Abstract
The mitochondria-bound adapter MAVS participates in IFN induction by recruitment of downstream partners such as members of the TRAF family, leading to activation of NF-κB, and the IRF3 pathways. A yeast two-hybrid search for MAVS-interacting proteins yielded the Polo-box domain (PBD) of the mitotic Polo-like kinase PLK1. We showed that PBD associates with two different domains of MAVS in both dependent and independent phosphorylation events. The phosphodependent association requires the phosphopeptide binding ability of PBD. It takes place downstream of the proline-rich domain of MAVS, within an STP motif, characteristic of the binding of PLK1 to its targets, where the central Thr234 residue is phosphorylated. Its phosphoindependent association takes place at the C terminus of MAVS. PLK1 strongly inhibits the ability of MAVS to activate the IRF3 and NF-κB pathways and to induce IFN. Reciprocally, depletion of PLK1 can increase IFN induction in response to RIG-I/SeV or RIG-I/poly(I)-poly(C) treatments. This inhibition is dependent on the phosphoindependent association of PBD at the C terminus of MAVS where it disrupts the association of MAVS with its downstream partner TRAF3. IFN induction was strongly inhibited in cells arrested in G2/M by nocodazole, which provokes increased expression of endogenous PLK1. Interestingly, depletion of PLK1 from these nocodazole-treated cells could restore, at least partially, IFN induction. Altogether, these data demonstrate a new function for PLK1 as a regulator of IFN induction and provide the basis for the development of inhibitors preventing the PLK1/MAVS association to sustain innate immunity.
The cellular innate immune response is triggered by a variety of microbial pathogens as an essential mechanism to limit the initial spread of these infections and involves the production of inflammatory cytokines and type I interferons (IFNα/β)6 (1). In the particular case of infections with RNA viruses, IFN-α/β induction occurs following interaction of viral RNAs with two cytosolic CARD-containing DE/xDH RNA helicases: RIG-I (retinoic acid-induced gene-I (2) or MDA-5 (melanoma differentiation-associated gene-5) (3, 4). RIG-I is activated upon recognition of single-stranded RNA (ssRNAs) with uncapped 5′-triphosphate ends, specific homopolymeric RNA motifs, or short dsRNA. In contrast, activation of MDA5 occurs upon recognition of long dsRNA (5–9). Through their N-terminal CARD domains, both RIG-I and MDA5 activate the same downstream partner, the mitochondria-bound protein MAVS (also termed interferon promoter stimulator 1 (IPS-1), virus-induced signaling adaptor (VISA), CARD adaptor-inducing IFN-β (Cardif)) (10–13). MAVS, in turn, activates the IKKαβγ complex and the TBK1/IKKϵ kinases, thus provoking activation of NF-κB and IRF3 transcription factors and, ultimately, IFN induction (14, 15).
The association between MAVS and RIG-I appears to be directed through homotypic interactions between their respective N-terminal CARD domains (16, 17). In contrast, the mechanisms involved in the association of MAVS with the downstream signaling pathways are more complex, because they require activation of several kinases such as IKKα, IKKβ, IKKϵ, and TBK1. MAVS associates with Fas-associated protein via death domain (FADD) and RIP (10–13, 18) and with proteins of the TRAF family, such as TRAF3 and TRAF6 (12, 19). Recruitment of FADD, RIP1, and TRAF3 to MAVS has been recently shown to occur in complex with TRAF family member-associated NF-κB activator (TANK) (20). In turn, TANK interacts with NEMO (NF-κB essential modulator), which bridges the nuclear factor-κB and the IRF3 signaling pathways (21, 22).
The two kinases TBK1 and IKKϵ present strong homologies (14, 15). They both phosphorylate IRF3 and can compensate each other for IFN induction in innate immune cells (23). Yet they differ in several aspects, among which is the privileged association of IKKϵ with MAVS at the mitochondria upon viral infection (18, 24, 25).
A systematic search for MAVS- and IKKϵ-interacting proteins in a yeast two-hybrid screen revealed that both MAVS and IKKϵ were interacting with the Polo-box domain (PBD) of the polo-like kinase PLK1. Here, we report the MAVS/PLK1 association. The IKKϵ/PLK1 association will be addressed in a separate study. PLK1 belongs to the family of mitotic serine/threonine Polo-like kinases that play critical roles in centrosome maturation, progression through mitosis, and the onset of cytokinesis (26). PLK1 overexpression is known to occur in tumors, and it is considered as a negative prognostic marker of progression to cancer (27). PLK1 usually associates with its cognate partners in a phosphodependent manner through a phosphopeptide binding motif present in its Polo-box domain (28, 29, 30). Here, we show that the Polo-box domain of PLK1 associates with two different domains of MAVS. While one of these associations requires a phosphorylation event and the phosphopeptide binding motif of PLK1, the second one is phosphorylation-independent. Importantly, this non phosphospecific mode of interaction between PLK1 and MAVS was able to strongly disrupt the association of MAVS with its downstream partner TRAF3 and to interfere with the IFN induction pathway. PLK1 thus represents a new negative regulator of the IFN induction pathway.
EXPERIMENTAL PROCEDURES
Reagents
Control (scrambled) siRNA and siRNA to PLK1 (nucleotides 1416–1438 (31) were synthesized by Dharmacon Research, Inc. (Lafayette, CO). Normal mouse IgG, anti-c-Myc (9E10), and anti-PLK1 (F-8) monoclonal antibodies (mAbs) were from Santa Cruz Biotechnology. The anti-MAVS polyclonal antibody was from ALEXIS. Anti-actin and anti-FLAG mAbs (M2) were from Sigma. Anti-V5 mAb was from Invitrogen and anti-HSP60 mAbs from BD. Aphidicolin, nocodazole, and poly(I)-poly(C) were from Sigma. Anti-IKKϵ was provided by John Hiscott.
Yeast Two-hybrid Analysis
Open-reading frames corresponding to MAVS (nucleotides 1–1539) were fused in-frame using the following primers : MAVS-forward (5′-GGGGACAACTTTGTACAAAAAAGTTGGCATGCCGTTTGCTGAAGACAAGA), MAVS-reverse (5′-GGGGACAACTTTGTACAAGAAAGTTGGTTATGAGGGCCTGTGGCATGG), with the coding sequence of the DNA binding domain of Gal4 using in vitro recombination (Gateway System, Invitrogen) as previously described (32). Then, the Gal4-DB-MAVS construct was transfected in AH109 yeast cells (Clontech) according to the manufacturer's procedure. The fusion protein did not induce transactivation. In parallel, a commercial human spleen cDNA library cloned in the Gal4-AD pPC86 two-hybrid vector (Invitrogen) was transfected in the Y187 yeast cells (Clontech). A mating strategy was used for two-hybrid screening of the human spleen cDNA library (33). Positive colonies were selected from ∼50 million diploids on synthetic medium missing histidine and supplemented with 10 mm 3-amino-triazole (Sigma-Aldrich). The positive colonies were submitted to three rounds of screening to avoid contamination by irrelevant DNA (34). Positive clones were analyzed by direct sequencing of PCR products obtained from yeast colonies. The cDNA from these clones was then recovered and inserted into pDONR vector (Invitrogen) to allow subsequent cloning in Gateway compatible acceptor plasmids, engineered by Y. Jacob.
Plasmids
The plasmid encoding FLAG-IKKϵ was described previously (14, 35). The pV5-IKKϵ was generated by restriction digestion from pFLAG-IKKϵ(WT) and subsequent subcloning in a pcDNA3.1(+)-V5 vector engineered in the laboratory. The pcDNA3.1(+) vector was from Invitrogen. The pcDNA3.0-c-Myc-MAVS (pMyc-MAVS) full-length (FL), 1–157 (N), and 156–540 (C) were described in Ref. 18. The Myc-MAVS constructs 364–540, 470–540, 1–180, 1–230, 1–280, 1–330, 1–360, 1–508, 156–513, 180–513, 230–513, 280–513, 330–513, Δ364–470, Δ364–470(T234A), T234A, T234D, T234E, 1–513(T234A), 1–513(T234D), 1–513(T234E), 1–513(S233DT234D), 1–513(S222DS233DT234D), ST233/234AA, SS257/258AA were generated by site-directed mutagenesis. pDEST GST-MAVS1–360 and pDEST GST-MAVS1–360 T234A were prepared as follows: the MAVS-(1–360) fragments (wild type or the threonine to alanine mutant) were amplified by PCR and cloned into pDEST27 Gateway mammalian expression vector (Invitrogen). The following PCR primers were used for the amplification of MAVS-(1–360) wild type and mutant: forward primer, 5′-ggggacaactttgtacaaaaaagttggcatgCCGTTTGCTGAAGACAAGA-3′ and reverse primer, 5′-ggggacaactttgtacaagaaagttggttaTGGCACCATGCCAGCACG-3′. The sequence of the PLK1 cDNA fragment recovered from the yeast positive colonies corresponded to its polo-box domain (PLK1-(323–603) for interaction with MAVS). The full-length Myc-PLK1 plasmid was obtained from Roy Golsteyn (36). pMyc-PLK1 T210D and T210V mutants were obtained by site-directed mutagenesis. Full-length PLK1, cloned in pDON207R (Invitrogen), was inserted in-frame 3′ of a triple FLAG epitope sequence into a translation optimized pCINeo vector (Promega) using the Gateway cloning procedure (Invitrogen) to generate pFLAG-PLK1. The PLK1-PBD fragment recovered from the yeast screen and subcloned in pDON207R was inserted in the triple FLAG pCINeo vector or pEGFP-C1-modified vector using the Gateway cloning procedure. They are referred to as pFLAG-PLK1-PBD and pEGFP-PLK1-PBD. The full-length sequence of MAVS was amplified from the pMyc-MAVS plasmid and cloned into the triple FLAG vector to generate pFLAG-MAVS. The pFLAG-PLK1 construct containing the mutation K82M was generated by site-directed mutagenesis. The PLK1-PBD constructs with the mutations H538A, L540M ± W414F, were generated by site-directed mutagenesis on the pFLAG-PLK1-PBD plasmid. The PLK1-PBD sequence and the full-length cDNA of MAVS were subcloned into the pM and pVP16 vectors (Clontech), respectively. The IFN-β firefly luciferase (IFN-β-pGL2) and β-galactosidase (pRSV-βGal) reporter plasmids were described previously (35). pGal4/Luc and pCMV Renilla luciferase reporter plasmids were obtained from Y. Jacob. pGal4/Luc was engineered from basic pGL3 vector (Promega). The FLAG-TRAF3 plasmid was generated by cloning human TRAF3 by reverse transcriptase-PCR assay and insertion into the pcDNA3 FLAG vector (a gift from R. Lin). The pEF-BOS FLAG- RIG-I plasmid was described in Ref. 2.
Cells and Virus
HEK 293T, Hela, and MCF7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). Huh7 cells were cultured as described previously (35). HEK 293T were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer. Infection with Sendaï virus (a gift from D. Garcin and D. Kolakofsky (37)) was performed as in Ref. 35.
Reporter Assays
For IFN-β luciferase reporter assays, 1.4 × 105 cells were transfected with 100 ng of IFN-β-pGL2 and 50 ng of pRSV-β Gal using Lipofectamine 2000 (Invitrogen). Indicated amounts of the PLK1 constructs were co-transfected with MAVS constructs or empty vector. siRNAs targeting PLK1 (final concentration 50 nm) were transfected 24 h before transfection of different amounts of MAVS. Cells were incubated for 24–48 h and were mock-treated or infected with Sendai virus. The reporter assays were performed as described in Ref. 35. Data are shown as the mean activity of luciferase activity (normalized to β-galactosidase activity) of triplicates ± S.E.
For Gal4 luciferase reporter assays, 1.4 × 105 cells were transfected with 100 ng of Gal4-luc and 20 ng of pCMV-Renilla using Lipofectamine 2000 (Invitrogen). 400 ng of pM-PLK1-PBD, containing the DNA binding domain of Gal4 fused in-frame with the Polo-box domain of PLK1, 400 ng of pVP16-MAVS, containing the full-length sequence of MAVS fused in-frame with the transactivator VP16, were transfected individually or co-transfected. The reporter assays were performed as recommended by the manufacturer. Data are shown as the mean normalized luciferase activity of triplicates ± S.E.
Immunoprecipitation and Immunoblot Analysis
Cells were washed once with phosphate-buffered saline and scraped in lysis buffer A (50 mm Tris-HCl, pH 7.5, 140 mm NaCl, 5% glycerol, 1% CHAPS) or buffer B (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100) containing cocktails of phosphatase and protease inhibitors (Complete, Roche Applied Science). During λ-phosphatase treatment (5 units/mg protein extract; 30 min at 30 °C), phosphatase inhibitors were omitted. The protein content was analyzed by immunoblot as described (35). Lysates were incubated at 4 °C overnight with the appropriate antibodies and then in the presence of A/G-agarose beads (Santa Cruz Biotechnology) for 60 min. The beads were washed three times, and the precipitated proteins were extracted using SDS-PAGE sample buffer at 95 °C. Lysate and immunoprecipitated material were resolved by SDS-PAGE, transferred onto polyvinylidene difluoride membrane (BioRad), and probed with antibodies directed against fused tags followed by appropriate secondary reagents. Bound antibodies were detected using the ECL Plus Western blotting Detection System (Amersham Biosciences).
In Vivo Phosphorylation
HEK 293T cells, plated at 3 × 106 cells/100 mm plate, were washed once in phosphate-free Dulbecco's Modified Eagle Medium (Invitrogen) and incubated for 10 h in this medium containing 10% fetal bovine serum, previously dialyzed against phosphate-buffer-saline, 800 μCi to 1 mCi of [32P]orthophosphoric acid (8500–9120 Ci (314–337TBq)/mmol PerkinElmer Life Sciences) and 20 μm MG132 (Sigma). After lysis in buffer A, protein extracts were submitted to immunoprecipitation with the appropriate antibodies. Following separation by SDS-PAGE, the proteins were transfered to a polyvinylidene difluoride membrane, and the presence of proteins in the complex was revealed by appropriate antibodies, while the phosphorylated proteins were revealed after exposure of the membrane to an intensifying screen followed by scanning on a PhosphorImager.
Real-time RT-PCR Analysis
Total cellular RNA was extracted by acid-guanidium thiocyanate-phenol chloroform using RNAble (Eurobio, Les Ulis, France) according to the manufacturer. The sequence of the primers used for the reverse transcription polymerase chain reaction (RT-PCR) of GAPDH, IFN-β, and ISG56 were described in Ref. 38. The sequences of the IL8 primers used for RT-PCR are: IL8-forward, 5′-AAGGGCCAAGAGAATATCCGAA-3′and IL8-reverse, 5′-ACTAGGGTTGCCAGATTTAACA-3′. The RT reactions were performed on 1 μg of total RNA using oligo dT primers and avian myeloblastosis virus (AMV) reverse transcriptase (Promega). Quantitative PCR was performed on a light cycler apparatus using SYBR Green, with 2 μl of the cDNA in a 10-μl reaction mixture containing 2 μl of Light Cycler FastStart Plus DNA master SYBR Green kit (Roche Applied Science) and 0.1 μm of each primer. Standard curves were established using 10-fold serial dilutions of plasmids containing amplicons (GAPDH, IFN-β, and ISG56) or purified PCR product (IL8). The measured amounts of each mRNA were normalized to the amounts of GAPDH mRNA. The amounts of RNA were expressed as number of copies/μg total.
RESULTS
Identification of the Polo-like Kinase PLK1 as Partner to MAVS
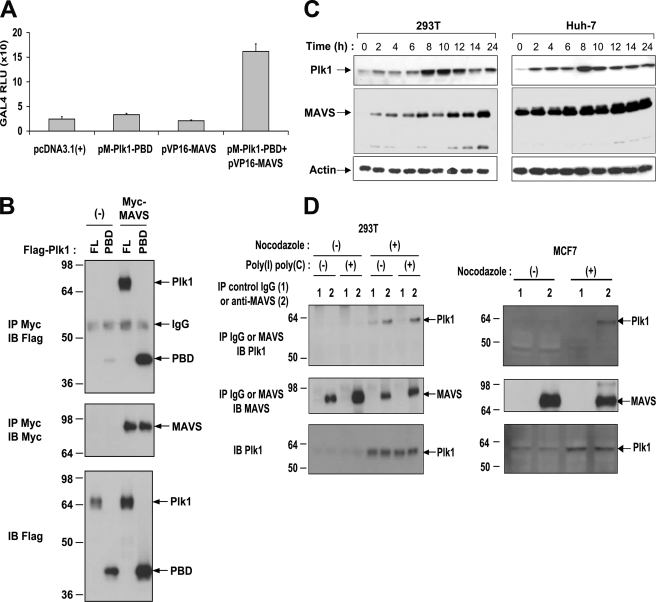
A high-throughput yeast two-hybrid screen (32) was performed to search for MAVS-interacting proteins. We used a MAVS construct deprived of its transmembrane domain (MAVS-(1–513)) to favor nuclear localization and efficient interaction with the prey library. Out of 5 × 107 transformants obtained with a human spleen cDNA library, 27 positive colonies were identified when MAVS was used as a bait. The mitotic serine/threonine Polo-like kinase PLK1 (39) was found as a major partner for MAVS along with two members of the tumor necrosis factor (TNF) receptor-associated factor family, TRAF2 and TRAF6. These data were thus in agreement with those obtained from a previous yeast two-hybrid screen using MAVS as bait (12). The sequence of the recovered PLK1 cDNA fragment (PLK1 amino acids 323–603) corresponds to its PBD. The interaction between MAVS and the PLK1-PBD was confirmed in human cells using a mammalian two-hybrid assay in which the PLK1-PBD sequence was fused in-frame with the DNA-binding domain of Gal4, while the MAVS sequence was fused to the transactivator VP16. Co-transfection of both constructs provoked an 8-fold transactivation of a Gal4-driven luciferase gene, thus showing the relevance of the interactions revealed by the yeast two-hybrid screen (Fig. 1A). Transient transfection and coprecipitation assays next showed that MAVS could interact with PLK1-PBD, as well as full-length PLK1 (Fig. 1B).
FIGURE 1.
Interaction of PLK1 with MAVS. A, HEK 293T cells were transfected with Gal4-luc and pCMV-Renilla reporter plasmids, together with an empty vector, pM-PLK1-PBD, containing the DNA binding domain of Gal4 fused in-frame with the Polo-box domain of PLK1, pVP16-MAVS, containing the full-length sequence of MAVS fused in-frame with the transactivator VP16, or both pM-PLK1-PBD and pVP16-MAVS. Firefly Gal4-luciferase activity was measured the day after and standardized to the Renilla luciferase activity. B, HEK 293T cells were transfected with pFLAG-PLK1 or pFLAG-PLK1-PBD together with either pcDNA3.1(+)(−) or pMyc-MAVS. 24 h after transfection, cell lysates (buffer B) were subjected to immunoprecipitation (IP) using c-Myc antibodies. The presence of PLK1 (FL), PLK1-PBD (PBD), or MAVS in the immunocomplexes was revealed by immunoblot using anti-FLAG or anti-Myc antibodies. Expression of PLK1, PLK1-PBD, and MAVS in total cell extracts was controlled with anti-FLAG or anti-Myc antibodies. C, HEK 293T and Huh-7 cells were plated at 60% of confluency and incubated after 24 h in presence of 1 μg/ml aphidicolin. After 14 h of incubation, the medium was removed, the cells washed three times in complete medium, and cell extracts were prepared at the indicated times. Expression of endogenous PLK1, MAVS, and actin was analyzed by immunoblot using antibodies directed against each of these proteins. D, HEK 293T or MCF7 cells were treated or not with 1 μg/ml aphidicolin for 14 h. In the case of HEK 293T, cells were also transfected with 3 μg of poly(I)-poly(C) 4 h before extraction. Cell lysates (buffer A) were immunoprecipitated with control IgG or anti-MAVS antibodies. The presence of MAVS and PLK1 in the immunoprecipitates and in the total extracts was analyzed by immunoblot using specific antibodies.
Endogenous Association of PLK1 with MAVS
Expression of PLK1 is known to be linked to the cell cycle (39). Therefore, to determine whether MAVS and PLK1 could associate at the endogenous level, we first established the kinetics of PLK1 and MAVS expression as a function of the cell cycle, in three different cell types: HEK 293T, Huh-7 (Fig. 1C), and Hela cells (supplemental Fig. S1), after growth arrest at G1/S interphase by aphidicolin treatment (26) or double thymidine block. These data confirmed the dependence of PLK1 expression upon the cell cycle with higher expression 8–12 h after block release, corresponding to the G2/M phase of the cycle. The expression of MAVS does not vary much during the cell cycle and a concomitant high expression of both proteins can be observed at the peak of PLK1 expression (Fig. 1C and supplemental Fig. S1). We then demonstrated that MAVS and PLK1 can interact at the endogenous level by using extracts from HEK 293T cells arrested at G2/M by nocodazole treatment as described (40). We also used the breast cancer cell line MCF7 (41), because PLK1 expression was reported to be constitutively elevated in tumor cells (42). Under our experimental conditions, however, the MCF7 behaved as the 293T cells with regards to endogenous levels of PLK1 and treatment with nocodazole was still necessary to increase these levels. Immunoprecipitation of endogenous MAVS pulled-down endogenous PLK1 from nocodazole-treated 293T and MCF7 cells in which expression of PLK1 was increased (Fig. 1D). Only a fraction of PLK1 could be pulled down by MAVS, which may reflect a competition between MAVS and the numerous (>600) PLK1 mitotic targets (40)). Interestingly, treatment of cells with poly(I)-poly(C), which activates endogenous MAVS through the dsRNA/RNA helicase pathway (4, 8), did not increase MAVS/PLK1 association (done with HEK293T cells; Fig. 1D left). This indicates that the association between MAVS and PLK1 may take place independently of MAVS activation.
PLK1-PBD Associates with Two Different Domains of MAVS
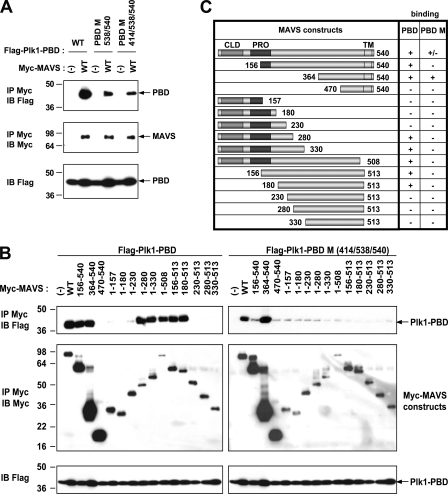
PLK1 recruits several of its targets by using a phosphopeptide binding motif located in its Polo-box domain. Through this motif, it can recognize phosphorylated threonine or serine residues, which have been previously phosphorylated by specific upstream kinases. This phosphopeptide binding ability of PLK1 is dependent on three residues His538, Lys540, and Trp414 (28, 29, 30). To determine the specificity of the association of PLK1 with MAVS, we generated the PLK1-PBD double mutant H538A/K540M and the PLK1-PBD triple mutant (PLK1-PBD W414F/H538A/K540M or PLK1-PBD M) and examined their ability to bind MAVS. Association of MAVS to both PLK1-PBD H538A/K540M and PLK1-PBD M mutants was reduced but still present, which suggested that the PLK1/MAVS association could be complex (Fig. 2A). By generating different MAVS deletion constructs, we found that PLK1-PBD binds MAVS at two different domains: an N-terminal 1–280 domain and a C-terminal 364–540 domain (Fig. 2, B and C). The MAVS N-terminal domain associates only to the PLK1-PBD construct with phosphopeptide binding ability. Comparison of the efficiency of binding of PLK1-PBD to different MAVS fragments then allowed us to restrict this region of MAVS to a deduced amino acid sequence encompassing the residues 180–280 (Fig. 2B). On the other hand, the MAVS C-terminal 364–540 domain binds PLK1-PBD, regardless of the mutations in the Trp414, His538, and Lys540 residues. With regard to this latter interaction, we could restrict it to the MAVS-(364–470) region (Fig. 2B). Two interesting observations provided more insights on this type of interaction. First, the MAVS constructs deleted of their transmembrane domain (514–540) and lacking up to the first 230 residues (MAVS-(230–513), -(280–513), and –(330–513)) were unable to bind PLK1-PBD M, although they contain the 364–470 domain. This indicates that binding of PLK1 to the C-terminal domain of MAVS depends on the transmembrane domain of MAVS and, probably, on the localization of MAVS at the mitochondria. Second, PLK1-PBD M does not bind to a MAVS-(155–540) construct despite the presence of both the 364–470 domain and the transmembrane domain. Only PLK1-PBD can bind this construct, in accord with its ability to specifically recognize the MAVS-(180–280) domain. This suggests that, in the absence of its first 155 residues, MAVS-(156–540) may adopt a conformation that does not allow PLK1-PBD M to have access to the 364–470 domain. Altogether, these results reveal that PLK1 uses its Polo-box domain to associate with two distinct domains of MAVS of ∼100 residues each, one near the proline-rich region of MAVS (180–280 amino acids) and one at the C-terminal domain of MAVS (367–470 amino acids) (summarized in Fig. 2C).
FIGURE 2.
PLK1-PBD interacts with two domains of MAVS. A, HEK 293T cells were transfected with pcDNA3.1(+)(−) or pMyc-MAVS (WT) together with pFLAG-PLK1-PBD either WT, or mutant (PBD M538/540 or PBD M 414/538/540). 24 h after transfection, cells lysates (buffer B) were immunoprecipitated (IP) with anti-c-Myc antibodies. The presence of the PLK1-PBD constructs and MAVS in the immunocomplexes and of PLK1-PBD in total cell extracts was revealed by immunoblot using anti-FLAG or anti-c-Myc antibodies. B, HEK 293T cells were transfected with pFLAG-PLK1-PBD or pFLAG-PLK1-PBD M 414/538/540 in presence of pcDNA3.1(+)(−) or different pMyc-MAVS constructs as indicated. 24 h after transfection, the cell lysates (buffer B) were immunoprecipitated (IP) with anti-c-Myc antibody. The presence of PLK1-PBD or PLK1-PBD M 414/538/540 and MAVS constructs in the immunocomplexes and of PLK1-PBD in total cell extracts was revealed by immunoblot using anti-FLAG and anti-c-Myc antibodies. C, schematic representation of the results shown in B. In MAVS, the region deliminating the CARD-like domain (CLD) is 1–77, the proline-rich region (PRO) is 103–173, and the transmembrane domain (TM) is 514–535. PBD M stands for PLK1-PBD M 414/538/540.
Association of MAVS with the Polo-box Domain of PLK1 Depends on Phosphorylation
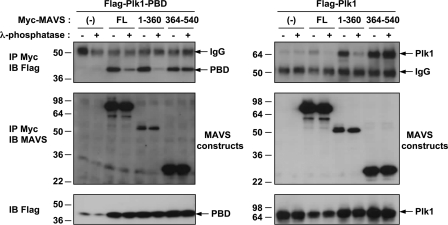
We next examined the dependence of MAVS phosphorylation on its interaction with PLK1. For this, we generated two MAVS constructs: MAVS-(1–360) and MAVS-(360–540), each containing one of the PLK1-interaction domains. These constructs were expressed in HEK 293T cells in the presence of PLK1 FL or PLK1-PBD, and the cell extracts were treated with λ phosphatase prior to MAVS immunoprecipitation. The results show that the association of MAVS-(1–360) or MAVS FL with PLK1 or PLK1-PBD was strongly inhibited by the phosphatase treatment. In contrast, association of the PLK1 constructs with MAVS-(360–540) was unaffected (Fig. 3). These data show that PLK1 associates with two separate domains of MAVS, one which depends on the phosphopeptide binding site of PLK1 and on a phosphorylation event, and one which is independent of both.
FIGURE 3.
Phosphorylation requirement for binding of PLK1 to MAVS. HEK 293T cells were transfected with pFLAG-PLK1-PBD (left panel) or pFLAG-PLK1 (right panel) together with one of the following plasmids: pcDNA3.1(+)(−), pMyc-MAVS (FL), pMyc-MAVS-(1–360), or pMyc-MAVS-(364–540). 24 h after transfection, cells were lysed in buffer B, treated or not with 5 units/μg of λ phosphatase, and submitted to immunoprecipitation (IP) with anti-c-Myc antibodies. The presence of PLK1-PBD, PLK1, and MAVS in the immunocomplexes and of PLK1 in the total cell extracts was revealed by immunoblot using anti-FLAG and anti-MAVS antibodies.
The Serine 233 and Threonine 234 Residues of MAVS Are Required for Its Association with the Phosphopeptide Binding Motif of PLK1-PBD
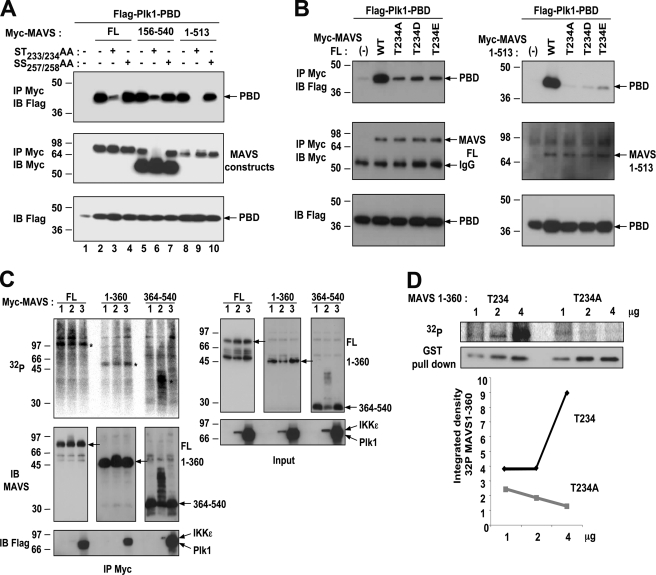
We have shown that the PBD of PLK1 associates specifically to the 180–280 domain of MAVS (Fig. 2B). This domain contains several threonine and serine residues that could serve as phosphorylation sites (supplemental Fig. S2). In particular, its 229PLARSTP235 motif resembles the PLK1 phosphopeptide binding consensus motif (P/F)-(Φ/P)-(Φ/A/Q)-(T/Q/H/M)-S-(pT/pS)-(P/X) (29). Introduction of the ST233/234AA substitution into full-length MAVS or MAVS-(156–540) strongly reduced their ability to bind PLK1-PBD, whereas the substitution SS257/258AA had no effect (Fig. 4A; lanes 3 and 6 compared with lanes 4 and 7). The remaining association depicted in lanes 3 and 6 is likely due to PLK1 interaction with the C-terminal domain of MAVS. Accordingly, the MAVS-(1–513) construct, which lost its ability to bind PLK1 through its C terminus, became unable to bind PLK1 when bearing the ST233/234AA substitution (Fig. 4A, lanes 9 and 10). Phosphorylation of the threonine residue within the motif STP has been previously shown to be required for the binding of the Polo-box domain, while the serine residue that precedes the phosphorylated Thr makes three hydrogen-bond interactions with PBD, through water molecules (29). MAVS constructs bearing the single T234A or S233A substitution were found to have a strongly reduced ability to bind PLK1-PBD (Fig. 4B and supplemental Fig. S3). However, substitution of the Thr234 residue to the phosphomimetic aspartic (D) or glutamic (E) acid residues could not restore the ability of PLK1-PBD to bind MAVS full-length or MAVS-(1–513) (Fig. 4B). It is possible that the D/E substitution is not able to act as a phosphomimetic and that authentic phosphorylation of Thr234 is necessary for recognition by PLK1-PBD. Altogether, these results clearly show that the Ser233 and Thr234 residues of MAVS play an important role in the recruitment of PLK1 to MAVS.
FIGURE 4.
Requirement of MAVS Ser233 and Thr234 for its specific binding with PLK1-PBD. A, HEK 293T cells were transfected with pFLAG-PLK1-PBD in the presence of pMyc-MAVS, either full-length (FL) or its fragments 156–540 or 1–513, in which the sequence was either unmodified (−) or carrying the mutations ST233/234A or SS257/258AA (+). B, HEK 293T cells were transfected with pFLAG-PLK1-PBD in the presence of pMyc-MAVS, either full-length (FL) or deleted of its transmembrane domain (1–513), in which the sequence was either unmodified (WT) or carrying the mutations T234A, T234D, or T234E. 24 h after transfection, cells lysates from experiments A and B (buffer B) were immunoprecipitated with anti-Myc antibodies. The presence of PLK1-PBD and of the MAVS constructs in the immunocomplexes and of PLK1-PBD in the total cell extracts was revealed by immunoblot using anti-FLAG or anti-Myc antibodies. C, HEK 293T cells were transfected with pMyc-MAVS full-length (FL) or its fragments 1–360 and 364–540 in presence of pcDNA3.1(+) (1), pFLAG-IKKϵ (2), or pFLAG-PLK1 (3). Six hours after transfection, the cells were incubated for 10 h in the presence of 1 mCi of [32P]orthophosphoric acid and 20 μm MG132. The cells lysates (buffer A) were immunoprecipitated with anti-c-Myc antibodies. The presence of MAVS constructs in the immunoprecipitates was first controlled by immunoblot using anti-MAVS antibodies, after which the membrane was exposed against an intensifying screen to reveal the 32P-labeled proteins through scanning on a phosphorimager. The membrane was then analyzed by immunoblot for the expression of PLK1 or IKKϵ using anti-FLAG antibodies. Expression of IKKϵ or PLK1 in the total cell extracts was controlled by immunoblots using anti-FLAG antibodies. D, HEK 293T cells were transfected with 1, 2, or 4 μg of pDEST GST-MAVS-(1–360) or pDEST GST-MAVS-(1–360) T234A and labeled with 1 mCi of [32P]orthophosphoric acid as above. The cells were lysed in buffer B minus glycerol and the GST-MAVS constructs were purified on glutathione-Sepharose. The presence of MAVS constructs and phospho-MAVS was revealed as above. The quantification of the phosphorylated bands versus the bands on the immunoblot was calculated using the ImageJ software.
In Vivo Phosphorylation of MAVS
Because PLK1 uses its phosphopeptide binding ability to bind MAVS, we set out to determine whether MAVS could be phosphorylated in vivo. We performed MAVS immunoprecipitation experiments after in vivo 32P labeling of cells expressing MAVS FL, or the MAVS-(1–360) or MAVS-(360–540) fragments. Expression of each of these constructs was performed alone or in the presence of Flag-PLK1 since PLK1 has been reported to phosphorylate some of its docking targets (43). As a control for phosphorylation, we used V5-IKKϵ, since we have observed that this kinase can phosphorylate MAVS (25). The results show that MAVS full-length was constitutively phosphorylated and that its phosphorylation was increased in the presence of IKKϵ (Fig. 4C). MAVS-(1–360) was also constitutively phosphorylated, whereas MAVS-(360–540) was not. Neither IKKϵ nor PLK1 could increase the phosphorylation of MAVS-(1–360). MAVS-(360–540) was not phosphorylated in the presence of PLK1 but became strongly phosphorylated in the presence of IKKϵ (Fig. 4C). The ability of IKKϵ to phosphorylate the C terminus of MAVS was beyond the scope of this investigation and will be addressed in a separate study. Finally, we compared the in vivo phosphorylation state of different concentrations of MAVS-(1–360) constructs that differ only by the presence of a threonine or an alanine residue at position 234. We have verified that the former binds PLK1-PBD and the other not (supplemental Fig. S3A). The results show a strong and specific increase in the phosphorylation state of MAVS-(1–360) expressing authentic Thr234, therefore indicating the importance of this residue in MAVS phosphorylation (Fig. 4D). Altogether, these results show for the first time that MAVS can be phosphorylated in vivo. In particular, the phosphorylation of the threonine 234 residue at the N-terminal domain of MAVS can provide the physiological conditions necessary to recruit PLK1 through the phosphopeptide binding motif.
PLK1 Inhibits MAVS-mediated IFNβ Stimulation
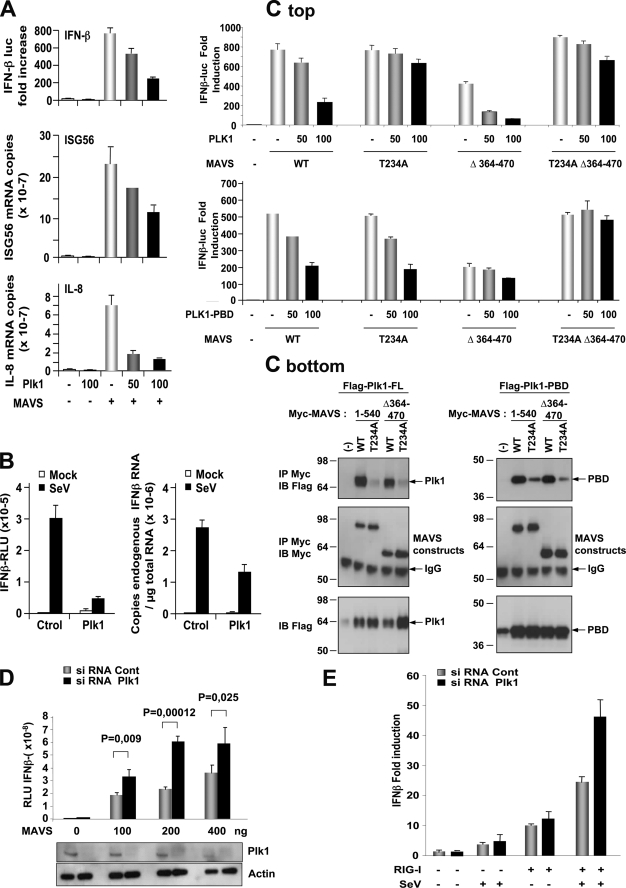
MAVS is an important factor of the IFN-inducing pathway. We, therefore, examined the effect of PLK1 on MAVS activity, using functional reporter assays (Fig. 5A). PLK1 alone had no intrinsic ability to induce IFN, in agreement with a previous report (12). However, when expressed in the presence of MAVS, PLK1 strongly inhibited the stimulation of the IFNβ promoter (60%), as well as the induction of the endogenous ISG56 (50%) and IL-8 (80%) genes, used as representatives of the IRF-3 and NF-κB signaling pathways, respectively (Fig. 5A). The effect of PLK1 on the IFN-inducing pathway was then examined in response to infection by Sendaï virus (SeV), one of the viruses activating the RIG-I/MAVS pathway (5). A 75-fold stimulation of the IFNβ promoter provoked by the SeV infection was strongly inhibited in the presence of overexpressed PLK1 (84%) (Fig. 5B, left). A similar inhibition was observed on endogenous IFNβ induced by SeV (Fig. 5B, right).
FIGURE 5.
PLK1 inhibits the MAVS-mediated IFN induction. A, HEK 293T cells were transfected with pMyc-MAVS (10 ng) alone or in the presence of pMyc-PLK1 (50 and 100 ng) and in the presence (top) or in the absence (middle and bottom) of IFN-β-pGL2 and pRSV-βgal. pMyc-PLK1 was also expressed individually. 24 h after transfection, luciferase activity was measured and normalized against the β-galactosidase activity (top panel). Expression levels of endogenous ISG56 (middle panel), and IL-8 (lower panel) were measured by RT-qPCR and normalized against endogenous GAPDH levels. Error bars represent the mean ± S.D. for triplicates. B, HEK 293T cells were transfected with 400 ng of an empty vector or pMyc-PLK1 in the presence (left) or in the absence (right) of IFNβ-pGL2 and pRSV-βgal. 24 h after transfection, cells were either mock-infected or infected with 20 HAU/ml of Sendaï virus. The stimulation of the IFNβ reporter plasmid or induction of the IFNβ endogenous gene were measured 18 h after infection, either by luciferase activity (left) or by RT-qPCR (right). Relative expression levels of IFNβ versus GAPDH is shown. Data are representative of two experiments run in triplicate. C top, HEK 293T cells were transfected with pMyc-MAVS (10 ng), either WT or with the mutation T234A, the deletion Δ364–470 or both (T234AΔ364–470) in the presence of 50 and 100 ng of pPLK1(top) or pPLK1-PBD (bottom) and in the presence of IFN-β-pGL2 and pRSV-βgal. 24 h after transfection, luciferase activity was measured and normalized against the β-galactosidase activity. The experiment was done in triplicate. Bottom, HEK 293T cells were transfected with pFLAG-PLK1-FL or pFLAG-PLK1-PBD in the presence of pMyc-MAVS, either full-length (1–540) or deleted of the 364–470 domain (Δ364–470), in which the sequence was either unmodified (WT) or with the mutation T234A. 24 h after transfection, the cells were lysates (buffer B) were immunoprecipitated with anti-Myc antibodies. The presence of the PLK1 and MAVS constructs in the immunoprecipitates and of the PLK1 constructs in the total cell extracts was revealed by immunoblot using anti-FLAG and anti-Myc antibodies. D, HEK 293T cells were first transfected with 50 nm Control (scrambled) or PLK1 siRNA and, after 24 h, with IFN-β-pGL2 and pRSV-βgal in presence of pMyc-MAVS (100, 200, or 400 ng). Luciferase activity was analyzed 24 h after the second transfection and normalized to the β-galactosidase activity. Error bars represent the mean ± S.D. for triplicates, and the p values were calculated according to the Student's t test. Cells lysates were analyzed by immunoblot using PLK1 and actin antibodies. E, HEK 293T cells were transfected with 50 nm Control or PLK1 siRNA and, after 24 h, with IFN-β-pGL2/pRSV-βgal and pRIG-I (400 ng). The cells were then infected with Sendaï virus (20 HAU/ml). After 6 h, luciferase activity was measured and normalized to the β-galactosidase activity.
We used MAVS mutants containing the T234A substitution and/or a deletion of the 364–470 domain to determine the relative importance of the two PLK1-binding domains on PLK1-mediated inhibition of MAVS. MAVS FL and the MAVS constructs MAVS T234A, MAVS Δ364–470 and the double mutant MAVS T234AΔ364–470 were expressed in the presence of PLK1 or PLK1-PBD and assessed for their ability to induce an IFNβ reporter vector (Fig. 5C, top). In parallel, these constructs were assayed for their ability to bind PLK1 or PLK1-PBD (Fig. 5C, bottom). The results confirmed efficient binding of both PLK1 and PLK1-PBD to MAVS FL (see Figs. 1, 2, and 4) and inhibition of MAVS activity by PLK1 (see Fig. 5, A and B). In addition, these results show that PLK1-PBD is just as efficient as PLK1 FL in inhibiting the activity of MAVS FL (Fig. 5C, top). PLK1 failed to bind MAVS T234A and MAVS T234AΔ364–470 and lost most of its ability to inhibit the activity of these MAVS constructs. In contrast, PLK1 was still efficient in binding MAVS Δ364–470 and could inhibit its activity. PLK1-PBD could bind MAVS T234A with reduced efficacy as compared with MAVS FL and was able to inhibit MAVS activity. PLK1-PBD was able also to bind MAVS Δ364–470 as efficiently as MAVS FL but could not inhibit it. Finally, PLK1-PBD was able to only poorly bind to MAVS T234AΔ364–470 and was not able to inhibit its IFN-inducing ability. Altogether, these results show that both PLK1-binding domains of MAVS are involved in the inhibition of MAVS by PLK1. A possible scenario of the MAVS/PLK1 interaction and effect on IFN induction is presented in the discussion (see also Fig. 8). The ability of PLK1 to inhibit the IFN induction pathway was further strengthened by the fact that depletion of endogenous PLK1 by siRNA resulted in an increase in MAVS activity (Fig. 5D). Finally, we analyzed the effect of the depletion of PLK1 on the RIG-I signaling pathway by transfecting limited amounts of RIG-I and analyzing stimulation of the IFN signaling pathway 6 h after infection with Sendaï virus. The results show that in conditions where the viral infection strongly activates RIG-I and IFN induction, depletion of PLK1 further potentiates this signaling pathway (Fig. 5E).
FIGURE 8.
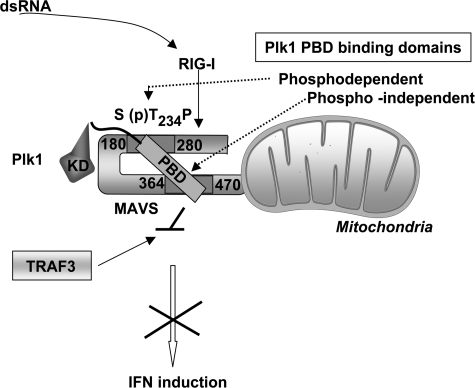
Model of interaction of PLK1 with MAVS. PLK1 interacts with the STP motif present in the MAVS-(180–280) domain through its phosphopeptide docking site contained within its Polo-box. This interaction is dependent on the phosphorylation of MAVS-Thr234. The PLK1 Polo-box interacts also with a region of MAVS at its C-terminal domain but does not use its phosphopeptide docking site for this. The latter interaction is responsible for the inhibition of the recruitment of TRAF3 to MAVS and for the inhibition of IFN induction.
PLK1 Inhibits IFN Induction by Disrupting the TRAF3/MAVS Association
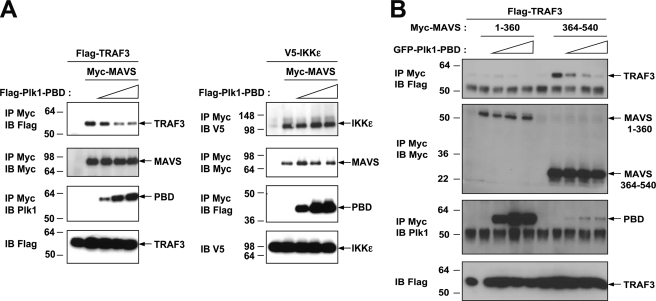
MAVS activity has been reported to depend on its association with members of the TRAF family, such as the E3-ubiquitin ligases TRAF2, TRAF6, and TRAF3 through TRAF-interacting motifs (TIM). In particular, TRAF3 is involved in the activation of the IRF3 pathway, which is essential for IFN induction (19). We thus determined the effect of PLK1-PBD on the MAVS/TRAF3 association. As a control, we compared it to the known association between MAVS and IKKϵ (18, 25). We performed co-precipitation studies of MAVS and TRAF3 in the presence of increasing amounts of PLK1-PBD. Increasing PLK1-PBD led to a strong reduction of the MAVS/TRAF3 association, whereas this had no effect on the MAVS/IKKϵ association (Fig. 6A). Recent reports show that MAVS dimerization leads to recruitment of downstream partners (44, 45). We showed that PLK1 was unable to prevent MAVS dimerization (supplemental Fig. S4), thus indicating that PLK1 can bind MAVS regardless of its state of activation. This is in accord with the data in Fig. 1 where we showed that binding of PLK1 to MAVS was not increased in the presence of poly(I)-poly(C). We then analyzed the effect of increasing concentrations of PLK1-PBD on the association of TRAF3 with the two MAVS-(1–360) and –(360–540) constructs, each containing one of the PLK1 binding domains. First, we found that TRAF3 associates with MAVS-(360–540) and not with MAVS-(1–360)7 in contrast with the data from Saha et al. (19). The difference between our results and those of Saha et al. may be related to the MAVS constructs used in each case. The role of the C-terminal TRAF3 binding domain of MAVS will be addressed in a separate study. Interestingly, PLK1-PBD was able to strongly abolish the interaction of TRAF3 to the MAVS-(360–540) domain (Fig. 6B). This was in accord with our functional reporter assay in which deletion of MAVS-(364–470) restores MAVS activity in presence of PLK1-PBD. These data demonstrate that one possible mechanism for the negative regulation of PLK1 in IFN induction is through the interaction of its PBD at the C-terminal domain of MAVS and involves deregulation of TRAF3.
FIGURE 6.
PLK1 inhibits binding of TRAF3 to MAVS. A, left, HEK 293T cells were transfected with pFLAG-TRAF3, pMyc-MAVS, and increasing amounts of pEGFP-PLK1-PBD. 24 h after transfection, cell lysates (buffer B) were immunoprecipitated (IP) using c-Myc antibodies. The presence of TRAF3 (upper panel), MAVS, and PLK1-PBD (middle panels) in the immunocomplexes was revealed by immunoblot using anti-FLAG, anti-c-Myc, or anti-PLK1 antibodies. Expression of TRAF3 was controlled in the total cell extracts using anti-FLAG antibodies (lower panel). A, right, HEK 293T cells were transfected with pV5-IKKϵ, pMyc-MAVS, and increasing amounts of pFLAG-PLK1-PBD. After 24 h, Myc-MAVS was immunoprecipitated as in A, left. The presence of IKKϵ (upper panel), MAVS and PLK1-PBD (middle panels) in the immunocomplexes was revealed by immunoblot using anti-V5, anti-c-Myc, or anti-FLAG antibodies. IKKϵ was measured in total cell extracts using anti-V5 antibodies (lower panel). B, HEK 293T cells were transfected with pFLAG-TRAF3 together with pMyc-MAVS-(1–360) or pMyc-MAVS-(364–540) and increasing amounts of pEGFP-PLK1-PBD. After 24 h, Myc-MAVS was immunoprecipitated as in A. The presence of TRAF3 (upper panel), MAVS and PLK1-PBD (middle panels) in the immunocomplexes was revealed by immunoblot using anti-FLAG, anti-c-Myc, or anti-PLK1 antibodies. TRAF3 was measured in total cell extracts using anti-FLAG antibodies (lower panel).
PLK1 Participates in the Inhibition of IFN Induction during Treatment with Antimicrotubule Drugs
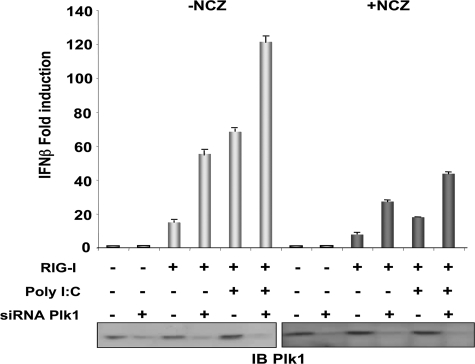
Because nocodazole treatment allows to detect PLK1/MAVS interaction (Fig. 1D), we were prompted to examine whether nocodazole could affect the MAVS-mediated innate immune response alone or in the presence of PLK1. To test this, HEK 293T cells, depleted or not from their endogenous PLK1, were submitted to nocodazole treatment and MAVS signaling was assayed after transfection with RIG-I and poly(I)-poly(C). The results first confirmed that depletion of PLK1 can increase the RIG-I-mediated IFN induction as shown in Fig. 5E where SeV was used as an activator for RIG-I. Here, poly(I)-poly(C) was used instead of SeV due to the concern that endocytosis of the virus might be impaired by the depolymerization of the microtubules upon nocodazole treatment. In those conditions, we observed that nocodazole treatment strongly increased the expression levels of PLK1 and led to a dramatic inhibition of IFN induction. Interestingly, depletion of PLK1 was found to partially restore IFN induction (Fig. 7). These data suggest that anticancer treatments using agents interfering with the dynamics of microtubules may provoke an unwanted increase in PLK1 that can interfere with the innate immune signaling pathways.
FIGURE 7.
PLK1 participates in the inhibition of IFN induction during treatment with nocodazole. HEK 293T cells were first transfected with 50 nm Control or PLK1 siRNA and, after 24 h, with IFN-β-pGL2/pRSV-βgal in the absence or presence of 400 ng of pRIG-I. 8 h after transfection, the cells were submitted to nocodazole treatment (50 ng/ml) for 20 h. 6 h before the end of treatment, cells were transfected with 100 ng/ml of poly(I)-poly(C). Luciferase activity was analyzed 6 h after the second transfection and normalized to the β-galactosidase activity. Error bars represent the mean ± S.D. for triplicates. Cell lysates were analyzed by immunoblot using PLK1 antibodies.
DISCUSSION
A search for partners to the IFN-inducing mitochondrial adapter MAVS by a yeast two-hybrid assay revealed the mitotic Polo-like kinase PLK1. We show that PLK1 interacts with MAVS through its Polo-box domain and interferes with induction of IFN.
One of the particularities of PLK1 is to bind its targets through its Polo-Box domain, in a phosphodependent manner, via a pSer/pThr-binding module that specifically recognizes Ser-(pSer/pThr)-Pro/X motifs and favors localization of PLK1 to specific subcellular structures (46). More that 600 potential targets for PLK1 have already been identified by affinity purification and mass spectrometry studies (40). Here, we showed that MAVS behaves as a target for PLK1 based on the fact that full association of MAVS with PLK1 requires the phosphopeptide binding motif of PLK1 (Fig. 2) and based on in vivo phosphorylation events (Fig. 3). We could link this specific association to the N-terminal end of MAVS, between residues 180 and 280 (Fig. 2). This region contains an Ser233-Thr234-Pro motif that could correspond to the phosphopeptide Ser-(pSer/pThr)-Pro recognized by PBD (29, 40), and indeed, we showed the importance of both MAVS residues Ser233 and Thr234 in its association with PLK1 because their substitution to alanine impairs the association (Fig. 4A). Substitution of Thr234 or Ser233 to the phosphomimetics D/E could not restore association of MAVS to PLK1-PBD (Fig. 4B and supplemental Fig. S3). This indicates that the authentic presence of a phosphate may be essential for MAVS to behave as a target for PLK1. Indeed, our experimental data showed that MAVS can be phosphorylated in vivo on its 1–360 domain and that the substitution T234A decreased its level of phosphorylation (Fig. 4D). We have observed that the phosphorylation of MAVS-(1–360) was not increased in the presence of PLK1. Priming kinases responsible for the phosphorylation of MAVS at its PLK1 binding STP motif are currently being searched.
We found that PLK1 was also able to bind to the C-terminal domain of MAVS, irrespective of its phosphopeptide binding ability (Fig. 2) and in a phosphoindependent manner (Fig. 3). This may involve direct binding of this MAVS domain to a different interface of PLK1-PBD or may involve other partner(s) related to the localization of MAVS at the mitochondria. Recently, a phosphorylation-independent binding of PLK1-PBD to the protein Bora has also been reported (47).
In view of the importance of MAVS in IFN induction, we examined the effect of PLK1 on this signaling pathway. PLK1 behaved as a strong inhibitor of the MAVS-mediated ability to induce IFN through the downstream NF-κB and IRF3 signaling pathways. PLK1 was also able to inhibit IFN induction in response to a viral stimulus, such as Sendaï virus and silencing of its expression could boost IFN induction in response to MAVS. MAVS mutated in its N-terminal PLK1 binding domain no longer binds PLK1 and is resistant to its inhibition. MAVS mutated in its C-terminal PLK1 binding domain still binds PLK1. These results indicate that the recruitment of PLK1 to the N-terminal domain of MAVS through its phosphopeptide docking site may occur first. Then PLK1 would bind the C terminus of MAVS through a different surface on its PBD (Fig. 5C). This would interfere with the recruitment of TRAF3 and subsequently inhibit MAVS activity. A model of the PLK1/MAVS association is presented in Fig. 8.
As expected from its negative role on MAVS activity, silencing of PLK1 was able to increase induction of IFN in response to ectopically expressed MAVS (Fig. 5D). In this respect, PLK1 belongs to the list of recently reported regulators of the IFN induction pathway at the level or immediately downstream of MAVS. For instance, the RNA helicase LGP2, similar to RIG-I and MDA5, but devoid of the CARD-like domain, binds a region close to the C-terminal domain of MAVS and can interfere with the binding of IKKϵ (16). NLRX1, a nucleotide-binding domain (NBD)- and leucine-rich repeat (LRR)-containing family member (NLR) localizes to the mitochondrial outer membrane, interacts with MAVS and inhibits MAVS activity (48). Deubiquitinating enzyme A (DUBA), an ovarian tumor domain-containing deubiquitinating enzyme, binds to and selectively cleaves the lysine 63-linked polyubiquitin chains on TRAF3 resulting in its dissociation from the downstream signaling complex containing TBK1 and behaves as a negative regulator of innate immune responses (49).
Silencing of PLK1 was found to increase MAVS activity, while its overexpression inhibited MAVS. PLK1 expression levels vary during the cell cycle. In the course of our study, we have shown that nocodazole treatment, which can increase the expression levels of PLK1 and favor its association with MAVS, can also strongly inhibit IFN induction. Depletion of PLK1 could partially restore IFN induction, confirming that at the endogenous level, PLK1 plays a role in the control of innate immunity. Nocodazole treatment is known to interfere with the polymerization of microtubules. PLK1 interacts through its kinase domain to all three components of microtubules, α-tubulin, β-tubulin, and γ-tubulin, regardless of their association in microtubules or not (50). It has been recently shown that mitochondria can move along the microtubule network (51). One attractive hypothesis is that, once bound to microtubules through its kinase domain, PLK1 may interact with MAVS through its Polo-box domain and control MAVS activity.
PLK1 overexpression has been characterized in many tumor cells as a marker of tumor progression (42). Several examples indicate a correlation between the dysregulation of PLK1 expression and/or activity during infection with some viruses that are known to provoke uncontrolled cell proliferation and drive the cells toward cancer. For example, CD4(+) T cells from simian immunodeficiency virus (SIV)-infected rhesus macaques with high viral loads exhibited both up-regulation of Plk1 and an aberrant activation-induced Plk1 response, indicating that SIV-related disease may contribute to aberrant T-cell responses and disease pathogenesis (52). Interestingly, in the case of HIV infection, Vpr can increase viral replication, arrest proliferating CD4+ T cells in the G2 phase of the cell cycle, and provoke cell division defects in fission yeast, Hela, and primary cells. In particular, it was shown to provoke mislocalization of the Plo1 yeast homolog of Polo kinase, resulting in multiple discrete cytoplasmic dots or concentration in the nucleus (53). The hypothesis that a reorganisation of PLK1 during HIV-I infection may favor its association with MAVS and control of innate immune response remains to be explored. The human papillomavirus type-16 E6/E7 proteins also provoked the deregulation of cellular genes such as Plk1, Aurora-A, cdk1, and Nek2, thus contributing to genomic instability and conversion to cancer cells, in addition to abrogating the normal functions of p53 and retinoblastoma (54).
Our results show an association between PLK1 and the C-terminal domain of MAVS and its control on MAVS activity. These data support the new notion that one of the aspects of the ability of PLK1 to drive the cells toward tumorigenesis is its ability to deregulate the innate immune control of the cells. We have shown that treatment of cells with antimicrotubule agents, such as nocodazole, can dramatically inhibit IFN induction but that some restoration of IFN induction could be achieved by depleting PLK1 from the cells. It has been recently reported that the treatment of breast cancer with siRNAs targeting PLK1 improved the sensitivity toward paclitaxel and induced an antiproliferative effect (55). It is possible that depleting PLK1 would prevent its negative control of MAVS and allow sufficient induction of IFN, which also has antiproliferative action. In addition to siRNA aimed at depleting PLK1 in tumoral cells, other strategies are aimed to inhibit the kinase activity of PLK1 through chemical inhibitors, such as the ATP-competitive dihydropteridinone derivative BI2536, (42, 56). The clinical potential of inhibitors that target the Polo-box domain to prevent access of PLK1 to its cellular partners is also being investigated. In this respect, the synthetic thymoquinone derivative Poloxin has been shown to cause PLK1 mislocalization, mitotic arrest and apoptosis in HeLa cells (57). Here, we have shown for the first time that the PLK1 inhibits IFN induction at the level of MAVS through its polo-box domain. Specific inhibitors that prevent binding of PLK1 to MAVS would therefore be of great interest for therapeutic intervention in viro-induced cancers, in conjunction with the use of antimicrotubule agents, to strengthen the ability of the cells to mount an innate immune response.
Supplementary Material
Acknowledgments
We thank Roy Golsteyn for the gift of the PLK1 plasmid, Olivera Grubisha for critical reading of the manuscript, and Robert Weil, Jesus Prieto, Damien Arnoult, and Erich Nigg for helpful discussions. The authors declare no conflict of interest.
This work was supported in part by grants from a Research Transversal Program (186), from the Pasteur Institute, and by the Agence Nationale pour la Recherche contre le SIDA (ANRS) (to E. F. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
S. Paz, M. Vilasco, and M. Arguello, unpublished observations.
- IFN
- interferon
- MAVS
- mitochondrial antiviral signaling
- CARD
- caspase activation and recruitment domain
- RIP
- receptor-interacting protein
- TRAF
- tumor necrosis factor receptor-associated factor
- TBK1
- TANK-binding kinase 1
- NF-κB
- nuclear factor-κB
- IκBα
- (inhibitor of κB) α
- IKK
- IκB (inhibitor of κB) kinase
- IRF3
- interferon regulatory factor-3
- IL-8
- interleukin-8
- ISG56
- interferon-stimulated gene 56
- dsRNA
- double-stranded RNA
- siRNA
- silencing RNA
- RSV
- Rous Sarcoma Virus
- CMV
- cytomegalovirus
- Gal4
- galactosidase transcription factor
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- FL
- full-length
- WT
- wild type
- GST
- glutathione S-transferase.
REFERENCES
- 1.Barton G. M., Medzhitov R. (2003) Science 300, 1524–1525 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 3.Kang D. C., Gopalkrishnan R. V., Wu Q., Jankowsky E., Pyle A. M., Fisher P. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e., Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 7.Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr. (2008) Nature 454, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito T., Gale M., Jr. (2008) J. Exp. Med. 205, 1523–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 11.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 12.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 13.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 14.Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 16.Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitour D., Meurs E. F. (2007) Sci STKE 2007, pe20. [DOI] [PubMed] [Google Scholar]
- 18.Lin R., Lacoste J., Nakhaei P., Sun Q., Yang L., Paz S., Wilkinson P., Julkunen I., Vitour D., Meurs E., Hiscott J. (2006) J. Virol. 80, 6072–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michallet M. C., Meylan E., Ermolaeva M. A., Vazquez J., Rebsamen M., Curran J., Poeck H., Bscheider M., Hartmann G., König M., Kalinke U., Pasparakis M., Tschopp J. (2008) Immunity 28, 651–661 [DOI] [PubMed] [Google Scholar]
- 21.Zhao T., Yang L., Sun Q., Arguello M., Ballard D. W., Hiscott J., Lin R. (2007) Nat. Immunol. 8, 592–600 [DOI] [PubMed] [Google Scholar]
- 22.Pomerantz J. L., Baltimore D. (1999) EMBO J. 18, 6694–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsui K., Kumagai Y., Kato H., Sato S., Kawagoe T., Uematsu S., Takeuchi O., Akira S. (2006) J. Immunol. 177, 5785–5789 [DOI] [PubMed] [Google Scholar]
- 24.Komuro A., Horvath C. M. (2006) J. Virol. 80, 12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paz S., Vilasco M., Arguello M., Sun Q., Lacoste J., Nguyen T. L., Zhao T., Shestakova E. A., Zaari S., Bibeau-Poirier A., Servant M. J., Lin R., Meurs E. F., Hiscott J. (2009) Mol. Cell. Biol. 29, 3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gumireddy K., Reddy M. V., Cosenza S. C., Boominathan R., Baker S. J., Papathi N., Jiang J., Holland J., Reddy E. P. (2005) Cancer Cell 7, 275–286 [DOI] [PubMed] [Google Scholar]
- 27.Reagan-Shaw S., Ahmad N. (2005) Faseb. J. 19, 611–613 [DOI] [PubMed] [Google Scholar]
- 28.Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N. (2003) EMBO J. 22, 5757–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 30.García-Alvarez B., de, Cárcer G., Ibañez S., Bragado-Nilsson E., Montoya G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spänkuch-Schmitt B., Wolf G., Solbach C., Loibl S., Knecht R., Stegmüller M., von Minckwitz G., Kaufmann M., Strebhardt K. (2002) Oncogene 21, 3162–3171 [DOI] [PubMed] [Google Scholar]
- 32.Walhout A. J., Sordella R., Lu X., Hartley J. L., Temple G. F., Brasch M. A., Thierry-Mieg N., Vidal M. (2000) Science 287, 116–122 [DOI] [PubMed] [Google Scholar]
- 33.Fromont-Racine M., Rain J. C., Legrain P. (1997) Nat. Genet. 16, 277–282 [DOI] [PubMed] [Google Scholar]
- 34.Vidalain P. O., Boxem M., Ge H., Li S., Vidal M. (2004) Methods 32, 363–370 [DOI] [PubMed] [Google Scholar]
- 35.Breiman A., Grandvaux N., Lin R., Ottone C., Akira S., Yoneyama M., Fujita T., Hiscott J., Meurs E. F. (2005) J. Virol. 79, 3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golsteyn R. M., Schultz S. J., Bartek J., Ziemiecki A., Ried T., Nigg E. A. (1994) J. Cell Sci. 107 ( Pt 6), 1509–1517 [DOI] [PubMed] [Google Scholar]
- 37.Strahle L., Garcin D., Kolakofsky D. (2006) Virology 351, 101–111 [DOI] [PubMed] [Google Scholar]
- 38.Vilasco M., Larrea E., Vitour D., Dabo S., Breiman A., Regnault B., Riezu J. I., Eid P., Prieto J., Meurs E. F. (2006) Hepatology 44, 1635–1647 [DOI] [PubMed] [Google Scholar]
- 39.Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 40.Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson R. B., Bates S. E., McManaway M. E., Lippman M. E. (1986) Cancer Res. 46, 1707–1713 [PubMed] [Google Scholar]
- 42.Strebhardt K., Ullrich A. (2006) Nat. Rev. Cancer 6, 321–330 [DOI] [PubMed] [Google Scholar]
- 43.Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E. A., Sillje H., Barr F. A. (2007) Nat. Cell Biol. 9, 436–444 [DOI] [PubMed] [Google Scholar]
- 44.Baril M., Racine M. E., Penin F., Lamarre D. (2009) J. Virol. 83, 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang E. D., Wang C. Y. (2009) J. Virol. 83, 3420–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 47.Seki A., Coppinger J. A., Du H., Jang C. Y., Yates J. R., 3rd, Fang G. (2008) J. Cell Biol. 181, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. (2008) Nature 451, 573–577 [DOI] [PubMed] [Google Scholar]
- 49.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 50.Feng Y., Hodge D. R., Palmieri G., Chase D. L., Longo D. L., Ferris D. K. (1999) Biochem. J. 339, 435–442 [PMC free article] [PubMed] [Google Scholar]
- 51.Huang B., Jones S. A., Brandenburg B., Zhuang X. (2008) Nature Methods 5, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bostik P., Dodd G. L., Villinger F., Mayne A. E., Ansari A. A. (2004) J. Virol. 78, 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang F., Re F., Sebastian S., Sazer S., Luban J. (2004) Mol. Biol. Cell 15, 1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel D., Incassati A., Wang N., McCance D. J. (2004) Cancer Res. 64, 1299–1306 [DOI] [PubMed] [Google Scholar]
- 55.Spänkuch B., Kurunci-Csacsko E., Kaufmann M., Strebhardt K. (2007) Oncogene 26, 5793–5807 [DOI] [PubMed] [Google Scholar]
- 56.Steegmaier M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., Grauert M., Adolf G. R., Kraut N., Peters J. M., Rettig W. J. (2007) Curr. Biol. 17, 316–322 [DOI] [PubMed] [Google Scholar]
- 57.Reindl W., Yuan J., Krämer A., Strebhardt K., Berg T. (2008) Chem Biol 15, 459–466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.