Abstract
Peroxisome proliferator activated receptor-γ co-activator-1α (PGC-1α) is a transcriptional co-activator that coordinately regulates the expression of distinct sets of metabolism-related genes in different tissues. Here we show that PGC-1α expression is reduced in skeletal muscles from mice lacking the sirtuin family deacetylase SIRT1. Conversely, SIRT1 activation or overexpression in differentiated C2C12 myotubes increased PGC-1α mRNA expression. The transcription-promoting effects of SIRT1 occurred through stimulation of PGC-1α promoter activity and were enhanced by co-transfection of myogenic factors, such as myocyte enhancer factor 2 (MEF2) and, especially, myogenic determining factor (MyoD). SIRT1 bound to the proximal promoter region of the PGC-1α gene, an interaction potentiated by MEF2C or MyoD, which also interact with this region. In the presence of MyoD, SIRT1 promoted a positive autoregulatory PGC-1α expression loop, such that overexpression of PGC-1α increased PGC-1α promoter activity in the presence of co-expressed MyoD and SIRT1. Chromatin immunoprecipitation showed that SIRT1 interacts with PGC-1α promoter and increases PGC-1α recruitment to its own promoter region. Immunoprecipitation assays further showed that SIRT1-PGC-1α interactions are enhanced by MyoD. Collectively, these data indicate that SIRT1 controls PGC-1α gene expression in skeletal muscle and that MyoD is a key mediator of this action. The involvement of MyoD in SIRT1-dependent PGC-1α expression may help to explain the ability of SIRT1 to drive muscle-specific gene expression and metabolism. Autoregulatory control of PGC-1α gene transcription seems to be a pivotal mechanism for conferring a transcription-activating response to SIRT1 in skeletal muscle.
Peroxisome proliferator activated receptor-γ co-activator-1α (PGC-1α)2 is a transcriptional co-activator that is recognized as a master controller of the expression of genes involved in metabolic regulation. PGC-1α exerts differential effects on metabolism in different tissues. In brown adipose tissue, PGC-1α increases the expression of uncoupling protein 1 and genes involved in mitochondrial oxidative pathways. In the liver, PGC-1α induces the expression of genes involved in gluconeogenesis and the metabolic response to starvation. In skeletal muscle, PGC-1α expression is rapidly induced by exercise in vivo (1), a response considered to be a mechanism for modulating metabolic flux in response to decreased ATP levels (2). Chronic exercise also increases PGC-1α expression in association with fiber-type switching toward the more oxidative and high endurance type IIa and type I fibers. The fiber-type switch promoted by PGC-1α is characterized by increased mitochondrial density and function, increased oxidative metabolism, increased expression of myofibrillar proteins characteristic of type I and type IIa muscle fibers and a switch in substrate fuel usage (3). Furthermore, greater levels of PGC-1α are found in oxidative fibers compared with glycolytic fibers, even in a rested state (3). Moreover, PGC-1α gene transcription is positively controlled by myogenic transcription factors, such as MyoD and MEF2 (4–6).
The action of PGC-1α is modulated by post-translation modifications (7, 8) such as acetylation (see below), or as a consequence of a strong transcriptional regulation of the PGC-1α gene by hormonal and metabolic stimuli that result in changes in PGC-1α levels, and therefore PGC-1α activity. Depending on the tissue and cell type, PGC-1α gene expression is regulated by cAMP-mediated pathways, peroxisome proliferator activated receptor-γ (PPARγ) and -β/δ (PPARβ/δ) activation, as well as the calcium-calmodulin pathway (7).
SIRT1 is a member of the sirtuin family of protein deacetylases, which have a major role in metabolic regulation. SIRT1 deacetylates histones as well as transcription factors and co-regulators and thereby controls regulatory protein activity and, ultimately, gene expression. The deacetylase activity of SIRT1 is controlled by the NAD+/NADH ratio, thus SIRT1 is considered to act as a sensor of the metabolic status of the cell and mediates adaptive gene expression in response to metabolic changes.
SIRT1 has been reported to exert negative control of myogenesis through deacetylation and subsequent repression of MyoD, particularly in association with the high NAD+ levels characteristic of myoblasts (9) and under conditions of glucose restriction (10). Moreover, SIRT1 represses the expression of metabolic genes such as uncoupling protein 3 (ucp3), which is a direct target of MyoD regulation (11, 12). Recent studies have also revealed that SIRT1 deacetylates PGC-1α in skeletal muscle, a modification that is associated with up-regulation of the fatty acid oxidation gene program in skeletal muscle (13). Treatment of mice with SIRT1 activators (resveratrol or SIRT1720) increases PGC-1α deacetylation, resulting in an increase in PGC-1α activity and transcription of PGC-1α target genes. In addition to deacetylating PGC-1α, SIRT1 activators have also been reported to increase PGC-1α levels (14, 15).
In the present study, we analyzed the role of SIRT1 in controlling PGC-1α gene expression in skeletal muscle cells. We report that SIRT1 increases PGC-1α gene transcription, an effect that involves PGC-1α autoregulatory processes and the interaction of SIRT1 with myogenic transcription factors. The direct control of PGC-1α gene expression, and thus PGC-1α levels and activity, by SIRT1 may be a key element in the regulatory mechanisms that links metabolic changes to adaptive gene expression in skeletal muscle.
EXPERIMENTAL PROCEDURES
mRNA Expression in SIRT1-null Mice
SIRT1-null mice, kindly provided by Frederick W. Alt (16) were cared and used in accordance with the European Community Council directive 86/609/EEC and approved by Committee for Ethics in Animal Experimentation, University of Barcelona. Thirty-day-old homozygous and heterozygous SIRT1-null mice were used, and wild-type littermates served as controls. After sacrifice, gastrocnemius, soleus, and tibialis anterior muscles were dissected, and total RNA was extracted using Tripure (Roche Applied Science). Reverse transcription was performed in a total volume of 20 μl using random hexamer primers (Applied Biosystems, Foster City, CA) and 0.5 μg of RNA. Real-time quantitative PCR was conducted in 25-μl reaction mixtures containing 1 μl of cDNA, 12.5 μl of TaqMan Universal PCR Master Mix (Applied Biosystems), 250 nm probes, and 900 nm primers from Assays-on-Demand Gene Expression Assay Mix (Applied Biosystems). TaqMan Gene Expression Assays primers for PGC-1α (Mm00447183_m1), UCP3 (Mm00494074_m1), and myogenin (Mm00446194_m1) were used. Each assay was performed in duplicate, and the mean value was used to calculate mRNA expression for the gene of interest and the housekeeping reference gene (β2-microglobulin). The amount of the gene of interest in each sample was normalized to that of the reference control using the comparative (2−ΔCT) method following the manufacturer's instructions.
Cell Culture
Mouse myoblastic C2C12 cells, obtained from the American Type Culture Collection (ATCC, Manassas, VA) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. C2C12 cells were induced to differentiate using Dulbecco's modified Eagle's medium supplemented with 2% horse serum. C2C12 myotubes were treated with 50 μm resveratrol (Calbiochem) and 10 mm nicotinamide (NAM, Sigma) on day 4 of differentiation. CV1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Adenoviral-mediated Gene Transfer of SIRT1, PGC-1α, and MyoD
The adenoviral expression vector for MyoD was obtained from Vector Biolabs (Philadelphia, PA) and that for PGC-1α was kindly provided by Dr. P. Puigserver. The adenoviral expression vector for murine SIRT1 has been previously described (11). C2C12 myotubes were infected with SIRT1, PGC-1α, MyoD, or AdCMV-GFP (control) adenoviral vectors at a multiplicity of infection of 50 for 3 h in Dulbecco's modified Eagle's medium. The efficiency of transduction was ∼90%, based on assessment of GFP fluorescence. Cells were subsequently cultured in C2C12 myotube differentiation medium as described above.
Construction of Promoter-Reporter Plasmids
The plasmid mPGC-1α-Luc containing the −2553 to +78 upstream region of the mouse PGC-1α gene linked to the promoterless firefly luciferase gene was a gift from Dr. B. Spiegelman. A deleted construct containing only the −140 to + 78 bp region (140bpPGC-1α-Luc) was obtained by digestion of the parental plasmid with KpnI and ZraI followed by re-ligation. The wild-type human PGC-1α-Luc plasmid (wt) and the E-boxes-mutated construct (Mt3) have previously been reported (4).
Transient Transfection Assays
C2C12 cells at 50% confluence were transiently transfected using the FuGENE Transfection Reagent (Roche Diagnostics, Barcelona, Spain) following the manufacturer's instructions. Each transfection point was assayed in triplicate in a 24-well plate and contained luciferase reporter vector (0.3 μg) and one or more of the following mammalian expression vectors (0.06 μg), as indicated in the text: pCMV-MyoD, pcCMV-MyoD-RRR (17), pcx-P/CAF or pcx-P/CAFΔHAT (18), pCMV-SIRT1 (Upstate Biotechnology, Inc., Lake Placid, NY), pCMV-SIRT1-H363Y (11), pCruzHA SIRT1 (19), pSV-PGC-1α (20), CMV-MEF2A, pCMV MEF2c and pcDNA I/A MEF2D (6), pCDNA3-FoxO1 (21), pSG5-PPARβ/δ (22), pcDNA3-e2F1, pCMV-DP1 and pTK-Luc (a luciferase expression vector driven by the thymidine kinase promoter) were kindly provided by Dr. Martínez-Balbas. The pRL-CMV expression vector for sea pansy (Renilla reniformis) luciferase was used as an internal transfection control (Promega, Madison, WI). Cells were incubated for 48 h after transfection and, treated with or without 10 μm GW501516 for 24 h before harvest as indicated in the text. Firefly luciferase and Renilla luciferase activities were measured in a Glomax 96 Microplate Luminometer using the Dual Luciferase Reporter assay system kit (Promega). Luciferase activity levels determined by PGC-1α promoter constructs were normalized to Renilla luciferase as a control for variation in transfection efficiency.
Chromatin Immunoprecipitation Assay
C2C12 cells were transfected with −140bpPGC-1α-Luc and expression vectors for MyoD, MEF2C, and/or HA-SIRT1 (which expresses an HA-SIRT1 fusion protein), as indicated. ChIP analyses of HA-SIRT1 recruitment used an anti-HA antibody (Roche Diagnostics) and Magna ChIP G (Upstate Biotechnology, Inc.), as recommended by the supplier.
ChIP analyses of PGC-1α recruitment were performed as above, but immunoprecipitations were performed using an anti-PGC-1α (H300) antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
The primers used to amplify the 177-bp fragment from −130 to +47 bp encompassing the proximal region of the mouse PGC-1α promoter were 5′-GTGCAGCAAGCTTGCACAGG-3′ (forward) and 5′-CAACTCCAATCCACTCTGACAC-3′ (reverse). The PCR products were electrophoresed on a 1.5% agarose gels, visualized by ethidium bromide staining, and quantified by densitometric analysis (Phoretics 1D Software, Phoretic International Ltd.).
Immunoprecipitation
Whole cell lysates were prepared by lysing C2C12 myotubes in 50 mm Tris-HCl, 0.1 m NaCl, 10 mm EDTA, 1% Nonidet P-40 (pH 7.4) containing a protease inhibitor mixture (Complete Mini, Roche Diagnostics). PGC-1α, SIRT1, or MyoD in the whole cell lysate fractions were immunoprecipitated with anti-PGC-1α, anti-SIRT1 (Upstate Biotechnology, Inc.) or anti-MyoD antibodies (Santa Cruz Biotechnology), respectively, using 500 μg of total protein, and collected with Trueblot anti-Rabbit Ig IP beads (eBioscience San Diego, CA) according to the manufacturer's instructions. Proteins were separated by SDS-PAGE on 8% or 10% gels, and transferred to Immobilon-P membranes (Millipore). To verify that the amounts of PGC-1α and SIRT1 protein in myogenic cells (input) were similar, we performed Western blots using equal amounts of protein extracts prior to immunoprecipitation.
RESULTS
Expression of the PGC-1α Gene in Skeletal Muscles from SIRT1-null Mice
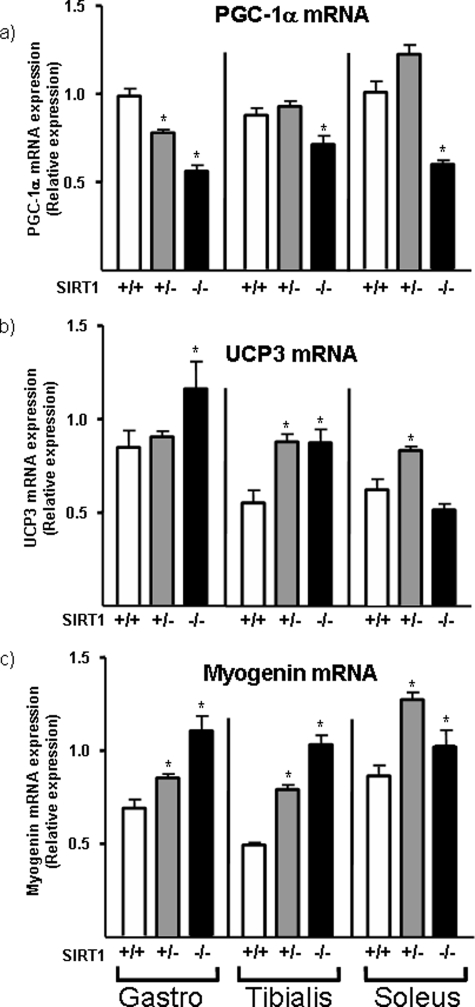
The effects of SIRT1 ablation on the expression of PGC-1α and other putative SIRT1 target genes in gastrocnemius muscle were determined in SIRT1 homozygous-null and SIRT1- heterozygous-null mice by quantitative reverse transcription-PCR, and compared with those seen in wild-type littermates (Fig. 1a). PGC-1α mRNA expression was significantly reduced in SIRT1 homozygous-null mice. The level of PGC-1α mRNA in SIRT1 heterozygous-null mice was intermediate between those in homozygous-null and wild-type mice. PGC-1α expression was also significantly reduced in the tibialis anterior and soleus muscles of SIRT1-null mice. To verify the specificity of the observed effects we assessed the expression of genes known to be negatively regulated by SIRT1 in myogenic cellular systems. UCP3 and myogenin genes, which are repressed by SIRT1 in vitro (9, 11) were consistently up-regulated in each of the skeletal muscles from SIRT1 homozygous-null mice and, in some cases, even in SIRT1 heterozygous-null mice (Fig. 1, b and c). Collectively, these observations indicate that SIRT1 is a potential positive regulator of PGC-1α gene expression in skeletal muscle.
FIGURE 1.
Expression of PGC-1α, UCP3, and myogenin mRNA in skeletal muscles from SIRT1-null mice. Gastrocnemius, tibialis anterior, and soleus muscles from 30-day-old SIRT1 homozygote-null mice (−/−), SIRT1 heterozygote-null mice (+/−), and wild-type littermates (+/+) were analyzed by quantitative reverse transcription-PCR. Data are presented as means ± S.E. of the relative abundance of transcript (8 mice/group). Statistically significant differences between SIRT1 −/− or SIRT1 +/− and SIRT1 +/+ are denoted by the asterisk (p < 0.05).
Effects of SIRT1 on PGC-1α Gene Expression in Myogenic Cells
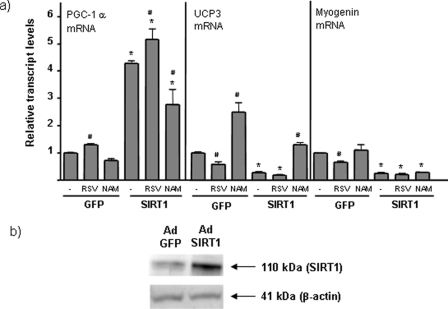
To determine the effects of SIRT1 on PGC-1α expression in myogenic cells, we transduced differentiated C2C12 myotubes with an adenoviral SIRT1 expression vector. In this experimental setting, SIRT1 was overexpressed ∼4- to 5-fold, as reported previously (Fig. 2b) (11). Overexpression of SIRT1 induced a marked increase in the expression of PGC-1α mRNA. In parallel, SIRT1 repressed UCP3 and myogenin gene expression, consistent with previous reports (9, 11). Treatment of C2C12 myotubes with resveratrol, an activator of SIRT1 activity, significantly increased PGC-1α gene expression and down-regulated UCP3 and myogenin expression (Fig. 2a). Resveratrol also increased the effects of adenoviral-mediated SIRT1 overexpression on PGC-1α levels. We also tested the effects of NAM, an SIRT1 inhibitor (23). NAM had no significant effect on basal expression of PGC-1α or myogenin expression, but attenuated SIRT1 overexpression induced PGC-1α up-regulation. Moreover, NAM increased basal levels of UCP3 mRNA and reduced the capacity of SIRT1 to inhibit UCP3 gene expression. Taken together, these findings indicate that SIRT1 exerts a positive effect on PGC-1α gene expression.
FIGURE 2.
Effects of SIRT1 overexpression and SIRT1 activity modulators on PGC-1α gene expression in muscle cells. C2C12 myotubes were transduced with Ad-SIRT1 or Ad-GFP (control) and treated with 50 μm resveratrol (resveratrol) or 10 mm nicotinamide (NAM) for 24 h before harvesting. a, data are presented as means ± S.E. of relative transcript levels from three to five independent experiments. Statistically significant differences (p < 0.05) are denoted by the asterisk (SIRT1 versus GFP) and # (drug treatment versus absence). b, representative immunoblot showing Ad-SIRT1-mediated overexpression of SIRT1.
SIRT1 Enhances PGC-1α Gene Transcription
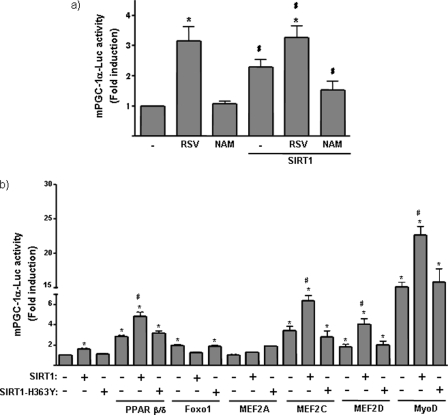
To determine whether the effects of SIRT1 on PGC-1α expression occur through induction of PGC-1α gene transcription, we transfected C2C12 myoblasts with a promoter-reporter construct containing 2 kb of the 5′ region of the mouse PGC-1α gene positioned upstream of the firefly luciferase gene (mPGC-1α-Luc). Tests of the effects of SIRT1-activity modulators showed that resveratrol increased basal PGC-1α promoter activity, whereas NAM had no effect under the basal conditions (Fig. 3a). Co-transfection of the PGC-1α promoter construct with an expression vector for SIRT1 increased PGC1-α promoter activity that was partially inhibited by NAM. These results indicate that SIRT1 acts on the 5′-upstream region of the PGC-1α gene to exert a positive effect on PGC-1α gene transcription.
FIGURE 3.
Effects of SIRT1 on PGC-1α gene promoter activity. C2C12 cells were transfected with a luciferase reporter construct driven by 2 kb of the mouse PGC-1α promoter region (PGC-1α -Luc). Data show the -fold induction of luciferase activity with respect to basal promoter values, and represent the means ± S.E. from at least three independent experiments performed in triplicate. a, cells were co-transfected with an expression vector for SIRT1 and treated with 50 μm resveratrol (RSV) or 10 mm NAM for 24 h, as indicated. Statistically significant differences (p < 0.05) due to drugs with respect to basal promoter values are denoted by the asterisk, and those due to the effects of SIRT1 are denoted by #. b, cells were co-transfected with expression vectors for PPARβ/δ, Foxo1, MEF2A, MEF2C, MEF2D, or MyoD, as indicated, as well as an expression vector for SIRT1 or the mutant form, SIRT1-H363Y. Cells transfected with PPARβ/δ were also treated with the specific agonist, 10 μm GW501516. Data show the -fold induction of luciferase activity with respect to basal promoter values, and represent the means ± S.E. from at least three independent experiments performed in triplicate. Statistically significant differences (p < 0.05) for each transfection condition with respect to basal promoter values are denoted by an asterisk, and those due to the effects of SIRT1 in the absence or presence of a given expression vector are denoted by #.
To gain insight into the mechanisms of SIRT1 action, we assessed the capacity of SIRT1 to co-activate known transcriptional activators of the PGC-1α promoter. To accomplish this, we co-transfected C2C12 myoblasts with expression vectors for these transcription factors, the PGC-1α promoter-reporter construct and expression vectors for SIRT1 or SIRT1-H363Y, a mutant form of SIRT1 that lacks deacetylase activity (24) (Fig. 3b). Previous reports have shown that PPAR β/δ activation increases PGC-1α promoter activity in muscle cells (25). Consistent with this, we found that SIRT1 acted as a co-activator of PPARβ/δ, increasing PGC-1α promoter activity. In contrast, the inactive SIRT1-H363Y mutant did not (Fig. 3b). Foxo1, which has also shown to act as a positive regulator at the PGC-1α promoter (4), was not co-activated by SIRT1, which instead tended to reduce PGC1α promoter activity in the presence of Foxo1.
We also studied the effects of SIRT1 on the action of MyoD and MEF2 isoforms, myogenic transcription factors known to act as positive regulators of the PGC-1α gene in muscle cells (4–6). Both MEF2D and MEF2C, but not MEF2A, increased PGC-1α promoter activity. As expected, MyoD was also a powerful activator. Co-transfection of SIRT1 co-activated these myogenic factors, increasing PGC-1α promoter activity. The SIRT1-H363Y mutant form lacked co-activator activity, indicating that this effect was dependent on SIRT1 deacetylase activity.
Involvement of Myogenic Factors in SIRT1 Effects on PGC-1α Gene Expression
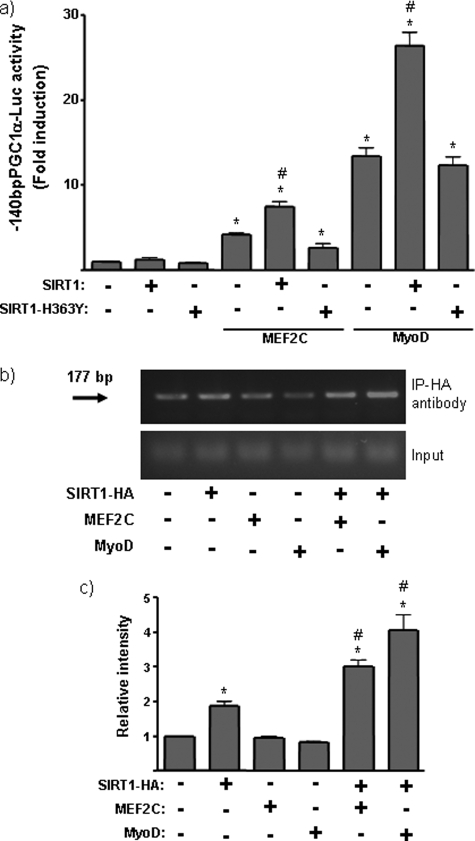
PGC-1α gene expression was further analyzed using a minimal construct of the mPGC-1α promoter (−140bpPGC-1α-Luc), which is devoid of most regions of the PGC-1α gene responsive to regulatory transcription factors but retains the region responsible for MyoD action (4) (Fig. 4a). The −140bpPGC-1α construct remained sensitive to MyoD trans-activation, as expected, and was also sensitive to MEF2C transactivation. SIRT1, again, enhanced the transactivation caused by these myogenic transcription factors.
FIGURE 4.
SIRT1 action on the myogenic factor-responsive region of the PGC-1α promoter. a, C2C12 cells were transfected with a promoter-reporter construct in which luciferase expression is driven by the proximal region (140 bp) of the PGC-1α gene promoter. Where indicated, cells were co-transfected with expression vectors for SIRT1, SIRT1-H363Y, MyoD, and/or MEF2C. Data show the -fold induction of luciferase activity with respect to basal promoter values and represent the means ± S.E. from at least three independent experiments performed in triplicate. Statistically significant differences (p < 0.05) for each transfection condition with respect to basal promoter values are denoted by an asterisk, and those due to the effects of SIRT1 in the absence or presence of a given expression vector are denoted by #. b, representative ChIP analysis of SIRT1 binding to the proximal region of the mouse PGC-1α promoter. Experiments were performed in the presence of a transfected 140-bp mPGC-1α-Luc construct and co-transfected expression vectors for HA-tagged SIRT1, MEF2C, and/or MyoD, as indicated. The arrow indicates the 177-bp PCR product from the mouse PGC-1α promoter. c, quantitative analysis of ChIP amplification. Data are expressed as the means ± S.E. of the -fold induction in relative intensity of the amplified PCR product from three independent experiments. Statistically significant differences (p < 0.05) are denoted by the asterisk (SIRT1-HA, MEF2C, or MyoD versus vector control) and # (MEF2c + SIRT1-HA or MyoD + SIRT1-HA versus SIRT1-HA, only).
We next used ChIP analysis to determine whether SIRT1 interacts directly with the PGC-1α gene promoter region. C2C12 cells were co-transfected with the PGC-1α promoter construct and an HA-tagged SIRT1 expression vector, together with expression vectors for MyoD or MEF2C, as indicated (Fig. 4, b and c). Unrelated immunoglobulin was used as a control. ChIP analysis using an anti-HA antibody revealed that a 177-bp PCR product of the PGC-1α promoter encompassing the proximal region was enriched in cells transfected with HA-SIRT1. The recruitment of SIRT1 to the PGC-1α promoter was further enhanced by expression of MEF2C or MyoD. This indicates that SIRT1 was specifically associated with a binding complex on the PGC-1α promoter in the presence of myogenic factors.
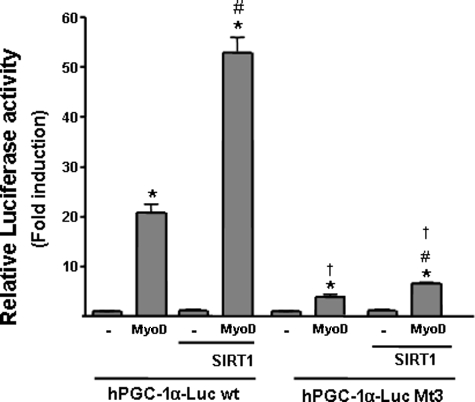
To further analyze the involvement of MyoD in the action of SIRT1 on PGC-1α gene transcription, we studied the responsiveness of the human PGC-1α promoter, in which the elements responsive to MyoD have been characterized previously (4). A region of the human minimal PGC-1α promoter (−49 bp) is practically identical to the corresponding mouse PGC-1α gene region and, as previously reported, retained MyoD responsiveness (Fig. 5). SIRT1 markedly increased the responsiveness of the PGC-1α promoter to MyoD. A PGC-1α promoter variant (Mt3) in which two MyoD-binding E boxes had been mutated (4) showed a marked reduction in MyoD-dependent promoter activity, as expected. In addition, SIRT1-dependent activity of this promoter in the presence of MyoD was substantially reduced (Fig. 5).
FIGURE 5.
Effects of SIRT1 on a MyoD response-defective mutant form of the human PGC-1α promoter. C2C12 cells were transfected with a luciferase construct containing 49 bp of the proximal human PGC-1α promoter core region (hPGC-1α-LUC wt) or a version in which the two E boxes (which mediate MyoD responsiveness) have been mutated (hPGC-1α-LUC Mt3). Cells were co-transfected with expression vectors for SIRT1 and/or MyoD as indicated. The results are expressed as -fold induction of luciferase activity with respect to basal promoter values, and represent the means ± S.E. of at least three independent experiments performed in triplicate. Statistically significant differences are denoted by the asterisk (MyoD versus vector control, p < 0.01), # (SIRT1 versus control vector, p < 0.01), and † (wt versus Mt3, p < 0.005).
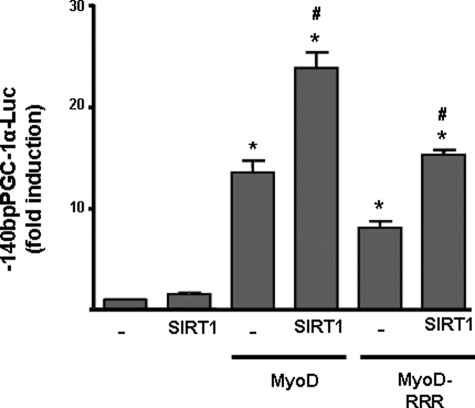
To assess whether SIRT1 exerted its co-activating effects on MyoD-dependent PGC-1α gene activation by altering MyoD acetylation status, we used an expression vector for a mutant form of MyoD in which lysine residues capable of being acetylated had been mutated to arginines (MyoD-RRR), and therefore could not be acetylated (17). We observed that MyoD-RRR-dependent transactivation of PGC-1α was reduced compared with that of wild-type MyoD and SIRT1 co-transfection did not restore full promoter activity as had been seen with the wild-type MyoD. However, SIRT1 induced an increase of the same magnitude in PGC-1α promoter activity in the presence of co-transfected wild-type MyoD or MyoD-RRR (Fig. 6).
FIGURE 6.
Effects of a non acetylable form of MyoD on mediating the responsiveness of the PGC-1α promoter to SIRT1. C2C12 cells were transfected with the 2-kb mouse PGC-1α-Luc promoter construct and co-transfected with expression vectors for MyoD or MyoD-RRR (a non acetylable mutant form of MyoD) and SIRT1, as indicated. Results are presented as means ± S.E. of the -fold induction of relative luciferase activity with respect to basal activity. Statistically significant differences (p < 0.05) are denoted by the asterisk (MyoD versus vector control) and # (SIRT1 versus vector control).
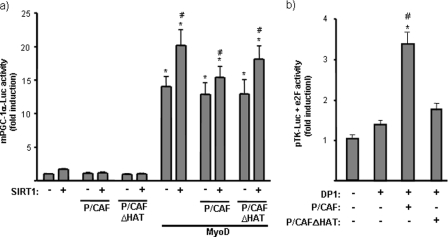
Previous reports have shown that SIRT1 interacts with MyoD via the acetyltransferase P/CAF, leading to repression of MyoD-dependent gene transcription (i.e. at the myogenin promoter) (9). To determine whether P/CAF could be involved in the action of SIRT1 on the PGC-1α gene, we co-transfected C2C12 myoblasts with the PGC-1α promoter-reporter construct and expression vectors for MyoD, SIRT1, and P/CAF or a mutant form of P/CAF without acetyltransferase activity (P/CAFΔHAT) (Fig. 7a). Neither P/CAF nor its mutant form modified the action of MyoD on the PGC-1α promoter, whereas, in parallel transfections, a positive P/CAF control yielded the expected result (Fig. 7b).
FIGURE 7.
Effects of P/CAF on the functional interaction of SIRT1 and MyoD on PGC-1α gene transcription. a, C2C12 cells were transfected with a luciferase reporter construct containing 2 kb of the mouse PGC-1α promoter region (mPGC-1α -Luc), and were co-transfected with expression vectors for MyoD, P/CAF, or P/CAFΔHAT (a mutated form of P/CAF devoid of acetyltransferase activity) and SIRT1, as indicated. Results are expressed as means ± S.E. of the -fold induction of relative luciferase activity with respect to basal activity. Statistically significant differences (p < 0.05) are denoted by the asterisk (MyoD versus vector control) and # (SIRT1 versus vector control). b, C2C12 cells were transfected with pTK-Luc (a luciferase expression vector driven by the thymidine kinase promoter) and co-transfected with pcDNA3-e2F1. DP1, P/CAF, and/or P/CAFΔHAT expression vectors were co-transfected, as indicated. Results are presented as means ± S.E. of at least three independent experiments. Statistically significant differences (p < 0.05) due to DP1 are denoted by the asterisk, and those due to the co-transfection of P/CAF expression vectors are denoted by #.
The Interaction of PGC1-α with Its Own Promoter Contributes to the Positive Regulation of PGC-1α Gene Transcription by SIRT1 Plus MyoD
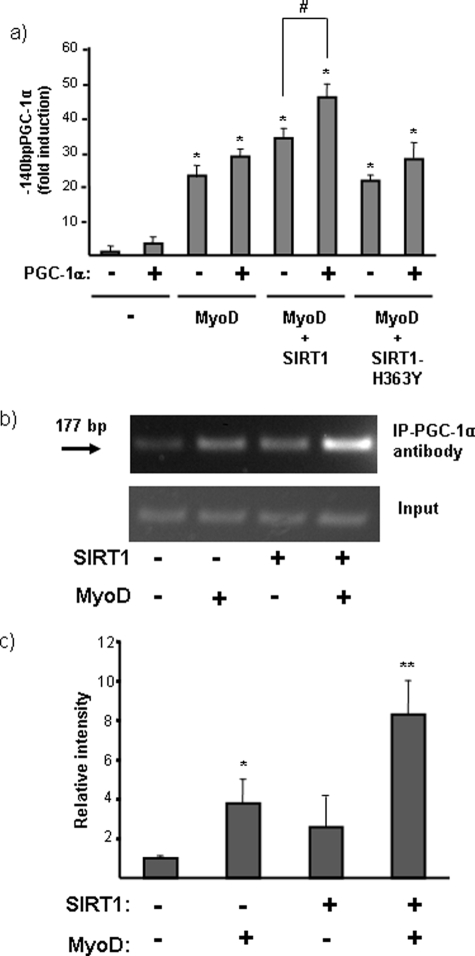
PGC-1α has been proposed to enhance its own transcription by an autoregulatory loop involving PPARs or the myogenic factor MEF2C (5, 6, 23). We thus sought to determine whether PGC-1α itself participated in the SIRT1-mediated stimulation of the PGC-1α gene promoter, and whether interactions with MyoD were involved. Using CV1 cells, which lack endogenous PGC-1α expression, were observed that exogenously expressed PGC-1α significantly increased its own promoter activity only in the presence of co-transfected SIRT1 and MyoD (Fig. 8a). To establish whether PGC-1α was recruited to its own promoter, we performed ChIP analyses (Fig. 8b). These assays showed that MyoD and SIRT1 expression individually enhanced the recruitment of PGC-1α to the minimal PGC-1α promoter, and the presence of both MyoD and SIRT1 induced maximal recruitment (Fig. 8c). We observed similar results for MEF2C (data not shown). These results indicate that PGC-1α may participate in the positive autoregulation of its own promoter via an interaction with SIRT1 and myogenic transcription factors.
FIGURE 8.
Effects of PGC-1α on the action of SIRT1 and MyoD at the PGC-1α gene promoter. a, CV1 cells were transfected with a promoter-reporter construct in which luciferase expression is driven by the proximal region (140 bp) of the PGC-1α gene promoter. Cells were co-transfected with expression vectors for PGC-1α, SIRT1, and/or MyoD, as indicated. Data are presented as -fold induction of luciferase activity with respect to basal promoter values, and represent the means ± S.E. of at least three independent experiments performed in triplicate. Statistically significant differences (p < 0.05) for each transfection condition with respect to basal promoter values are denoted by the asterisk, and those due to the effects of PGC-1α in the absence or presence of a given expression vector are denoted by #. b, representative ChIP analysis of PGC-1α binding to the proximal region of its own promoter. Experiments were performed in the presence of a transfected 140bpPGC-1α-Luc construct and co-transfected expression vectors for PGC-1α, SIRT1, and/or MyoD, as indicated. A representative ChIP analysis is shown. The arrow indicates the 177-bp PCR product from the mouse PGC-1α promoter. c, quantitative analysis of ChIP amplification. Data are expressed as the means ± S.E. of the -fold induction in relative intensity of the amplified PCR product from three independent experiments. Statistical significance of differences is denoted by: *, p < 0.05; **, p < 0.005.
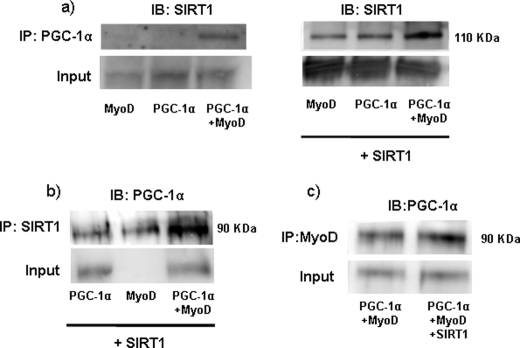
The Interaction of SIRT1 with PGC-1α Is Enhanced by MyoD
To determine whether these observations indicated that MyoD promoted the interaction between SIRT1 and PGC-1α, we performed co-immunoprecipitation assays in C2C12 cells in which MyoD, PGC-1α, or both had been overexpressed by adenoviral-mediated gene transfer. Endogenous SIRT1 was barely detectable in PGC-1α immunoprecipitates from cells MyoD or PGC-1α alone (Fig. 9a, left), but expression levels were markedly increased in cells that co-expressed both MyoD and PGC-1α (Fig. 9a, right). We also assessed the presence of PGC-1α in SIRT1 immunoprecipitates (Fig. 9b) and found that the highest levels of PGC-1α were also detected in cells that overexpressed both SIRT1 and MyoD. Finally, when an anti-MyoD antibody was used to immunoprecipitate cell extracts, we found substantial levels of PGC-1α in the immunoprecipitated material, indicating that these two proteins interact. Moreover, this interaction was enhanced by the presence of SIRT1 (Fig. 9c). Taken together, these data indicate that the presence of MyoD enhances the formation of SIRT1-PGC-1α complexes, and strongly support the participation of these three partners in building the highly active transcriptional complexes responsible for increasing PGC-1α gene transcription in response to SIRT1.
FIGURE 9.
Effect of MyoD on the interaction of PGC-1α with SIRT1. a, immunoprecipitation analysis of the interaction of PGC-1α with SIRT1. C2C12 cells were transduced with adenoviral vectors for PGC-1α and/or MyoD (left panel), and SIRT1 (right panel), as indicated. Cell extracts were immunoprecipitated (IP) using an antibody directed against PGC-1α and immunoblotted (IB) with an anti-SIRT1 antibody (top panel). Whole cell extracts were probed with the anti-SIRT1 antibody to confirm equivalent SIRT1 input before immunoprecipitation (bottom panels). b, C2C12 cells were transduced with adenoviral vectors for PGC-1α and/or MyoD, and SIRT1, as indicated. Cell extracts were immunoprecipitated using an antibody directed against SIRT1 and immunoblotted with an anti-PGC-1α antibody (top panel). Whole cell extracts were probed with the anti-PGC-1α antibody to confirm equivalent PGC-1α input before immunoprecipitation (bottom panel). c, C2C12 cells were transduced with adenoviral vectors for PGC-1α, MyoD, with or without co-transduction with the SIRT1 vector, as indicated. Cell extracts were immunoprecipitated using an antibody directed against MyoD and immunoblotted with an anti-PGC-1α antibody (top panel). Whole cell extracts were probed with the anti-PGC-1α antibody to confirm equivalent PGC-1α input before immunoprecipitation (bottom panel).
DISCUSSION
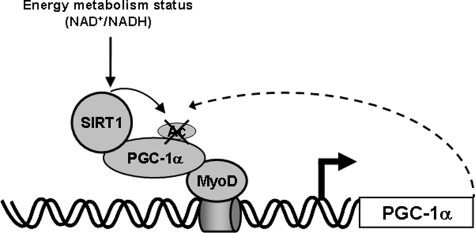
In this study we have identified a novel pathway in which SIRT1 action positively regulated PGC-1α gene transcription in skeletal muscle. We propose a model in which SIRT1 interacts with and deacetylates PGC-1α, increasing its activity on its own promoter through interactions with MyoD. The result is a SIRT1-enhanced positive autoregulatory loop (Fig. 10).
FIGURE 10.
Schematic representation of the possible mechanism-of-action of SIRT1 on PGC-1α gene transcription. Schematic overview of the interaction of SIRT1 with MyoD and involvement of the PGC-1α positive autoregulatory loop.
In addition to their importance in the control of muscle metabolism, the observed effects of SIRT1 on PGC-1α gene expression reveal several novel mechanistic aspects of muscle gene transcription regulation by SIRT1.
The observation that SIRT1 positively regulates PGC-1α gene expression in conjunction with MyoD is not a common feature of MyoD-sensitive genes. SIRT1 usually represses the action of MyoD on its target genes, as shown by myogenin (9). Thus, in the specific case of the PGC-1α gene, the positive effect of SIRT1 is likely to involve a unique architecture of the MyoD-containing transcriptional complex and the existence of positive autoregulatory processes elicited by PGC-1α itself. Our present findings indicate that P/CAF, which has been reported to mediate repressive effects of SIRT1 on MyoD-dependent gene transcription (9), does not have an important role in the control of the PGC-1α gene promoter. The existence of a PGC-1α-positive autoregulatory loop and the fact that PGC-1α activity is increased by deacetylation by SIRT1 (13) are consistent with the positive effect of SIRT1 on the transcriptional regulatory complex containing MyoD and PGC-1α. Indeed, the transcription of another MyoD target gene, UCP3, in which PGC-1α does not exert positive effects (11), is repressed by SIRT1. Moreover, the PGC-1α-positive autoregulation of its own gene promoter has been previously reported and shown to be mediated by PPARs (25, 26) or the myogenic factor MEF2C (5, 6), as also found in the present study. In light of the present findings, it could be hypothesized that other muscle genes that are targets of positive activation by MyoD and PGC-1α could be positively regulated by SIRT1. Our preliminary data support this proposal. For instance, SIRT1 seems to activate the gene encoding carnitine palmitoyl-transferase 1-β,3 a positive target of myogenic factors and PGC-1α (13, 27).
The transcriptional control of PGC-1α gene expression by SIRT1 might mediate adaptive gene expression in response to metabolic stimuli (NAD+/NADH ratio) by inducing PGC-1α expression and, therefore, PGC-1α target genes. This mechanism would be additive to the action reported by Gerhart-Hines et al. (2007) in which SIRT1 has shown to activate PGC-1α target genes by deacetylating PGC-1α, thereby increasing its activity. SIRT1 (and the molecules that influence its activity) can be expected to act through both mechanisms, enhanced expression and enhanced activity of PGC-1α, to control a plethora of genes in skeletal muscle, promote oxidative metabolism, and contribute to the enrichment of highly oxidative fiber types in muscle. Reports that the expression levels of PGC-1α (3, 28) and SIRT1 (29) are increased in highly oxidative muscle fibers relative to glycolytic fibers are consistent with our present findings. On the other hand, calorie restriction has been reported to ameliorate the age-related decline in skeletal muscle oxidative function (30, 31). The induction of PGC-1α by SIRT1 and subsequent promotion of mitochondrial oxidative metabolism could contribute to this positive effect of calorie restriction. In conclusion, we report that SIRT1 controls PGC-1α gene expression in skeletal muscle and MyoD is a key mediator of this action. The involvement of myogenic factors in SIRT1 action on PGC-1α may provide a molecular mechanism to help explain how SIRT1 drives muscle-specific adaptive changes in gene expression and metabolism.
Acknowledgments
We thank Drs. M. Martinez-Balbas, B. Spiegelman, T. Finkel, and P. Puigserver for kindly supplying expression vectors, and F. Alt for supplying SIRT1-null mice.
This work is supported by Grant SAF2008-01896 from the Ministerio de Ciencia e Innovación, Spain.
R. Amat and F. Villarroya, unpublished observations.
- PGC-1α
- peroxisome proliferator activated receptor-γco-activator-1α
- UCP1
- uncoupling protein 1
- UCP3
- uncoupling protein 3
- NAM
- nicotinamide
- ChIP
- chromatin immunoprecipitation
- MyoD
- myogenic determining factor
- PPARγ
- peroxisome proliferator activated receptor-γ
- GFP
- green fluorescent protein
- CMV
- cytomegalovirus
- MEF2
- myocyte enhancer factor 2.
REFERENCES
- 1.Pilegaard H., Saltin B., Neufer P. D. (2003) Diabetes 52, 657–662 [DOI] [PubMed] [Google Scholar]
- 2.Russell A. P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C. A., Bell D. R., Kralli A., Giacobino J. P., Dériaz O. (2003) Diabetes 52, 2874–2881 [DOI] [PubMed] [Google Scholar]
- 3.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 4.Chang J. H., Lin K. H., Shih C. H., Chang Y. J., Chi H. C., Chen S. L. (2006) Endocrinology 147, 3093–3106 [DOI] [PubMed] [Google Scholar]
- 5.Handschin C., Rhee J., Lin J., Tarr P. T., Spiegelman B. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czubryt M. P., McAnally J., Fishman G. I., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 8.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. (2003) Mol. Cell 12, 51–62 [DOI] [PubMed] [Google Scholar]
- 10.Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amat R., Solanes G., Giralt M., Villarroya F. (2007) J. Biol. Chem. 282, 34066–34076 [DOI] [PubMed] [Google Scholar]
- 12.Solanes G., Pedraza N., Iglesias R., Giralt M., Villarroya F. (2000) FASEB J. 14, 2141–2143 [DOI] [PubMed] [Google Scholar]
- 13.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 15.Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 16.Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solanes G., Pedraza N., Iglesias R., Giralt M., Villarroya F. (2003) Mol. Endocrinol. 17, 1944–1958 [DOI] [PubMed] [Google Scholar]
- 18.Blanco J. C., Minucci S., Lu J., Yang X. J., Walker K. K., Chen H., Evans R. M., Nakatani Y., Ozato K. (1998) Genes Dev. 12, 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 20.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 21.Nadal A., Marrero P. F., Haro D. (2002) Biochem. J. 366, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amri E. Z., Bonino F., Ailhaud G., Abumrad N. A., Grimaldi P. A. (1995) J. Biol. Chem. 270, 2367–2371 [DOI] [PubMed] [Google Scholar]
- 23.Landry J., Slama J. T., Sternglanz R. (2000) Biochem. Biophys. Res. Commun. 278, 685–690 [DOI] [PubMed] [Google Scholar]
- 24.Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 25.Hondares E., Pineda-Torra I., Iglesias R., Staels B., Villarroya F., Giralt M. (2007) Biochem. Biophys. Res. Commun. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 26.Hondares E., Mora O., Yubero P., Rodriguez de la Concepción M., Iglesias R., Giralt M., Villarroya F. (2006) Endocrinology 147, 2829–2838 [DOI] [PubMed] [Google Scholar]
- 27.Baldán A., Relat J., Marrero P. F., Haro D. (2004) Nucleic Acids Res. 32, 4742–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 29.Suwa M., Nakano H., Radak Z., Kumagai S. (2008) Metabolism 57, 986–998 [DOI] [PubMed] [Google Scholar]
- 30.Hepple R. T., Baker D. J., Kaczor J. J., Krause D. J. (2005) FASEB J. 19, 1320–1322 [DOI] [PubMed] [Google Scholar]
- 31.Baker D. J., Betik A. C., Krause D. J., Hepple R. T. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61, 675–684 [DOI] [PubMed] [Google Scholar]