Abstract
Redox-active copper is implicated in the pathogenesis of Alzheimer disease (AD), β-amyloid peptide (Aβ) aggregation, and amyloid formation. Aβ·copper complexes have been identified in AD and catalytically oxidize cholesterol and lipid to generate H2O2 and lipid peroxides. The site and mechanism of this abnormality is not known. Growing evidence suggests that amyloidogenic processing of the β-amyloid precursor protein (APP) occurs in lipid rafts, membrane microdomains enriched in cholesterol. β- and γ-secretases, and Aβ have been identified in lipid rafts in cultured cells, human and rodent brains, but the role of copper in lipid raft amyloidogenic processing is presently unknown. In this study, we found that copper modulates flotillin-2 association with cholesterol-rich lipid raft domains, and consequently Aβ synthesis is attenuated via copper-mediated inhibition of APP endocytosis. We also found that total cellular copper is associated inversely with lipid raft copper levels, so that under intracellular copper deficiency conditions, Aβ·copper complexes are more likely to form. This explains the paradoxical hypermetallation of Aβ with copper under tissue copper deficiency conditions in AD.
Imbalance of metal ions has been recognized as one of the key factors in the pathogenesis of Alzheimer disease (AD).2 Aberrant interactions between copper or zinc with the β-amyloid peptide (Aβ) released into the glutamatergic synaptic cleft vicinity could result in the formation of toxic Aβ oligomers and aggregation into plaques characteristic of AD brains (reviewed in Ref. 1). Copper, iron, and zinc are highly concentrated in extracellular plaques (2, 3), and yet brain tissues from AD (4–6) and human β-amyloid precursor protein (APP) transgenic mice (7–10) are paradoxically copper deficient compared with age-matched controls. Elevation of intracellular copper levels by genetic, dietary, and pharmacological manipulations in both AD transgenic animal and cell culture models is able to attenuate Aβ production (7, 9, 11–15). However, the underlying mechanism is at present unclear.
Abnormal cholesterol metabolism is also a contributing factor in the pathogenesis of AD. Hypercholesterolemia increases the risk of developing AD-like pathology in a transgenic mouse model (16). Epidemiological and animal model studies show that a hypercholesterolemic diet is associated with Aβ accumulation and accelerated cognitive decline, both of which are further aggravated by high dietary copper (17, 18). In contrast, biochemical depletion of cholesterol using statins, inhibitors of 3-hydroxy-3-methyglutaryl coenzyme A reductase, and methyl-β-cyclodextrin, a cholesterol sequestering agent, inhibit Aβ production in animal and cell culture models (19–25).
Cholesterol is enriched in lipid rafts, membrane microdomains implicated in Aβ generation from APP cleavage by β- and γ-secretases. Recruitment of BACE1 (β-secretase) into lipid rafts increases the production of sAPPβ and Aβ (23, 26). The β-secretase-cleaved APP C-terminal fragment (β-CTF), and γ-secretase, a multiprotein complex composed of presenilin (PS1 or PS2), nicastrin (Nct), PEN-2 and APH-1, colocalize to lipid rafts (27). The accumulation of Aβ in lipid rafts isolated from AD and APP transgenic mice brains (28) provided further evidence that cholesterol plays a role in APP processing and Aβ generation.
Currently, copper and cholesterol have been reported to modulate APP processing independently. However, evidence indicates that, despite tissue copper deficiency, Aβ·Cu2+ complexes form in AD that catalytically oxidize cholesterol and lipid to generate H2O2 and lipid peroxides (e.g. hydroxynonenal and malondialdehyde), which contribute to oxidative damage observed in AD (29–35). The underlying mechanism leading to the formation of pathological Aβ·Cu2+ complexes is unknown. In this study, we show that copper alters the structure of lipid rafts, and attenuates Aβ synthesis in lipid rafts by inhibition of APP endocytosis. We also identify a paradoxical inverse relationship between total cellular copper levels and copper distribution to lipid rafts, which appear to possess a privileged pool of copper where Aβ is more likely to interact with Cu2+ under copper-deficiency conditions to form Aβ·Cu2+ complexes. These data provide a novel mechanism by which cellular copper deficiency in AD could foster an environment for potentially adverse interactions between Aβ, copper, and cholesterol in lipid rafts.
EXPERIMENTAL PROCEDURES
Cell Culture
SH-SY5Y human neuroblastoma cells were cultured in RPMI media 1640 with GLUTAMAXTM-I (Invitrogen) supplemented with 20% fetal calf serum. The SH-SY5Y (swAPP695) cell line was generated by transfecting SH-SY5Y cells with an APP cDNA containing the Swedish mutation (swAPP695) cloned into the pIRESpuro2 vector (36). Cells were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Stably transfected cells were selected and maintained using 2 μg/ml puromycin (Sigma).
Animals
All the experimental procedures involving animals were performed in accordance with guidelines established by the animal ethics committee at the University of Melbourne, according to the National Health and Medical Research Council of Australia. Four human Swedish mutant APP expressing Tg2576 transgenic (37) and three non-transgenic littermate brain hemispheres were harvested from 15-month-old female mice. To minimize metal contamination of samples, all tubes were pre-soaked in 1% HNO3 and rinsed in MilliQ water prior to use. Mice were sacrificed by anesthetization with 250 μl of 65 mg/ml sodium pentobarbitone (Lethabarb; Virbac) followed by transcardial perfusion with phosphate-buffered saline (pH 7.4). Brains were dissected and snap frozen on dry ice and stored at −80 °C until use.
Antibodies
The following antibodies were used in this study: anti-flotillin-2 (BD Transduction Laboratories), anti-PS1 N-terminal 20 amino acids (98/1) (38), anti-nicastrin (Sigma), anti-Aβ1–16 (W0-2) (39), anti-FLAG M2 (Sigma), and anti-Na,K-ATPase (mouse monoclonal antibody (464.6), Abcam).
Treatment of Cells with Metals
Confluent cells were preincubated with 50 μg/ml cycloheximide in serum-free Hanks' balanced salt solution (HBSS; Sigma) for 20 min. Cells were then incubated in the presence of 50 μg/ml cycloheximide in HBSS with copper chelators (50 μm bathocuproinedisulfonic acid (BCS) and 50 μm d-penicillamine), CuCl2 (2 μm or 5 μm), FeCl3 (2 μm), or ZnCl2 (2 μm) for 3 h.
Western Blot Analysis
Protein samples were separated by SDS-PAGE using 4–12% BisTris gradient gels (Invitrogen), using either MOPS or MES running buffer (Invitrogen). After separation, proteins were transferred to a 0.2-μm nitrocellulose membrane (Bio-Rad). Blots for detection of Aβ were boiled for 5 min in phosphate-buffered saline prior to blocking. Blots were blocked with Tris-buffered saline containing 0.05% Tween® 20 (Sigma) and 5% skim milk, and probed with appropriate primary antibodies diluted in the blocking buffer. For detection of protein by chemiluminescence, appropriate secondary IgG-horseradish peroxidase-conjugated antibodies were used. Blots were developed using an ECLTM Western blotting detection system (Amersham Biosciences), and visualized using the LAS-3000 Imaging System (Fujifilm). Densitometry quantification of immunoreactive signals was performed using MultiGauge (version 3; Fujifilm).
Cell Surface Biotinylation Assay
Cell surface proteins were isolated from confluent SH-SY5Y cells cultured in 75-cm2 flasks post-copper treatments using the Pierce® Cell Surface Protein Isolation Kit according to the manufacturer's instructions (Pierce) with some modifications. Briefly, for post-copper treatments, each flask of cells were labeled with 10 ml of Sulfo-NHS-SS-Biotin/phosphate-buffered saline for 30 min. at 4 °C. Unreacted biotin was quenched with 500 μl of Quenching Solution supplied in the kit. Each flask of cells was harvested by scraping, and processed individually. After cell lysis, total protein concentration in the cell lysate was determined by the BCA protein assay (Pierce), and total protein of 150 μg from each flask was incubated with 125 μl of NeutrAvidin-agarose to precipitate biotinylated surface proteins. Unbound proteins were removed by washes in Wash Buffer supplied in the kit supplemented with CompleteTM EDTA-free protease inhibitor mixture (Roche). Biotinylated surface proteins were eluted from NeutrAvidin with 30 μl of 1× NuPAGE LDS sample buffer (Invitrogen) containing 50 mm dithiothreitol, and analyzed by Western blotting.
Isolation of Lipid Rafts
Lipid rafts were isolated from confluent SH-SY5Y cells cultured in three 175-cm2 flasks and prepared as described previously (27) with some modifications. Briefly, after metal treatments as described above, cells were resuspended in 0.5 ml of lysis buffer containing 25 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, and 0.5% Brij-58 (Sigma) supplemented with CompleteTM EDTA-free protease inhibitor mixture (Roche). Cells were homogenized by passages through a 25-gauge needle. Total protein concentration in homogenates was determined by the BCA protein assay (Pierce), and homogenates containing 4 mg of total protein were adjusted to a 45% final concentration of sucrose (final volume, 1 ml). This was layered sequentially with 35% sucrose (2 ml) and 5% sucrose (2 ml) to form a discontinuous sucrose gradient. The samples were subjected to ultracentrifugation at 46,600 × g at 4 °C for 19 h in a Beckman SW50.1 rotor. Ten 0.5-ml fractions were collected from the top of the gradient. For the detection of Aβ, lipid rafts were isolated from SH-SY5Y (swAPP695) cells. For γ-secretase activity assay, sucrose gradient fractions, diluted with lysis buffer, were concentrated by ultracentrifugation at 55,000 × g at 4 °C for 70 min in a Beckman TLA55 rotor.
Lipid rafts were isolated from mice brains as described previously (28) with modifications. Briefly, brain homogenates containing 10 mg of total protein were adjusted to a 45% final concentration sucrose (final volume, 4 ml). The discontinuous sucrose gradient was formed by sequential layering of 35% sucrose (4 ml) and 5% sucrose (4 ml). The samples were subjected to ultracentrifugation at 39,000 × g at 4 °C for 19 h in a Beckman SW40 rotor. Twelve 1-ml fractions were collected from the top of the gradient. Equal volumes (10 μl) of sucrose gradient fraction samples were analyzed for protein by Western blotting.
Metal Analysis
SH-SY5Y cells and mice brain homogenates were pre-digested overnight in 50 μl of concentrated HNO3 (Aristar Grade, BDH), followed by heating to 90 °C for 20 min. The digested homogenate and sucrose gradient fraction samples were diluted with 1% HNO3 and metal levels (copper, iron, and zinc) were measured by inductively coupled plasma mass spectrometry (Ultramass 700, Varian, Australia). The instrument was calibrated using certified inductively coupled plasma mass spectrometry standard solutions (AccuStandard) containing 0, 10, 50, and 100 ppb of all metals measured in 1% HNO3.
Cholesterol Analysis
Cholesterol was measured using the fluorometric Amplex Red cholesterol assay (Molecular Probes), according to the manufacturer's instructions. Fluorescence was detected with excitation wavelength at 560 ± 5 nm and emission at 590 ± 5 nm. Cholesterol levels in lipid raft fractions were expressed as a percentage of the total cellular or brain cholesterol.
In Vitro γ-Secretase Activity Assay
In vitro γ-secretase activity in sucrose gradient fractions were determined as the generation of ϵ-CTF from a recombinant APP substrate, as described previously (40). Membranes from the sucrose gradient fractions sedimented by ultracentrifugation were pooled, fractions 3–5 were designated lipid rafts and fractions 8–10, non-lipid rafts. Pooled lipid raft and non-lipid raft membrane pellets were resuspended in 18 μl of buffer containing 20 mm Hepes (pH 7.0), 5 mm MgCl2, 5 mm CaCl2, 0.15 m KCl, and 0.5% CHAPSO. To start the reaction, 1 mm ATP and phospholipids (phosphatidylcholine and phosphatidylethanolamine; 2.5 μg/ml each) were added to the reaction mixture, followed by 0.3 μg of purified MC99–3FLAG peptide. Assays were performed in parallel in the presence of the γ-secretase inhibitor, L-685,458 (1 μm; Merck). Reactions were incubated at 37 °C for 16 h, and stopped by the addition of SDS sample buffer. Following Western blotting, ϵ-CTF was detected using the anti-FLAG M2 antibody. Gene Tools (version 3.07; SynGene) was used to quantify the density of ϵ-CTF immunoreactivity. Data were normalized to protein concentrations in the fraction pools. To account for nonspecific release of ϵ-CTF, γ-secretase activity for each sample was determined by subtracting ϵ-CTF immunoreactivity in the presence of L-685,458 from that without immunoreactivity.
Statistical Analysis
Quantified data represent the mean ± S.E. of at least three independent experiments. Statistical analyses were performed using GraphPad PrismTM Software for Windows. For SH-SY5Y cell data, statistical significance was determined by one-way analysis of variance with Bonferroni or linear trend post hoc tests; and for mice brain tissue data, statistical significance was determined by t test. Significance was defined as *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
RESULTS
Copper Induces Microdomain Redistribution of Flotillin-2, a Lipid Raft Marker
Amyloidogenic processing of APP occurs in lipid rafts (23, 26–28, 41). To investigate the effect of copper on lipid raft APP processing, we isolated lipid rafts from SH-SY5Y human neuroblastoma cells exposed to differential copper conditions. It has been reported that the transcription and translation of APP are under metal regulation (42–44), and that copper regulates genes in the cholesterol biosynthetic pathway (45, 46). To eliminate confounding metal-mediated changes in APP expression and in cholesterol biosynthetic genes expression, all cell culture experiments were performed in the presence of cycloheximide, an inhibitor of protein synthesis. We manipulated the intracellular copper levels by exposing SH-SY5Y cells for 3 h to serum-free HBSS medium containing: 1) copper chelators (50 μm BCS and 50 μm d-penicillamine) to induce copper deficiency; 2) 2 μm CuCl2; or 3) 5 μm CuCl2. The rationale for performing the in vitro SH-SY5Y cell culture experiments in the absence of serum was based on the observation that copper released from hippocampal neurons exists freely in solution, not bound to any protein (47). Also, the use of serum-free HBSS removes additional factors present in serum that could affect our data interpretation. Physiological concentrations of copper were selected based on the serum concentration of “free” non-ceruloplasmin bound copper in AD being 2.6 ± 4.6 μm (48), and the estimation of synaptic cleft copper concentration being 14 ± 4 μm (49). In addition, exposure of SH-SY5Y cells to medium containing 2 μm FeCl3 or ZnCl2 were included as metal specificity controls.
Metal analysis by inductively coupled plasma mass spectrometry showed that copper chelation selectively reduced intracellular copper levels and did not affect iron and zinc levels (supplemental Fig. S1, a–c). However, cellular copper deficiency induced by chelators was statistically insignificant compared with untreated control, and this was possibly due to partial metal deficiency as the result of serum-free HBSS usage. Intracellular copper levels were responsive to elevations in extracellular copper concentrations, and exogenous iron and zinc (2 μm) did not have an effect on intracellular copper levels (supplemental Fig. S1a). Similarly, intracellular iron and zinc levels were responsive only to changes in extracellular iron and zinc conditions, respectively (supplemental Fig. S1, b and c).
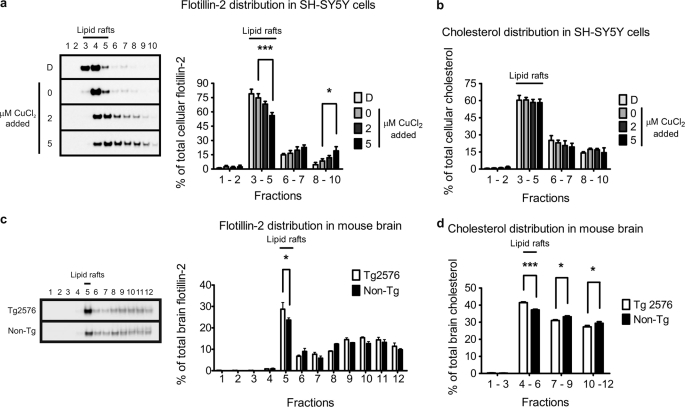
Lipid rafts are recognized as detergent-insoluble membrane structures primarily composed of cholesterol and sphingolipids (50). We isolated lipid rafts, according to established protocols, as low-density buoyant detergent-insoluble membranes on a discontinuous sucrose gradient. Flotillin-2, a commonly used lipid raft protein marker (51), was localized predominantly to the low-density fraction 4 of the untreated control cells with moderate immunoreactivity in fractions 3 and 5 (Fig. 1a). Smaller amounts of flotillin-2 immunoreactivity were also detected in heavier fractions 6–10. Fractions 3–5 were confirmed to contain lipid rafts by cholesterol analysis (Fig. 1b), which indicated that they were most enriched in cholesterol. On this basis, fractions 3–5 were designated as the lipid raft fractions.
FIGURE 1.
Copper induces flotillin-2 redistribution from lipid rafts. Lipid rafts were isolated from SH-SY5Y cells exposed for 3 h to differential copper conditions, or from brain tissues of copper-deficient 15-month-old female Tg2576 (n = 4) and age-matched non-Tg control (n = 3) mice on a discontinuous sucrose gradient by ultracentrifugation as described under “Experimental Procedures.” Lipid raft fractions were identified by Western blot analysis as flotillin-2 positive (a) fractions 3–5 in SH-SY5Y untreated control, and (c) fraction 5 in non-Tg mice brains. Cholesterol analysis further confirmed these flotillin-2-positive lipid raft fractions by their concentrated cholesterol content (b and d). D, copper-deficient (50 μm BCS and 50 μm d-penicillamine). Densitometry analysis of flotillin-2 levels in the indicated fractions were expressed as a percentage of total cellular or brain tissue flotillin-2. Cholesterol data were expressed as a percentage of total cellular or brain tissue cholesterol. Data represent the results of at least three independent experiments (mean ± S.E.). *, p ≤ 0.05; ***, p ≤ 0.001 compared with untreated SH-SY5Y cells or non-Tg control.
Extracellular copper manipulations (across a low physiological concentration range) markedly altered flotillin-2 distribution. Copper depletion increased flotillin-2 recruitment to the lipid raft fractions (Fig. 1a). Conversely, copper elevation induced redistribution of flotillin-2 into higher density sucrose fractions, generally considered to be non-lipid raft fractions (Fig. 1a). This copper-induced diffusion of flotillin-2 localization was analogous to lipid raft disruption and flotillin-1 dispersion as the result of cholesterol depletion by methyl-β-cyclodextrin and statins (52). In our study, this effect was specific to copper, with no significant change in flotillin-2 lipid raft distribution observed in SH-SY5Y cells incubated with iron or zinc (data not shown).
Brain copper levels in AD and APP transgenic (e.g. Tg2576) mice brains are significantly reduced compared with age-matched control brains (reviewed in Ref. 4). To investigate whether decreased brain copper is associated with flotillin-2 lipid raft redistribution in vivo, we isolated lipid rafts from 15-month-old female Tg2576 and non-transgenic (non-Tg) littermate mice brains. Consistent with our previous reports (8, 10), the brains of Tg2576 mice were significantly deficient in copper and zinc compared with the age-matched non-transgenic controls (supplemental Fig. S2, a–c). In these samples, fraction 5 was identified as the lipid raft fraction by maximum flotillin-2 immunoreactivity (Fig. 1c), and confirmed by cholesterol analysis (Fig. 1d). Unlike SH-SY5Y cells (Fig. 1b), there was a small but significant redistribution of cholesterol toward lipid rafts in the Tg2576 brain tissues (Fig. 1d). This could be attributed to changes in copper status in SH-SY5Y cells being more acute than in the living animal. Nevertheless, increased flotillin-2 distribution to lipid rafts in copper-deficient Tg2576 brain (Fig. 1c) was consistent with the effect of copper deficiency in SH-SY5Y cells (Fig. 1a).
Increased Copper Is Associated with Decreased APP Processing in Lipid Rafts
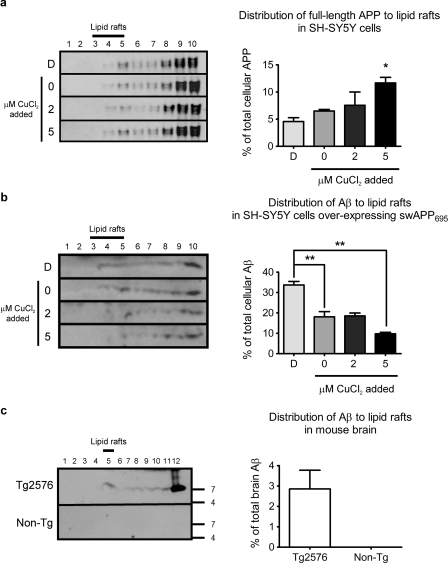
APP processing is modulated by flotillin-2 (53). To assess the impact of copper-stimulated flotillin-2 redistribution on APP processing in lipid rafts, we analyzed APP and Aβ present in lipid rafts under differential copper conditions by Western blotting. Under conditions of elevated copper, which distributes flotillin-2 out of lipid rafts (Fig. 1a), there was a significant increase in full-length APP within lipid rafts (Fig. 2a). However, endogenous Aβ in lipid rafts isolated from SH-SY5Y cells was below the limit of detection, therefore we prepared lipid rafts from SH-SY5Y cells overexpressing the same human APP695 isoform carrying the familial AD Swedish mutation (swAPP695) as that present in the transgenic Tg2576 mice. We found that under elevated copper conditions, concomitant with the increase in full-length APP, there was significantly less Aβ distributed to lipid rafts (Fig. 2b).
FIGURE 2.
Copper exposure reduced Aβ levels within lipid rafts. Western blot analysis of (a) full-length APP in lipid rafts isolated from SH-SY5Y cells and (b) Aβ in lipid rafts isolated from SH-SY5Y cells overexpressing swAPP695, exposed to differential copper conditions for 3 h. c, Western blot analysis of Aβ in lipid rafts isolated from brain tissues of copper-deficient 15-month-old female Tg2576 (n = 4) and age-matched non-Tg control (n = 3) mice. APP and Aβ were detected using W0-2 antibody. Aβ was not detected in brain tissues of non-Tg control mice. D, copper-deficient (50 μm BCS and 50 μm d-penicillamine). Densitometry analyses of APP and Aβ in lipid raft fractions were expressed as a percentage of total cellular APP and Aβ, respectively. Cell data represent the results from at least three independent experiments (mean ± S.E.). *, p ≤ 0.05; **, p ≤ 0.01 compared with untreated SH-SY5Y cells or non-Tg control.
Dimeric Aβ has been reported to be the predominant species present in lipid rafts isolated from Tg2576 mice and AD brain (28). We also found apparently dimeric Aβ to be the primary species present in our lipid raft preparations from brains of Tg2576 mice (Fig. 2c). Aβ was not detectable by the W0-2 antibody on Western blot in non-transgenic mice brain tissue.
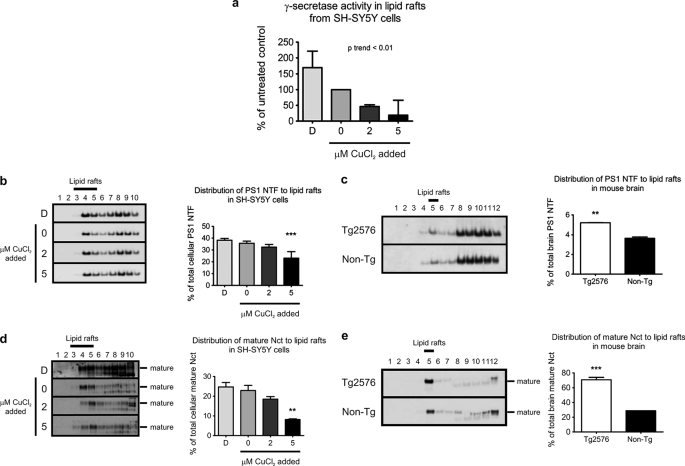
Increased Copper Disperses Lipid Raft γ-Secretase Proteins and Inhibits Activity
To further evaluate the impact of copper on lipid raft APP processing, we analyzed lipid raft distribution and activity of the γ-secretase, the protease complex involved in the cleavage of APP β-CTF to generate Aβ (reviewed in Ref. 54). We measured γ-secretase activity in lipid rafts isolated from SH-SY5Y cells incubated under differential copper conditions in vitro, by the release of ϵ-CTF from purified MC99–3FLAG substrate (40). We found a strong inverse relationship (p < 0.01) between γ-secretase activity in lipid rafts and cellular copper levels (Fig. 3a). Under conditions of copper deficiency, there was ∼70% increase in lipid raft γ-secretase activity. Conversely, exposure of cells to as little as 2 μm exogenous copper resulted in ∼50% decrease in activity compared with the untreated control (Fig. 3a). These results are consistent with the impact of copper modulation on Aβ generation that we observed in SH-SY5Y cells (Fig. 2b).
FIGURE 3.
Copper alters distribution of γ-secretase to lipid rafts. a, γ-secretase activity in lipid rafts isolated from SH-SY5Y cells exposed to differential copper conditions for 3 h was determined by the release of ϵ-CTF from MC99–3FLAG peptide. γ-Secretase activity in lipid rafts was expressed as a percentage of the untreated control. Presenilin 1 N-terminal fragment (PS1 NTF) in lipid rafts isolated from (b) SH-SY5Y cells exposed to differential copper conditions for 3 h, and (c) brain tissues of 15-month-old female Tg2576 (n = 4) and age-matched non-Tg control (n = 3) mice was detected using an anti-PS1 N terminus (98/1) antibody by Western blotting. Nct in lipid rafts isolated from (d) SH-SY5Y cells exposed to differential copper conditions for 3 h, and (e) mouse brain tissue was detected using an anti-nicastrin antibody by Western blotting. The mature Nct was the predominant species present in lipid rafts. D, copper-deficient (50 μm BCS and 50 μm d-penicillamine). Densitometry analyses of PS1 NTF and Nct levels in lipid raft fractions were expressed as a percentage of total cellular or brain tissue PS1 and Nct levels, respectively. SH-SY5Y cell data represent the results from at least three independent experiments (mean ± S.E.). **, p ≤ 0.01; ***, p ≤ 0.001 compared with untreated SH-SY5Y cells or non-Tg control.
To determine the basis for this change, we assayed two core components of the γ-secretase, PS1 and Nct, by Western blotting. PS1 or PS2 forms the catalytic core of γ-secretase, and genetic mutations in presenilins cause familial AD (55, 56). In concordance with reduced γ-secretase activity under elevated copper conditions (Fig. 3a), a decrease in PS1 distribution to lipid rafts was also observed, which reached significance when SH-SY5Y cells were exposed to medium supplemented with 5 μm CuCl2 (Fig. 3b). In the brain tissue of Tg2576 mice we found an increased distribution of PS1 to lipid rafts (Fig. 3c), consistent with the lower copper levels in these brain tissues (supplemental Fig. S2a).
Nct is the substrate receptor that recruits the proteolytically processed C-terminal fragments of type I transmembrane proteins such as the APP β-CTF into the γ-secretase complex (57). Analysis of lipid raft-associated Nct isolated from SH-SY5Y cells and mice brains showed that active mature Nct was the predominant species present in lipid rafts (Fig. 3, d and e). The distribution of mature Nct to lipid rafts paralleled PS1 distribution, with these two proteins increased in lipid rafts under copper deficiency conditions in both SH-SY5Y cells (Fig. 3d) and Tg2576 brain (Fig. 3e). Therefore, the decrease in lipid raft γ-secretase activity induced by copper (Fig. 3a) may be at least partially explained by redistribution of PS1 and Nct out of the lipid raft fractions.
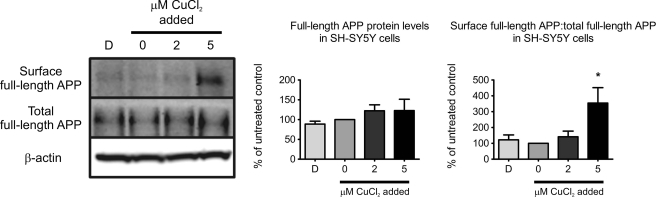
Elevated Copper Promotes Cell Surface Localization of APP
β- Cleavage of APP occurs in endosomes where BACE1 is concentrated (58), therefore, APP needs to be internalized from the cell surface by endocytosis for this event to occur. To evaluate the effect of copper on APP surface localization, we performed surface biotinylation after cells were exposed to differential copper conditions. Consistent with reduced Aβ production in lipid rafts in the presence of elevated cellular copper (Fig. 2b), there was a significant retention of APP at the cell surface (Fig. 4). Copper had no effect on Na,K-ATPase, a plasma membrane resident protein (supplemental Fig. S3). Therefore, copper inhibits APP endocytosis.
FIGURE 4.
Copper promotes retention of APP at cell surface. Surface proteins biotinylated using sulfo-NHS-SS-biotin post-copper treatments were precipitated with NeutrAvidin-agarose, and analyzed by Western blotting. APP was detected using W0-2 antibody. Total APP were normalized to β-actin as a loading control, and expressed as a percentage of the untreated control. The ratio of surface APP to normalized total APP was expressed as a percentage of the untreated control. Data represent the results of two independent experiments (mean ± S.E.). *, p ≤ 0.05 compared with untreated SH-SY5Y cells.
Copper Distribution to Lipid Rafts Is Inversely Associated with Cellular Copper Levels
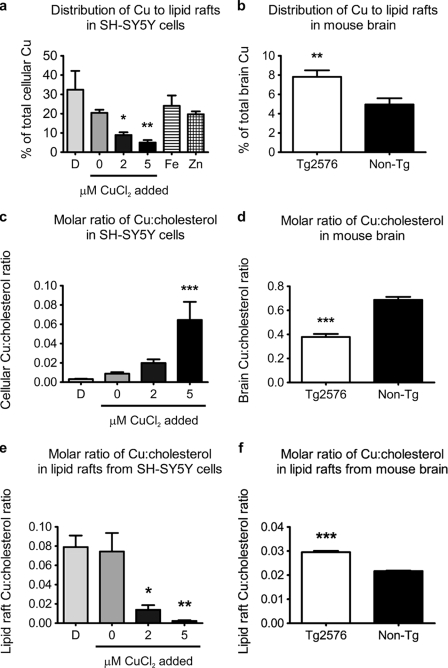
The concentration of metals in lipid rafts has not, to our knowledge, been determined previously. To examine the relationship between total cellular and lipid raft copper levels, we measured the metal contents in sucrose gradient fractions of SH-SY5Y cells treated under differential copper conditions. Surprisingly, copper distribution to lipid rafts under these cellular copper conditions (Fig. 5a) was inversely associated with total cellular copper (supplemental Fig. S1a). In other words, as cellular copper levels fall, lipid raft copper levels are preserved. There was no significant changes in iron or zinc distributions to lipid rafts under these metal treatment conditions (supplemental Fig. S4, a and b).
FIGURE 5.
Copper distribution to lipid rafts associates inversely with total cellular copper levels. Analysis of copper levels in lipid rafts isolated from (a) SH-SY5Y cells exposed to differential metal conditions for 3 h, and from (b) brain tissues of copper-deficient 15-month-old female Tg2576 (n = 4) and age-matched non-Tg control (n = 3) mice. Lipid raft copper levels were expressed as a percentage of total cellular or brain tissue copper (mean ± S.E.). Molar ratio of total cellular copper:cholesterol in (c) SH-SY5Y cells exposed to differential copper conditions, and (d) mouse brain tissues. Copper:cholesterol molar ratio in lipid rafts isolated from (e) SH-SY5Y cells exposed to differential copper conditions, and (f) mouse brain tissues. Data represent the results of at least three independent experiments (mean ± S.E.). D, copper-deficient (50 μm BCS and 50 μm d-penicillamine). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 compared with untreated SH-SY5Y cells or non-Tg control.
The inverse relationship between cellular copper levels and lipid raft copper distribution was consistent with findings from Tg2576 and non-transgenic control mice brains. Compared with samples from non-transgenic control mice, there was significantly less total copper in the brain tissues of Tg2576 mice (supplemental Fig. S2a), whereas copper distribution to lipid raft fractions was significantly greater in these brain tissues (Fig. 5b). There was no change in iron and zinc distributions to lipid rafts isolated from Tg2576 brains (supplemental Fig. S4, c and d).
Since copper elevation reduces the relative distribution of copper to lipid rafts (Fig. 5a), but does not perturb the proportional cholesterol distribution to lipid rafts (Fig. 1a), we calculated the molar ratios of copper:cholesterol in these compartments. As expected, the molar ratio of total cellular copper:cholesterol in SH-SY5Y cells increased with elevation of intracellular copper levels (Fig. 5c). This association was reflected in vivo in the copper-deficient Tg2576 mouse brain, where the molar ratio of copper:cholesterol was, as expected, significantly reduced compared with the non-transgenic control brain tissue (Fig. 5d). However, for lipid rafts, the molar ratio of copper:cholesterol was inverted compared with the total cellular copper:cholesterol molar ratio. The molar ratio of copper:cholesterol in lipid rafts actually increased under conditions of total cellular copper deficiency in SH-SY5Y cells and was ∼6- and 40-fold greater than the lipid raft copper:cholesterol ratio of cells exposed to 2 and 5 μm CuCl2, respectively (Fig. 5e). This paradoxical association of copper with cholesterol in lipid rafts was recapitulated in the increased molar ratio of copper:cholesterol in lipid rafts isolated from copper-deficient Tg2576 brain compared with lipid rafts from control brains (Fig. 5f). This is important because cellular copper deficiency, by paradoxically elevating the stoichiometry of copper:cholesterol (Fig. 5, e and f) and Aβ in lipid rafts (Fig. 2, b and c), must increase the probability of Aβ·Cu2+ complex formation. Aβ·Cu2+ complexes in turn oxidize cholesterol and lipid, forming oxysterols and lipid peroxidation products that characterize brain pathology in AD and APP transgenic mice (29–32, 34, 35, 59).
DISCUSSION
Copper and cholesterol have been reported independently in the literature as modulating factors of APP processing and Aβ generation, of relevance to the pathogenesis of AD. In the present study, using SH-SY5Y cells as an in vitro cell model and Tg2576 mouse as an in vivo AD animal model, we showed that copper modulates flotillin-2 lipid raft association, which consequently affects APP endoctyosis and processing in cholesterol-rich lipid rafts. We also found that cellular copper levels are inversely associated with lipid raft copper levels. Therefore, under the copper deficiency conditions present in AD brain tissues, Aβ, copper, and cholesterol are concentrated in lipid rafts. This increases the probability of pro-oxidant Aβ·Cu2+ complexes catalytically oxidizing cholesterol and long chain fatty acids, as observed in brain tissue of AD and APP transgenic mice (29–35).
In this study, we found that copper induced redistribution of flotillin-2 away from cholesterol-rich lipid rafts (Fig. 1), but to the best of our knowledge, flotillin-2 is not known to be a copper binding or copper-sensitive protein. This redistribution is similar to the effects of pharmacological depletion of cholesterol and lipid raft disruption (52). However, the proportional distribution of cholesterol in the raft fractions did not alter under differential copper conditions (Fig. 1b), indicating that flotillin-2 redistribution is not a consequence of changes in lipid raft density. Therefore, flotillin-2 may not be a perfect marker for lipid rafts because our data show that it can be perturbed by copper to redistribute into non-raft fractions, whereas the density and cholesterol content of the raft fractions are unchanged.
Endocytosis of APP to BACE1-rich endosomes is required to initiate β-cleavage of APP (58), and this event is sensitive to flotillin-2 and cholesterol depletion (53). Flotillin-2 and cholesterol are both enriched in lipid rafts, which have been proposed to partition a subset of total cellular APP undergoing amyloidogenic processing (23). Consistent with this mechanism, we found elevated copper reduced flotillin-2 association with lipid rafts (Fig. 1a), decreased lipid raft processing (Fig. 2a) and endocytosis (Fig. 4) of APP. Therefore, our data suggest that the mechanism underlying copper attenuation of Aβ generation is the inhibition of flotillin-2-mediated APP endocytosis via lipid rafts.
The redox-active property of copper makes it an essential cofactor for many proteins involved in diverse cellular processes, but paradoxically, the redox activity of copper also makes it potentially cytotoxic (60). Under normal physiological conditions, the uptake, distribution, and efflux of copper are tightly regulated. It is therefore intriguing to observe under differential cellular copper conditions that the copper level is maintained in lipid rafts in both SH-SY5Y cells and Tg2576 brain tissues (Fig. 5). Intracellular copper does not usually exist as a free ion (61), therefore its association with lipid rafts could be due to complexes formed by cuproproteins. Lipid rafts are implicated in various cellular processes including signaling, and protein trafficking (50). The preservation of copper in lipid rafts suggests that lipid rafts might serve an important, as yet unidentified function in copper homeostasis, which is worthy of further investigation.
Aβ oligomers are implicated in the pathogenesis of AD, affecting cognitive function, memory, synaptic function, and long-term potentiation (62, 63). Cholesterol-dependent interactions of Aβ with gangliosides in lipid rafts (64, 65) and interactions of Aβ with copper (66–68) both promote Aβ aggregation. We and others have previously reported that Aβ·Cu2+ complexes can catalytically oxidize cholesterol to generate H2O2 (29–32). Our current data indicate that lipid rafts provide a favorable environment for the co-enrichment of copper and Aβ, especially under conditions of copper deficiency. We hypothesize that in aging and AD, where there is intracellular copper deficiency (5, 6, 69), the concomitant enrichment of Aβ and copper within lipid rafts promotes the formation of redox-active Aβ·Cu2+ complexes, fostering the catalytic oxidation of cholesterol, lipid, and the generation of neurotoxic H2O2. This further creates a vulnerable environment for Aβ to cross-link, forming SDS-resistant oligomers characteristic of Aβ extracted from AD brains (67, 68). This also explains the paradoxical enrichment of copper and cholesterol within amyloid plaques, whereas the neighboring tissue is copper-deficient, as well as the benefit of raising the brain copper levels to relieve amyloid pathology in transgenic mice (7, 9, 11, 14), and the potential therapeutic benefits of copper ionophores for treating AD (70).
Acknowledgment
We thank Dr. A. F. Hill (Dept. of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Australia) for the gift of swAPP695 cDNA in the pIRESpuro2 expression vector.
This work was supported by an Alzheimer's Association Zenith Award (to A. I. B.) and grants (to A. I. B.) from the American Health Assistance Foundation, Australian Research Council, Bethlehem Griffiths Research Foundation, and National Health and Medical Research Council (Australia).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- AD
- Alzheimer disease
- APP
- β-amyloid precursor protein
- BCS
- bathocuproinedisulfonic acid
- Nct
- nicastrin
- CTF
- C-terminal fragment
- PS
- presenilin
- HBSS
- Hanks' balanced salt solution
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- MES
- 4-morpholineethanesulfonic acid
- Tg
- transgenic.
REFERENCES
- 1.Bush A. I., Tanzi R. E. (2008) Neurotherapeutics 5, 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovell M. A., Robertson J. D., Teesdale W. J., Campbell J. L., Markesbery W. R. (1998) J. Neurol. Sci. 158, 47–52 [DOI] [PubMed] [Google Scholar]
- 3.Miller L. M., Wang Q., Telivala T. P., Smith R. J., Lanzirotti A., Miklossy J. (2006) J. Struct. Biol. 155, 30–37 [DOI] [PubMed] [Google Scholar]
- 4.Adlard P. A., Bush A. I. (2006) J. Alzheimers Dis. 10, 145–163 [DOI] [PubMed] [Google Scholar]
- 5.Magaki S., Raghavan R., Mueller C., Oberg K. C., Vinters H. V., Kirsch W. M. (2007) Neurosci. Lett. 418, 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Religa D., Strozyk D., Cherny R. A., Volitakis I., Haroutunian V., Winblad B., Naslund J., Bush A. I. (2006) Neurology 67, 69–75 [DOI] [PubMed] [Google Scholar]
- 7.Bayer T. A., Schäfer S., Simons A., Kemmling A., Kamer T., Tepest R., Eckert A., Schüssel K., Eikenberg O., Sturchler-Pierrat C., Abramowski D., Staufenbiel M., Multhaup G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14187–14192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard C. J., Cappai R., Volitakis I., Cherny R. A., White A. R., Beyreuther K., Masters C. L., Bush A. I., Li Q. X. (2002) J. Biol. Chem. 277, 44670–44676 [DOI] [PubMed] [Google Scholar]
- 9.Phinney A. L., Drisaldi B., Schmidt S. D., Lugowski S., Coronado V., Liang Y., Horne P., Yang J., Sekoulidis J., Coomaraswamy J., Chishti M. A., Cox D. W., Mathews P. M., Nixon R. A., Carlson G. A., St. George-Hyslop P., Westaway D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14193–14198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maynard C. J., Cappai R., Volitakis I., Cherny R. A., Masters C. L., Li Q. X., Bush A. I. (2006) J. Inorg. Biochem. 100, 952–962 [DOI] [PubMed] [Google Scholar]
- 11.Adlard P. A., Cherny R. A., Finkelstein D. I., Gautier E., Robb E., Cortes M., Volitakis I., Liu X., Smith J. P., Perez K., Laughton K., Li Q. X., Charman S. A., Nicolazzo J. A., Wilkins S., Deleva K., Lynch T., Kok G., Ritchie C. W., Tanzi R. E., Cappai R., Masters C. L., Barnham K. J., Bush A. I. (2008) Neuron 59, 43–55 [DOI] [PubMed] [Google Scholar]
- 12.Borchardt T., Camakaris J., Cappai R., Masters C. L., Beyreuther K., Multhaup G. (1999) Biochem. J. 344, 461–467 [PMC free article] [PubMed] [Google Scholar]
- 13.Cater M. A., McInnes K. T., Li Q. X., Volitakis I., La Fontaine S., Mercer J. F., Bush A. I. (2008) Biochem. J. 412, 141–152 [DOI] [PubMed] [Google Scholar]
- 14.Cherny R. A., Atwood C. S., Xilinas M. E., Gray D. N., Jones W. D., McLean C. A., Barnham K. J., Volitakis I., Fraser F. W., Kim Y., Huang X., Goldstein L. E., Moir R. D., Lim J. T., Beyreuther K., Zheng H., Tanzi R. E., Masters C. L., Bush A. I. (2001) Neuron 30, 665–676 [DOI] [PubMed] [Google Scholar]
- 15.White A. R., Du T., Laughton K. M., Volitakis I., Sharples R. A., Xilinas M. E., Hoke D. E., Holsinger R. M., Evin G., Cherny R. A., Hill A. F., Barnham K. J., Li Q. X., Bush A. I., Masters C. L. (2006) J. Biol. Chem. 281, 17670–17680 [DOI] [PubMed] [Google Scholar]
- 16.Refolo L. M., Malester B., LaFrancois J., Bryant-Thomas T., Wang R., Tint G. S., Sambamurti K., Duff K., Pappolla M. A. (2000) Neurobiol. Dis. 7, 321–331 [DOI] [PubMed] [Google Scholar]
- 17.Morris M. C., Evans D. A., Tangney C. C., Bienias J. L., Schneider J. A., Wilson R. S., Scherr P. A. (2006) Arch. Neurol. 63, 1085–1088 [DOI] [PubMed] [Google Scholar]
- 18.Sparks D. L., Schreurs B. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11065–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons M., Keller P., De Strooper B., Beyreuther K., Dotti C. G., Simons K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6460–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buxbaum J. D., Cullen E. I., Friedhoff L. T. (2002) Front. Biosci. 7, a50–59 [DOI] [PubMed] [Google Scholar]
- 21.Won J. S., Im Y. B., Khan M., Contreras M., Singh A. K., Singh I. (2008) J. Neurochem. 105, 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan N. B., Siegel G. J., Feinstein D. L. (2004) Neurochem. Res. 29, 1897–1911 [DOI] [PubMed] [Google Scholar]
- 23.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch C., Eckert G. P., Mueller W. E. (2003) Biochem. Pharmacol. 65, 843–856 [DOI] [PubMed] [Google Scholar]
- 25.Fassbender K., Simons M., Bergmann C., Stroick M., Lutjohann D., Keller P., Runz H., Kuhl S., Bertsch T., von Bergmann K., Hennerici M., Beyreuther K., Hartmann T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordy J. M., Hussain I., Dingwall C., Hooper N. M., Turner A. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11735–11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vetrivel K. S., Cheng H., Lin W., Sakurai T., Li T., Nukina N., Wong P. C., Xu H., Thinakaran G. (2004) J. Biol. Chem. 279, 44945–44954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawarabayashi T., Shoji M., Younkin L. H., Wen-Lang L., Dickson D. W., Murakami T., Matsubara E., Abe K., Ashe K. H., Younkin S. G. (2004) J. Neurosci. 24, 3801–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson T. J., Alkon D. L. (2005) J. Biol. Chem. 280, 7377–7387 [DOI] [PubMed] [Google Scholar]
- 30.Opazo C., Huang X., Cherny R. A., Moir R. D., Roher A. E., White A. R., Cappai R., Masters C. L., Tanzi R. E., Inestrosa N. C., Bush A. I. (2002) J. Biol. Chem. 277, 40302–40308 [DOI] [PubMed] [Google Scholar]
- 31.Puglielli L., Friedlich A. L., Setchell K. D., Nagano S., Opazo C., Cherny R. A., Barnham K. J., Wade J. D., Melov S., Kovacs D. M., Bush A. I. (2005) J. Clin. Invest. 115, 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimoto N., Tasaki M., Shimanouchi T., Umakoshi H., Kuboi R. (2005) J. Biosci. Bioeng. 100, 455–459 [DOI] [PubMed] [Google Scholar]
- 33.Smith D. P., Smith D. G., Curtain C. C., Boas J. F., Pilbrow J. R., Ciccotosto G. D., Lau T. L., Tew D. J., Perez K., Wade J. D., Bush A. I., Drew S. C., Separovic F., Masters C. L., Cappai R., Barnham K. J. (2006) J. Biol. Chem. 281, 15145–15154 [DOI] [PubMed] [Google Scholar]
- 34.Jiang D., Men L., Wang J., Zhang Y., Chickenyen S., Wang Y., Zhou F. (2007) Biochemistry 46, 9270–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray I. V., Liu L., Komatsu H., Uryu K., Xiao G., Lawson J. A., Axelsen P. H. (2007) J. Biol. Chem. 282, 9335–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharples R. A., Vella L. J., Nisbet R. M., Naylor R., Perez K., Barnham K. J., Masters C. L., Hill A. F. (2008) FASEB J. 22, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 37.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 38.Culvenor J. G., Evin G., Cooney M. A., Wardan H., Sharples R. A., Maher F., Reed G., Diehlmann A., Weidemann A., Beyreuther K., Masters C. L. (2000) Exp. Cell Res. 255, 192–206 [DOI] [PubMed] [Google Scholar]
- 39.Ida N., Hartmann T., Pantel J., Schröder J., Zerfass R., Förstl H., Sandbrink R., Masters C. L., Beyreuther K. (1996) J. Biol. Chem. 271, 22908–22914 [DOI] [PubMed] [Google Scholar]
- 40.Hoke D. E., Tan J. L., Ilaya N. T., Culvenor J. G., Smith S. J., White A. R., Masters C. L., Evin G. M. (2005) FEBS J. 272, 5544–5557 [DOI] [PubMed] [Google Scholar]
- 41.Cordy J. M., Hooper N. M., Turner A. J. (2006) Mol. Membr. Biol. 23, 111–122 [DOI] [PubMed] [Google Scholar]
- 42.Bellingham S. A., Lahiri D. K., Maloney B., La Fontaine S., Multhaup G., Camakaris J. (2004) J. Biol. Chem. 279, 20378–20386 [DOI] [PubMed] [Google Scholar]
- 43.Rogers J. T., Randall J. D., Cahill C. M., Eder P. S., Huang X., Gunshin H., Leiter L., McPhee J., Sarang S. S., Utsuki T., Greig N. H., Lahiri D. K., Tanzi R. E., Bush A. I., Giordano T., Gullans S. R. (2002) J. Biol. Chem. 277, 45518–45528 [DOI] [PubMed] [Google Scholar]
- 44.Armendariz A. D., Gonzalez M., Loguinov A. V., Vulpe C. D. (2004) Physiol. Genomics 20, 45–54 [DOI] [PubMed] [Google Scholar]
- 45.Huster D., Purnat T. D., Burkhead J. L., Ralle M., Fiehn O., Stuckert F., Olson N. E., Teupser D., Lutsenko S. (2007) J. Biol. Chem. 282, 8343–8355 [DOI] [PubMed] [Google Scholar]
- 46.Svensson P. A., Englund M. C., Markström E., Ohlsson B. G., Jernås M., Billig H., Torgerson J. S., Wiklund O., Carlsson L. M., Carlsson B. (2003) Atherosclerosis 169, 71–76 [DOI] [PubMed] [Google Scholar]
- 47.Schlief M. L., Craig A. M., Gitlin J. D. (2005) J. Neurosci. 25, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Squitti R., Ventriglia M., Barbati G., Cassetta E., Ferreri F., Dal Forno G., Ramires S., Zappasodi F., Rossini P. M. (2007) J. Neural. Transm. 114, 1589–1594 [DOI] [PubMed] [Google Scholar]
- 49.Hopt A., Korte S., Fink H., Panne U., Niessner R., Jahn R., Kretzschmar H., Herms J. (2003) J. Neurosci. Methods 128, 159–172 [DOI] [PubMed] [Google Scholar]
- 50.Simons K., Ikonen E. (1997) Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 51.Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. (1997) J. Biol. Chem. 272, 13793–13802 [DOI] [PubMed] [Google Scholar]
- 52.Urano Y., Hayashi I., Isoo N., Reid P. C., Shibasaki Y., Noguchi N., Tomita T., Iwatsubo T., Hamakubo T., Kodama T. (2005) J. Lipid Res. 46, 904–912 [DOI] [PubMed] [Google Scholar]
- 53.Schneider A., Rajendran L., Honsho M., Gralle M., Donnert G., Wouters F., Hell S. W., Simons M. (2008) J. Neurosci. 28, 2874–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steiner H., Fluhrer R., Haass C. (2008) J. Biol. Chem. 283, 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherrington R., Rogaev E. I., Liang Y., Rogaeva E. A., Levesque G., Ikeda M., Chi H., Lin C., Li G., Holman K., Tsuda T., Mar L., Foncin J.-F., Bruni A. C., Montesi M. P., Sorbi S., Rainero I., Pinessi L., Nee L., Chumakov I., Pollen D., Brookes A., Sanseau P., Polinsky R. J., Wasco W., Da Silva H. A. R., Haines J. L., Pericak-Vance M. A., Tanzi R. E., Roses A. D., Fraser P. E., Rommens J. M., St George-Hyslop P. H. (1995) Nature 375, 754–760 [DOI] [PubMed] [Google Scholar]
- 56.Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 57.Shah S., Lee S. F., Tabuchi K., Hao Y. H., Yu C., LaPlant Q., Ball H., Dann C. E., 3rd, Südhof T., Yu G. (2005) Cell 122, 435–447 [DOI] [PubMed] [Google Scholar]
- 58.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 59.Smith D. G., Cappai R., Barnham K. J. (2007) Biochim. Biophys. Acta 1768, 1976–1990 [DOI] [PubMed] [Google Scholar]
- 60.Peña M. M., Lee J., Thiele D. J. (1999) J. Nutr. 129, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 61.Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O'Halloran T. V. (1999) Science 284, 805–808 [DOI] [PubMed] [Google Scholar]
- 62.Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 63.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 64.Kakio A., Nishimoto S., Yanagisawa K., Kozutsumi Y., Matsuzaki K. (2002) Biochemistry 41, 7385–7390 [DOI] [PubMed] [Google Scholar]
- 65.Kim S. I., Yi J. S., Ko Y. G. (2006) J. Cell. Biochem. 99, 878–889 [DOI] [PubMed] [Google Scholar]
- 66.Atwood C. S., Moir R. D., Huang X., Scarpa R. C., Bacarra N. M., Romano D. M., Hartshorn M. A., Tanzi R. E., Bush A. I. (1998) J. Biol. Chem. 273, 12817–12826 [DOI] [PubMed] [Google Scholar]
- 67.Atwood C. S., Perry G., Zeng H., Kato Y., Jones W. D., Ling K. Q., Huang X., Moir R. D., Wang D., Sayre L. M., Smith M. A., Chen S. G., Bush A. I. (2004) Biochemistry 43, 560–568 [DOI] [PubMed] [Google Scholar]
- 68.Barnham K. J., Haeffner F., Ciccotosto G. D., Curtain C. C., Tew D., Mavros C., Beyreuther K., Carrington D., Masters C. L., Cherny R. A., Cappai R., Bush A. I. (2004) FASEB J. 18, 1427–1429 [DOI] [PubMed] [Google Scholar]
- 69.Strozyk D., Launer L. J., Adlard P. A., Cherny R. A., Tsatsanis A., Volitakis I., Blennow K., Petrovitch H., White L. R., Bush A. I. (2009) Neurobiol. Aging 30, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lannfelt L., Blennow K., Zetterberg H., Batsman S., Ames D., Harrison J., Masters C. L., Targum S., Bush A. I., Murdoch R., Wilson J., Ritchie C. W. (2008) Lancet Neurol. 7, 779–786 [DOI] [PubMed] [Google Scholar]