Abstract
Facile control of targeted intracellular protein degradation has many potential uses in basic science and biotechnology. One promising approach to this goal is to redesign adaptor proteins, which can regulate proteolytic specificity by tethering substrates to energy-dependent AAA+ proteases. Using the ClpXP protease, we have probed the minimal biochemical functions required for adaptor function by designing and characterizing variant substrates, adaptors, and ClpX enzymes. We find that substrate tethering mediated by heterologous interaction domains and a small bridging molecule mimics substrate delivery by the wild-type system. These results show that simple tethering is sufficient for synthetic adaptor function. In our engineered system, tethering and proteolysis depend on the presence of the macrolide rapamycin, providing a foundation for engineering highly specific degradation of target proteins in cells. Importantly, this degradation is regulated by a small molecule without the need for new adaptor or enzyme biosynthesis.
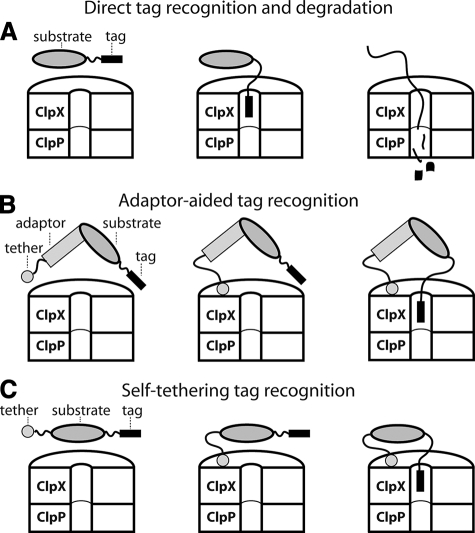
Targeted proteolytic degradation plays important roles in protein quality control and in regulating cellular circuitry in organisms ranging from bacteria to humans (1–4). In some instances, substrates are recognized directly by a protease enzyme via a degradation tag (Fig. 1A) (see Refs. 5 and 6). In other cases, adaptor proteins or multiple types of substrate sequences are also required to ensure efficient degradation (Fig. 1, B and C) (see Refs. 7 and 8).
FIGURE 1.
A, the ClpX component of the ClpXP protease recognizes some substrates via a degradation tag, denatures the substrate, and then translocates the unfolded protein into ClpP for degradation. B, adaptor-assisted binding of a substrate to ClpXP. C, self-tethering of a substrate to ClpXP.
Experimentally induced degradation can be used as a tool to probe the role of specific proteins in cellular processes. For example, a protein that is normally stable can be modified to make its degradation conditionally dependent on the presence of an adaptor, allowing studies of the consequences of depletion after induction of adaptor synthesis (7, 9). Such systems complement methods, such as RNA interference, that rely upon repressing biosynthesis of the target protein but offer significant advantages when rapid depletion of otherwise long-lived proteins is the goal (10–12). We are interested in engineering synthetic adaptor systems to control targeted intracellular degradation.
ClpXP is a AAA+ protease present in bacteria and mitochondria that consists of two components, ClpX and ClpP. Hexamers of ClpX recognize degradation tags in specific substrate proteins, unfold them in a reaction that requires ATP hydrolysis, and then use additional cycles of ATP hydrolysis to translocate the unfolded polypeptide into an interior chamber of ClpP, where proteolysis takes place (see Fig. 1A). The simplest way in which an adaptor could stimulate degradation is by tethering a specific substrate to a protease, thereby increasing its effective concentration and facilitating proteolysis (see Fig. 1B; for review, see Ref. 3). The SspB adaptor, for example, appears to function by this mechanism. SspB enhances ClpXP degradation of certain substrates, including N-RseA and proteins bearing the ssrA-degradation tag (2, 8, 13). ClpXP degrades these substrates in the absence of SspB, but Km for degradation is substantially lower when this adaptor is present. Two features of SspB are consistent with a tethering mechanism. It has a substrate-binding domain with a groove that binds a portion of the ssrA tag or a sequence in N-RseA, and it contains a flexible C-terminal extension terminating with a peptide motif (XB) that binds to the N-terminal domain of ClpX (14–19). Mutations that prevent SspB binding to ClpX or block substrate binding to SspB eliminate stimulation of degradation (13, 16, 20).
It has not been rigorously established, however, that tethering per se is sufficient for the activity of any adaptor. Based on biochemical experiments, for instance, Thibault et al. (21) proposed that the adaptor activity of SspB is mediated, in part, by its ability to direct the movement of the N-terminal domains of ClpX, and thereby to regulate the delivery of tagged substrates to ClpXP. For some adaptors, tethering of the substrate to the protease is not sufficient for degradation. For example, the ClpS adaptor tethers N-end rule substrates to the AAA+ ClpAP protease (22–24), but some ClpS mutants mediate efficient substrate tethering to ClpAP without facilitating degradation (25). In such cases, more complicated transactions between the adaptor and the protease appear to be needed to ensure that the substrate is properly delivered to the protease. Moreover, in some instances, adaptors play roles in substrate delivery but are also required for assembly of the active protease (26).
The studies reported here were motivated by two major goals. First, we wished to test if a completely synthetic adaptor system could be used to regulate substrate degradation. Second, we sought to design a proteolysis system that could be controlled by the presence or absence of a small molecule. To define the minimal biochemical properties required for adaptor-protein function, we engineered and characterized synthetic variants of adaptors, substrates, and the ClpXP protease. We reasoned that if specialized interactions between SspB and the N-terminal domain of ClpX were a requisite part of substrate delivery, then replacing either component would preclude efficient degradation. By contrast, we found that rapid degradation of an otherwise poor substrate was possible in the absence of SspB and the N-domain as long as substrate·enzyme tethering was maintained by other interaction domains. These results show that tethering alone is sufficient for synthetic- adaptor function. We were also able to control degradation in vitro and in vivo using systems in which a small molecule, rapamycin, drives assembly of tethered proteolytic complexes. Thus, targeted degradation can be engineered to depend, in a conditional fashion, on the presence of a small molecule. In principle, degradation under small molecule control has many of the advantages of chemical genetics (27), but should be even simpler and more widely applicable as a method of functional inhibition. In addition, controlling degradation in this fashion is possible even when biosynthesis of new macromolecules is precluded.
EXPERIMENTAL PROCEDURES
Buffers
LB1 buffer (pH 7.6) contained 20 mm HEPES, 400 mm NaCl, 100 mm KCl, 20 mm imidazole, 10% glycerol, and 10 mm 2-mercaptoethanol. LB2 buffer (pH 8.0) contained 100 mm NaH2PO4, 10 mm Tris·HCl, 6 m guanidine HCl, and 10 mm imidazole. EB1 buffer (pH 7.6) contained 20 mm HEPES, 400 mm NaCl, 100 mm KCl, 200 mm imidazole, 10% glycerol, and 10 mm 2-mercaptoethanol. QB1 buffer (pH 7.0) contained 50 mm NaPO4 and 100 mm NaCl. GF1 buffer (pH 7.6) contained 50 mm Tris·HCl, 1 mm dithiothreitol, 300 mm NaCl, 0.1 mm EDTA, and 10% glycerol. PD-1 buffer (pH 7.6) contained 25 mm HEPES KOH, 5 mm MgCl2, 10% glycerol, and 200 mm KCl. YEG media contained 0.5% yeast extract, 1% NaCl, and 0.4% glucose. 1.5× YT broth (pH 7.0) contained 1.3% Tryptone, 0.75% yeast extract, and 0.75% NaCl.
Plasmids and Strains
XB-tail-Arc-DAS+4 was cloned into a pET24d vector and consisted of the following sequences from the N to the C terminus: (M)GDDRGGRPALRVVK (XB motif underlined); residues 113–154 of Escherichia coli SspB; a His6 tag; phage P22 Arc repressor; the st11 sequence H6KNQHD; and a DAS+4 tag (AANDENYSENYADAS). Arc-DAS+4 lacks the XB-tail sequence but is otherwise identical to XB-tail-Arc-DAS+4. Arc-ssrA is identical to Arc-DAS+4 but has a wild-type ssrA tag (AANDENYLAA). FKBP-linker-ClpXΔN and FKBP-linker-[ClpXΔN]3 were cloned in pACYC vectors and consisted of the N-terminal human FKBP12 protein, followed by residues 139–165 of E. coli SspB, a His6 tag, and either by E. coli ClpXΔN (residues 61–424) or the covalently linked [ClpXΔN]3 trimer (28). For studies in vivo, FKBP-linker-[ClpXΔN]3 was placed under control of a constitutive promoter and ribosome-binding site (j23101 and b0032, respectively)2 and expressed from a plasmid bearing a ColE1 origin of replication. The gene for SspBcore-FRB was generated in a pET vector by fusing the coding sequence for residues 1–113 of E. coli SspB to the FRB domain of rat mTor (residues 2015–2114) with a connecting linker sequence of H6RGS. Plasmids encoding λO-DHFRII-FRB and λO-titin-I27-FRB were generated by replacing the SspBcore/His6 cassette in the SspBcore-FRB construct with a fragment encoding (M)TNTAKILNFGRS-(DHFRII/titin-I27)-GGSEH6GS. Standard techniques were used to replace the λO tag in λO-titin-I27-FRB with the sequence MD6. The GFP-DAS+4 and titin-I27-DAS+4 substrates were expressed from pET vectors with N-terminal H6ID2LG tags for ease of purification. All growth and degradation experiments in vivo were performed in E. coli strain X90 (F′lacIqlac′ pro′/ara Δ(lac-pro) nalA argE(am) rifRthi-1 clpX−, recA−).
Protein Expression and Purification
E. coli ClpP was expressed and purified as described previously (30). Unless noted, all other proteins were overexpressed from isopropyl 1-thio-β-d-galactopyranoside-inducible promoters in E. coli strain BLR (BL21 recA− λ(DE3)). Briefly, cells were grown to A600 0.7 at 37 °C in 1–2 liters of 1.5× YT broth, the cells were chilled to 18 °C, and expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested 4 h after induction, resuspended in LB1 buffer (15 ml/liter of culture), and frozen at −80 °C until purification. To aid lysis, cells were subjected to two rounds of freezing and thawing before the addition of 1 mm phenylmethylsulfonyl fluoride, 0.1 mg/ml lysozyme, and 250 units of benzonase nuclease. After a 30-min incubation at 4 °C, lysates were centrifuged at 8000 × g in a Sorvall SA600 rotor for 40 min, and the supernatant was decanted and saved.
For purification of ClpX and its variants, the lysate supernatant was incubated with 1 ml of Ni2+-NTA3 resin equilibrated with LB1 buffer for 5 min. After two bulk washes with 30 column volumes (CV) of lysis buffer, the slurry was poured into a column, washed with 20 CVs of LB1 buffer, and the protein was eluted with EB1 buffer (8 × 0.5 CV elutions). Fractions were pooled based on Bradford assays, concentrated using Amicon Ultracel 10k filters, and chromatographed on a Superdex S200 gel-filtration column equilibrated in GF1 buffer. Fractions were analyzed by SDS-PAGE, concentrated, and stored frozen in GF1 buffer at −80 °C.
The GFP-DAS+4, titin-I27-DAS+4, and λO-DHFRII-FRB proteins were purified by Ni2+-NTA chromatography as described above, and stored frozen in EB1 buffer at −80 °C. XB-tail-Arc-DAS+4 was lysed in denaturing buffer LB2. After centrifugation as described above, the soluble fraction was applied to a Ni2+-NTA column, washed with 10 CVs of LB2, 20 CVs of LB1, and eluted as described above. Fractions were pooled based on Bradford analysis, exchanged into QB1 buffer using a spin column, and loaded onto an Amersham Biosciences 5/50 GL MonoQ column equilibrated in QB1 buffer. The column was washed with 5 CVs of QB1 buffer and a 10-ml gradient from 0.1 to 1 m NaCl in QB1 buffer was applied. Appropriate fractions were concentrated, pooled, and stored frozen at −80 °C. For each protein, matrix-assisted laser desorption ionization time-of-flight mass spectrometry indicated that the full-length protein without truncations had been purified.
Biochemical Assays
Assays were performed in PD-1 buffer at 30 °C using a NADH-coupled colorimetric assay and a plate reader for ATPase assays (31) or an ATP-regeneration system using creatine phosphate for degradation assays (32). Degradation assays of XB-tail-Arc-DAS+4 (10 μm) by 0.3 μm ClpX6 and 0.9 μm ClpP14 were quenched by boiling in SDS and monitored by SDS-PAGE. GFP degradation was monitored by loss of fluorescence (excitation 467 nm; emission 511 nm). Degradation of titin-I27-DAS+4 was monitored by release of radioactive peptides soluble in trichloroacetic acid (5). Log-phase growth rates at 30 °C in YEG media were measured using a plate reader to monitor A600; the plasmid expressing FKBP-linker-[ClpXΔN]3 was maintained using ampicillin (100 μg/ml), and the plasmid expressing λO-DHFRII-FRB was maintained using tetracycline (10 μg/ml). Degradation assays in vivo were performed by centrifuging 1 ml of 0.7 A600 cultures, resuspending the pellet in 8 m urea, normalizing each sample by total protein content using a Bradford assay, running SDS-PAGE, transferring by electroblotting to a polyvinylidene difluoride membrane, and probing using a polyclonal anti-DHFRII antibody (a gift from Dr. Elizabeth Howell, University of Tennessee, Knoxville, TN).
RESULTS
Tethering-dependent Degradation with No Adaptor
SspB uses one part of its structure to bind a substrate and another part (the XB peptide) to bind to the N-domain of ClpX (14–17, 19, 33). For a substrate that normally requires delivery by SspB, we reasoned that the need for the substrate-binding portion of this adaptor might be obviated by fusing the XB peptide and the flexible tail of SspB directly to a substrate. In principle, this design would allow the protein to tether itself to the N-domain of ClpX. Efficient ClpXP degradation of such a “substrate” would support a passive tethering model, whereas poor degradation would suggest that the core substrate-binding domain of SspB plays a more active role in delivery.
We constructed and purified a protein consisting of the XB peptide of SspB, the flexible tail of SspB, the Arc repressor protein, and the DAS+4 degradation tag, arranged from the N to C terminus (XB-tail-Arc-DAS+4). The DAS+4 tag (AANDENSENYADAS) is a variant of the ssrA tag that binds SspB normally but binds ClpX with dramatically reduced affinity (7). Proteins containing the DAS+4 tag are degraded inefficiently by ClpXP, except at very high substrate concentrations or in the presence of SspB. In degradation assays monitored by SDS-PAGE, ClpXP degraded a 33-fold excess of the XB-tail-Arc-DAS+4 protein to near completion over the course of 60 min (Fig. 2A). By contrast, an otherwise identical variant lacking the XB peptide and tail (Arc-DAS+4) was degraded much more slowly under identical conditions (Fig. 2B).
FIGURE 2.
SDS-PAGE assays of protein degradation by the ClpXP or ClpXΔN/ClpP proteases (300 nm ClpX or ClpXΔN; 900 nm ClpP). The XB-tail-Arc-DAS+4 substrate (10 μm) required autotethering to the N-terminal domain of ClpX for efficient degradation. Hence, this substrate was not degraded if the N-domain of ClpX was deleted or if excess XB peptide was present in the proteolysis reaction. Degradation of the Arc-ssrA substrate (10 μm) did not require the ClpX N-domain and was not inhibited by XB peptide.
Two additional control experiments confirmed that degradation of XB-tail-Arc-DAS+4 depended on tethering of the XB region of the substrate to the N-terminal domain of ClpX. First, ClpXΔN, a truncated enzyme lacking the N-domain, failed to support ClpP degradation of XB-tail-Arc-DAS+4 (Fig. 2C) but degraded a tethering-independent substrate (Arc-ssrA) rapidly (Fig. 2D). Second, addition of free XB peptide substantially slowed degradation of the XB-tail-Arc-DAS+4 by wild-type ClpXP (Fig. 2E) but had no effect on degradation of Arc-ssrA (Fig. 2F).
Taken together, these results suggest that the role of SspB in enhancing substrate degradation is largely one of tethering the substrate to the N-terminal domain of ClpX and increasing its local concentration. If interactions between SspB and the N-domain are essential for facilitating degradation, then these contacts must be limited to the flexible tail and XB peptide, which comprise the C-terminal 41 residues of SspB.
Artificial Tethering Supports Substrate Delivery
Do contacts between the XB peptide and the N-domain of ClpX serve functions other than simple tethering? To address this question, we designed a new binding interface that involved neither the N-domain of ClpX nor the XB peptide. First, we constructed a ClpX variant containing the human FKBP12 protein at the N terminus, a linker region, and ClpXΔN at the C terminus (FKBP-linker-ClpXΔN; Fig. 3A). Second, we fused a SspB variant containing the substrate-binding core domain but lacking the flexible tail and XB motif to the N terminus of the FRB domain from rat mTor (SspBcore-FRB). The FKBP12 protein and the FRB domain bind to each other with high affinity only in the presence of the small molecule rapamycin (34). Thus, adaptor-mediated tethering in this system is predicted to be rapamycin dependent.
FIGURE 3.
A, schematic depictions of hexamers formed by the FKBP-linker-ClpXΔN or FKBP-linker-[ClpXΔN]3 enzyme variants. B, dependence of ATP hydrolysis rates on enzyme concentration. The rate of ATP hydrolysis by FKBP-linker-[ClpXΔN]3 was a linear function of enzyme concentration (r > 0.999; average rate = 82 ATP min−1 enzyme−1), indicating that this protein forms a stable hexamer at low protein concentrations. ATP hydrolysis by the FKBP-linker-ClpXΔN enzyme, by contrast, was highly nonlinear in a fashion that suggested hexamer dissociation at concentrations below 100 nm. C, the rate of ATP hydrolysis by FKBP-linker-[ClpXΔN]3 (0.3 μm pseudo-hexamer) was slowed by the presence of ClpP (0.9 μm) and enhanced by the presence of an unfolded substrate (1.4 μm), the carboxymethylated titin-I27-VP15-ssrA protein (37). D, Michaelis-Menten plot of the substrate dependence of the steady-state rate of degradation of the GFP-ssrA substrate by FKBP-linker-[ClpXΔN]3 (100 nm pseudo-hexamer) and ClpP14 (300 nm). The solid line is a nonlinear least-squares fit of the experimental data (Km = 3.6 μm; Vmax = 1.1 min−1 enzyme−1).
The FKBP-linker-ClpXΔN enzyme displayed rates of ATP hydrolysis that increased non-linearly at low protein concentrations (Fig. 3B). Because N-domain dimerization normally helps stabilize the active hexameric form of ClpX (33, 35), replacing this domain with FKBP12 probably resulted in weaker hexamerization. To stabilize the active enzyme, FKBP12 and the linker were fused to the N terminus of a trimeric form of ClpXΔN in which the subunits were connected with a flexible linker (Fig. 3A) (28). Purified FKBP-linker-[ClpXΔN]3 exhibited linear ATP hydrolysis rates at concentrations ranging from 50 to 500 nm (Fig. 3B), consistent with stable pseudo-hexamer formation at low enzyme concentrations. As observed for wild-type ClpX6 (36, 37), ATP hydrolysis by the linked FKBP-linker-[ClpXΔN]3 enzyme was slightly repressed by ClpP binding and stimulated by addition of an unfolded substrate (Fig. 3C). Importantly, FKBP-linker-[ClpXΔN]3 and ClpP degraded GFP-ssrA with a Km of 3.6 μm and a Vmax of 1.1 min−1 enzyme−1 (Fig. 3D). These steady-state kinetic parameters are similar to those reported for wild-type ClpXP (30), showing that the presence of the FKBP12 domain in the linked enzyme does not interfere with degradation of ssrA-tagged substrates.
Next, we tested if the artificial SspBcore-FRB adaptor could deliver substrates to FKBP-linker-[ClpXΔN]3 in a rapamycin-dependent fashion (Fig. 4A). In the presence of this adaptor/enzyme pair, ClpP and rapamycin, two different DAS+4-tagged substrates were degraded with Km values near 1 μm and Vmax values expected based on the resistance of these native proteins to ClpX unfolding (Fig. 4, B and C) (30, 37). In the absence of rapamycin, degradation of both substrates was extremely slow (Fig. 4, B and C). Similarly, no degradation of untagged GFP or titin-I27 was observed in the presence of SspBcore- FRB, rapamycin, FKBP-linker-[ClpXΔN]3, and ClpP (data not shown). These experiments indicate that artificial tethering mediated by the FRB·rapamycin·FKBP12 complex results in efficient adaptor-dependent degradation. We conclude that simple tethering of substrates to ClpX is sufficient to explain adaptor-mediated enhancement of degradation.
FIGURE 4.
A, schematic showing delivery of DAS+4-tagged substrate to FKBP-linker-[ClpXΔN]3 by the synthetic SspBcore-FRB adaptor and rapamycin. B, substrate dependence of GFP-DAS+4 degradation by FKBP-linker-[ClpXΔN]3 (100 nm pseudo hexamer) and ClpP14 (300 nm) in the presence of SspBcore-FRB (200 nm) and the presence (1 μm) or absence of rapamycin. The line for the plus rapamycin curve is a fit to the Michaelis-Menten equation (Km = 0.51 μm; Vmax = 1.4 min−1 enzyme−1). The line for the no rapamycin data is a linear fit. C, Michaelis-Menten plots for titin-I27-DAS+4 degradation by FKBP-linker-[ClpXΔN]3 (100 nm pseudo hexamer) and ClpP14 (300 nm) in the presence of SspBcore-FRB (200 nm) and the presence (1 μm) or absence of rapamycin. The fit to the plus rapamycin data gave Km = 2.2 μm and Vmax = 0.17 min−1 enzyme−1. The line for the no rapamycin data is a linear fit.
Rapamycin-dependent Degradation
Artificial tethering mediated by the FRB-rapamycin-FKBP12 interaction requires the interaction of three molecular components. To test the kinetics of assembly, we monitored the fluorescence of the GFP-DAS+4 substrate mixed with SspBcore-FRB, FKBP-linker-[ClpXΔN]3, and ClpP (Fig. 5A). When rapamycin was added, degradation reached an enhanced steady-state rate within the dead time of the experiment (∼20 s; Fig. 5A). Thus, FRB·rapamycin·FKBP12 binding and subsequent degradation occurs on the time scale of many biological responses. We also tested the response of the system to rapamycin concentrations and found that the degradation rate was unchanged at rapamycin concentrations above the enzyme concentration (Fig. 5B). This result indicates that near stoichiometric quantities of the small molecule with respect to FKBP-linker-[ClpXΔN]3 and SspBcore-FRB saturate the system. Thus, the response of the system to rapamycin is rapid and sensitive.
FIGURE 5.
A, rapamycin addition results in rapid degradation. The GFP-DAS+4 substrate (0.5 μm), SspBcore-FRB (1 μm), FKBP-linker-[ClpXΔN]3 (0.5 μm pseudo-hexamer), and ClpP14 (1.5 μm) were preincubated and slow steady-state degradation was observed by loss of substrate fluorescence. At the time indicated by the arrow, rapamycin (2 μm) was added. Within the dead-time of the experiment (≈20 s), degradation of GFP-DAS+4 reached a new and much faster steady-state rate. B, changing rapamycin concentration over the range shown had almost no effect on the steady-state rate of degradation of 35S-titin-I27-DAS+4 (10 μm) by the FKBP-linker-[ClpXΔN]3 (100 nm pseudo hexamer), ClpP14 (300 nm), and SspBcore-FRB (200 nm). C, degradation of 35S-titin-I27-DAS+4 (10 μm) by FKBP-linker-[ClpXΔN]3 (200 nm pseudo-hexamer) and ClpP14 (600 nm) was assayed as a function of the SspBcore-FRB adaptor concentration in the presence of rapamycin (1 μm).
To evaluate the effects of the adaptor concentration, we titrated increasing quantities of the SspBcore-FRB adaptor against fixed concentrations of FKBP-linker-[ClpXΔN]3, ClpP, and rapamycin and assayed degradation of titin-I27-DAS+4. Degradation initially increased linearly and reached a maximal value at a ratio of one SspBcore-FRB dimer for each FKBP-linker-[ClpXΔN]3 pseudo-hexamer. For the wild-type proteins, it is known that one substrate-bound SspB dimer binds to one ClpX hexamer (38). At high concentrations of the SspBcore-FRB adaptor (10 μm), the degradation rate was reduced to ∼25% of its maximal value (Fig. 5C). Under these conditions, free adaptor molecules probably compete with ClpX-bound adaptors for substrate binding.
Tethering-dependent Delivery of a λO-tagged Substrate
The results presented so far show that alternative mechanisms of tethering can facilitate ClpXP proteolysis of substrates with C-terminal degradation tags. Degradation of proteins bearing the N-terminal λO tag normally requires the N-domain of ClpX (39), which is missing from the FKBP-linker-[ClpXΔN]3 variant. To test the versatility of the artificial tethering system, we constructed and purified a substrate with an N-terminal λO tag (NH2-TNTAKILNFGR) (40), followed by the titin-I27 domain, and then the FRB domain. This λO-titin-I27-FRB substrate contains no sequences from the SspB adaptor but can tether itself via rapamycin to the FKBP-linker-[ClpXΔN]3 enzyme.
The purified λO-titin-I27-FRB fusion protein was degraded by FKBP-linker-[ClpXΔN]3 and ClpP when rapamycin was present but not when it was absent (Fig. 6A). As a control, we constructed an otherwise identical fusion protein in which the λO tag was replaced by the sequence MD6 (MDDDDDD), which we hypothesized would not be recognized by ClpX. No degradation of this protein was observed in the presence or absence of rapamycin (Fig. 6A). We conclude that an N-terminal λO tag can serve as a degradation signal for substrates that can tether themselves to ClpX.
FIGURE 6.
A, schematic showing rapamycin-dependent degradation of a substrate with an N-terminal λO tag and C-terminal FRB domain by FKBP-linker-[ClpXΔN]3 and ClpP. The gel below the schematic shows that variants of the titin-I27 protein with an N-terminal λO tag and C-terminal FRB domain were degraded in a rapamycin-dependent fashion in vitro as assayed by SDS-PAGE. Replacing the λO tag with a MD6 sequence blocked degradation. All reactions contained substrate (10 μm), FKBP-linker-[ClpXΔN]3 (300 nm pseudo-hexamer), and ClpP14 (900 nm). B, relative growth rates in M9 minimal medium of clpX− E. coli strains expressing λO-DHFRII-FRB as a function of rapamycin (10 μm, when present), trimethoprim (100 μg/ml, when present), and a plasmid expressing FKBP-linker-[ClpXΔN]3. C, Western blotting shows intracellular degradation of λO-DHFRII-FRB after addition of rapamycin (10 μm) but not after a mock addition in clpX− E. coli strains expressing FKBP-linker-[ClpXΔN]3. Protein synthesis was not blocked in this experiment. The sample volume in each lane was adjusted to yield the same amount of total cellular protein.
Rapamycin-dependent Degradation in Vivo
For studies of degradation in E. coli, we constructed a substrate with an N-terminal λO tag, followed by the DHFRII enzyme and the FRB domain (λO-DHFRII-FRB). This substrate was degraded in a rapamycin-dependent manner by FKBP-linker-[ClpXΔN]3 and ClpP in vitro (data not shown). Expression of DHFRII in E. coli results in resistance to trimethoprim, an antibiotic that inhibits the endogenous dihydrofolate reductase enzyme (41–45). When we expressed λO-DHFRII-FRB in a clpX− strain containing FKBP-linker-[ClpXΔN]3, the cells grew well in the presence of trimethoprim (Fig. 6B). When rapamycin was added, however, these cells became trimethoprim sensitive and growth slowed substantially (Fig. 6B). This result was not caused by rapamycin toxicity, as rapamycin did not affect growth in media lacking trimethoprim or in a strain without FKBP-linker-[ClpXΔN]3 (Fig. 6B). Thus, the FKBP-linker-[ClpXΔN]3 enzyme and ClpP appear to degrade λO-DHFRII-FRB in a rapamycin-dependent fashion in vivo. To confirm this inference, we prepared cell lysates at different times after treatment with rapamycin or a mock addition and subjected them to SDS-PAGE and Western blotting using an anti-DHFRII antibody. The steady-state level of λO-DHFRII-FRB was reduced rapidly upon addition of rapamycin (Fig. 6C), even though protein synthesis was not blocked in this experiment. Hence, rapamycin-dependent tethering of FRB fusion substrates to FKBP-linker-[ClpXΔN]3 can successfully control degradation in the cell.
DISCUSSION
The results presented here demonstrate that tethering of a substrate to a AAA+ protease is sufficient for adaptor function. We found, for example, that the normal tethering function of the SspB adaptor protein could be transferred directly to a substrate by fusing the ClpX-binding peptide and tail of SspB to an otherwise poor substrate for the ClpXP protease. This result shows that the substrate-binding domain of SspB is dispensable for adaptor function. Moreover, we were able to engineer a completely artificial tethering system, in which the N-domain of ClpX was replaced with the FKBP12 protein and the normal tail and ClpX-binding peptide of the SspB adaptor was replaced with the FRB domain. In the presence of rapamycin, a small bridging molecule, substrates bearing a weak degradation tag were degraded efficiently in the presence of this artificial adaptor and re-engineered enzyme. Finally, we established a synthetic rapamycin-dependent system in which substrates bearing an N-terminal λO-degradation tag and the FRB domain were efficiently degraded by a FKBP-ClpX variant and ClpP. It is important to note that this substrate contained no parts from the SspB adaptor and ClpX lacked its N-domain, which is required for normal adaptor-mediated delivery of substrates to wild-type ClpX. Because the functions mediated by SspB and the ClpX N-domain can effectively be replaced by other tethering elements, it is possible that the wild-type adaptor system stimulates proteolysis simply by tethering substrates to ClpXP.
Although tethering of substrates to ClpX appears to be sufficient for adaptor function, there clearly must also be other geometric and structural criteria that need to be met (46). For example, a potential substrate that is tethered too rigidly and therefore could not reach the translocation pore would presumably not be degraded efficiently. In the studies reported here, we tried to account for this factor by engineering a flexible linker between the FKBP12 and ClpX portions of our synthetic enzyme. In some cases, however, it may also be necessary to design flexibility into the adaptor or substrate as well.
Efficient ClpXP degradation mediated by the λO tag normally requires both a multimeric substrate and the N-domain of ClpX. For example, the tetrameric λO protein is degraded well by wild-type ClpXP but is not degraded efficiently by ClpXΔN and ClpP (33, 39). Moreover, Km for ClpXP degradation of a dimeric λO-tagged substrate was substantially lower than that for a monomeric λO-tagged protein (47). Nevertheless, we found that a monomeric substrate, λO-titin-I27-FRB, was degraded efficiently by FKBP-linker-[ClpXΔN]3 and ClpP. This result suggests that degradation of λO-tagged substrates does not require substrate multimerization or the ClpX N-domain if alternative mechanisms of tethering to ClpX are available. We propose that λO-tagged substrates normally require multimerization for efficient degradation, because one λO tag in a multimer tethers the substrate to ClpX via the N-domain, allowing a second λO tag to be engaged by the translocation channel of ClpX to initiate degradation. Indeed, this dual function tag model is supported by reports that both the isolated N-domain of ClpX and ClpXΔN itself bind to the λO protein (33, 39).
Multivalent recognition of degradation signals is likely to facilitate the targeted proteolysis of many substrates at low concentrations. For example, monomeric substrates might contain more than one type of degradation tag. Indeed, Flynn et al. (40) reported that roughly 25% of the identified cellular substrates for ClpXP contained more than one degradation motif. For multimeric substrates, the same sequence signal in two different subunits could be recognized by different parts of the protease as proposed above. We found, for example, that clpX+ strains containing λO-DHFRII-FRB were trimethoprim sensitive (data not shown), suggesting that wild-type ClpXP can degrade this substrate. Because DHFRII is a tetramer, it is likely that a λO tag from one subunit tethers the substrate to the N-domain of ClpX, allowing the λO tag from a second subunit to be engaged by ClpX for degradation. Recent studies indicate that ClpX disassembly of the tetrameric MuA transposase also involves recognition of multiple classes of sequence elements (29).
Furthermore, our results demonstrate that adaptor-mediated degradation can be placed under small molecule control both in vitro and in vivo. In the cell, the kinetics of rapamycin uptake and the subsequent assembly of the FRB·rapamycin· FKBP12 complex are sufficiently rapid to ensure that degradation starts within minutes of addition of the small molecule. The system described here may not be optimal for many desired uses in controlling intracellular degradation. Nevertheless, our results suggest that it should be possible to design improved systems that combine small molecule control, fast temporal responses, and the exquisite specificity of a genetically encoded degradation tag. We expect such systems to be useful in the study of protein products, including those encoded by essential genes, for which the phenotype of depletion on a short time scale needs to be determined. In principle, such a system could be used with either N- or C-terminal tethering-dependent tags, greatly extending their applicability.
Acknowledgments
We thank Kathleen McGinness, Andreas Martin, Kevin Griffith, Eyal Gur, Peter Chien, Shankar Sundar, and Elizabeth Howell for helpful discussions and materials.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-16892 and GM-48224.
Registry of Standard Biological Parts.
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- CV
- column volume
- GFP
- green fluorescent protein
- DHFR
- dihydrofolate reductase.
REFERENCES
- 1.Gottesman S. (2003) Annu. Rev. Cell Dev. Biol. 19, 565–587 [DOI] [PubMed] [Google Scholar]
- 2.Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., Neher S. B., Oakes E. S., Siddiqui S. M., Wah D. A., Baker T. A. (2004) Cell 119, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T. A., Sauer R. T. (2006) Trends Biochem. Sci. 31, 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 5.Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neher S. B., Flynn J. M., Sauer R. T., Baker T. A. (2003) Genes Dev. 17, 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinness K. E., Baker T. A., Sauer R. T. (2006) Mol. Cell 22, 701–707 [DOI] [PubMed] [Google Scholar]
- 8.Flynn J. M., Levchenko I., Sauer R. T., Baker T. A. (2004) Genes Dev. 18, 2292–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffith K. L., Grossman A. D. (2008) Mol. Microbiol. 70, 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998) Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- 11.Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., Wandless T. J. (2006) Cell 126, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janse D. M., Crosas B., Finley D., Church G. M. (2004) J. Biol. Chem. 279, 21415–21420 [DOI] [PubMed] [Google Scholar]
- 13.Levchenko I., Seidel M., Sauer R. T., Baker T. A. (2000) Science 289, 2354–2356 [DOI] [PubMed] [Google Scholar]
- 14.Dougan D. A., Weber-Ban E., Bukau B. (2003) Mol. Cell 12, 373–380 [DOI] [PubMed] [Google Scholar]
- 15.Wah D. A., Levchenko I., Rieckhof G. E., Bolon D. N., Baker T. A., Sauer R. T. (2003) Mol. Cell 12, 355–363 [DOI] [PubMed] [Google Scholar]
- 16.Park E. Y., Lee B. G., Hong S. B., Kim H. W., Jeon H., Song H. K. (2007) J. Mol. Biol. 367, 514–526 [DOI] [PubMed] [Google Scholar]
- 17.Levchenko I., Grant R. A., Wah D. A., Sauer R. T., Baker T. A. (2003) Mol. Cell 12, 365–372 [DOI] [PubMed] [Google Scholar]
- 18.Levchenko I., Grant R. A., Flynn J. M., Sauer R. T., Baker T. A. (2005) Nat. Struct. Mol. Biol. 12, 520–525 [DOI] [PubMed] [Google Scholar]
- 19.Song H. K., Eck M. J. (2003) Mol. Cell 12, 75–86 [DOI] [PubMed] [Google Scholar]
- 20.Bolon D. N., Wah D. A., Hersch G. L., Baker T. A., Sauer R. T. (2004) Mol. Cell 13, 443–449 [DOI] [PubMed] [Google Scholar]
- 21.Thibault G., Tsitrin Y., Davidson T., Gribun A., Houry W. A. (2006) EMBO J. 25, 3367–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougan D. A., Reid B. G., Horwich A. L., Bukau B. (2002) Mol. Cell 9, 673–683 [DOI] [PubMed] [Google Scholar]
- 23.Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D. A., Bukau B. (2006) Nature 439, 753–756 [DOI] [PubMed] [Google Scholar]
- 24.Wang K. H., Sauer R. T., Baker T. A. (2007) Genes Dev. 21, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou J. Y., Sauer R. T., Baker T. A. (2008) Nat. Struct. Mol. Biol. 15, 288–294 [DOI] [PubMed] [Google Scholar]
- 26.Kirstein J., Schlothauer T., Dougan D. A., Lilie H., Tischendorf G., Mogk A., Bukau B., Turgay K. (2006) EMBO J. 25, 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh D. P., Chang Y. T. (2006) Chem. Rev. 106, 2476–2530 [DOI] [PubMed] [Google Scholar]
- 28.Martin A., Baker T. A., Sauer R. T. (2005) Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
- 29.Abdelhakim A. H., Oakes E. C., Sauer R. T., Baker T. A. (2008) Mol. Cell 30, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y. I., Burton R. E., Burton B. M., Sauer R. T., Baker T. A. (2000) Mol. Cell 5, 639–648 [DOI] [PubMed] [Google Scholar]
- 31.Nørby J. G. (1988) Methods Enzymol. 156, 116–119 [DOI] [PubMed] [Google Scholar]
- 32.Martin A., Baker T. A., Sauer R. T. (2008) Mol. Cell 29, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtyra U. A., Thibault G., Tuite A., Houry W. A. (2003) J. Biol. Chem. 278, 48981–48990 [DOI] [PubMed] [Google Scholar]
- 34.Choi J., Chen J., Schreiber S. L., Clardy J. (1996) Science 273, 239–242 [DOI] [PubMed] [Google Scholar]
- 35.Grimaud R., Kessel M., Beuron F., Steven A. C., Maurizi M. R. (1998) J. Biol. Chem. 273, 12476–12481 [DOI] [PubMed] [Google Scholar]
- 36.Kim Y. I., Levchenko I., Fraczkowska K., Woodruff R. V., Sauer R. T., Baker T. A. (2001) Nat. Struct. Biol. 8, 230–233 [DOI] [PubMed] [Google Scholar]
- 37.Kenniston J. A., Baker T. A., Fernandez J. M., Sauer R. T. (2003) Cell 114, 511–520 [DOI] [PubMed] [Google Scholar]
- 38.Wah D. A., Levchenko I., Baker T. A., Sauer R. T. (2002) Chem. Biol. 9, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 39.Singh S. K., Rozycki J., Ortega J., Ishikawa T., Lo J., Steven A. C., Maurizi M. R. (2001) J. Biol. Chem. 276, 29420–29429 [DOI] [PubMed] [Google Scholar]
- 40.Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. (2003) Mol. Cell 11, 671–683 [DOI] [PubMed] [Google Scholar]
- 41.Fleming M. P., Datta N., Gruneberg R. N. (1972) Br. Med. J. 1, 726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattishall K. H., Acar J., Burchall J. J., Goldstein F. W., Harvey R. J. (1977) J. Biol. Chem. 252, 2319–2323 [PubMed] [Google Scholar]
- 43.Stone D., Smith S. L. (1979) J. Biol. Chem. 254, 10857–10861 [PubMed] [Google Scholar]
- 44.Smith S. L., Stone D., Novak P., Baccanari D. P., Burchall J. J. (1979) J. Biol. Chem. 254, 6222–6225 [PubMed] [Google Scholar]
- 45.Krahn J. M., Jackson M. R., DeRose E. F., Howell E. E., London R. E. (2007) Biochemistry 46, 14878–14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGinness K. E., Bolon D. N., Kaganovich M., Baker T. A., Sauer R. T. (2007) J. Biol. Chem. 282, 11465–11473 [DOI] [PubMed] [Google Scholar]
- 47.Farrell C. M., Baker T. A., Sauer R. T. (2007) Mol. Cell 25, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]