged printor were subjected to immunoprecipitation with anti-HA antibody (12CA5). Immunoprecipitates were analyzed by immunoblotting (IB) with anti-HA and anti-Myc antibodies. The asterisks indicate potential degradation products of printor. B, co-immunoprecipitation of printor with full-length torsinA. Lysates from HeLa cells expressing HA-tagged printor or HA vector and C-terminal FLAG-tagged torsinA or FLAG vector were subjected to immunoprecipitation (IP) with anti-FLAG antibody. The immunoprecipitates were analyzed by immunoblotting with anti-FLAG and anti-HA antibodies. C, association of endogenous torsinA and printor in the mouse brain. Homogenates from mouse cerebellum were subjected to immunoprecipitation using anti-printor antibody or the preimmune serum, followed by immunoblot analysis with anti-printor and anti-torsinA antibodies.
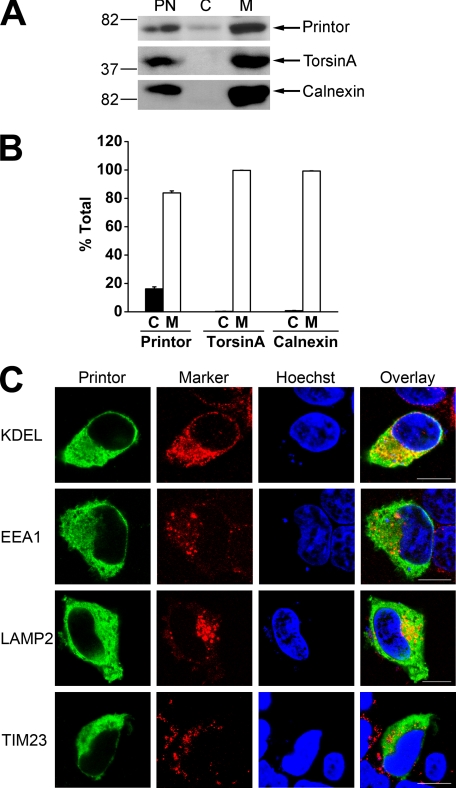
Printor Exists in Both Cytosolic and Membrane-associated Pools and Associates with the ER
To investigate the intracellular distribution of endogenous printor protein, we performed subcellular fractionation experiments to separate the postnuclear supernatant from SH-SY5Y cells into cytosol and membrane fractions. Western blot analysis of these fractions revealed that printor was present in both the cytosol and membrane fractions (Fig. 5A). Quantification analysis revealed that 16.2 ± 1.4% of printor is in the cytosolic fraction and 83.8 ± 1.4% of printor in the membrane-associated pool (Fig. 5B). This is in contrast to torsinA or calnexin, which are exclusively found in the membrane fraction (Fig. 5A). To determine the membrane compartment(s) that printor is associated with, we performed double labeling immunofluorescence confocal microscopic analysis to compare the intracellular distribution of printor with various markers of intracellular organelles in SH-SY5Y cells (Fig. 5C). We found that the distribution of printor exhibited a significant overlap with that of the ER marker KDEL, but not the early endosomal marker EEA1, lysosomal marker LAMP2, or mitochondrial marker TIM23, suggesting that printor is associated with the ER compartment.
FIGURE 5.
Printor is found in both cytosolic and membrane-associated fractions. A, post-nuclear supernatant (PN) from SH-SY5Y cells was separated into cytosol (C) and membrane (M) fractions. Aliquots representing an equal percentage of each fraction were analyzed by immunoblotting with anti-printor, anti-torsinA, and anti-calnexin antibodies. B, the level of the indicated proteins in each fraction was quantified using NIH Scion Image and shown as a percentage of the total level of the indicated protein. Data represent mean ± S.E. from at least three independent experiments. C, SH-SY5Y cells expressing Myc-tagged printor were immunostained with anti-Myc and anti-KDEL, anti-EEA1, anti-LAMP2, or anti-TIM23 primary antibodies followed by detection with secondary antibodies conjugated to Texas Red (marker proteins, red) or FITC (printor, green). Hoechst stain was used to visualize the nucleus. Scale bars, 10 μm.
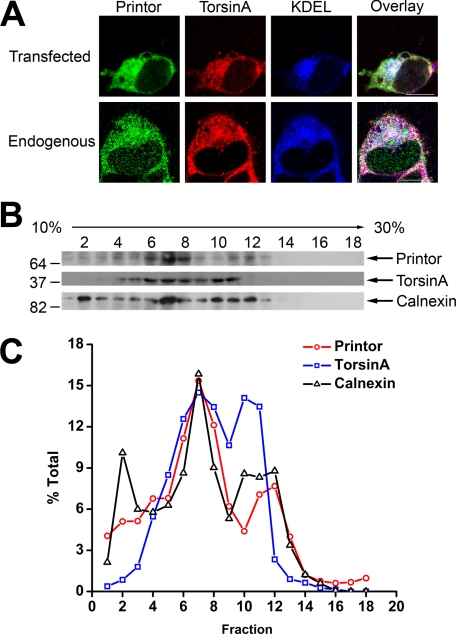
Printor Co-localizes with TorsinA Mainly at the ER Rather Than the NE
To determine whether printor and torsinA co-localized in the ER, we first performed triple labeling immunofluorescence confocal microscopic analysis in SH-SY5Y cells expressing C-terminal HA-tagged torsinA and Myc-tagged printor with anti-HA, anti-Myc, and anti-KDEL antibodies. We observed a significant overlap in the three immunostaining patterns (Fig. 6A), suggesting that printor and torsinA co-localize at the ER. We then performed triple labeling analysis with anti-printor, anti-torsinA, and anti-KDEL antibodies and found that endogenous printor and torsinA co-localize at the ER (Fig. 6A). To complement our immunocytochemistry data, we performed density gradient fractionation experiments to determine whether endogenous torsinA and printor associate with the ER membrane compartment. Upon fractionation of SH-SY5Y post-nuclear supernatant on a 10–30% linear Opti-Prep gradient, a clear co-fractionation of printor with torsinA and the ER integral membrane protein calnexin was observed in fractions 4–12, providing additional evidence supporting the association of printor with torsinA at the ER membrane compartment (Fig. 6B). Quantification analysis revealed that these three proteins co-fractionate in two membrane pools that peaked in fractions 7 and 11 (Fig. 6C), which may represent smooth ER and rough ER membranes, respectively.
FIGURE 6.
Co-localization of printor and torsinA in the ER. A, SH-SY5Y cells expressing Myc-tagged printor and C-terminal HA-tagged torsinA (top) were immunostained with primary antibodies against Myc, HA, and KDEL, followed by detection with secondary antibodies conjugated to Texas Red (torsinA, red), FITC (printor, green), or Cy5 (KDEL, blue). Untransfected SH-SY5Y cells (bottom) were immunostained with primary antibodies against printor, torsinA, and KDEL, followed by detection with secondary antibodies conjugated to Texas Red (torsinA, red), FITC (printor, green), or Cy5 (KDEL, blue). Scale bars, 10 μm. B, post-nuclear supernatant from SH-SY5Y cells was fractionated on a 10–30% Opti-Prep gradient into 18 fractions, with fraction 1 corresponding to the top of the gradient. Equal volumes of each fraction were analyzed by SDS-PAGE followed by immunoblotting using anti-printor, anti-torsinA, and anti-calnexin antibodies. C, the level of the indicated protein in each fraction was quantified using NIH Scion Image and shown as a percentage of the total level of the indicated protein. Data are representative of at least three independent experiments.
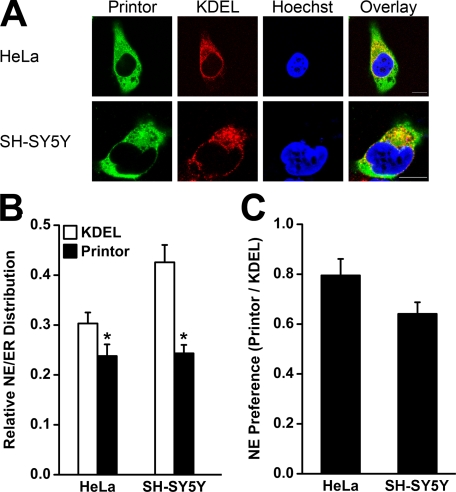
Given that the ER membrane is in structural continuity with the NE membrane (55), we next performed quantitative double labeling analysis as described (16) to compare the relative distribution of printor and the ER marker KDEL between the ER and NE subdomains in the non-neuronal cell line HeLa (Fig. 7A, top) and the dopaminergic neuronal cell line SH-SY5Y (Fig. 7A, bottom). The results revealed that in both cell types, the relative NE/ER distribution of printor is significantly lower than that of KDEL, indicating that printor is preferentially localized to the ER compared with KDEL (Fig. 7B). To control for cell-type variation in the relative NE/ER distribution of KDEL, we determined the NE preference of printor by normalizing the NE/ER ratio of printor to the corresponding NE/ER ratio of KDEL in the same cells (Fig. 7C). We found that the NE preference of printor in both cell types was less than 1, consistent with an enhanced ER preference. This result is in direct contrast to our previous finding that torsinA exhibits neuronal cell type-specific NE preference (16) and suggests that the main site of co-localization between printor and torsinA is at the ER rather than the NE.
FIGURE 7.
Printor displays ER preference in both HeLa and SH-SY5Y cells. A, HeLa (top) or SH-SY5Y (bottom) cells expressing Myc-tagged printor were stained with primary antibodies against Myc and ER marker KDEL, followed by detection with secondary antibodies conjugated to Texas Red (KDEL, red) or FITC (printor, green). Hoechst stain was used to visualize the nucleus. Scale bars, 10 μm. B, quantification shows the relative distribution of printor and KDEL in the NE versus the ER. Data represent mean ± S.E. (error bars) from at least three independent experiments. *, significantly different from the NE/ER ratio of KDEL (p < 0.05). C, NE preference of printor was determined by normalizing the NE/ER ratio of printor in HeLa or SH-SY5Y cells to the corresponding NE/ER ratio of KDEL in the same cells. Data represent mean ± S.E. from at least three independent experiments.
Printor Shows Reduced Co-localization with the ATP-bound Form of TorsinA
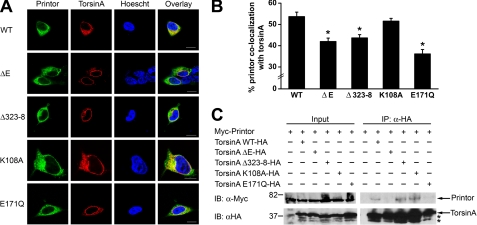
Because torsinA contains a AAA+ ATPase domain and is predicted to be a chaperone protein that binds a substrate(s) in an ATPase cycle-dependent manner (11, 12), we examined the co-localization of printor with the ATP binding-deficient mutant torsinA K108A, which would be unable to bind the substrate(s), and the “substrate trap” ATP hydrolysis-deficient mutant torsinA E171Q, which would bind tightly to the substrate(s). Double labeling immunofluorescence confocal microscopic analysis of SH-SY5Y cells co-expressing Myc-tagged printor and C-terminal HA-tagged torsinA WT, torsinA K108A, or torsinA E171Q revealed that the ATP-free form of torsinA K108A, like torsinA WT, visibly co-localized with printor, but the ATP-bound form of torsinA E171Q appeared to have reduced co-localization with printor (Fig. 8A). Quantification analysis showed that 51.5 ± 1.3% of printor co-localizes with torsinA K108A, which is similar to the extent of printor co-localization with torsinA WT (53.7 ± 2.1%). In contrast, only 36.1 ± 2.1% of printor co-localized with torsinA E171Q, indicating a significant (p < 0.005) reduction (Fig. 8B). Reciprocal quantification analysis showed that 62.4 ± 1.4% of torsinA K108A co-localizes with printor, which is similar to the extent of torsinA WT co-localization with printor (63.7 ± 1.7%). In contrast, torsinA E171Q exhibited a significantly (p < 0.005) decreased level of co-localization with printor (52.9 ± 2.1%) compared with torsinA WT.
FIGURE 8.
Printor interaction and co-localization with torsinA is disrupted by torsinA ΔE and E171Q mutations. A, co-localization between printor and WT or mutant torsinA. SH-SY5Y cells co-expressing Myc-tagged printor and C-terminal HA-tagged torsinA WT, torsinA ΔE, torsinA Δ323–328, torsinA K108A, or torsinA E171Q were stained with primary antibodies against HA and Myc, followed by detection with secondary antibodies conjugated to Texas Red (torsinA, red) or FITC (printor, green). Hoechst stain was used to visualize the nucleus. Scale bars, 10 μm. B, quantification shows the percentage of printor protein that co-localizes with WT or mutant torsinA. Data represent mean ± S.E. (error bars) from at least three independent experiments. *, significantly different from the percentage of printor co-localization with torsinA WT (p < 0.005). C, extracts from SH-SY5Y cells expressing Myc-tagged printor and C-terminal HA-tagged torsinA WT, torsinA ΔE, torsinA Δ323–328, torsinA K108A, or torsinA E171Q were subjected to immunoprecipitation (IP) with anti-HA antibody followed by immunoblotting (IB) with anti-HA and anti-Myc antibodies.
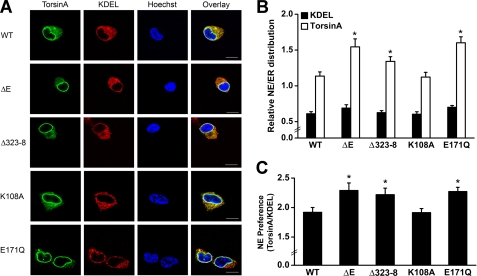
To determine whether the observed reduction in the co-localization between printor and the ATP-bound form of torsinA E171Q was due to a change in torsinA localization, we performed additional double labeling immunofluorescence microscopic analysis to compare the intracellular distribution of C-terminal HA-tagged torsinA WT, torsinA K108A, and torsinA E171Q in SH-SY5Y cells (Fig. 9A). Consistent with our previous report (16), we found the NE/ER ratio of torsinA WT was significantly greater than the NE/ER ratio of KDEL in SH-SY5Y cells (Fig. 9B), indicating a preferential localization of torsinA WT to the NE compared with the ER marker KDEL. The NE/ER ratio of torsinA K108A was similar to that of torsinA WT, but torsinA E171Q showed a significantly higher NE/ER ratio compared with torsinA WT (Fig. 9B). After normalization to the NE/ER ratio of KDEL in the same cells, torsinA E171Q had an NE preference of 2.27 ± 0.07 compared with 1.92 ± 0.08 for torsinA WT and 1.91 ± 0.07 for torsinA K108A (Fig. 9C). Together, these results suggest that ATP binding induces translocation of torsinA from the ER to the NE, leading to reduced co-localization of torsinA with printor.
FIGURE 9.
TorsinA E171Q mutation and dystonia-associated mutations promote translocation of torsinA from the ER to NE. A, SH-SY5Y cells expressing C-terminal HA-tagged torsinA WT, torsinA ΔE, torsinA Δ323–328, torsinA K108A, or torsinA E171Q were stained with primary antibodies against HA and ER marker KDEL, followed by detection with secondary antibodies conjugated to Texas Red (KDEL, red) or FITC (torsinA, green). Hoechst stain was used to visualize the nucleus. Scale bars, 10 μm. B, quantification shows the relative distribution of torsinA and KDEL in the NE versus the ER. Data represent mean ± S.E. (error bars) from at least three independent experiments. *, significantly different from the NE/ER ratio of torsinA WT (p < 0.05). C, NE preference of torsinA was determined by normalizing the NE/ER ratio of torsinA to the corresponding NE/ER ratio of KDEL in the same cells. Data represent mean ± S.E. from at least three independent experiments. *, significantly different from torsinA WT (p < 0.05).
Dystonia-associated Mutations Reduce the Co-localization between Printor and TorsinA
To determine the effects of dystonia-associated mutations on torsinA co-localization with printor, we performed double labeling immunofluorescence confocal microscopic analysis to examine the co-localization of C-terminal HA-tagged torsinA WT, torsinA ΔE, or torsinA Δ323–328 with Myc-tagged printor in SH-SY5Y cells. We found that torsinA WT visibly co-localized with printor, but torsinA ΔE and Δ323–328 mutants appeared to have reduced co-localization with printor (Fig. 8A). Quantification analysis revealed that, whereas 53.7 ± 2.1% of printor co-localized with torsinA WT, only 42.0 ± 1.6 and 43.7 ± 1.5% of printor co-localized with torsinA ΔE and the torsinA Δ323–328 mutant, respectively (Fig. 8B), indicating that both dystonia-associated mutations cause a significant (p < 0.005) reduction in the co-localization of printor with torsinA. Reciprocal quantification analysis showed that 56.4 ± 2.0% of torsinA ΔE and 58.0 ± 1.9% of torsinA Δ323–328 mutant proteins co-localized with printor, both of which represent a significant (p < 0.05) decrease compared with the extent of torsinA WT co-localization with printor (63.7 ± 1.7%). As we have shown previously (16), both torsinA ΔE and Δ323–328 mutations promoted translocation of torsinA from the ER to the NE (Fig. 9). Together, these data suggest that the reduced co-localization of printor with torsinA ΔE and torsinA Δ323–328 mutants may result from the mislocalization of dystonia-associated mutants to the NE.
TorsinA ΔE and E171Q Mutations Disrupt the Interaction of TorsinA with Printor
To determine the effects of various torsinA mutations on the ability of torsinA to bind printor, we performed co-immunoprecipitation analysis in SH-SY5Y cells co-expressing HA-tagged printor and either C-terminal Myc-tagged torsinA WT, torsinA K108A, torsinA E171Q, torsinA ΔE, torsinA Δ323–328, or the Myc vector control. We found that, like torsinA WT, torsinA K108A, an ATP binding-deficient mutant, which cannot bind the substrate, was able to interact with printor. In contrast, the substrate trap ATP hydrolysis-deficient mutant torsinA E171Q was incapable of interacting with printor (Fig. 8C). These data argue against the possibility of printor being a torsinA substrate and suggest that printor may be a cofactor of torsinA. Our co-immunoprecipitation analysis revealed that the rare dystonia mutation torsinA Δ323–328 had no significant effect on the interaction of torsinA with printor. In contrast, DYT1 dystonia-associated mutation torsinA ΔE completely abolished the ability of torsinA to bind printor (Fig. 8C).
DISCUSSION
TorsinA ΔE mutation is the major cause for DYT1 dystonia, but little is known about torsinA function and its regulation in cells and how torsinA ΔE mutation leads to the disease pathogenesis. In the present study, we identified and characterized a novel protein, printor, that interacts and co-localizes with torsinA. Printor is a novel member of the BTB-BACK-Kelch (BBK) protein family (51, 56, 57) and contains an N-terminal BTB homology region, a BACK domain, and six kelch repeats. Several BBK proteins have been implicated in neuronal development (58–60) and protein ubiquitination (50, 57, 61–63), and mutations in at least one BBK protein, gigaxonin, lead to neuronal dysfunction (64). A major difference between printor and other BBK proteins is that the printor BTB homology region contains a 48-amino acid P/Q-rich intervening sequence, making it difficult to predict whether this region of printor represents a functional BTB domain. The six kelch repeats of printor are predicted to form a β-propeller structure with multiple protein binding sites (53, 56). Thus, printor could potentially interact with multiple proteins or participate in the formation of multiprotein complexes. The finding that printor orthologues are encoded by the genomes of chordate animals, together with our biochemical data, indicates that the importance of the printor-torsinA association is widespread but previously unrecognized.
Our immunohistochemical and immunoblot analyses reveal that, like torsinA, printor is expressed in the brain, where it is enriched in neurons but not glia. The presence of printor in neuronal cell bodies and processes suggests that printor may participate in neuronal and synaptic function. The abundant expression of printor and its co-distribution with torsinA in brain regions thought to be affected in dystonia (e.g. cerebellum) supports a role for printor in the pathogenesis of dystonia. However, similar to the expression pattern of torsinA, printor is not confined to the dystonia-affected brain regions, but rather, it is widely distributed throughout the brain and is also expressed in many other tissues, suggesting that printor, like torsinA, may have a functional role important to many cell types, including neurons.
TorsinA contains a well conserved AAA+ ATPase domain and is believed to have a chaperone-like function in promoting conformational remodeling of substrate proteins (13, 14, 65). Several binding partners of torsinA have been reported, including LAP1 and its homologous protein LULL1 (21), nesprins (22), snapin (27), and dopamine transporter (33). These torsinA binding partners have been proposed to be substrates of torsinA (20–22, 27, 33), and some of these proteins have been shown to preferentially bind to the ATP-bound form of torsinA (21, 22, 33). In contrast, we found that printor binds to torsinA WT and the ATP binding-deficient mutant torsinA K108A, but not to the substrate trap ATP hydrolysis-deficient mutant torsinA E171Q. Our results support a role for printor as a cofactor rather than a substrate of torsinA. Increasing evidence indicates that AAA+ proteins utilize cofactors to extend and regulate their binding repertoire and achieve functional specificity (66–68). For example, the AAA+ protein N-ethylmaleimide-sensitive factor cofactor, α-SNAP, recruits N-ethylmaleimide-sensitive factor to the SNARE complex and stimulates the ATPase activity of N-ethylmaleimide-sensitive factor for disassembly of the SNARE complex (11, 12). Cdc48/p97 AAA+ ATPase regulates membrane fusion through interaction with its cofactor p47, and participates in the ER-associated degradation process by forming a complex with its cofactors, Ufd1 and Npl4 (68). Given that torsinA is a 332-amino acid protein with a 220-amino acid AAA+ ATPase homology domain and 40- amino acid hydrophobic regions (69), it seems particularly plausible that torsinA ATPase would use co-factor(s) to extend its binding repertoire for substrate recognition and function regulation. Printor, with its multiple protein-protein interaction domains, could either directly recruit torsinA substrates or act as a scaffold for organizing a multicomponent torsinA-chaperone complex. Alternatively, printor could act as a regulator of torsinA, either modulating torsinA ATPase activity or regulating the torsinA protein expression level or localization. Further studies are needed to determine the exact role of the printor-torsinA interaction.
Our finding of an endogenous printor-torsinA complex and the co-localization of printor with torsinA mainly at the ER rather than the NE suggests that printor may act as a cofactor for mediating the ER function of torsinA, participating in substrate recognition and processing. Printor contains two putative transmembrane domains at its C terminus and two potential N-linked glycosylation sites in the N-terminal region (Fig. 1). This supports a lumenal orientation for the N terminus of printor (Fig. 1D) and suggests that the lumen of the ER is the likely location of the torsinA-printor interaction. Consistent with this possibility, our subcellular fractionation studies reveal that a majority of endogenous printor are membrane-associated and co-fractionate with endogenous torsinA and the ER membrane on the Opti-Prep density gradient. Together, our results raise the possibility that torsinA and printor may function together as a chaperone complex for regulating protein folding in the ER lumen or protein trafficking through the ER.
In addition to the membrane-associated pool, our subcellular fractionation analysis indicates that there is a small cytosolic pool of endogenous printor in SH-SY5Y cells. Although torsinA is generally thought to be a ER lumenal protein, a small population of torsinA has been observed in the cytosol (10, 70) and a significant pool of torsinA has been shown to have the AAA+ domain facing the cytoplasm (20). Moreover, torsinA has been reported to bind several cytosolic proteins (20, 27, 71). Thus, it is possible that a small pool of printor may interact with torsinA in the cytosol and regulate cytosolic protein folding.
Although torsinA ΔE mutation accounts for most cases of DYT1 dystonia, the pathological mechanism by which the torsinA mutation causes dystonia remains elusive. To understand torsinA function in normal physiology and its dysfunction in dystonia, a major research effort in the field has been directed toward the search for binding partners of torsinA. Thus far, this effort has resulted in the isolation of several torsinA binding partners (20–22, 27). However, none of these interactions is impaired by the dystonia-associated torsinA ΔE mutation. In contrast, our study reveals, for the first time, an interaction between torsinA and its putative cofactor printor that is completely disrupted by the torsinA ΔE mutation. Our data supports a loss-of-function pathogenic mechanism for the torsinA ΔE mutation in dystonia and underscores the importance of the printor-torsinA interaction in normal physiology.
In addition to the torsinA ΔE mutation, torsinA Δ323–328 mutation has also been associated with dystonia, although it is unclear whether torsinA Δ323–328 is pathogenic or not because of its co-occurrence with a mutation in another dystonia-related protein, ϵ-sarcoglycan (6). The structural and functional consequences of the torsinA Δ323–328 mutation remain mostly unknown. Our results reveal that torsinA Δ323–328 mutation, like the torsinA ΔE mutation, promotes translocation of torsinA from the ER to the NE, leading to reduced co-localization of torsinA with printor. However, unlike the torsinA ΔE mutation, the torsinA Δ323–328 mutation does not affect the interaction of torsinA with printor. These data suggest that the printor binding site involves the second α-helix in the torsinA C-terminal region where glutamate residues 302 and 303 reside, but not the extreme α-helix in the torsinA C-terminal tail where amino acids Phe323–Tyr328 reside (72). Thus, deletion of a single glutamate residue at positions 302 or 303 (torsinA ΔE) would alter the helical register and disrupt the ability of torsinA to bind printor, whereas deletion of amino acids Phe323–Tyr328 (torsinA Δ323–328) would not affect the ability of torsinA to bind printor.
In conclusion, our study identifies a novel protein, printor, which interacts and co-localizes with torsinA, and provides evidence supporting a role for printor as a cofactor rather than a substrate of torsinA. Our findings suggest that disruption of the printor-torsinA interaction by the torsinA ΔE mutation could contribute to the pathophysiology of DYT1 dystonia.
Acknowledgments
We are grateful to Paul Worley (Johns Hopkins University) for providing the rat hippocampal/cortical cDNA library and William T. Dauer (Columbia University) for providing pcDNA3.1/V5-His-TOPO torsinA E171Q and torsinA K108A. We thank Marla Gearing (Emory University) and Bruce H. Wainer (University of Florida) for advice on immunohistochemical analysis. The confocal imaging analysis was performed at the Emory Neuroscience Core Facility supported in part by National Institutes of Health Grant P30 NS055077.
This work was supported, in whole or in part, by National Institutes of Health Grants NS054334 (to L. M. G.), NS050650 (to L.-S. C.), and ES015813 and GM082828 (to L. L.).
- ER
- endoplasmic reticulum
- NE
- nuclear envelope
- WT
- wild type
- FITC
- fluorescein isothiocyanate
- HA
- hemagglutinin
- AAA+
- ATPases associated with a variety of cellular activities
- MAP
- microtubule-associated protein
- LAP1
- lamina-associated polypeptide 1
- BTB
- broad complex, tramtrack, and bric a brac.
REFERENCES
- 1.Bressman S. B., Fahn S., Ozelius L. J., Kramer P. L., Risch N. J. (2001) Arch. Neurol. 58, 681–682 [DOI] [PubMed] [Google Scholar]
- 2.Walker R. H., Brin M. F., Sandu D., Good P. F., Shashidharan P. (2002) Neurology 58, 120–124 [DOI] [PubMed] [Google Scholar]
- 3.Rostasy K., Augood S. J., Hewett J. W., Leung J. C., Sasaki H., Ozelius L. J., Ramesh V., Standaert D. G., Breakefield X. O., Hedreen J. C. (2003) Neurobiol. Dis. 12, 11–24 [DOI] [PubMed] [Google Scholar]
- 4.Ozelius L. J., Hewett J. W., Page C. E., Bressman S. B., Kramer P. L., Shalish C., de Leon D., Brin M. F., Raymond D., Corey D. P., Fahn S., Risch N. J., Buckler A. J., Gusella J. F., Breakefield X. O. (1997) Nat. Genet. 17, 40–48 [DOI] [PubMed] [Google Scholar]
- 5.Leung J. C., Klein C., Friedman J., Vieregge P., Jacobs H., Doheny D., Kamm C., DeLeon D., Pramstaller P. P., Penney J. B., Eisengart M., Jankovic J., Gasser T., Bressman S. B., Corey D. P., Kramer P., Brin M. F., Ozelius L. J., Breakefield X. O. (2001) Neurogenetics 3, 133–143 [DOI] [PubMed] [Google Scholar]
- 6.Klein C., Liu L., Doheny D., Kock N., Müller B., de Carvalho Aguiar P., Leung J., de Leon D., Bressman S. B., Silverman J., Smith C., Danisi F., Morrison C., Walker R. H., Velickovic M., Schwinger E., Kramer P. L., Breakefield X. O., Brin M. F., Ozelius L. J. (2002) Ann. Neurol. 52, 675–679 [DOI] [PubMed] [Google Scholar]
- 7.Kock N., Naismith T. V., Boston H. E., Ozelius L. J., Corey D. P., Breakefield X. O., Hanson P. I. (2006) Hum. Mol. Genet. 15, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 8.Clarimon J., Asgeirsson H., Singleton A., Jakobsson F., Hjaltason H., Hardy J., Sveinbjornsdottir S. (2005) Ann. Neurol. 57, 765–767 [DOI] [PubMed] [Google Scholar]
- 9.Liu Z., Zolkiewska A., Zolkiewski M. (2003) Biochem. J. 374, 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewett J., Ziefer P., Bergeron D., Naismith T., Boston H., Slater D., Wilbur J., Schuback D., Kamm C., Smith N., Camp S., Ozelius L. J., Ramesh V., Hanson P. I., Breakefield X. O. (2003) J. Neurosci. Res. 72, 158–168 [DOI] [PubMed] [Google Scholar]
- 11.Patel S., Latterich M. (1998) Trends Cell Biol. 8, 65–71 [PubMed] [Google Scholar]
- 12.Hanson P. I., Whiteheart S. W. (2005) Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 13.McLean P. J., Kawamata H., Shariff S., Hewett J., Sharma N., Ueda K., Breakefield X. O., Hyman B. T. (2002) J. Neurochem. 83, 846–854 [DOI] [PubMed] [Google Scholar]
- 14.Caldwell G. A., Cao S., Sexton E. G., Gelwix C. C., Bevel J. P., Caldwell K. A. (2003) Hum. Mol. Genet. 12, 307–319 [DOI] [PubMed] [Google Scholar]
- 15.Shashidharan P., Kramer B. C., Walker R. H., Olanow C. W., Brin M. F. (2000) Brain Res. 853, 197–206 [DOI] [PubMed] [Google Scholar]
- 16.Giles L. M., Chen J., Li L., Chin L. S. (2008) Hum. Mol. Genet. 17, 2712–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Alegre P., Paulson H. L. (2004) J. Neurosci. 24, 2593–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodchild R. E., Dauer W. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naismith T. V., Heuser J. E., Breakefield X. O., Hanson P. I. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7612–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamm C., Boston H., Hewett J., Wilbur J., Corey D. P., Hanson P. I., Ramesh V., Breakefield X. O. (2004) J. Biol. Chem. 279, 19882–19892 [DOI] [PubMed] [Google Scholar]
- 21.Goodchild R. E., Dauer W. T. (2005) J. Cell Biol. 168, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nery F. C., Zeng J., Niland B. P., Hewett J., Farley J., Irimia D., Li Y., Wiche G., Sonnenberg A., Breakefield X. O. (2008) J. Cell Sci. 121, 3476–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao J., Gong S., Zhao Y., LeDoux M. S. (2004) Brain Res. Dev. Brain Res. 152, 47–60 [DOI] [PubMed] [Google Scholar]
- 24.Augood S. J., Keller-McGandy C. E., Siriani A., Hewett J., Ramesh V., Sapp E., DiFiglia M., Breakefield X. O., Standaert D. G. (2003) Brain Res. 986, 12–21 [DOI] [PubMed] [Google Scholar]
- 25.Konakova M., Pulst S. M. (2001) Brain Res. 922, 1–8 [DOI] [PubMed] [Google Scholar]
- 26.Walker R. H., Brin M. F., Sandu D., Gujjari P., Hof P. R., Warren Olanow C., Shashidharan P. (2001) Brain Res. 900, 348–354 [DOI] [PubMed] [Google Scholar]
- 27.Granata A., Watson R., Collinson L. M., Schiavo G., Warner T. T. (2008) J. Biol. Chem. 283, 7568–7579 [DOI] [PubMed] [Google Scholar]
- 28.Grundmann K., Reischmann B., Vanhoutte G., Hübener J., Teismann P., Hauser T. K., Bonin M., Wilbertz J., Horn S., Nguyen H. P., Kuhn M., Chanarat S., Wolburg H., Van der Linden A., Riess O. (2007) Neurobiol. Dis. 27, 190–206 [DOI] [PubMed] [Google Scholar]
- 29.Balcioglu A., Kim M. O., Sharma N., Cha J. H., Breakefield X. O., Standaert D. G. (2007) J. Neurochem. 102, 783–788 [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., DeCuypere M., LeDoux M. S. (2008) Exp. Neurol. 210, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shashidharan P., Sandu D., Potla U., Armata I. A., Walker R. H., McNaught K. S., Weisz D., Sreenath T., Brin M. F., Olanow C. W. (2005) Hum. Mol. Genet. 14, 125–133 [DOI] [PubMed] [Google Scholar]
- 32.Goodchild R. E., Kim C. E., Dauer W. T. (2005) Neuron 48, 923–932 [DOI] [PubMed] [Google Scholar]
- 33.Torres G. E., Sweeney A. L., Beaulieu J. M., Shashidharan P., Caron M. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15650–15655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 35.Kirk E., Chin L. S., Li L. (2006) J. Cell Sci. 119, 4689–4701 [DOI] [PubMed] [Google Scholar]
- 36.Chin L. S., Nugent R. D., Raynor M. C., Vavalle J. P., Li L. (2000) J. Biol. Chem. 275, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 37.Kwong J., Roundabush F. L., Hutton Moore P., Montague M., Oldham W., Li Y., Chin L. S., Li L. (2000) J. Cell Sci. 113, 2273–2284 [DOI] [PubMed] [Google Scholar]
- 38.Chin L. S., Raynor M. C., Wei X., Chen H. Q., Li L. (2001) J. Biol. Chem. 276, 7069–7078 [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Chin L. S., Levey A. I., Li L. (2002) J. Biol. Chem. 277, 28212–28221 [DOI] [PubMed] [Google Scholar]
- 40.Gearing M., Wilson R. W., Unger E. R., Shelton E. R., Chan H. W., Masters C. L., Beyreuther K., Mirra S. S. (1993) J. Neuropathol. Exp. Neurol. 52, 22–30 [DOI] [PubMed] [Google Scholar]
- 41.Lee J. T., Wheeler T. C., Li L., Chin L. S. (2008) Hum. Mol. Genet. 17, 906–917 [DOI] [PubMed] [Google Scholar]
- 42.Olzmann J. A., Brown K., Wilkinson K. D., Rees H. D., Huai Q., Ke H., Levey A. I., Li L., Chin L. S. (2004) J. Biol. Chem. 279, 8506–8515 [DOI] [PubMed] [Google Scholar]
- 43.Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007) J. Cell Biol. 178, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Chin L. S., Weigel C., Li L. (2001) J. Biol. Chem. 276, 40824–40833 [DOI] [PubMed] [Google Scholar]
- 45.Chin L. S., Vavalle J. P., Li L. (2002) J. Biol. Chem. 277, 35071–35079 [DOI] [PubMed] [Google Scholar]
- 46.Liang J., Yin C., Doong H., Fang S., Peterhoff C., Nixon R. A., Monteiro M. J. (2006) J. Cell Sci. 119, 4011–4024 [DOI] [PubMed] [Google Scholar]
- 47.Webber E., Li L., Chin L. S. (2008) J. Mol. Biol. 382, 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagase T., Kikuno R., Ishikawa K. I., Hirosawa M., Ohara O. (2000) DNA Res. 7, 65–73 [DOI] [PubMed] [Google Scholar]
- 49.Chen W., Zollman S., Couderc J. L., Laski F. A. (1995) Mol. Cell. Biol. 15, 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. (2003) Mol. Cell 12, 783–790 [DOI] [PubMed] [Google Scholar]
- 51.Stogios P. J., Privé G. G. (2004) Trends Biochem. Sci. 29, 634–637 [DOI] [PubMed] [Google Scholar]
- 52.Robinson D. N., Cooley L. (1997) J. Cell Biol. 138, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams J., Kelso R., Cooley L. (2000) Trends Cell Biol. 10, 17–24 [DOI] [PubMed] [Google Scholar]
- 54.Way M., Sanders M., Garcia C., Sakai J., Matsudaira P. (1995) J. Cell Biol. 128, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellenberg J., Siggia E. D., Moreira J. E., Smith C. L., Presley J. F., Worman H. J., Lippincott-Schwartz J. (1997) J. Cell Biol. 138, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prag S., Adams J. C. (2003) BMC Bioinformatics 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005) Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandez M. C., Andres-Barquin P. J., Martinez S., Bulfone A., Rubenstein J. L., Israel M. A. (1997) J. Neurosci. 17, 3038–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim T. A., Lim J., Ota S., Raja S., Rogers R., Rivnay B., Avraham H., Avraham S. (1998) J. Cell Biol. 141, 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soltysik-Espanola M., Rogers R. A., Jiang S., Kim T. A., Gaedigk R., White R. A., Avraham H., Avraham S. (1999) Mol. Biol. Cell 10, 2361–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., Hannink M. (2005) J. Biol. Chem. 280, 30091–30099 [DOI] [PubMed] [Google Scholar]
- 62.van den Heuvel S. (2004) Curr. Biol. 14, R59–R61 [PubMed] [Google Scholar]
- 63.Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 64.Bomont P., Cavalier L., Blondeau F., Ben Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B., Landrieu P., Hentati F., Koenig M. (2000) Nat. Genet. 26, 370–374 [DOI] [PubMed] [Google Scholar]
- 65.Caldwell G. A., Cao S., Gelwix C. C., Sexton E. G., Caldwell K. A. (2004) Adv. Neurol. 94, 79–85 [PubMed] [Google Scholar]
- 66.Dougan D. A., Mogk A., Zeth K., Turgay K., Bukau B. (2002) FEBS Lett. 529, 6–10 [DOI] [PubMed] [Google Scholar]
- 67.White S. R., Lauring B. (2007) Traffic 8, 1657–1667 [DOI] [PubMed] [Google Scholar]
- 68.Schuberth C., Buchberger A. (2008) Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giles L. M., Li L., Chin L. S. (2009) Autophagy 5, 82–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Callan A. C., Bunning S., Jones O. T., High S., Swanton E. (2007) Biochem. J. 401, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hewett J. W., Zeng J., Niland B. P., Bragg D. C., Breakefield X. O. (2006) Neurobiol. Dis. 22, 98–111 [DOI] [PubMed] [Google Scholar]
- 72.Zhu L., Wrabl J. O., Hayashi A. P., Rose L. S., Thomas P. J. (2008) Mol. Biol. Cell 19, 3599–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]