Abstract
Chondrosarcoma is a primary bone tumor with a dismal prognosis; most patients with this disease develop fatal pulmonary metastases, suggesting the need for a better systemic treatment. Anti-angiogenesis treatment may be useful, because angiogenesis is critical for both tumor growth and metastasis. Vascular endothelial growth factor (VEGF) is the most potent pro-angiogenic factor and is regulated by pathways related to the normal physiologic response to hypoxia and genetic alterations related to the malignant phenotype. Our prior work has shown that VEGF is overexpressed in high grade chondrosarcoma and chondrosarcoma cell lines. Working on the premise that developmental pathways giving a selective growth advantage are often recapitulated in tumors, we investigated the regulation of VEGF by HDAC4 and Runx2 in chondrosarcoma. We tested the hypothesis that there is dysregulation of HDAC4/Runx2/VEGF gene expression and that decreased HDAC4 expression accounts for at least some of the increased VEGF expression seen in chondrosarcoma. We show that reduced expression of HDAC4 in chondrosarcoma cells increases expression of Runx2 leading to increased expression of VEGF and in vitro angiogenesis. Thus, both hypoxia and dysregulated expression of a developmental pathway are causes of increased VEGF expression in chondrosarcoma.
Chondrosarcomas are mesenchymal tumors in which the primary tissue is cartilage; they include 20% of primary bone tumors and occur in patients of all ages (1, 2). Chondrosarcomas are difficult tumors to cure, because they are unresponsive to the standard adjuvant treatment, chemotherapy (3) and radiation therapy (4), resulting in cure rates of less than 10% (5, 6), with the vast majority of patients succumbing to pulmonary metastases. Angiogenesis is critical for both tumor growth and development of metastases, and inhibiting angiogenesis has become a therapeutic strategy. We have demonstrated that grade II and III chondrosarcomas have more microvascularity than benign or grade I tumors (7) and that these tumors overexpress vascular endothelial growth factor (VEGF)2 (8). Because VEGF is the most important proangiogenic molecule and is overexpressed in high grade chondrosarcoma, we have focused on the regulation of VEGF in this tumor.
VEGF expression is determined by normal physiologic hypoxia-related pathways and genetic abnormalities. We know that both of these broad categories of gene regulation are operational in chondrosarcomas. Included in genetic abnormalities are epigenetic phenomena such as DNA methylation and histone modification that regulate chromatin structure and gene expression (9). Both histone acetylases and histone deacetylases (HDACs) are key enzymes that catalyze the reversible acetylation/deacetylation of core histone tails, which is an essential mechanism of the epigenetic control of gene expression (10). HDACs function as transcriptional co-repressors. HDACs can deacetylate DNA binding transcription factors, thereby decreasing their binding affinity, localization, and half-life (11). The activity of HDACs is affected by their phosphorylation state, thereby linking them to cell signaling pathways (12). The mammalian HDACs fall into three classes based on their structural and biochemical characteristics (13). Recent studies show that the class II HDACs are involved in cellular differentiation and developmental processes, and their dysregulation may be involved in carcinogenesis. HDAC4 along with -5, -7, and -9 compose the class IIa HDACs. Class I HDACs are ubiquitously expressed, whereas class II HDACs have tissue-specific expression and regulate cell differentiation. HDAC4 is expressed in muscle, brain, and cartilage.
Targets of HDAC4 include Runx1 and Runx2. Runx2 is important for skeletal development. There has been no direct link established between HDAC4 and angiogenesis; however, in the growth plate, HDAC4 is expressed in prehypertrophic chondrocytes and regulates chondrocyte hypertrophy and endochondral bone formation by binding and inhibiting the activity of Runx2/Cbfa1 (11) and induces cell death in a caspase-9-dependent manner (14). Runx2 is a transcription factor that is necessary for chondrocyte hypertrophy and endochondral ossification (11). HDAC4 expression decreases in the more mature hypertrophic chondrocytes, releasing Runx2 activity, and endochondral ossification ensues. Runx2 is known to up-regulate VEGF expression during endochondral bone formation, and both changes in HDAC4 and Runx2 expression are necessary for this process to occur (15). The functions of HDAC4 and Runx2 in the growth plate have been demonstrated in HDAC4 knock-out and Runx2 gain of function mice in which there is premature ossification of the growth plate in both models. Overexpression of Runx2 in fibroblasts induces an increase in their VEGF mRNA level and protein production (15). In chondrosarcoma, overexpression of Runx2 has been detected in high grade tumors, although its significance has been heretofore unknown (16, 17). In lung cancer patients, reduced expression of class II HDACs is associated with poor progression (18). Taken together, these studies highlight the importance of the class II HDACs in control of Runx2 transcription in normal chondrocyte differentiation and suggest Runx2 may play an important role in angiogenesis and chondrosarcoma development.
In this study, we tested the hypothesis that decreased HDAC4 expression in chondrosarcoma cell lines accounts for some of the increased VEGF expression and angiogenic activity in these cells. We also investigated the mechanism of HDAC4 regulation of Runx2 and VEGF transcription.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Human chondrocytes isolated from normal adult articular cartilage were cultured in complete medium (40% Dulbecco's modified Eagle's medium, 40% minimum Eagle's medium, 20% F-12 medium) with 10% fetal bovine serum (19). Chondrosarcoma (CS) cell line JJ (a gift from Dr. Joel Block, Rush Medical School, Chicago) was also cultured in complete medium (19), and CS-1 (a gift from Dr. Francis Hornicek, Harvard Medical School, Boston) was cultured in RPMI 1640 medium with 10% fetal bovine serum. All cells were cultured in a humidified incubator under 5% CO2. JJ was derived from a human grade II chondrosarcoma and CS-1 from a human grade III chondrosarcoma (20–22).
Recombinant DNA and Transfections
The following constructs were provided by the individuals listed: pcDNA3-HDAC4-FLAG, Eric A. Miska, University of Cambridge, Cambridge, UK (23); pCMV/Runx2, Renny Franceshi, University of Michigan, Ann Arbor, MI (24); pGL3-Runx2-LUC (0.6 kb of rat RUNX2 promoter), Gary Stein, University of Massachusetts Medical School (25); human VEGF promoter constructs pGL3-VEGF3.3-LUC (−2362/+955) and pGL3-VEGF1.7-LUC (−794/+955), Dr. Lee Ellis, University of Texas M.D. Anderson Cancer Center, Houston, TX (26); and HA-CBP, Eugene Chin, Rhode Island Hospital, Providence, RI. Cells were transiently transfected with DNA plasmids using FuGENE 6 (Roche Applied Science) after seeding for 18 h. After transfection, cells were incubated for 48 h and harvested for the following experiments.
Site-directed Mutagenesis
Site-directed mutagenesis of the two putative Runx2-binding sites in the VEGF promoter (pGL3-VEGF1.7-LUC (−794/+955)) was performed to change the core-binding motif TGG/ACC (27) to TCT/AGA using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). First, the putative Runx-binding site mutation between nucleotides −559 and −553 (Fig. 4C) was generated by amplifying the promoter fragment using the forward primer carrying the mutation site indicated in capital letters, 5′-ggggcgctcggccaGAacagggaagctg-3′ and the reverse primer 5′-cagcttccctgttctggccgagcgcccc-3′. The PCR product was digested with DpnI to destroy the parental DNA, followed by transformation in XL1-Blue competent cells (Stratagene, La Jolla, CA). The second mutation between +293 and +287 bp was then generated by using the single mutant promoter as the template with forward primer 5′-tctacttccccaaatcactgtACattttggaaaccagcagaaag-3′ and reverse primer 5′-ctttctgctggtttccaaaatgtacagtgatttggggaagtaga-3′. Incorporation of the substitution mutations was confirmed by sequencing (Brown University, Providence, RI).
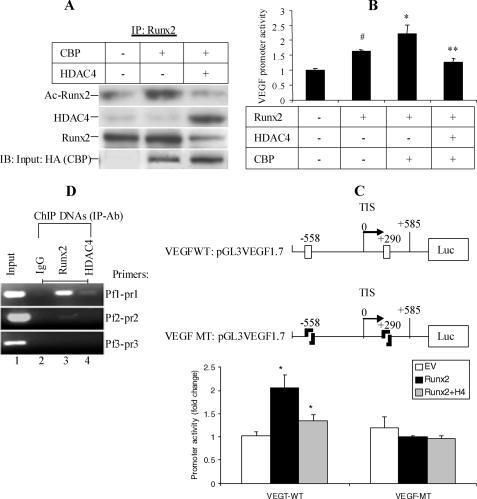
FIGURE 4.
Functional relationship of HDAC4, Runx2, and VEGF. A, interaction of HDAC4 and Runx2 and effect of HDAC4 on Runx2 protein. CS-1 cells were transfected with CBP and/or HDAC4 or empty vector. After 48 h, cell lysates were used for immunoprecipitation with Runx2 antibody and protein A/G beads. Western blot was performed with anti-acetylated lysine antibody, Runx2 antibody, and HDAC4 antibody, sequentially. Last row is Western blot of nonimmunoprecipitated cell lysate with anti-hemagglutinin (HA) antibody showing the input CBP amount. The results shown were representative of two independent experiments. IP, immunoprecipitated; IB, immunoblot. B, effect of HDAC4 on Runx2 protein activity. CS-1 cells were co-transfected with VEGF-luciferase, pRL-SV40, and 0.2 μg of expression plasmids for HDAC4, Runx2, or CBP. After 48 h, cells were analyzed for dual luciferase activity by normalizing to Renilla luciferase. Data represent the mean of triplicate samples ± S.D. (n = 3). #, p < 0.05, significant difference from the EV control (column 1). *, p < 0.01, significant difference from column 2. **, p < 0.05, significant difference from column 3. C, no effect of HDAC4 or Runx2 on VEGF promoter activity after mutation of Runx2-binding motifs. Schematic representation of VEGF promoter reporter construct indicating positions of Runx2-binding motifs at −558 and +290 relative to the transcription initiation sites (TIS) that were mutated using site-directed mutagenesis. The wild type (WT) or mutant (MT) VEGF promoters (0.5 μg) were co-transfected with 0.5 μg of EV, Runx2, or HDAC4 expression constructs in JJ cells. The luciferase (Luc) activity was determined 2 days after transfection and normalized to Renilla luciferase values used as an internal control. The relative fold change data were obtained by comparing Runx2 or Runx2 with HDAC4 to EV (n = 3). Data are the mean ± S.D. (n = 3). *, p < 0.05, compared with empty vector. D, ChIP analysis of endogenous Runx2 binding to the VEGF promoter in CS cells. CS cells were cross-linked with formaldehyde when cells are 80% confluent for ChIP analysis. Three independent immunoprecipitation reactions were carried out with Runx2 antibody (lane 3), HDAC4 antibody (Ab) (lane 4), and normal rabbit IgG (lane 2), respectively. The PCR products obtained with ChIP DNAs using the VEGF promoter-primers (Pf1-pr1, Pf2-pr2, Pf3-pr3) were run on 2% agarose gel containing ethidium bromide. The experiment was repeated three times with similar results.
Real Time RT-PCR (qPCR)
RNA was isolated from chondrocytes and transfected and nontransfected CS cell lines with RNAqueous kit (Ambion, Austin, TX). After treatment with TURBO DNase (Ambion), 1 μg of RNA was reverse-transcribed with random hexamers to obtain first-strand cDNA using iScript cDNA kit (Bio-Rad). The quantification of mRNA for HDAC4, Runx2, and VEGF was performed by two-step real time quantitative RT-PCR (Qiagen, Valencia, CA). Primers for HDAC4 were (5′ to 3′) as follows: forward, gag aga ctc acc ctt ccc g, and reverse, ccg gtc tgc acc aac caa g. Runx2 primers were as follows: forward, ggc agg cac agt ctt ccc, and reverse, ggc cca gtt ctg aag cac c, and can detect the three Runx2 transcript variants. The primers used for VEGF and 18 S rRNA (18 S) were reported previously (19). 18 S was used as an internal control gene because it has been shown to be the optimal reference gene (28). Amplification conditions were as follows: 2 min of preincubation at 50 °C, 10 min at 95 °C for enzyme activation, and 40 cycles at 95 °C denaturation for 10 s, 55 °C annealing for 30 s, and 72 °C extension for 30 s. The comparative threshold cycle (Ct) method, i.e. 2−ΔΔCt method, was used for the calculation of fold amplification (29). Each experiment was evaluated with three PCRs, and each experiment was repeated three times, the sample size necessary to maintain power at 0.80 to detect a 50% decrease with an α of 0.025 (one-tailed t test). The expression of HDAC4, Runx2, and VEGF in each tumor cell line was compared with chondrocytes using a one-tailed Student's t test to take advantage of the directional hypothesis.
Luciferase Assay
After subculture for 18 h, CS cells in 12-well plates were 70% confluent and then transiently transfected with reporter plasmid and co-transfected with gene expression vector, or empty vector, or different concentrations of HDAC4 vector as indicated using FuGENE 6 (Roche Applied Science), according to the manufacturer's instructions. The total amount of transfected DNA (1.5 μg/well) for each experiment was kept constant by addition of EV when needed. Cells were lysed 48 h after transfection. Luciferase activity was measured to assess reporter gene expression in a luminometer (Turner Design, Sunnyvale, CA) using Dual-Luciferase® Reporter Assay System (Promega, Madison, WI). In this system, cells were co-transfected with 0.02 μg of pRL-SV40 (Renilla luciferase driven by a full SV40 promoter) to control for transfection efficiency. Luciferase values were normalized for transfection efficiency (firefly/Renilla ratios). The relative fold induction or inhibition refers to the ratio of luciferase activity measured in expression vector-transfected cells relative to the activity obtained in the empty vector-transfected cells. Each measurement was made in duplicate.
Western Blot
Whole cell lysates containing 20 μg of protein were heated and separated by 4–20% SDS-polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). Membranes were blocked with 5% nonfat milk/PBST for 1 h and subsequently incubated overnight with primary antibodies (anti-VEGF and anti-HDAC4 polyclonal antibody, Santa Cruz Biotechnology; anti-Runx2 antibody, CeMines, Inc., Golden, CO). Signals were detected using a horseradish peroxidase-conjugated secondary antibody and an enhanced chemiluminescence detection kit (Amersham Biosciences). Bands were quantified using Image Acquisition and Analysis Software (UVP Biosystems, Upland, CA) and normalized to actin. Because of heterogeneity of Western blot data, normalized data were natural logarithm-transformed and analyzed with Student's t test with Bonferroni correction.
Co-immunoprecipitation
CS cells in 10-cm2 dishes were transfected with 4 μg of HA-CBP and/or FLAG-HDAC4. 48 h later, cells were lysed, and the lysates were incubated with 2 μg of anti-Runx2 antibody (CeMines, Inc., Golden, CO) with shaking at 4 °C overnight, and then with 30 μl of prewashed protein A/G Plus-agarose immunoprecipitation reagent (Santa Cruz Biotechnology) for 2 h at 4 °C. Beads were washed four times in 1× wash buffer (Actif Motif, Carlsbad, CA), resuspended in 1 volume of Western loading buffer, and boiled for 10 min at 95 °C. The presence of immunoprecipitated proteins (acetylated Runx2, total Runx2, and HDAC4) was detected by SDS-PAGE and immunoblotting by using anti-acetylated lysine antibody (Cell Signaling Technology, Beverly, MA), anti-Runx2 antibody (CeMines, Inc.), and anti-HDAC4 polyclonal antibody (Santa Cruz Biotechnology).
Knockdown of Runx2 and HDAC4 via RNA Interference
Cells in 6-well plates were transfected with 5 nm HDAC4 siRNA (MU-003497-02, Dharmacon, Lafayette, CO) or Runx2 siRNA (Hs_RUNX2_3_HP siRNA, target sequence CAGCACGCTATTAAATCCAAA, Qiagen) per ml by HiPerFect transfection reagent (Qiagen). mRNA and protein were obtained from cell lysates as described above 72 h after transfection.
ELISA
Soluble VEGF was detected in the conditioned media (CM) obtained from CS cells transfected with HDAC4 expression plasmid (different concentrations) or empty plasmid or siRNA specific for Runx2 or scramble siRNA for control using a commercially available assay kit (R & D Systems, Minneapolis, MN) according to manufacturer's instructions. VEGF levels in the CM were measured two times for each condition and normalized for the number of cells at the end of the culture period. Immunodepletion of VEGF in CM obtained from transfected CS cells as described above was performed by incubating either with 1 μg of anti-VEGF goat IgG (R & D Systems, Minneapolis, MN) or with control normal goat IgG per 1 ml of CM for 2 h at 4 °C with rotation; then 30 μl of protein A/G-agarose (Santa Cruz Biotechnology) was added and incubated for 2 h more at 4 °C with rotation. The resulting immune complexes were removed by centrifugation, and then VEGF levels were remeasured in the CM by ELISA.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP experiments were performed following the protocol provided with the ChIP-IT kit (Active Motif, Carlsbad, CA) using JJ and CS-1 cells. Cells were treated with formaldehyde to cross-link DNA-bound protein and subsequently subjected to sonication in lysis buffer containing protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin). Immunoprecipitation was performed with 2 μg of the anti-Runx2 antibody or anti-HDAC4 antibody (Santa Cruz Biotechnology) for each ChIP reaction. Following immunoprecipitation, proteins were removed by treatment with proteinase K, and the DNA was purified by the DNA mini-columns supplied with the kit. PCR was performed using primers for three regions of the human VEGF promoter (5′ to 3′) as follows: Pf1, gcttgccattccccacttgaatc; Pr1, tgtctgtctgtctgtccgtcag; Pf2, gtgtgcagacggcagtcactag; Pr2, gatctattggaatcctggagtgacc; Pf3, ctacagacgttccttagtgctgg; Pr3, caccaagtttgtggagctgagaac.
HUVEC Proliferation Assay
CM from CS cells were tested for angiogenic activity in the HUVEC assay. HUVECs in EGM-2 medium (Cambrex, East Rutherford, NJ) were plated at 5 × 103 cells per well in 24-well plates and starved of extrinsic VEGF and basic fibroblast growth factor for 24 h. HUVECs were then cultured in the different CM for 24 h followed by 18 h with [3H]thymidine (1 μCi/well). Proliferation rate was measured from radioactivity incorporated into total DNA using a Topcount NXT scintillation counter (Packard Instrument Co.) and calculated as a ratio of experimental to control. Results are an average of three independent experiments with four data points for each condition. Similarly, we tested the effect of VEGF-immunodepleted CM on HUVEC proliferation in the presence or absence of rhVEGF165 (R & D Systems).
HUVEC Tube Formation in Co-culture Assay
To study the effects of co-culturing HUVEC with JJ cells on HUVEC tube formation, we used a three-dimensional Matrigel system. In this system, each well of 4-well chamber slides was first divided into two equal parts by a thin strip of agarose gel (30). 100 μl of growth factor-reduced Matrigel (BD Biosciences) was added into one side of the wells and allowed to solidify for 30 min at 37 °C. JJ cells were then plated on the Matrigel, and 2 days after transfection, the other side of the wells were coated with Matrigel onto which HUVECs were plated. JJ cells and HUVECs were initially cultured in their own media. After 2 h, the wells were covered with enough of the HUVEC medium (2% fetal bovine serum, HUVEC medium without VEGF and fibroblast growth factors) so that the agarose divider no longer separated the two wells. After 1 day of co-culture, images of HUVEC were taken using an SpotII digital camera (Diagnostic Instruments, Sterling Heights, MI) attached to a Nikon microscope. Eight different fields were evaluated for each treatment, each image being selected on the basis of the optimal focal plane that had the majority of cells in focus. The tubular structures were traced, and the total length, area, and number of junctions were analyzed using Elements software (Nikon, Inc, Mellville, NY). The experiment was repeated three times.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism version 3.0 (GraphPad Software, San Diego). ELISA, luciferase activity, and HUVEC proliferation and tube formation results were analyzed with one-way analysis of variance. Post test comparisons were made with Bonferroni correction. Experiments with two groups were analyzed with the Student's t test. The null hypothesis of no difference was rejected at a significance level of 5%.
RESULTS
HDAC4, Runx2, and VEGF Expression in Chondrosarcoma Cell Lines
As a first step in evaluating the potential role of HDAC4, Runx2, and VEGF as dysregulated pathways in chondrosarcoma, we analyzed expression of these genes and proteins in chondrocyte and chondrosarcoma cell lines using quantitative real time PCR and Western blotting (Fig. 1). We found that HDAC4 gene expression was decreased to 17 and 39% in chondrosarcoma cells compared with chondrocytes (Fig. 1A), whereas Runx2 was increased 4.1- and 15.6-fold (Fig. 1B), and VEGF was increased 5.9- and 8.5-fold (Fig. 1C). All chondrosarcoma gene expressions were significantly different from chondrocytes (p < 0.01). Protein levels of Runx2 and VEGF showed analogous changes (Fig. 1D). Quantification of Western blots showed HDAC4 was decreased to 47% in chondrosarcoma cells compared with chondrocytes, and Runx2 and VEGF were increased 3.4- and 3.1-fold compared with chondrocytes.
FIGURE 1.
Expression of HDAC4, Runx2, and VEGF in chondrocytes and chondrosarcoma cells. A–C, endogenous mRNA level of HDAC4, Runx2, and VEGF. Gene expression in chondrocytes (CH) and chondrosarcoma cells JJ and CS-1 was quantified by qPCR and normalized to 18 S. The data are the mean ± S.D. (n = 3); *, p < 0.01, chondrosarcoma cells versus chondrocytes. D, representative Western blot of HDAC4, Runx2, and VEGF. Whole cell lysates prepared from chondrocytes, JJ, and CS-1 were used for SDS-PAGE and Western blot detection of HDAC4, Runx2, VEGF, and actin using specific antibodies. E, quantification of Western blots of four independent experiments, which were quantitated by normalizing to actin; *, p < 0.02; #, p < 0.01.
VEGFa mRNA and Protein Levels Are Regulated by HDAC4
To determine whether the decreased expression of HDAC4 is a cause, either directly or indirectly, of increased VEGF expression, chondrosarcoma cells were transfected with either HDAC4 expression construct or siRNA directed against HDAC4. We tested if increased HDAC4 expression would decrease VEGF expression. Real time PCR showed that HDAC4 transfection repressed the endogenous VEGFa transcription to 34% in JJ cells and to 29% in CS-1 cells compared with the control (EV) (p < 0.01) (Fig. 2A). Fig. 2, B and C, shows high level of HDAC4 expression after HDAC4 transfection. Western blotting also showed that endogenous VEGFa protein level decreased after HDAC4 transfection (Fig. 2C). Thus, VEGF expression was repressed by overexpression of HDAC4. The opposite effect was seen with HDAC4 knockdown. As can be seen from Fig. 2, E and F, siRNA effectively decreased the expression of endogenous HDAC4 mRNA and protein levels, whereas VEGFa mRNA increased 4.1-fold (p < 0.01) (Fig. 2D) as did protein levels (Fig. 2F). Similar results were obtained with JJ cells (data not shown). To understand the mechanism of the regulation of VEGF expression by HDAC4, we did the following studies.
FIGURE 2.
Overexpression of HDAC4 decreases and HDAC4 knockdown increases VEGF expression in chondrosarcoma cell lines. A and B, VEGF and HDAC4 mRNA level after transfection with HDAC4. B, expression after EV transfection defined as 1. Chondrosarcoma cells JJ and CS-1 in 10-cm2 dishes were transfected with 7 μg of HDAC4 expression construct (H4) or empty vector (EV). After 48 h, VEGF and HDAC4 gene expression was quantified by qPCR and normalized to 18 S. *, p < 0.01, HDAC4 versus EV transfection. C, VEGF and HDAC4 protein levels after transfection with HDAC4. Whole cell lysates from cells transfected with EV or HDAC4 expression vector (H4) were used to detect VEGF and HDAC4 protein level with specific antibodies by Western blot. Representative Western blot results are shown from one of three independent experiments. Actin was used as the loading control. D and E, VEGF and HDAC4 mRNA level after knockdown of HDAC4. CS-1 cells were treated with HDAC4 siRNA (H4-siRNA) or nontargeting control siRNA (Con-siRNA) at 5 nm/ml for 72 h. VEGF and HDAC4 gene expression was quantified by qPCR and normalized to 18 S. *, p < 0.01. F, HDAC4 and VEGF protein expression after HDAC4 knockdown. Protein levels in cell lysates were analyzed by Western blot. Representative Western blot results are shown from one of three independent experiments. Actin was used as the loading control.
HDAC4 Inhibits VEGFa Expression by Inhibiting Runx2 Expression
We used VEGFa and Runx2 reporter constructs to investigate the effects of HDAC4 and Runx2, alone and in combination, on the VEGF and Runx2 promoters. The results of luciferase activity analysis show that transfection of HDAC4 expression construct repressed VEGFa promoter-driven luciferase activity to 54% in JJ cells (p < 0.05) and to 35% in CS-1 (p < 0.02) (Fig. 3A). Because Runx2 is known to be a target for HDAC4, and because Runx2 is known to regulate endochondral ossification, which requires VEGF, we analyzed the role of Runx2 in the HDAC4 regulation of VEGF expression. When CS-1 was transfected with the HDAC4 expression construct and a Runx2-luciferase reporter construct, promoter activity decreased to 38% of control (Fig. 3B). HDAC4 also decreased Runx2 mRNA in JJ and CS-1 (p < 0.01) (Fig. 3C).
FIGURE 3.
HDAC4 regulates VEGF through Runx2. A, VEGF promoter activity after HDAC4 transfection. Chondrosarcoma cells JJ and CS-1 were transiently transfected with HDAC4 expression construct (0.6 μg/well), VEGF-luciferase, pRL-SV40, or EV. The amount of transfected DNA was kept at 1.5 μg/well by addition of the appropriate amount of empty vector. After 48 h, the cells were harvested for dual luciferase assays. The relative promoter activity (fold) was the ratio of luciferase value (Firefly/Renilla) measured in expression vector-transfected cells relative to that obtained in the EV-transfected cells as described under “Experimental Procedures.” Data represent the mean of three experiments ± S.D. (n = 3). *, p < 0.05 compared with empty vector. B, Runx2 promoter activity after HDAC4 transfection. CS-1 cells were transfected with a Runx2/luciferase reporter construct, pRL-SV40, and HDAC4 expression construct 0.6 μg/ml or EV. After 48 h, cells were analyzed for dual luciferase activity by normalizing to Renilla luciferase. Data represent the mean ± S.D. (n = 3); *, p < 0.01. C, effect of HDAC4 on Runx2 mRNA. Chondrosarcoma cells were transfected with HDAC4 expression construct 3 μg/ml or EV. Runx2 gene expression was quantified by qPCR and normalized to 18 S. *, p < 0.01. D and E, effect of Runx2 knockdown on VEGF mRNA and protein levels. CS cells were treated with Runx2 siRNA (R2-siRNA) or nontargeting control siRNA (Con-siRNA) at 5 nm/ml for 72 h. VEGF gene expression was quantified by qPCR and normalized to 18 S. The data are the mean ± S.D. (n = 3); *, p < 0.01 compared with control-siRNA. Protein levels of VEGF and Runx2 in cell lysates were analyzed by Western blot. The data are representative of three experiments. F, dose-response effect of HDAC4 transfection on VEGF promoter activated by Runx2. CS-1 was transfected with Runx2 expression construct (0.3 μg/well) and HDAC4 concentrations of 0.05, 0.1, 0.2, or 0.6 μg/well. Control transfections were performed with EV. Total DNA used for transfection was kept constant by adding empty vector. Cells were also co-transfected with VEGF-luciferase, pRL-SV40. After 48 h, cells were analyzed for dual luciferase activity by normalizing to Renilla luciferase. Data are mean ± S.D., n = 3. *, p < 0.01, versus EV (column 1); #, p < 0.05, versus Runx2 transfection alone (column 2). G and H, effect of co-transfection of Runx2 siRNA and HDAC4 construct on VEGF mRNA and protein. Runx2 was knocked down in CS-1 cells with Runx2 siRNA (R2-si) or nontargeting control siRNA (Con-si) at 5 nm/ml for 72 h. After 24 h, some cells were transfected with HDAC4 (H4) 1.0 μg/ml or empty vector. VEGF mRNA was measured with qRT-PCR. *, p < 0.001 compared with empty vector- control siRNA. Secreted VEGF into CM was determined with ELISA; *, p < 0.05; #, p < 0.01.
When Runx2 expression was knocked down using siRNA (Fig. 3E), similar effects were seen on VEGF expression as with HDAC4 transfection. VEGF mRNA decreased to 33% of control (p < 0.01) (Fig. 3D), and the protein level also decreased (Fig. 3E).
When CS-1 was transfected with a Runx2 expression construct, VEGF promoter activity increased 2.9-fold compared with EV (p < 0.01) (Fig. 3F). The effect of Runx2 on the VEGF promoter activity could be counteracted by HDAC4 transfection in a dose-dependent manner (Fig. 3F) (p < 0.05 for 0.1–0.6 μg).
To assess if HDAC4 has a direct effect on VEGF expression, we performed separate and combination transfections with HDAC4 and Runx2 siRNA. HDAC4 transfection decreased VEGF mRNA to 35% of control (p < 0.001) and secreted VEGF into conditioned media from 311 to 202 pg/ml (p < 0.05). Runx2 knockdown had a similar effect as follows: VEGF mRNA decreased to 29% of control (p < 0.001), and VEGF protein decreased to 107 pg/ml (p < 0.01). HDAC4 transfection after Runx2 knockdown had no additional effect on VEGF mRNA (27% of control, p > 0.05 compared with Runx2 siRNA) or secreted protein (102 pg/ml, p > 0.05) (Fig. 3, G and H). The results are consistent with Runx2 being the mediator of HDAC4 regulation of VEGF.
HDAC4 Deacetylates Runx2 and Reverses the Runx2-mediated Activation of VEGF Promoter
To show more definitively that the effect of HDAC4 on the VEGF promoter activity is a result of the HDAC4 effect on Runx2, we did co-immunoprecipitation and luciferase assays with different combinations of HDAC4, Runx2, and CBP transfections as well as ChIP analysis of Runx2 and HDAC4 binding to the VEGF promoter. CBP acetylates Runx2 and increases its transcriptional activity and half-life (31, 32). Fig. 4A shows that after Runx2 pulldown, HDAC4 is present, and therefore endogenous HDAC4 and Runx2 proteins interacted in vivo (columns 1 and 2). HDAC4 transfection reduced total and acetylated Runx2, completely reversing the effect of CBP on Runx2 protein levels.
We next evaluated these transfection combinations on VEGF promoter activity. Runx2 increased the activity of the VEGF promoter, which was further increased by co-transfection with CBP. Co-transfection of HDAC4 with Runx2/CBP reversed the effect of CBP on VEGF promoter activity (Fig. 4B).
To show that there is no direct effect of HDAC4 on the VEGF promoter, the two Runx2-binding sites flanking the transcription initiation site were mutated. In the mutated promoter, there was no effect of Runx2 or HDAC4 transfection on promoter activity (Fig. 4C).
To evaluate endogenous Runx2 binding to the VEGF promoter, ChIP was performed. In ChIP analysis, Runx2 binds to a predicted Runx2-binding site (Pf1-pr1). HDAC4 pulldown showed the weak presence of the Runx2-binding site, potentially from protein-protein interaction with Runx2 (Fig. 4D).
HDAC4 Expression Decreases Angiogenic Activity of Chondrosarcoma Cells
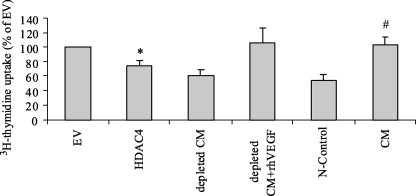
To confirm that alteration in HDAC4 expression has a phenotypic effect on angiogenic activity, we utilized the HUVEC proliferation assay. Restoration of HDAC4 expression in chondrosarcoma cells should decrease angiogenic activity through its effect on Runx2 and VEGF. Therefore, conditioned media from experiments in which HDAC4 was overexpressed (Fig. 2) and in which Runx2 was knocked down (Fig. 3) were compared and tested for VEGF content with ELISA and for their stimulatory effect on HUVECs. HDAC4 transfection decreased thymidine incorporation in HUVECs by 25% compared with EV, p < 0.05 (Fig. 5). VEGF depletion by immunoprecipitation resulted in no detectable VEGF by ELISA and decreased incorporation by 39%, which was not statistically different from HDAC4 transfection or unconditioned media, p > 0.05. Conditioned media and EV had similar biologic activity as 10 ng/ml rhVEGF165. Similar results were obtained with Runx2 knockdown (data not shown). The results suggest that the majority of the stimulatory effect in CM is from VEGF and that HDAC4 transfection prevents the majority of thymidine incorporation by CM. To show this more definitively, VEGF content in CM was measured by ELISA. HDAC4 transfection reduced VEGF concentration from 382 to 226 pg/ml. The results were similar as direct Runx2 knockdown with siRNA, which decreased VEGF from 426 to 223 pg/ml (Table 1). Furthermore, HDAC4 transfection showed a dose-response effect on VEGF content in CM and on HUVEC proliferation (Table 2). Similar inhibition rates on HUVECs were obtained from the condition medium collected from CS-1 transfected by HDAC4 or Runx2 siRNA (data not shown).
FIGURE 5.
HDAC4 overexpression decreases HUVE cell proliferation. Conditioned media from untransfected JJ cells and EV or HDAC4 transfection were tested in the HUVEC assay. 50–70% confluent JJ cells were transfected with HDAC4 expression construct or EV. HUVECs were serum-deprived for 24 h and then conditioned media from JJ cells were added for 48 h. Tritiated thymidine (1 μC/ml) was added for the last 18 h. Cells were washed and lysed, and incorporated tritium was measured. Data are mean ± S.D., n = 3. The groups were statistically different, one-way analysis of variance, p < 0.001; *, p < 0.05 compared with EV. #, p < 0.001 compared with N-control. CM was VEGF depleted by immunoprecipitation with VEGF goat polyclonal antibody and then rescued with addition of 10 ng/ml rhVEGF165. N-control is culture media for chondrosarcoma cells.
TABLE 1.
HDAC4 expression or Runx2 silencing decreases VEGF in conditioned media
50–70% confluent JJ cells were transfected with HDAC4 expression construct or EV or Runx2 siRNA or nontargeting control siRNA. After 48–72 h, cell lysates were analyzed with qPCR and Western blotting (Fig. 2C and Fig. 3E), and CM was used for ELISA. Secreted VEGF into conditioned media was determined with ELISA. Data are mean ± S.D., n = 3.
| Treatment (CM) | EV | HDAC4 | Control-siRNA | Runx2-siRNA |
|---|---|---|---|---|
| VEGF (pg/ml/105 cells) | 382.6 ± 38.1 | 226.2 ± 21.5a | 426.0 ± 33.4 | 222.9 ± 27.9b |
a p < 0.01 versus EV.
b p < 0.001 versus control-siRNA.
TABLE 2.
HDAC4 dose-response effect on VEGF content in CM and HUVEC proliferation
JJ cells were transfected with increasing amounts of HDAC4 expression construct or empty vector (0 μg/well HDAC4). CM was collected after 48 h. Soluble VEGF was measured by ELISA (mean ± S.D., n = 3). HUVECs were serum-deprived for 24 h, and then conditioned media were added for 48 h. Tritiated thymidine (1 μC/ml) was added for the last 18 h. Cells were washed and lysed, and incorporated tritium was measured (mean ± S.D., n = 3).
| Treatment (CM) | HDAC4 (μg/well) |
|||
|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | |
| VEGF (pg/ml/105 cells) | 453.1 ± 9.4 | 397.3 ± 19.5 | 361.4 ± 40.7a | 242.2 ± 26.9b |
| [3H]Thymidine uptake(% of control) | 100 | 85.8 ± 3.4 | 80.1 ± 2.8a | 74.4 ± 7.4b |
a p < 0.01.
b p < 0.001 versus 0 μg of HDAC4 transfection.
To confirm the angiogenic activity of CS cells, JJ-HUVEC three-dimensional co-culture was performed, and tube formation by HUVEC was evaluated. Runx2 silencing decreased tube length by 25% (p < 0.01), tube area by 36% (p < 0.002), and tube junctions by 40% (p < 0.001), similar to the effect of VEGF blocking antibody. Taken together, the results indicate that HDAC4 inhibits Runx2 activity by deacetylation of Runx2 and by decreasing Runx2 transcription, which in turn results in decreased VEGF, thereby linking the effect of decreased HDAC4 to angiogenesis in chondrosarcoma (Fig. 6).
FIGURE 6.
Runx2 silencing in chondrosarcoma cells reduces HUVEC tubulogenesis in co-culture. JJ cells were cultured in Matrigel and transfected with control siRNA or Runx2 siRNA for 3 days, and then HUVECs were added to the culture with EGM2 medium without VEGF and basic fibroblast growth factor for 1 day. HUVECs were photographed, and tube formation was quantified with image analysis software as described under “Experimental Procedures.” Tube length, area, and junctions were quantified in eight 10× fields per condition, graphed as the mean ± S.D. (n = 8). *, p < 0.01 compared with control siRNA. Scale bar, 100 μm. Representative data are shown in the upper right panel, and results of quantification from one of three experiments are summarized in the lower left panels and upper and lower right panels. A, unconditioned medium; B, JJ cells without transfection; C, control siRNA; D, Runx2 siRNA; E, JJ cells transfected with control siRNA and then 10 ng/ml VEGFa antibody added to medium.
DISCUSSION
Little is known about the mechanisms of angiogenesis in chondrosarcoma. Previous studies have shown that grades II and III chondrosarcomas have higher amounts of microvessel density (7), VEGF expression (8), hypoxia-inducing factor 1α (HIF-1α) expression (8), and Runx2 expression (16, 17). In cell culture we have shown that hypoxia and the associated increased expression of HIF-1α increases VEGF expression (19), but we also know that even under normoxic conditions, chondrosarcoma cells express and secrete higher levels of VEGF than chondrocytes in the absence of HIF-1α expression. VEGF is a known mediator of angiogenesis in many tumors. The report on expression of JNK2, p-JNK2, c-Jun, pc-Jun, c-Fos, p-ERK, Runx2, and VEGF in chondrosarcoma utilizing immunohistochemistry (16) showed that Runx2 and VEGF were expressed at higher levels in high grade chondrosarcoma. The authors postulated that the JNK/ERK-AP-1/-Runx2 signal transduction “network” is associated with induction of VEGF and chondrosarcoma development; however, no functional interactions between these factors were analyzed. Our results are consistent with this study in that the chondrosarcoma cell lines have the same expression profile of Runx2 (16, 17) and VEGF (8) as the primary tumors. Our results also show a functional relationship between these differences in gene expression and that at least some of the increased Runx2 and VEGF expression is related to reduced HDAC4 expression in the tumor cells.
The mechanism of HDAC4 repression of VEGF promoter activity is mediated, at least in part, through inhibiting Runx2 promoter activity and the interaction of HDAC4 with Runx2 protein to deacetylate Runx2, a transcription factor regulating the VEGF promoter. We have shown that HDAC4 decreases Runx2 expression and activity and that decreased Runx2 expression leads to decreased VEGF expression. We also showed that endogenous Runx2 binds to the VEGF promoter. HDAC4 and Runx2 are known to be necessary for growth plate maturation, a process that culminates in endochondral ossification and angiogenesis. In one study, HDAC4 was shown to bind to Runx2 and inhibit Runx2 transcriptional activity. HDAC4 homozygous knock-out mice had premature mineralization of endochondral bones that was similar to Runx2 gain of function mice (11). In another model, transforming growth factor-β-mediated repression of osteoblast differentiation requires HDAC4 and -5 acting as co-repressors of Runx2 transcription (33). In a study of Runx2 regulation of osteocalcin, it was found that HDAC3 binds to the amino terminus of Runx2 and decreases its ability to up-regulate osteocalcin expression (34). In a model of osteoblast maturation, BMP-2 signaling causes a net increase in Runx2 acetylation which thereby increases Runx2 transcriptional activity (35). In pro-osteoblast scenarios, which would include endochondral ossification in the growth plate, Runx2 expression and activity are increased, whereas in situations where osteoblast maturation is inhibited, Runx2 activity is inhibited by a variety of mechanisms that include HDACs. In our study of chondrosarcoma cells, endogenous HDAC4 expression is reduced, leading to increased Runx2 expression and transcriptional activity, whereas enhanced HDAC4 expression could decrease the VEGF expression mRNA level and protein production and decrease VEGF content and angiogenic activity of conditioned media from chondrosarcoma cells. The functional interactions between HDAC4, Runx2, and VEGF in chondrosarcoma are analogous to those found in the growth plate.
The processes of migration, invasion, metastasis, angiogenesis, and proliferation are phenotypes shared by cells in a growing organism and tumor cells. The recapitulation of developmental pathways can give a selective growth advantage to tumor cells by facilitating these processes. This concept has resulted in a renewed appreciation of developmental pathways as potential therapeutic targets. Some of the factors involved in developmental pathways that are re-expressed in tumors include Indian hedgehog, bone morphogenic protein, Wnt/β-catenin, and notch (36, 37). Our results show that a pathway expressed in the growth plate, which is the normal embryologic equivalent of chondrosarcoma, is re-expressed in the malignant chondrocytes.
Angiogenesis is the result of the net effect of pro- and anti-angiogenic factors expressed or secreted by tumor and stromal cells. Runx2 is known to regulate multiple processes related to angiogenesis. Knockdown of Runx2 in human bone marrow endothelial cells results in inhibition of endothelial cell phenotype with decreased migration, invasion, tube formation, and differentiation (38). In Runx2 heterozygous knock-out mice, there is loss of vascularization and VEGF expression in hypertrophic growth plate chondrocytes resulting in loss of endochondral ossification, whereas in fibroblasts, Runx2 transfection increases VEGF promoter activity and gene and protein expression (15). In chondrosarcoma cells, we also found that Runx2 mediates expression of VEGF leading to increased angiogenic activity. In the HUVEC assays, we showed that overexpression of HDAC4 or silencing Runx2 decreased the stimulatory effect of conditioned media from chondrosarcoma cells on HUVEC proliferation and tube formation by decreasing expression of VEGF. Some of the angiogenic activity could be from other angiogenic factors. Data presented in this study support the notion that enhancing HDAC4 expression or blocking Runx2 expression or function could be useful strategies to decrease angiogenesis in chondrosarcoma.
In summary, our results show that the expression of HDAC4 is reduced in chondrosarcoma cells when compared with normal chondrocytes, whereas Runx2 and VEGF were overexpressed in the tumor cells. Decreased HDAC4 expression is shown to result in increased VEGF expression through its effect on Runx2. HDAC4 decreases total and acetylated Runx2 by its deacylating and transcriptional repressor activities. Reduced HDAC4 results in higher levels of Runx2, and Runx2 increases transcription of VEGF, ultimately culminating in increased VEGF protein levels and angiogenic activity of the chondrosarcoma cells. Of course there are many factors that regulate angiogenesis besides VEGF and many other pathways that may regulate expression of VEGF. Our long term goal is to provide the molecular underpinnings of angiogenesis in chondrosarcoma so as to identify the relevant molecular targets. Our results show that in addition to the hypoxia-mediated expression of VEGF that is present in most normal tissues and tumors, which is probably dominant, abnormalities in expression of genes not related to hypoxia also drive the expression of VEGF. Targeting only the hypoxia-mediated pathways may not be efficacious in light of the dysregulated expression of HDAC4/Runx2 that we have described.
Acknowledgments
We thank Jack Wands, M.D., for a critical review of the manuscript and Jason Machon, Ph.D., for statistical support.
This work was supported, in whole or in part, by National Institutes of Health Grant AG014399 and Grant 1P20RR024484-01 from the NCRR. This work was also supported by the RIH Orthopaedic Foundation.
- VEGF
- vascular endothelial growth factor
- Runx2
- runt-related transcription factor 2
- HDAC
- histone deacetylase
- qPCR
- real time RT-PCR
- CM
- conditioned media
- HUVEC
- human umbilical vein endothelial cell
- HIF-1α
- hypoxia inducing factor 1-alpha
- EV
- empty vector
- H4
- HDAC4
- ChIP
- chromatin immunoprecipitation
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- ELISA
- enzyme-linked immunosorbent assay
- CS
- chondrosarcoma
- CBP
- cAMP-response element-binding protein (CREB)-binding protein.
REFERENCES
- 1.Huvos A. G. (1991) Bone Tumors Diagnosis, Treatment and Prognosis, 2nd Ed., pp. 343–365, W. B. Saunders Co., Philadelphia [Google Scholar]
- 2.Landis S. H., Murray T., Bolden S., Wingo P. A. (1999) CA Cancer J. Clin. 49, 8–31, 1 [DOI] [PubMed] [Google Scholar]
- 3.Terek R. M., Schwartz G. K., Devaney K., Glantz L., Mak S., Healey J. H., Albino A. P. (1998) J. Orthop. Res. 16, 585–590 [DOI] [PubMed] [Google Scholar]
- 4.Moussavi-Harami F., Mollano A., Martin J. A., Ayoob A., Domann F. E., Gitelis S., Buckwalter J. A. (2006) Biochem. Biophys. Res. Commun. 346, 379–385 [DOI] [PubMed] [Google Scholar]
- 5.Lee F. Y., Mankin H. J., Fondren G., Gebhardt M. C., Springfield D. S., Rosenberg A. E., Jennings L. C. (1999) J. Bone Joint Surg. Am. 81, 326–338 [DOI] [PubMed] [Google Scholar]
- 6.Capanna R., Bertoni F., Bettelli G., Picci P., Bacchini P., Present D., Giunti A., Campanacci M. (1988) J. Bone Joint Surg. 70, 60–69 [PubMed] [Google Scholar]
- 7.McGough R. L., Aswad B. I., Terek R. M. (2002) Clin. Orthop. 397, 76–82 [DOI] [PubMed] [Google Scholar]
- 8.McGough R. L., Lin C., Meitner P., Aswad B. I., Terek R. M. (2002) Clin. Orthop. 397, 62–69 [DOI] [PubMed] [Google Scholar]
- 9.Rountree M. R., Bachman K. E., Herman J. G., Baylin S. B. (2001) Oncogene 20, 3156–3165 [DOI] [PubMed] [Google Scholar]
- 10.Tsukiyama T., Wu C. (1997) Curr. Opin. Genet. Dev. 7, 182–191 [DOI] [PubMed] [Google Scholar]
- 11.Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 12.Zheng Q., Sebald E., Zhou G., Chen Y., Wilcox W., Lee B., Krakow D. (2005) Am. J. Hum. Genet. 77, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verdin E., Dequiedt F., Kasler H. G. (2003) Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 14.Paroni G., Mizzau M., Henderson C., Del Sal G., Schneider C., Brancolini C. (2004) Mol. Biol. Cell 15, 2804–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelzer E., Glotzer D. J., Hartmann C., Thomas D., Fukai N., Soker S., Olsen B. R. (2001) Mech. Dev. 106, 97–106 [DOI] [PubMed] [Google Scholar]
- 16.Papachristou D. J., Papachristou G. I., Papachristou G. J., Papaefthimiou O. A., Agnantis N. J., Basdra E. K., Papavassiliou A. G. (2005) Histopathology 47, 565–574 [DOI] [PubMed] [Google Scholar]
- 17.Park H. R., Park Y. K. (2007) Pathol. Oncol. Res. 13, 32–37 [DOI] [PubMed] [Google Scholar]
- 18.Osada H., Tatematsu Y., Saito H., Yatabe Y., Mitsudomi T., Takahashi T. (2004) Int. J. Cancer 112, 26–32 [DOI] [PubMed] [Google Scholar]
- 19.Lin C., McGough R., Aswad B., Block J. A., Terek R. (2004) J. Orthop. Res. 22, 1175–1181 [DOI] [PubMed] [Google Scholar]
- 20.Block J. A., Inerot S. E., Gitelis S., Kimura J. H. (1991) J. Bone Joint Surg. 73, 647–658 [PubMed] [Google Scholar]
- 21.Morioka H., Weissbach L., Vogel T., Nielsen G. P., Faircloth G. T., Shao L., Hornicek F. J. (2003) Clin. Cancer Res. 9, 1211–1217 [PubMed] [Google Scholar]
- 22.Shao L., Kasanov J., Hornicek F. J., Morii T., Fondren G., Weissbach L. (2003) Biochem. Pharmacol. 66, 2381–2395 [DOI] [PubMed] [Google Scholar]
- 23.Miska E. A., Langley E., Wolf D., Karlsson C., Pines J., Kouzarides T. (2001) Nucleic Acids Res. 29, 3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao G., Jiang D., Ge C., Zhao Z., Lai Y., Boules H., Phimphilai M., Yang X., Karsenty G., Franceschi R. T. (2005) J. Biol. Chem. 280, 30689–30696 [DOI] [PubMed] [Google Scholar]
- 25.Drissi H., Luc Q., Shakoori R., Chuva De Sousa , Lopes S., Choi J. Y., Terry A., Hu M., Jones S., Neil J. C., Lian J. B., Stein J. L., Van Wijnen A. J., Stein G. S. (2000) J. Cell Physiol. 184, 341–350 [DOI] [PubMed] [Google Scholar]
- 26.Zhang L., Yu D., Hu M., Xiong S., Lang A., Ellis L. M., Pollock R. E. (2000) Cancer Res. 60, 3655–3661 [PubMed] [Google Scholar]
- 27.Javed A., Barnes G. L., Jasanya B. O., Stein J. L., Gerstenfeld L., Lian J. B., Stein G. S. (2001) Mol. Cell. Biol. 21, 2891–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asp J., Brantsing C., Lövstedt K., Benassi M. S., Inerot S., Gamberi G., Picci P., Lindahl A. (2005) Int. J. Oncol. 27, 1577–1582 [PubMed] [Google Scholar]
- 29.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 30.Wang I. E., Shan J., Choi R., Oh S., Kepler C. K., Chen F. H., Lu H. H. (2007) J. Orthop. Res. 25, 1609–1620 [DOI] [PubMed] [Google Scholar]
- 31.Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., Yao T. P. (2002) EMBO J. 21, 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grönroos E., Hellman U., Heldin C. H., Ericsson J. (2002) Mol. Cell 10, 483–493 [DOI] [PubMed] [Google Scholar]
- 33.Kang J. S., Alliston T., Delston R., Derynck R. (2005) EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 35.Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 36.Bailey J. M., Singh P. K., Hollingsworth M. A. (2007) J. Cell. Biochem. 102, 829–839 [DOI] [PubMed] [Google Scholar]
- 37.Velcheti V., Govindan R. (2007) J. Thorac. Oncol. 2, 7–10 [DOI] [PubMed] [Google Scholar]
- 38.Sun L., Vitolo M., Passaniti A. (2001) Cancer Res. 61, 4994–5001 [PubMed] [Google Scholar]