Abstract
The action of Clostridium difficile toxins A and B depends on inactivation of host small G-proteins by glucosylation. Cellular inositol hexakisphosphate (InsP6) induces an autocatalytic cleavage of the toxins, releasing an N-terminal glucosyltransferase domain into the host cell cytosol. We have defined the cysteine protease domain (CPD) responsible for autoprocessing within toxin A (TcdA) and report the 1.6 Å x-ray crystal structure of the domain bound to InsP6. InsP6 is bound in a highly basic pocket that is separated from an unusual active site by a β-flap structure. Functional studies confirm an intramolecular mechanism of cleavage and highlight specific residues required for InsP6-induced TcdA processing. Analysis of the structural and functional data in the context of sequences from similar and diverse origins highlights a C-terminal extension and a π-cation interaction within the β-flap that appear to be unique among the large clostridial cytotoxins.
Clostridium difficile is a Gram-positive, spore-forming anaerobe that infects the colon and causes a range of disorders, including diarrhea, pseudomembranous colitis, and toxic megacolon (1, 2). Two large toxins, TcdA2 and TcdB (308 and 270 kDa, respectively) are recognized as the main virulence factors of C. difficile, although their relative importance is the subject of on-going study (3, 4). These proteins belong to a class of homologous toxins called large clostridial toxins (LCTs) and have been classified more broadly as AB toxins, wherein a B moiety is involved in the delivery of an enzymatic A moiety into the cytosol of a target cell. In LCTs, the A subunit is an N-terminal glucosyltransferase that inactivates small G-proteins, such as Rho, leading to cell rounding and apoptosis of the intoxicated cell (5, 6). The B subunit corresponds to the remainder of the toxin and is responsible for binding the target cell through a C-terminal receptor-binding domain (7–9) and forming the membrane pore needed for translocation of the A subunit (10, 11). Unlike other known AB toxins, the glucosyltransferase A domains of LCTs are released from the B subunits by an autoproteolytic cleavage event (12). Cleavage is triggered by host inositol phosphates and the reducing environment of the cytosol (12).
In LCTs, autoproteolysis has been attributed to a cysteine protease activity located within the N-terminal region of the B subunit (13). This region was identified based on homology with the cysteine protease domain (CPD) found in the multifunctional autoprocessing repeats in toxins (MARTX) toxins from Gram-negative bacteria (14). Autoprocessing in the MARTX toxin from Vibrio cholera (VcRTx) is also stimulated by InsP6 (15). A recent crystal structure of VcRTx CPD bound to InsP6 suggests a novel mechanism of InsP6-induced allosteric activation (16). The CPDs of TcdA and VcRTx share only 19% sequence identity. To gain insight into the mechanistic commonalities between these entirely different toxins and to delineate the LCT-specific modes of InsP6-induced processing, we performed structural and functional analyses on the cysteine protease from TcdA.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Point Mutants
The nucleotide sequence coding for amino acids 543–809 of TcdA (TcdA CPD) was amplified from C. difficile strain 10463 genomic DNA. The DNA was cloned into a modified pET27 vector such that the resulting protein contains an N-terminal His10 tag followed by a 3C protease cleavage site. The sequence preceding the TcdA CPD sequence is MGSSHHHHHHHHHHGSSLEVLFQGPGS. Following 3C cleavage, only non-native residues GPGS remain. The extended construct, TcdA g-CPD, encodes amino acids 510–809. This plasmid was constructed similarly, except the last two residues of the leader sequence are VD rather than GS. Point mutants were generated by QuikChange site-directed mutagenesis.
Protein Expression and Purification
Transformed Escherichia coli BL21(DE3) cells were grown in Terrific Broth containing 50 mg/liter kanamycin. Ten ml of overnight culture was used to inoculate 1 liter of medium, and the culture was placed at 37 °C and 230 rpm. When the cultures reached A600 = 0.6, the temperature was changed to 16 °C, and expression was induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. After 16 h, the cells were harvested by centrifugation and resuspended in 100 mm NaCl, 20 mm Tris, pH 8.0. Following French Press lysis, the lysates were centrifuged at 48,000 × g for 20 min. Protein was purified from the supernatant by nickel affinity chromatography. For crystallization trials, the His10 tag was cleaved from TcdA CPD by 3C protease overnight at 4 °C. The mixture was then run back over an Ni2+-nitrilotriacetic acid column to remove the cleaved His10 tag and the His-tagged 3C protease. Cleaved TcdA CPD was further purified by gel filtration chromatography in 50 mm NaCl, 20 mm Tris, pH 8.0. Selenomethionine-substituted TcdA CPD was prepared using E. coli BL843(DE3) in minimal medium containing 40 mg/liter l-selenomethionine following the same procedure, except 5 mm methionine and 1 mm dithiothreitol were added to the buffers to prevent oxidation of the selenium. TcdA g-CPD and mutants used for cleavage assays were expressed as above and purified by nickel affinity and ion exchange chromatography. The His10 tag was not cleaved for these proteins.
Crystallization
TcdA CPD was concentrated to 60–80 mg/ml in 50 mm NaCl, 20 mm Tris, pH 8.0. InsP6 (Sigma) was added to 10 mm. TcdA CPD was crystallized by the hanging drop method at 21 °C with a 1:1 ratio of protein and mother liquor containing 100 mm Tris, pH 8.0–9.0, 25–30% polyethylene glycol 8000, and 200 mm guanadinium chloride. Crystals were mounted on cryo loops and flash-cooled in liquid nitrogen. No cryoprotectants were required.
Structure Determination and Refinement
X-ray data were collected from single crystals on LS-CAT beamlines 21-ID-D and 21-ID-G at the Advanced Photon Source (Argonne, IL). Diffraction data were indexed, integrated, and scaled using HKL2000 (17). Phases were determined from anomalous scattering data using autoSHARP (18). The model was built using Coot (19) and refined using Phenix (20) with riding hydrogens and seven TLS groups per chain. The final model contains two CPD molecules (consisting of residues 547–570 and 575–801 for chain A and 548–570, 578–655, and 661–803 for chain B), two InsP6 molecules, and 649 water molecules.
InsP6-induced Cleavage Assays
TcdA g-CPD and g-CPD mutant proteins were exchanged into 60 mm NaCl, 250 mm sucrose, 20 mm Tris, pH 7.5. 100 μl of protein at 5 μm was mixed with 1 μl of buffer or InsP6 stock solution and incubated at 37 °C. After 2 h, the reaction was stopped by the addition of loading buffer and heating. For the time course reactions, 1 ml of 5 μm protein was treated with 10 μl of 500 μm InsP6. At the indicated time points, 50-μl aliquots were removed, SDS loading buffer was added, and the samples were heated for 5 min. The samples were analyzed by SDS-PAGE with Coomassie staining. Band intensities were quantified using the Kodak 1D 3.6 software. The percentage of cleavage was calculated as the intensity of the CPD band divided by the sum of the intensities of the g-CPD and CPD bands.
NMR Spectroscopy
TcdA CPD was expressed and purified as described for crystallography. The protein was then exchanged into a 50 mm NaCl, 50 mm potassium phosphate, pH 7.0 buffer by gel filtration chromatography and concentrated to 0.6 mm ± 5 mm InsP6. NMR data were recorded at 25 °C on a Bruker DRX600 spectrometer with a 5 mm TXI-Z cryoprobe and processed with Topspin 2.0b (Bruker).
RESULTS
Defining the TcdA Cysteine Protease Domain
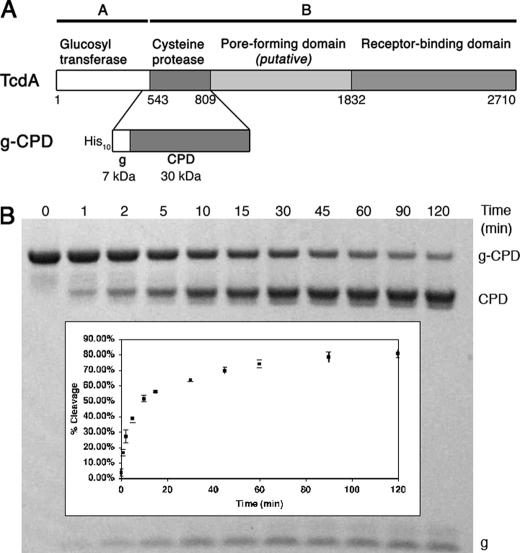
TcdA and TcdB holotoxins undergo autoproteolysis in the presence of InsP6 and dithiothreitol to release an N-terminal glucosyltransferase domain (residues 1–542 for TcdA) (21). In TcdB, the autoproteolytic activity and InsP6 binding have been mapped to fragments corresponding to residues 1–955 and residues 544–955, respectively (22). Sequence alignment with VcRTx suggests domain boundaries of 543–769 (TcdA) and 543–767 (TcdB) (13). To define a structural domain required for TcdA autoproteolysis, we designed a construct for the recombinant expression of TcdA 510–809. The 33 residues from the glucosyltransferase domain C terminus were included to provide a substrate for the enzyme and to permit visualization of cleavage by SDS-PAGE (Fig. 1A). Incubation of this 37-kDa protein, g-CPD, with InsP6 causes proteolysis, resulting in two cleavage products of 30 kDa (CPD) and 7 kDa (g). When InsP6 and TcdA g-CPD were mixed in equal molar ratios, proteolysis occurred quickly, with 50% of the protein cleaved in about 10 min (Fig. 1B). A shorter construct corresponding to TcdA residues 510–769 did not undergo cleavage, even when InsP6 was added in excess (data not shown).
FIGURE 1.
Defining the TcdA CPD. A, TcdA is an AB toxin with four functional domains. The CPD is defined in this work as residues 543–809 based on the InsP6-induced activity of g-CPD, residues 510–809. B, time course of g-CPD (5 μm) cleavage in the presence of 5 μm InsP6 shows 50% cleavage within 10 min. Inset, the band intensities of cleaved and uncleaved proteins were analyzed by densitometry to calculate the percentage of cleavage. The values represent the mean ± S.D. of three independent experiments.
Structure of the TcdA Cysteine Protease Domain
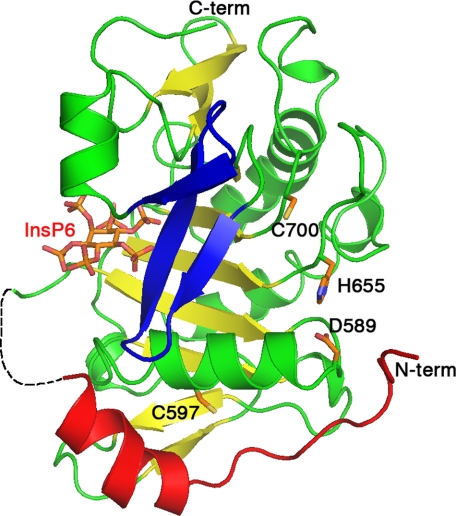
TcdA amino acids 543–809, henceforth TcdA CPD, was crystallized in the presence of InsP6, and the structure was determined at 1.6 Å (Table 1). The asymmetric unit contained two CPD molecules, which align with an root mean square deviation of 0.236 Å based on the backbone α-carbon positions. For the remainder of this paper, we refer to chain A. The TcdA CPD is composed of a nine-stranded β-sheet flanked by five α-helices (Fig. 2). The closest structural homolog is that of the VcRTx CPD (Protein Data Bank code 3eeb-A) (16), which aligns with an root mean square deviation of 2.9 Å over 187 residues (supplemental Fig. S1), whereas the next closest homolog is caspase-7 (Protein Data Bank code 1SHJ-B) (23), aligning with a root mean square deviation of 4.8 Å over 135 residues. In both the VcRTx and TcdA CPD structures, InsP6 binds on one face of the β-sheet and is separated from the proposed active site (Cys700, His655, and Asp589 in TcdA) by a three-stranded β-hairpin structure termed the β-flap (Fig. 2 and supplemental Fig. S1). The N terminus of the protein wraps around the exterior of the domain with the most N-terminal of the resolved residues (Gly547) near the proposed catalytic site (Fig. 2). The TcdA CPD contains about 50 more residues than the VcRTx CPD, which leads to additional β-strands at the top and bottom of the β-sheet and two additional α-helices (supplemental Figs. S1 and S2). The two cysteines of the TcdA CPD domain (Cys597 and Cys700) are more than 20 Å apart, consistent with the observation that for g-CPD, reductants are not needed for InsP6-induced auto-processing (Figs. 1B and 2). The fact that reducing agents enhance the autoprocessing of holotoxin (13) suggests the presence of a disulfide involving at least one cysteine from another domain.
TABLE 1.
X-ray data collection and refinement statistics
Outer resolution bin statistics are given in parentheses.
| Native | Selenium | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9784 | 0.9784 |
| Resolution (outer shell) (Å) | 50-1.6 (1.66-1.60) | 50-1.8 (1.86-1.80) |
| Rmerge (%)a | 4.3 (26.8) | 5.5 (26.6) |
| Mean I/σI | 23.5 (2.4) | 24.9 (6.7) |
| Completeness (%) | 96.2 (81.9) | 99.6 (96.5) |
| Redundancy | 4.8 (2.8) | 7.6 (6.9) |
| Unique observations | 68,137 (5,753) | 47,174 (4,525) |
| Anomal. phasing power | 1.226 | |
| Refinement | ||
| Rcryst/Rfree (%)b | 16.54/19.67 | |
| No. of protein atoms | 4,007 | |
| No. of ligand atoms | 84 | |
| No. of solvent waters | 649 | |
| Bond length root mean square deviation (Å) | 0.011 | |
| Bond angle root mean square deviation (degrees) | 1.469 | |
| Average protein B (Å2) | 39 | |
| Ramachandran plot (%)c | ||
| Most favored | 417 (92.3%) | |
| Allowed | 33 (7.3%) | |
| Generously allowed | 1 (0.2%) | |
| Disallowed | 1 (0.2%) | |
aRmerge = Shkl(Si|Ihkl,i − 〈Ihkl〉))/Shkl,i〈Ihkli〉, where Ihkl,i, is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and 〈Ihkl〉 is the mean intensity of that reflection.
b Rcryst = S‖Fo,hkl| − |Fc,hkl‖/|Fo,hkl|, where |Fo,hkl| and |Fc,hkl| are the observed and calculated structure factor amplitudes. Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
c Calculated with the program PROCHECK ((29)).
FIGURE 2.
Structure of the TcdA CPD bound to InsP6. TcdA CPD is shown as a ribbon diagram with InsP6 and the side chains of Cys700, His655, Asp589, and Cys597 shown as sticks. The InsP6 binding site and catalytic site are separated by a three-stranded β-flap (blue). The central β-sheet is shown in yellow. The N terminus (red) wraps around the domain with the most N-terminal residues near the catalytic site.
TcdA CPD Active Site
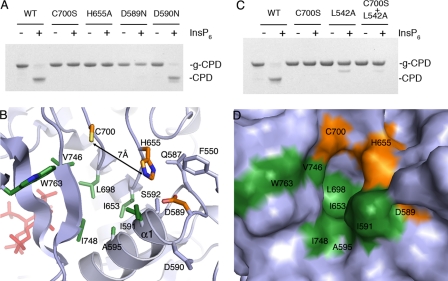
In cysteine proteases, the nucleophilicity of the active site cysteine is typically enhanced by hydrogen bonding to an adjacent histidine. Often, an additional negatively charged residue, such as aspartic acid, is present to stabilize the positive charge of the histidine. TcdA Cys700, His655, and Asp589 align to residues that have been implicated as catalytic residues in TcdB CPD (13, 22) and VcRTx CPD (15). To address the role of these residues in autoproteolysis of TcdA, each was mutated within g-CPD, and the proteins were tested for InsP6-inducible cleavage. Mutation of Cys700, His655, or Asp589 abrogates InsP6-inducible cleavage (Fig. 3A). The adjacent Asp590 cannot substitute for Asp589, and mutation of Asp590 does not cause a defect in protease activity (Fig. 3A). Although these results are consistent with a catalytic triad, the structure indicates that although His655 and Asp589 are in hydrogen bonding contact, Cys700 is >6 Å from His655 and >10 Å from Asp589 (Fig. 3B). This arrangement, which we also observe in the CPD structure of VcRTx, suggests that the mechanism of catalysis in this class of self-cleaving proteases may be different from one in which a catalytic triad is in place.
FIGURE 3.
Catalytic site of the TcdA CPD. A, wild-type (WT), C700S, H655A, D589N, and D590N g-CPD at 5 μm were incubated with 100 μm InsP6 for 2 h at 37 °C. In contrast to wild-type and D590N, the C700S, H655A, and D589N mutants were unable to undergo InsP6-inducible cleavage. B, ribbon diagram of the catalytic site of CPD (gray) with residue side chains of the catalytic site (orange) and hydrophobic pocket (green) shown as sticks. Other side chains within the active site are colored gray. C, L542A g-CPD (cleavage site mutant) does not undergo cleavage upon the addition of InsP6. When C700S and L542A g-CPD are mixed, no InsP6-dependent cleavage is observed. D, surface representation of the catalytic site reveals a likely substrate-binding pocket. The Asp589, His655, and Cys700 residues are colored orange, and the hydrophobic residues are colored green. The orientation is identical to that in B.
Intramolecular Cleavage by CPD
To test whether InsP6-induced cleavage in TcdA is inter- or intramolecular, we mutated the conserved P1 residue from leucine to alanine (L542A) in g-CPD. This mutation prevented InsP6-inducible cleavage at the normal cleavage site (Fig. 3C). When this mutant was mixed with the catalytic cysteine mutant, g-CPD C700S, no cleavage was observed. The g-CPD L542A was unable to cleave the normal cleavage site of g-CPD C700S, suggesting that intermolecular cleavage does not occur for TcdA. In the crystal structure of TcdA CPD, residues 543–546 are unstructured. In TcdA CPD containing an intact cleavage site, the unstructured residues 543–546 probably extend into the catalytic site. The structure of the TcdA CPD suggests that Leu542 may bind in a hydrophobic pocket located between the catalytic cysteine and the β-flap (Fig. 3D). This pocket is made up of the residues Ile591, Ala595, Ile653, Leu698, Val746, Ile748, and Trp763 (shown in green in Fig. 3, B and D), which are mostly conserved among LCTs (supplemental Fig. S3).
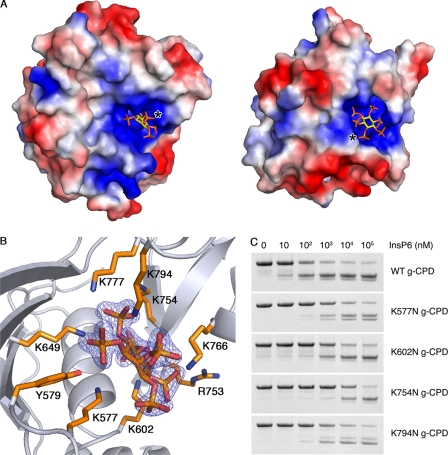
Binding of InsP6
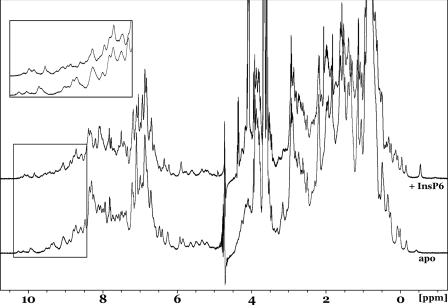
The InsP6 molecule is highly charged, bearing six negatively charged phosphate groups and, accordingly, binds within a positively charged pocket of the TcdA CPD (Fig. 4A). Binding of InsP6 results in a significant stabilization of TcdA CPD, as reflected by an increased resistance to digestion by chymotrypsin (supplemental Fig. S3). Efforts to obtain crystals of the TcdA CPD in the absence of InsP6 were unsuccessful, so NMR spectroscopy was used to assess the nature of InsP6-induced structural changes. The one-dimensional 1H NMR spectrum of apo-TcdA shows excellent dispersion of the signals indicative of a well folded globular domain (see Fig. 6, lower trace). The addition of InsP6 into the sample resulted in a substantial perturbation of the signals with an even greater dispersion (Fig. 6, upper trace). This is most evident in the very high field methyl region (−0.4 to +0.4 ppm) and the very low field amide region (8.4–10.4 ppm; inset) of the spectrum. These data indicate that the TcdA CPD is folded in the absence of InsP6 and imply that InsP6 binding induces significant conformational reorganization and structural stability within TcdA CPD.
FIGURE 4.
InsP6 binding pocket. A, the TcdA CPD (left) and VcRTx CPD (right) surfaces colored by electrostatic potential (negatively charged surfaces are red, positively charged regions are blue, and neutral regions are shown in white) demonstrate that InsP6, shown as a stick model, binds in positively charged pockets. The protein molecules were aligned and are shown in identical views, but the orientation of the InsP6 molecules and the position of the axial phosphate groups (denoted with an asterisk) differs. B, InsP6 and the side chains of residues that directly interact with InsP6 are shown as sticks. The orientation is identical to that in A. The displayed electron density is a 2Fo − Fc map contoured at 5σ in which InsP6 was omitted from the phase calculations. C, wild type (WT), K577N, K602N, K754N, and K794N g-CPD at 5 μm were tested for autoproteolysis over a range of InsP6 concentrations. Each mutant required 10–1000-fold more InsP6 in order to undergo autoproteolysis at levels comparable with wild type.
FIGURE 6.
InsP6-induced structural changes. One-dimensional 1H NMR spectra of the TcdA CPD in the presence (upper trace) and absence (lower trace) of InsP6 show clear dispersion of peaks indicative of folded proteins. The inset shows major changes in the peaks, suggesting that significant structural changes are induced by InsP6 binding.
Insights into the effects of InsP6 binding on TcdA CPD are provided by the high resolution structure of the complex. The TcdA CPD has nine residues that make direct contacts with the InsP6 (Fig. 4B). These include one arginine (Arg753), one tyrosine (Tyr579), and seven lysines (Lys577, Lys602, Lys649, Lys754, Lys766, Lys777, and Lys794). Thus, binding of InsP6 involves residues spanning the entire domain, consistent with the significant reorganization of the structure upon binding. Lys794, which is conserved among all LCTs except TpeL of C. perfringens (supplemental Fig. S4), coordinates the axial phosphate group of the InsP6. This residue is located on the C-terminal β-strand, which is entirely absent from the VcRTx CPD. Thus, the position of the axial phosphate group and the orientation of the InsP6 in the VcRTx CPD structure are markedly different from that in TcdA CPD (Fig. 4A).
In VcRTx CPD, Lys602, Lys754, Lys766, Lys777, and Arg753 are conserved and important for binding InsP6 and proteolysis (supplemental Fig. S2) (16). To test the importance of both conserved and unique InsP6-binding interactions in TcdA autoproteolysis, lysine residues 577, 602, 754, and 794 were mutated to asparagine in the g-CPD protein background and tested for InsP6-inducible cleavage (Fig. 4C). The mutant proteins required 10–1,000 times higher concentrations of InsP6 to achieve an equivalent amount of cleavage as wild type g-CPD, confirming the significance of each of these InsP6 ligands.
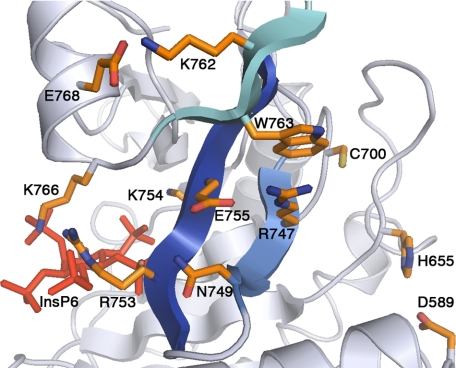
The β-Flap
In the structural analysis of the VcRTx CPD, Lupardus et al. (16) propose the β-flap as a link between InsP6 binding and the catalytic site. In TcdA, the β-flap contains a network of amino acid side chains that may be involved in transmitting InsP6-induced structural changes to the active site (Fig. 5). The active site side of the β-flap includes Val746, Ile748, and Trp763, residues that line the hydrophobic pocket thought to be involved in binding the P1 residue of the substrate (Leu542) (Fig. 3D). On the InsP6 side of the β-flap, the conserved Arg753-Lys754 pair and Lys766 directly coordinate the InsP6 (Figs. 4B and 5). Lys766 is located at the beginning of α4, a structural element not found in the structure of VcRTx CPD. In addition to interacting with InsP6, the α4 helix stabilizes the β-flap with an electrostatic interaction between Glu768 and Lys762 (Fig. 5). Arg747 also stabilizes the β-flap structure. This is through an electrostatic interaction with Glu755 and a π-cation interaction with Trp763 (Fig. 5). Although Trp763 is strictly conserved in the broad family of CPD sequences, the Arg747 and Glu757 residues are only conserved within the LCTs (supplemental Fig. S4). In VcRTx and other homologs from Gram-negative bacteria, the residue corresponding to Arg747 is an alanine. Efforts to analyze a TcdA CPD R747A mutant failed due to protein instability, implying a role for Arg747 in the absence of InsP6. Thus, in TcdA (and probably all LCTs), the β-flap is an important structural element both in separating the InsP6-binding and substrate-binding sites and in contributing to the global stability of the fold.
FIGURE 5.
β-flap. A close up of the β-flap (blue) reveals a network of interactions that link InsP6 (red) to the catalytic site (Cys700 and His655).
DISCUSSION
TcdA and VcRTx represent two classes of large toxins that target the eukaryotic cytoskeleton through very different mechanisms. The proteins contain a single region of homology, the CPD, which is involved in proteolytic processing of the toxins. Comparison of the atomic structures from these highly divergent domains provides an opportunity to gain insight into the selective pressures needed for function in both systems.
The CPD from TcdA is larger than that of VcRTx and contains a C-terminal extension that was not readily apparent through sequence analysis. The observation that Lys777 and Lys794 are involved in coordination of InsP6 provides a ready explanation for the lack of activity in the 510–769 construct we made based on sequence alignment with VcRTx. The altered conformation of InsP6 within the active site reflects the contributions from this C-terminal extension as well as other structural differences in the TcdA and VcRTx InsP6 binding sites. Lys766 is presented by the α4 helix, not present in VcRTx, and the loop containing Lys577 is positioned very differently in the two molecules (supplemental Fig. S1). The capacity to evolve differences in how InsP6 (and related molecules) are bound has been noted in the family of RNA editing enzymes (24) and could be relevant in a newly identified and largely uncharacterized family of CPD homologs from bacteria and eukaryotes (25).
The VcRTx and TcdA CPD structures were obtained in the presence of InsP6. The dispersion of peaks observed in the apo- and InsP6-bound proteins by NMR suggest that both proteins are folded but that InsP6 binding confers stability and induces significant structural change (Fig. 6). This is not unexpected given the energetically unfavorable accumulation of positive charges that one would predict in the absence of InsP6 (Fig. 4A). We propose that the energetic gains of complementing these positive charges with InsP6 allow the molecule to overcome the energetic barrier needed to access its active conformation. An example of such an allosteric transition in a single-domain protein has been documented in the phosphorylation of the NtrC response regulator (26). In TcdA, the InsP6-induced conformational changes are likely to involve the organization of the substrate-binding pocket through stabilization of the β-flap.
A trio of Asp, His, and Cys residues has been shown to be essential in the autoproteolytic processing of VcRTx (15), TcdB (13), and TcdA (this work). The structures of the TcdA and VcRTx CPDs, however, indicate that these residues do not adopt a conventional catalytic triad arrangement. Instead, the cysteine and histidine residues are separated by a large cavity, which is likely to serve as the substrate-binding pocket (Fig. 3, B and D). In TcdA, this pocket is ∼11 Å long, ∼7 Å wide, and ∼7 Å deep and bounded by α1, the β-flap, and the loop containing His655. One end of the cavity is hydrophobic and believed to represent a binding site for the P1 leucine residue of the substrate (Fig. 3D). Although His655 is on a flexible loop and could reorient in the presence of substrate, the structural changes needed to account for the observed importance of Asp589 would be substantial and require rearrangement of α1 and occlusion of the substrate binding cavity (Fig. 3B). Thus, despite biochemical evidence consistent with a catalytic triad, the TcdA and VcRTx CPD crystal structures indicate that the His and Asp residues are not positioned to increase the nucleophilicity of the Cys. Rather, His655 may be acting as an oxyanion hole with Asp589 important in its capacity to orient His655. TcdA and VcRTx CPDs may employ a primitive mechanism of proteolysis wherein the need to “activate” the nucleophile is obviated by the intramolecular and/or single turnover nature of the reaction. Mutant serine proteases in which His and Asp have been removed from the catalytic triad still catalyze reactions with ∼10,000-fold rate enhancements (27), explained largely by their ability to constrain substrate and stabilize the developing oxyanion (28).
In summary, we have shown that the CPD of TcdA is able to use InsP6 to activate an intramolecular autoproteolytic processing event. The structural analysis reveals striking similarities in the mechanisms of TcdA and VcRTx InsP6-inducible cleavage but also shows differences in the conformation of InsP6 and the organization of the β-flap. These observations should aid the elucidation of unique and conserved molecular features required for virulence in TcdA, the LCTs, and the larger family of CPDs from MARTX toxins.
Supplementary Material
Acknowledgment
We gratefully acknowledge Tara Archuleta for help with protein purification.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM075156 (to W. J. C.). This work was also supported by a Cellular and Molecular Microbiology Training Grant (to R. N. P.), the Germs and Defense Summer Science Academy (to M. C.), and Vanderbilt University Medical Center development funds. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (code 3HO6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TcdA and -B
- C. difficile toxin A and B, respectively
- InsP6
- inositol hexakisphosphate
- CPD
- cysteine protease domain
- LCT
- large clostridial toxin
- MARTX
- multifunctional autoprocessing repeats in toxins
- VcRTx
- V. cholera RTx toxin
- g-CPD
- residues 510–809 of TcdA.
REFERENCES
- 1.Lyerly D. M., Krivan H. C., Wilkins T. D. (1988) Clin. Microbiol. Rev. 1, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFarland L. V., Stamm W. E. (1986) Am. J. Infect. Control 14, 99–109 [DOI] [PubMed] [Google Scholar]
- 3.Lyras D., O'Connor J. R., Howarth P. M., Sambol S. P., Carter G. P., Phumoonna T., Poon R., Adams V., Vedantam G., Johnson S., Gerding D. N., Rood J. I. (2009) Nature 458, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyerly D. M., Saum K. E., MacDonald D. K., Wilkins T. D. (1985) Infect. Immun. 47, 349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 6.Gerhard R., Nottrott S., Schoentaube J., Tatge H., Olling A., Just I. (2008) J. Med. Microbiol. 57, 765–770 [DOI] [PubMed] [Google Scholar]
- 7.Frisch C., Gerhard R., Aktories K., Hofmann F., Just I. (2003) Biochem. Biophys. Res. Commun. 300, 706–711 [DOI] [PubMed] [Google Scholar]
- 8.Greco A., Ho J. G., Lin S. J., Palcic M. M., Rupnik M., Ng K. K. (2006) Nat. Struct. Mol. Biol. 13, 460–461 [DOI] [PubMed] [Google Scholar]
- 9.von Eichel-Streiber C., Sauerborn M. (1990) Gene 96, 107–113 [DOI] [PubMed] [Google Scholar]
- 10.Barth H., Pfeifer G., Hofmann F., Maier E., Benz R., Aktories K. (2001) J. Biol. Chem. 276, 10670–10676 [DOI] [PubMed] [Google Scholar]
- 11.Qa'Dan M., Spyres L. M., Ballard J. D. (2000) Infect. Immun. 68, 2470–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C. (2007) Nature 446, 415–419 [DOI] [PubMed] [Google Scholar]
- 13.Egerer M., Giesemann T., Jank T., Satchell K. J., Aktories K. (2007) J. Biol. Chem. 282, 25314–25321 [DOI] [PubMed] [Google Scholar]
- 14.Sheahan K. L., Cordero C. L., Satchell K. J. (2007) EMBO J. 26, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prochazkova K., Satchell K. J. (2008) J. Biol. Chem. 283, 23656–23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupardus P. J., Shen A., Bogyo M., Garcia K. C. (2008) Science 322, 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 19.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 21.Rupnik M., Pabst S., Rupnik M., von Eichel-Streiber C., Urlaub H., Söling H. D. (2005) Microbiology 151, 199–208 [DOI] [PubMed] [Google Scholar]
- 22.Egerer M., Giesemann T., Herrmann C., Aktories K. (2009) J. Biol. Chem. 284, 3389–3395 [DOI] [PubMed] [Google Scholar]
- 23.Hardy J. A., Lam J., Nguyen J. T., O'Brien T., Wells J. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macbeth M. R., Schubert H. L., Vandemark A. P., Lingam A. T., Hill C. P., Bass B. L. (2005) Science 309, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei J., Lupardus P. J., Garcia K. C., Grishin N. V. (2009) Protein Sci. 18, 856–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volkman B. F., Lipson D., Wemmer D. E., Kern D. (2001) Science 291, 2429–2433 [DOI] [PubMed] [Google Scholar]
- 27.Carter P., Wells J. A. (1988) Nature 332, 564–568 [DOI] [PubMed] [Google Scholar]
- 28.Hilvert D. (2000) Annu. Rev. Biochem. 69, 751–793 [DOI] [PubMed] [Google Scholar]
- 29.Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.