Abstract
Prion propagation involves a conformational transition of the cellular form of prion protein (PrPC) to a disease-specific isomer (PrPSc), shifting from a predominantly α-helical conformation to one dominated by β-sheet structure. This conformational transition is of critical importance in understanding the molecular basis for prion disease. Here, we elucidate the conformational properties of a disulfide-reduced fragment of human PrP spanning residues 91–231 under acidic conditions, using a combination of heteronuclear NMR, analytical ultracentrifugation, and circular dichroism. We find that this form of the protein, which similarly to PrPSc, is a potent inhibitor of the 26 S proteasome, assembles into soluble oligomers that have significant β-sheet content. The monomeric precursor to these oligomers exhibits many of the characteristics of a molten globule intermediate with some helical character in regions that form helices I and III in the PrPC conformation, whereas helix II exhibits little evidence for adopting a helical conformation, suggesting that this region is a likely source of interaction within the initial phases of the transformation to a β-rich conformation. This precursor state is almost as compact as the folded PrPC structure and, as it assembles, only residues 126–227 are immobilized within the oligomeric structure, leaving the remainder in a mobile, random-coil state.
Prion diseases, such as Creutzfeldt-Jacob and Gerstmann-Sträussler-Scheinker in humans, scrapie in sheep, and bovine spongiform encephalopathy in cattle, are fatal neurological disorders associated with the deposition of an abnormally folded form of a host-encoded glycoprotein, prion (PrP)2 (1). These diseases may be inherited, arise sporadically, or be acquired through the transmission of an infectious agent (2, 3). The disease-associated form of the protein, termed the scrapie form or PrPSc, differs from the normal cellular form (PrPC) through a conformational change, resulting in a significant increase in the β-sheet content and protease resistance of the protein (3, 4). PrPC, in contrast, consists of a predominantly α-helical structured domain and an unstructured N-terminal domain, which is capable of binding a number of divalent metals (5–12). A single disulfide bond links two of the main α-helices and forms an integral part of the core of the structured domain (13, 14).
According to the protein-only hypothesis (15), the infectious agent is composed of a conformational isomer of PrP (16) that is able to convert other isoforms to the infectious isomer in an autocatalytic manner. Despite numerous studies, little is known about the mechanism of conversion of PrPC to PrPSc. The most coherent and general model proposed thus far is that PrPC fluctuates between the dominant native state and minor conformations, one or a set of which can self-associate in an ordered manner to produce a stable supramolecular structure composed of misfolded PrP monomers (3, 17). This stable, oligomeric species can then bind to, and stabilize, rare non-native monomer conformations that are structurally complementary. In this manner, new monomeric chains are recruited and the system can propagate.
In view of the above model, considerable effort has been devoted to generating and characterizing alternative, possibly PrPSc-like, conformations in the hope of identifying common properties or features that facilitate the formation of amyloid oligomers. This has been accomplished either through PrPSc-dependent conversion reactions (18–20) or through conversion of PrPC in the absence of a PrPSc template (21–25). The latter approach, using mainly disulfide-oxidized recombinant PrP, has generated a wide range of novel conformations formed under non-physiological conditions where the native state is relatively destabilized. These conformations have ranged from near-native (14, 26, 27), to those that display significant β-sheet content (21, 23, 28–33). The majority of these latter species have shown a high propensity for aggregation, although not all are on-pathway to the formation of amyloid. Many of these non-native states also display some of the characteristics of PrPSc, such as increased β-sheet content, protease resistance, and a propensity for oligomerization (28, 29, 31) and some have been claimed to be associated with the disease process (34).
One such PrP folding intermediate, termed β-PrP, differs from the majority of studied PrP intermediate states in that it is formed by refolding the PrP molecule from the native α-helical conformation (here termed α-PrP), at acidic pH in a reduced state, with the disulfide bond broken (22, 35). Although no covalent differences between the PrPC and PrPSc have been consistently identified to date, the role of the disulfide bond in prion propagation remains disputed (25, 36–39). β-PrP is rich in β-sheet structure (22, 35), and displays many of the characteristics of a PrPSc-like precursor molecule, such as partial resistance to proteinase K digestion, and the ability to form amyloid fibrils in the presence of physiological concentrations of salts (40).
The β-PrP species previously characterized, spanning residues 91–231 of PrP, was soluble at low ionic strength buffers and monomeric, according to elution volume on gel filtration (22). NMR analysis showed that it displayed radically different spectra to those of α-PrP, with considerably fewer observable peaks and markedly reduced chemical shift dispersion. Data from circular dichroism experiments showed that fixed side chain (tertiary) interactions were lost, in contrast to the well defined β-sheet secondary structure, and thus in conjunction with the NMR data, indicated that β-PrP possessed a number of characteristics associated with a “molten globule” folding intermediate (22). Such states have been proposed to be important in amyloid and fibril formation (41). Indeed, antibodies raised against β-PrP (e.g. ICSM33) are capable of recognizing native PrPSc (but not PrPC) (42–44). Subsequently, a related study examining the role of the disulfide bond in PrP folding confirmed that a monomeric molten globule-like form of PrP was formed on refolding the disulfide-reduced protein at acidic pH, but reported that, under their conditions, the circular dichroism response interpreted as β-sheet structure was associated with protein oligomerization (45). Indeed, atomic force microscopy on oligomeric full-length β-PrP (residues 23–231) shows small, round particles, showing that it is capable of formation of oligomers without forming fibrils (35). Notably, however, salt-induced oligomeric β-PrP has been shown to be a potent inhibitor of the 26 S proteasome, in a similar manner to PrPSc (46). Impairment of the ubiquitin-proteasome system in vivo has been linked to prion neuropathology in prion-infected mice (46).
Although the global properties of several PrP intermediate states have been determined (30–32, 35), no information on their conformational properties on a sequence-specific basis has been obtained. Their conformational properties are considered important, as the elucidation of the chain conformation may provide information on the way in which these chains pack in the assembly process, and also potentially provide clues on the mechanism of amyloid assembly and the phenomenon of prion strains. As the conformational fluctuations and heterogeneity of molten globule states give rise to broad NMR spectra that preclude direct observation of their conformational properties by NMR (47–50), here we use denaturant titration experiments to determine the conformational properties of β-PrP, through the population of the unfolded state that is visible by NMR. In addition, we use circular dichroism and analytical ultracentrifugation to examine the global structural properties, and the distribution of multimeric species that are formed from β-PrP.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Preparation
The protein construct used in this study comprised residues 91–231 of human prion protein (huPrP91–231). Expression and purification of 15N- and 13C/15N-labeled protein for NMR study was carried out as described previously (6). Samples were stored in 6 m guanidinium hydrochloride, 100 mm EDTA, pH 8.0 at −20 °C, prior to use. Protein samples were converted from the normal α-helical (α-PrP) conformation to the β-sheet conformation (β-PrP), by reducing the protein disulfide bond and refolding at acidic pH as previously described (22). Briefly, purified α-PrP was denatured by the addition of guanidinium hydrochloride to 6 m in the presence of 100 mm DTT, refolded under reducing conditions by dialysis against 10 mm sodium acetate, 2 mm DTT, pH 4, and subject to ultracentrifugation at 150,000 × g for 4 h. The yield for conversion to β-PrP was 88%, calculated prior to ultracentrifugation, and 27% after.
13C/15N-labeled protein samples for the backbone assignment of the urea-denatured unfolded state of PrP were buffer exchanged by repeated concentration/dilution in Amicon 50 pressure cells into 10 mm sodium acetate, 2 mm sodium azide, pH 4.0, 2 mm DTT (10% D2O (v/v)) containing 5 m urea. Sample protein concentrations were 1.1 mm.
NMR Spectroscopy
NMR spectra were acquired at 298 K on Bruker DRX-600 and DRX-800 spectrometers equipped with 5-mm 13C/15N/1H triple-resonance probes. Proton chemical shifts were referenced to 1 mm TSP added to the samples. 15N and 13C chemical shifts were calculated relative to TSP, using the gyromagnetic ratios of 15N, 13C, and 1H (15N/1H = 0.101329118, 13C/1H = 0.25144953). NMR data were processed and analyzed on Linux Workstations using Felix 2004 (Accelrys, San Diego, CA) software.
Backbone Resonance Assignments of the Unfolded States of Prion Protein
Backbone resonance assignments were achieved using the standard suite of triple resonance experiments (HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH) (51–54). Peak picking was performed semimanually. The backbone assignment methodology used the “asstools” set of assignment programs (55). The default option and values within the program were used, with the standard table of Cα and C′ chemical shifts replaced with a table containing ranges of 0.5 ppm from random coil values. The long 13C′ and 15N T2 relaxation times allowed long 13C′ (180 ms) and 15N (69 ms) acquisition times, giving good resolution in these dimensions in the HNCO and HN(CA)CO spectra. Cα and Cβ matches were used mainly for residue-type and preceding residue-type identification. All peaks were assigned apart from residues 146–148, 207–208, and 211–212.
Urea Titration of Prion Protein Monitored by NMR Spectroscopy
For the measurement of backbone 13C chemical shifts at varying urea concentrations, 500 μm samples of 13C/15N-labeled β-PrP in 10 mm sodium acetate, 2 mm sodium azide, 1 mm TSP, 2 mm DTT, pH 4.0 (10% D2O (v/v)), were titrated with aliquots of freshly prepared 9 m urea in the same buffer. Following each addition of denaturant, samples were equilibrated for 60 min at 298 K, before being placed in the magnet. Cα and C′ chemical shifts were determined through HN(CO)CA and HNCO experiments, respectively (51, 53, 54), acquired at 298 K. As urea hydrolyzes, resulting in the production of ammonia, the pH of samples was found to increase over the course of the acquisition of triple resonance spectra (2 to 4 days). (It was not possible to significantly increase the buffering capacity of the sample as β-PrP is prone to aggregation at high ionic strength.) Consequently, to minimize the change in sample pH over the range of urea concentrations monitored, the titration was carried out in three separate titrations, with Cα and C′ chemical shifts were determined at the following urea concentrations: 0.61, 0.79, 1.10, and 1.51 m; 1.53, 2.00, 2.50, 3.01, and 3.50 m; and 3.48, 3.98, 4.53, and 5.23 m.
As the acquisition of 1H-15N HSQC spectra is considerably faster than that of triple resonance spectra it was possible to acquire HN and 15N chemical shifts independently in single 1H-15N HSQC titrations. For these, samples of 50 μm and 500 μm 15N-labeled β-PrP in the same buffer described above were titrated in an analogous manner to the 13C/15N titrations. 1H-15N HSQC spectra were acquired using sensitivity enhanced pulsed field gradient coherence selection (54, 56) with acquisition times of 273 and 128 ms in the 1H and 15N dimensions, respectively. The intensities of resolved HSQC resonances were measured and normalized to the intensity of the backbone resonance of residue Gly-94, which is unstructured in α-PrP and β-PrP, and whose intensity remained approximately constant throughout the titration. The modulus of the 1H/15N chemical shift differences (Dmod) was calculated in the following way; Dmod = sqrt ((Ddhn)2 + (Ddn/10)2), where Ddhn and Ddn are the chemical shift differences in 1H and 15N, respectively.
Sample Preparation for Concurrent NMR/CD/Analytical Ultracentrifugation (AUC) Experiments
β-PrP samples for simultaneous CD and AUC analysis were prepared in various concentrations of urea (0.0–5.0 m) in 10 mm sodium acetate, 2 mm DTT, 2 mm sodium azide, pH 4.0, by dilution of the PrP stock solution with buffer A (10 mm sodium acetate, pH 4.0, 2 mm DTT, 2 mm sodium azide) and buffer B (9 m urea in 10 mm sodium acetate, pH 4.0, 2 mm DTT, 2 mm sodium azide) as appropriate. Samples were then concentrated to 50 μm PrP using Vivaspin 6 spin concentrators (Sartorius), and the flow-through used as the matched buffer reference for CD and AUC experiments.
CD Spectroscopy
Far-UV CD spectra were acquired on a JASCO J-715 spectropolarimeter using 0.1-mm path length cuvettes and typically 10 accumulations, with a bandwidth of 1 nm and integration 1 s−1. CD spectra were acquired both before and after the ultracentrifugation, and at 50 μm protein concentration in various concentrations of urea, as described below.
Equilibrium Denaturation Data Monitored by CD
The amide CD absorption of 50 μm β-PrP in 10 mm sodium acetate, 2 mm sodium azide, 2 mm DTT, pH 4.0, was recorded in varying concentrations of urea as previously described (6, 13). The mean residue ellipticity signal ([θ]r) at 215 nm was converted to the proportion of molecules in the unfolded state, αU, according to the relationship,
where [θ]r0M and [θ]r5M are the mean residue ellipticity values for the beginning and end of the urea denaturation transition (i.e. 0 and 100%, respectively). The normalized data were then fitted to a two-state unfolding equation,
where m represents the sensitivity of the unfolding transition to denaturant, K(N/U) represents the equilibrium constant between the native and unfolded states, and D is the denaturant concentration.
The two-state unfolding model is not formally correct for the observed β-PrP CD unfolding transition (see “Results”), as the data represent a transition between monomeric species at high urea concentration and multimeric species at lower urea concentrations. The form of the CD transition does nevertheless follow that for two-state unfolding and therefore the data are fitted to the two-state unfolding model to derive minimum and maximum values for [θ]r, representing the signal of folded and unfolded species, regardless of association state. These values were then used to derive the proportion of folded species at each urea concentration.
Analytical Ultracentrifugation
Sedimentation velocity AUC experiments were carried out using a Beckman Optima XL-1 analytical ultracentrifuge. Samples prepared for parallel CD and sedimentation velocity analysis were loaded into Beckman AUC sample cells with 12-mm optical path two-channel centerpieces, with the matched buffer in the reference sector. Cells were loaded into a AnTi-50 rotor and spun at 50,000 rpm; scans were acquired using both interference and absorbance optics (280 nm) at 10-min intervals over a period of 16 h. The sedimentation profiles were analyzed using the software SEDFIT (version 11.3) (57). Partial specific volume (v̄) for huPrP91-231 was calculated from the amino acid sequence using SEDNTERP software. Buffer densities and viscosities were measured using an Anton Paar DMA5000 density meter and an Anton Paar AMVn automated microviscometer, respectively. Sedimentation velocity data were analyzed using the c(s) method of distribution (57) to characterize the sedimentation coefficient distribution of all species present in solution. The proportion of the monomer in each sample was calculated by integration of the peak between 1 and 2 Svedbergs in the c(s) distribution.
RESULTS
General Characteristics of β-PrP NMR Spectra
In the absence of denaturant there is a substantial loss of both signal intensity and chemical shift dispersion observed in the NMR spectra of β-PrP (Fig. 1), when compared with α-PrP (58). Under native conditions only 38 of the 137 possible resonances of β-PrP are observable; features consistent with the formation of a partially folded molten globule intermediate (47–50). Previously, we assigned the visible NMR resonances of β-PrP to the region that is unstructured in α-PrP (residues 91–126 and 229–231); the signals from the remainder of the protein being too broad to be observed (58). Thus, any structural rearrangements that have occurred in β-PrP are within the region of the protein that forms the folded core of both α-PrP and PrPC. The N-terminal residues of β-PrP show little chemical shift dispersion and display chemical shift values close to those expected for random coil; these residues also show chemical shift values that are highly similar to those observed in native α-PrP (Fig. 1B). The intensities of N-terminal residue peaks in β-PrP are also similar to those of α-PrP. From 15N studies on α-PrP (6, 59), we can conclude that, as in α-PrP, these residues are predominantly unstructured (58). This is in contrast to full-length β-PrP (residues 23–231), where residues 105–210 appear to be engaged in intra- and/or intermolecular interactions (35). The NMR characteristics of β-PrP appear similar to other partially folded molten globule and oligomeric states of PrP (30, 32, 33), which also contain a predominantly disordered N-terminal region (residues 91–126) and a line broadened, unobservable region corresponding to the folded domain of α-PrP. The latter intermediates are, however, unlike β-PrP in that they are formed with the native PrPC disulfide bond intact; also they have been shown to be off-pathway with respect to the formation of amyloid from native α-PrP (28, 29).
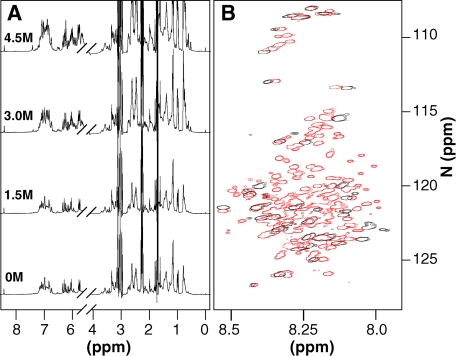
FIGURE 1.
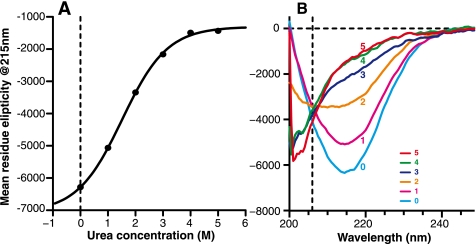
Equilibrium denaturation of β-PrP. A, one-dimensional NMR spectra of 500 μm 15N-labeled β-PrP at increasing urea concentrations. B, 1H-15N HSQC spectra of 15N-labeled β-PrP in the absence of denaturant (black) and the denatured state in 5.2 m urea (red). In the absence of denaturant, the majority of resonances of β-PrP are not visible due to extensive line broadening (only 38 of the possible 137 resonances). Those that are visible arise from the N and very C terminus (residues 91–126, 229–231), which are predominantly unstructured in β-PrP.
Assignment of Urea-denatured State
To determine the structural characteristics of β-PrP on a per residue basis, it is necessary to recover the line-broadened signals, a task that is often readily achieved through the addition of a chemical denaturant (50, 60). Previous experiments demonstrated that β-PrP is prone to aggregation at high ionic strength and therefore urea was used here as an alternative to guanidine hydrochloride, to minimize any aggregation (58). One-dimensional 1H and two-dimensional 1H-15N HSQC spectra of β-PrP were recorded at urea concentrations ranging from 0 to 5.2 m (Fig. 1). By 5.0 m urea all the amide resonances expected for the fully denatured protein are observable and, therefore, the unfolded state is judged to be the predominant species. A backbone and Cβ NMR assignment (Cα, Cβ, C′ (carbonyl), HN, NH) was obtained for the denatured state using a standard suite of triple-resonance NMR experiments (14, 51–54). Sequential assignments were achieved mainly through linking of preceding and intraresidue carbonyl (C′) chemical shifts, utilizing the greater chemical shift dispersion of the 13C′ and 15N resonances (61), with Cα and Cβ matches used mainly for residue-type and preceding residue-type identification. The chemical shifts of the Cα, Cβ, and C′ resonances were found to be within the ranges previously reported for random coils (62) and, therefore, indicated that the protein was predominantly unfolded.
Structured Regions within β-PrP
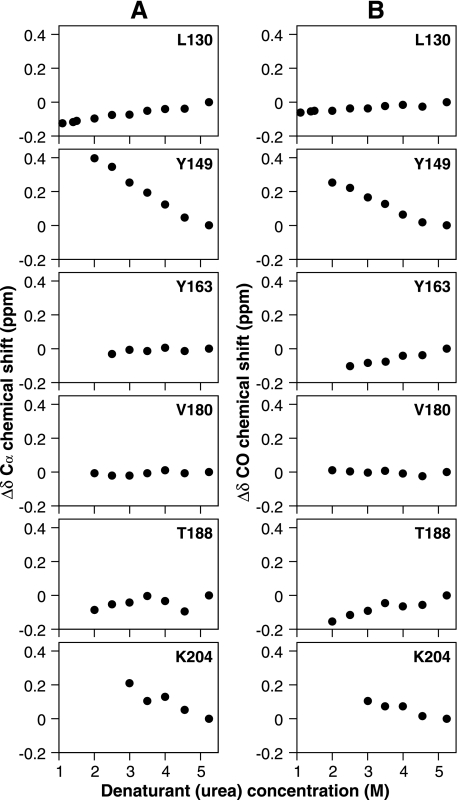
To determine the conformational properties of β-PrP in the absence of denaturant, the chemical shifts of Cα, C′, HN, and NH resonances were determined at concentrations of urea between 0 and 5.2 m. It is possible to follow the movement in NMR resonance frequencies between denaturant concentrations, as the conformational transitions taking place are fast relative to the NMR chemical shift differences, leading to averaging of the NMR signals (the fast exchange regime) (63). By tracking the change in chemical shift and disappearance of individual resonances as a function of the urea concentration (and the consequent change in the relative populations of the unfolded state and β-PrP), it was possible to identify structural regions in β-PrP, as 13C (in particular Cα and C′) chemical shifts provide a reliable indication of the structural preference of secondary structure elements (62, 64). Fig. 2 shows the denaturant concentration dependence of Cα and C′ chemical shift changes observed for representative residues from the region of β-PrP that are unobservable in the absence of denaturant. The residues displayed are found within regions of well defined secondary structure in α-PrP, with Leu-130 and Tyr-163 present in the short anti-parallel β-sheet, and the remainder found in its three α-helices. As can be seen in Fig. 2, the residues exhibit wide differences in both the direction and magnitude of chemical shift changes for both Cα and C′ resonances. For instance, Tyr-149 and Lys-204 (which are in helices I and III of α-PrP) both experience strong downfield (positive) shifts as the denaturant is decreased. Such downfield chemical shift changes for Cα and C′ indicate that these regions of β-PrP exhibit a preference for backbone torsional angles (ϕ and ψ) in the α-helical region (62, 65). In contrast Val-180, found in α-helix II of α-PrP, exhibits very little change in chemical shift throughout the course of the titration, whereas Leu-130 and Tyr-163 (situated in the α-PrP β-sheet) and Thr-188 (at the end of helix II in α-PrP) show small upfield (negative) shifts.
FIGURE 2.
Chemical shift changes (Δδ) of representative β-PrP peaks in HNCO and HN(CO)CA spectra. Cα (A) and C′ (carbonyl) (B) chemical shift changes (ppm) as a function of urea. Rapid loss of peak intensity (as illustrated in Fig. 8) precluded measurement of data for certain residues at lower denaturant concentrations. A downfield (positive) chemical shift change is indicative of formation of α-helical structure, with upfield (negative) shifts being indicative of β-sheet.
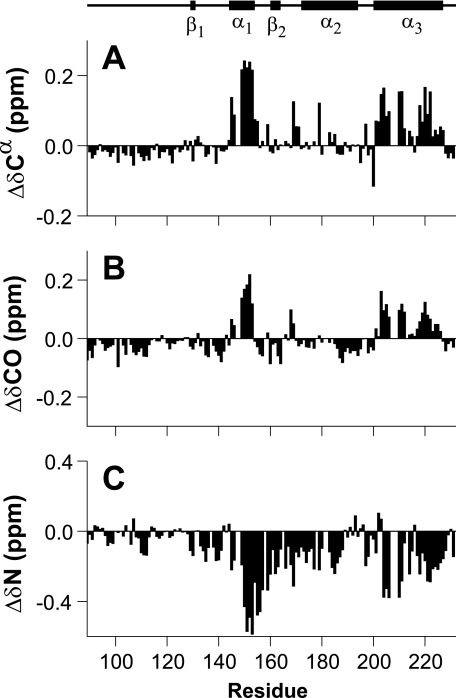
A comparison of the changes in Cα and C′ chemical shift from throughout the protein are shown in Fig. 3. The changes shown are those occurring between 3.5 and 5.2 m urea, as below 3.5 m urea a significant number of resonances (≈19 in the triple resonance spectra) became unobservable through line broadening. Hence, the chosen range of urea concentrations is a compromise between the magnitude of the observed chemical shift changes and the number of observable residues. (The denaturant-dependent loss of peak intensities is discussed in greater detail later.) The consistent sign and distribution of the chemical shift changes as the population of β-PrP increases (Fig. 3) indicate that many of the residues that adopt an α-helical conformation in the native structure of α-PrP also adopt a helical conformation in β-PrP (residues 144–154 and 200–228). These regions of the protein correspond to helices I and III of α-PrP (Fig. 4). Intriguingly, the region corresponding to helix II of α-PrP (residues 172–194) shows little indication of adopting an α-helical conformation in β-PrP. This is particularly interesting as helix III, to which helix II is disulfide-bonded in α-PrP, still exhibits distinct helix propensity. These observations are consistent with data showing that the biochemical properties of ovine PrPSc correlate with the ease of unwinding of helix II (66), and are also consistent with the observed high intrinsic helix propensity of residues within helix I (residues 144–154), which is suggestive of stability against environmental changes, and therefore a lower likelihood of initiating the transition from PrPC into PrPSc (67).
FIGURE 3.
Chemical shift changes of backbone resonances of β-PrP between 5.2 and 3.5 m urea. Cα (A) and carbonyl (C′) (B) chemical shift changes. 13C chemical shifts accurately indicate secondary structure elements (14, 64, 83). The regions that exhibit the largest chemical shift changes are those that are found in helices I and III of α-PrP, and which retain their α-helicity in β-PrP. A significant number of residues (∼19) are unobservable below 3.5 m urea because of line broadening of resonances, precluding measurement of data. C, 15N chemical shift changes. Although not as diagnostic of secondary structure as 13C chemical shifts, the degree of 15N chemical shift change reflects the change in the structural ensemble, and confirms 13C chemical shift data that the conformational changes occurring in the region comprising helix II of α-PrP are less than the other helical regions.
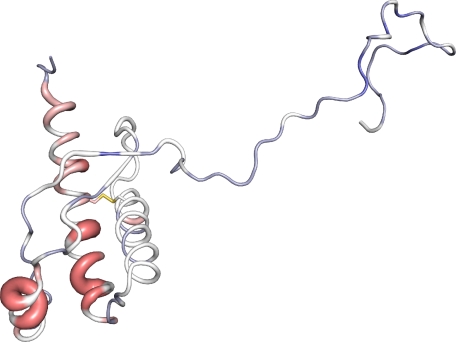
FIGURE 4.
Cα chemical shift changes in β-PrP between 5.2 and 3.5 m urea displayed on the three-dimensional structure of PrPC (6, 76). Residues that show downfield Cα chemical shift changes, and thus retain α-helicity within β-PrP are colored red, whereas those that are in a more extended conformation are colored from white to blue. The magnitude of the chemical shift changes are also shown by the width of the backbone representation. This figure was prepared using PyMOL (84).
For these α-helical regions, the 13C chemical shift changes observed are in the range of 0.1–0.25 ppm for Cα resonances, and 0.05–0.23 ppm for C′ resonances (Fig. 3). In contrast, the chemical shift changes observed in these regions following the unfolding of α-PrP are 3.0–5.0 ppm for Cα, and 1.7–3.5 ppm for C′ resonances (14). Thus, the chemical shift changes observed in the unfolding of these regions of β-PrP between 3.5 and 5.2 m urea are ∼5% of the α-PrP native to denatured state unfolding transition. The low denaturant dependence for these changes (Fig. 2) indicate that the free energy and m-value changes associated with the folding of monomeric β-PrP are relatively small, and that the gain in structure is not associated with a concerted large scale burial of hydrophobic groups. This is consistent with the formation of a partly folded state. Indeed, the chemical shift changes observed show an approximately linear dependence with denaturant, indicating that the transition between the partially folded and denatured state ensembles is not fully resolved, and is incomplete in the range of accessible denaturant concentrations. Thus it is not possible to use the chemical shift changes to extrapolate to the end point of the transition. However, by extrapolating the observed linear chemical shifts to 0 m urea this would indicate that in the absence of denaturant, helices I and III in β-PrP would retain at least ∼30% of their native α-helicity (14) (Fig. 2) in zero denaturant. In comparison, the native-like pH 4 molten-globule state of apomyoglobin retains between 50 and 80% native structure for its four most structured helices (68), and 10–20% native structure in its pH 2.3 acid-denatured state (69).
For the remainder of the line-broadened region of β-PrP, Cα chemical shift changes provide little consistent information on the preferred conformational distribution, whereas the majority of C′ shift changes are of a similar sign and magnitude to those observed for the second strand of the α-PrP β-sheet (Fig. 3). Of interest are the C′ chemical shift changes observed for residues 186–195, which show small, but consistent C′ upfield shift movements, suggesting a small preference for an extended conformation. Given the small chemical shift changes observed it is not possible to conclude whether this region is overwhelmingly dominated by a β-type conformation, but as this region comprises the C terminus of helix II of α-PrP (5, 6), it thus displays a more extended conformation in β-PrP compared with that seen in α-PrP (Fig. 4). This region of the protein provides the hinge in a three-dimensional domain-swapped dimer of human PrP (39), and appears to undergo a marked conformational change in ovine PrPSc (66), suggesting a possible role for this region of the protein in the conformational change to PrPSc. It is also interesting to observe that the residues immediately preceding helix I show similar upfield C′ chemical shifts. Peptides that contain residues 138–141 but not helix I, form fibrils more readily than peptides that are devoid of this region (70), and it has been suggested that this region of the protein may provide a nucleus for the oligomerization (67).
Also of note are the measurable 13C shift changes in the unstructured N terminus of the protein (residues 91–126). This region of the protein is predominantly unstructured, as indicated by 15N relaxation rates (6, 14). However, previous studies have indicated that this region of the protein may be exchanging between more structured or collapsed states, as it exhibits differential peak intensities (14), and residues 105–126 appear to be capable of forming intra- and/or intermolecular interactions in full-length β-PrP oligomers (35). The chemical shift changes observed here suggest some change in conformational preferences for this region of the protein, the small Cα and C′ chemical shift changes indicating that it may be sampling more extended conformations, within β-PrP. This region becomes partially secluded in PrPSc (71), and undergoes a conformational change that is essential in prion propagation (72, 73).
Comparison of CD Signal and NMR Chemical Shift Changes
To obtain an overall view of secondary structure changes in β-PrP as a function of denaturant concentration, the dependence of the CD spectrum was measured over the same urea concentration range as the NMR spectra (Fig. 5). In the absence of denaturant the CD response is dominated by a β-sheet-rich species, and is similar to a soluble oligomeric form of PrP that was induced by heat treatment of α-PrP in the presence of phospholipids (31). At 5 m urea, however, the CD spectrum corresponds to a largely unfolded species, in line with the NMR data at high urea concentrations. The CD response at intermediate urea concentrations showed that, in contrast to α-PrP, the denaturation of β-PrP demonstrated little evidence of a steep unfolding transition. Instead, there was a gradual loss of secondary structure signal observed as the denaturant concentration was increased (Fig. 5A), with a midpoint for the unfolding transition at ∼1.8–2.5 m urea. The lack of a steep unfolding transition is in keeping with the idea that β-PrP in the absence of denaturant is a molten globule that lacks significant tertiary structure (47–50). The unfolding of β-PrP, however, retains characteristics of a two-state transition, as indicated by the apparent isodichroic point at 206 nm in the far-UV CD denaturation (Fig. 5B), implying that the primary CD-observable species are the β-sheet-rich species and the unfolded state.
FIGURE 5.
Equilibrium denaturation of β-PrP monitored by far-UV circular dichroism. A, equilibrium unfolding profile, monitored by mean residue ellipticity at 215 nm. The solid line shows a non-linear least squares fit to a two-state model for unfolding. This is for illustrative purposes, as the CD signal is reporting a transition between a monomeric and oligomeric species (see “Experimental Procedures”). B, far-UV CD spectra of β-PrP at increasing urea concentrations (0–5 m). Note the isodichroic point at 206 nm, which is strongly indicative of an unfolding transition involving two states.
Analysis of Sample Homogeneity
The lack of pronounced NMR chemical shift changes consistent with a high proportion of extended β-sheet structure within β-PrP is at variance with the strong β-sheet signal reported by CD. Additionally, the range of urea concentrations over which chemical shift changes can be reliably determined for many resonances corresponds to concentrations at which greater than 90% of the β-sheet signal observed by CD has been lost (Fig. 5A). The line broadening of signals arising from the C terminus of β-PrP is consistent with β-PrP being a molten globule; however, line broadening could also be due to aggregation or multimerization of the molecule (45), resulting in a molecule of high molecular weight and, thus, long rotational correlation time (63). To differentiate between these possibilities, sedimentation velocity AUC was employed to identify species present during the β-PrP denaturation. The advantage of this technique over other commonly used techniques for identifying multimeric species, such as size exclusion chromatography and the measurement of hydrodynamic radius by pulsed-field gradient NMR, is that all species present within a sample can be observed (74), and complications arising from subtly different solution conditions or calibrations are avoided.
For this experiment a single 50 μm sample of β-PrP was prepared using the previously established β-PrP preparation protocol (58), and NMR, CD, and AUC denaturation data acquired concurrently, within 24 h of sample preparation, to minimize time-dependent aggregation. In this way, the correlation between aggregation state, CD response, and NMR chemical shift change could be unambiguously established.
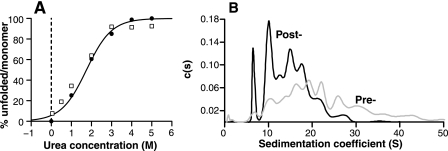
The CD response and denaturant-dependent loss of NMR signal of this sample was consistent with previously prepared samples, with a minimum at 215 nm in the absence of denaturant and a midpoint for the unfolding transition of ∼1.8–2.5 m urea (Fig. 6A). The sedimentation coefficient distributions showed that β-PrP did not sediment as a single species, but as a mixture of oligomers in the absence of denaturant, the majority (>98%) of β-PrP being multimeric (Fig. 6B). A broad distribution of species was found between 1 and ∼30 S, the largest species being of the order of 2 MDa. The high speed centrifugation spin, which is part of the β-PrP purification protocol (see “Experimental Procedures”), was found to remove species greater than ∼30 S (see Fig. 6B). The β-PrP sample could therefore be said to consist primarily of soluble oligomers, the large molecular size of which resulting in NMR signals broadening to such an extent that they are unobservable. A small (<2%) proportion of protein was found to have a sedimentation coefficient 1.4 S, however, the CD spectrum of this species could not be determined unambiguously given its small proportion within the sample. As the denaturant concentration is increased, the population of high molecular weight oligomers decreased, and a concomitant increase in the proportion of the 1.4 S species was observed; by 3 m urea ∼90% of the sample was composed of the 1.4 S species (Fig. 6A). This indicates that this species is the monomer. Additionally, as the value of its sedimentation coefficient does not change significantly with increasing urea concentration, this shows that it has a conformation in the absence of denaturant not dissimilar to that found under denaturing conditions. Comparison with the CD denaturation profile indicated that the denaturant-dependent loss of oligomers was co-incident with the loss of the CD signal of a β-sheet-rich species (Fig. 6A) and, thus, the oligomeric species give rise to the majority of the β-sheet CD response.
FIGURE 6.
A, percentage of monomeric species populated in the equilibrium denaturation of β-PrP, as monitored by sedimentation velocity AUC (black circles). The close association of the proportion of monomer with the loss of CD signal (open squares) indicates that the β-sheet secondary structure is lost before or is co-incident with the dissociation into monomer. B, continuous c(s) distribution characterizing the sedimentation coefficient of all species in a 50 μm sample of 15N-labeled β-PrP, prior to, and following the high speed centrifugation spin used in β-PrP purification (see “Experimental Procedures”).
The form of the CD and AUC transitions follow that for a two-state unfolding with low cooperativity, despite the change in the association state of the protein, with apparently solely the oligomeric (β-sheet-containing), and monomeric, unfolded species populated. The apparent two-state nature of the CD unfolding transition, as shown by the isodichroic point at 206 nm (Fig. 5B), indicates that the CD absorption spectrum of the lower molecular weight oligomers is indistinguishable from that of the larger scale oligomers and, thus, these species adopt very similar extended secondary structures. This conclusion is supported by the observation that the very high molecular weight species that are removed by the high speed spin used in the β-PrP purification exhibit a very similar CD spectrum to the soluble β-PrP (data not shown). Hence, it appears that there is a very similar β-sheet structure adopted throughout the molecular weight range of all the oligomeric species formed. Some indication of the nature of the β-sheet structure can be gleaned from Fourier transform infrared spectra of soluble β-PrP, albeit from mouse, which are similar to those of ordered fibrils but with a less pronounced 1622 cm−1 maximum characteristic of β-amyloid (40), rather than the amide I peak in the region of 1630 to 1643 cm−1, which is typical of natively folded β-sheet proteins (75). This suggests that the β-sheet contacts in the soluble, oligomeric forms of β-PrP are non-native and amyloid-like, and that the β-PrP oligomeric species are direct fibril precursors.
Structured Molten Globule Intermediate within the Denatured-state Ensemble
In contrast to the apparent simplicity of the CD transition, the NMR data indicate that the unfolded state reported by CD displays significant non-uniform behavior as a function of urea concentration, and constitutes a more broadly populated ensemble than expected for a fully denatured protein. Furthermore, the NMR data indicate a distribution of species in the denatured state ensemble containing a sizable population of an intermediate species, which, whereas not apparently containing a substantial β-sheet structure, has a conformation that is radically different than that of native α-PrP. The observed changes in NMR chemical shifts reflect a change in the conformational distribution of the monomeric component of β-PrP as it unfolds, rather than a change in association of the protein, as there are no discernible differences in chemical shift changes observed at reduced (50 versus 500 μm) protein concentration (Fig. 7).
FIGURE 7.
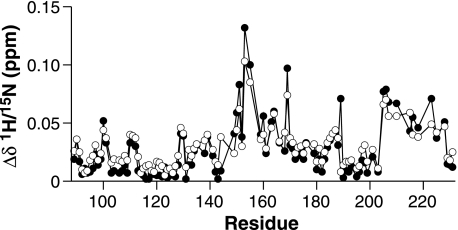
Chemical Shift differences (Δδ) of β-PrP HN and NH resonances between 4.5 and 2.5 m urea, at 500 (black circles) and 50 (open circles) μm β-PrP. A weighted average of both the 1H and 15N chemical shift changes are shown, taking into account the difference in the gyromagnetic ratios of 1H and 15N (see “Experimental Procedures”). No significant differences between the two protein concentrations are observed, thus the changes in chemical shift are not due to a change in protein association but rather due to a change in the conformation of the monomeric protein.
Some indication of the change in the conformational distribution of the denatured state ensemble can be obtained from AUC data, which show that there is an increase in hydrodynamic radius (Rh) of the monomeric species as the denaturant concentration is increased, well beyond the mid-point of the observed unfolding transition. The observed radii were 29, 32, and 34 Å at 3.0, 4.0, and 5.0 m urea, respectively. The Rh value obtained for β-PrP at 3 m urea correlates well with values obtained for both the disulfide-free (28 Å) (45) and disulfide-oxidized (28.3 Å) forms of α-PrP (32). Thus, the Rh for the denatured state ensemble is not significantly larger than that of the native state, and considerably smaller than the calculated Rh of 41.3 Å for the fully unfolded and disulfide-reduced state (32). This is suggestive of a population of a collapsed, partially structured intermediate state within the denatured state ensemble.
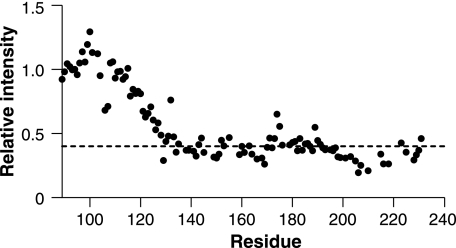
This conclusion is supported by changes in the NMR relaxation properties of nuclei of the line-broadened region of β-PrP (residues 127–228). As the population of β-PrP increases with reduced denaturant, there is a marked loss of NMR signal intensities, with different regions of the protein experiencing markedly different sensitivities to the lowering of denaturant. For example, although the majority of resonances from the line-broadened region of β-PrP are visible at urea concentrations greater than 2.0–2.5 m (approximately the mid-point of the CD unfolding transition), resonances for residues in the middle of helix III rapidly lose intensity, with a number broadening beyond detection by 3.0 m urea (Fig. 8). Broadening of NMR signals, resulting in less intense peaks, can arise in heterogeneous systems due to insufficient time averaging of sampled chemical shifts, because of restricted mobility (typically because of conformational fluctuations on a millisecond to microsecond time scale) (63). This is a common observation in molten globule states, and indicates that this region of the protein is undergoing complex motions on an intermediate time scale. The lack of sensitivity of NMR chemical shift changes to protein concentration show that this phenomenon is not due to a change in the association state of the protein. Furthermore, the line broadening of resonances showed no substantial correlation with the 1H, 15N, or the modulus of the 1H/15N chemical shift changes, indicating that the observed loss of NMR peak intensities in this range of urea concentrations reflect the increased population of a structured intermediate state (within the denatured state ensemble). The non-linearity of the resonance intensity with population, however, make quantification of the population unachievable.
FIGURE 8.
Denaturant dependence of signal intensities. The ratio of backbone (1H/15N) β-PrP resonances in the 1H-15N HSQC spectra acquired at 2.5 and 4.5 m urea are shown, with intensities normalized on the intensity of resonance Gly-94, which is in the unstructured region of β-PrP, and whose intensity and chemical shift changes showed negligible denaturant dependence. The dotted line shows the average signal loss for residues 129–231.
DISCUSSION
The data presented here establish that, under the original conditions in which a reduced and acidified form of recombinant PrP comprising residues 91–231 (termed β-PrP because of its high apparent content of β-sheet structure as measured by circular dichroism), is composed of a mixture of species. Whereas, originally, gel filtration of β-PrP at low ionic strength gave an elution position almost indistinguishable from the PrPC-like α-PrP (the predominant conformation when the disulfide bond is intact and the pH neutral), sedimentation velocity analytical ultracentrifugation reveal that in the absence of denaturant, β-PrP consists predominantly of a broad distribution of soluble oligomeric species, with a majority sedimenting between 6 and 20 Svedbergs. The distribution between oligomers and monomeric species shifts toward the latter on addition of urea as a denaturant.
The lack of a well defined oligomeric species within 91–231 β-PrP contrasts with the distinct spherical oligomers, which were observed following the similar reduction of the disulfide bond and refolding of full-length huPrP (residues 23–231) (35). Furthermore, we observe that residues 91–127 are predominantly unstructured and visible in the NMR spectra of 91–231 β-PrP, in contrast to full-length β-PrP, where residues 105–231 were found to be engaged in intra- and/or intermolecular interactions. This result is intriguing, suggesting a role for the predominantly unstructured peptide segment 23–90 in the conformational transition to a distinct oligomeric species. This is not without precedent, as Lührs et al. (31) have observed that peptide segment 105–120 is required for successful conversion of α-PrP into a soluble oligomeric species, which displays similar CD spectra to that observed for β-PrP, and there is evidence for some residues of the unfolded region of PrP transiently contacting the folded region of PrP (27, 76).
The monomeric species is an ensemble in fast conformational exchange comprising the unfolded state of the protein and a collapsed, molten globule state, the latter indicated by the differential loss of peak intensities at intermediate urea concentrations, a low degree of cooperativity in the unfolding transition, and a compact hydrodynamic radius. The sign and magnitude of the chemical shift changes between the unfolded state and the monomeric molten globule state indicate that two of the three α-helices found in α-PrP retain their preference for an α-helical conformation in the monomeric ensemble, but residues in helix II of α-PrP are in a more extended, random coil-like conformation. A similar study, which used pressure to dissociate oligomers of full-length β-PrP (35), identified the resultant semi-stable monomers with PrPC*, a rare metastable form of non-reduced PrP stabilized at high pressure (27). The most stable region of this I2 intermediate was in the region of the disulfide bond linking helices II and III, which corresponded well with hydrogen exchange data on oxidized PrP (13). Although it is difficult at this stage to correlate these dissociated monomeric species our data suggest that the loss of the disulfide bond in β-PrP has a significant effect on the stability the region surrounding the disulfide bond, and in particular on the stability of helix II.
The lack of stability of helical conformation within helix II fits with a previously proposed model for PrPSc formation (39), where dimerization and domain swapping involving this region of PrP occur early in the process. However, whereas the crystal structure of the α-helical domain-swapped dimer of PrP can be accommodated in β-PrP fibril subunits, the lack of significant α-helicity observed in the far-UV CD spectra of β-PrP oligomers and fibrils is inconsistent with this conformation constituting the fibril subunits (40). Furthermore, EPR measurements of recombinant PrP fibrils are consistent with a parallel, in-register β-sheet arrangement for the monomeric units dominating the conformations within the fibrils (77). Nevertheless, the disordered nature of the β-PrP precursor state parallels the behavior of the precursor states of a number of other amyloidogenic proteins (60, 78–80), where the fluctuating, partially structured nature of these precursors appears to allow more readily the formation of specific intermolecular contacts required for the formation of higher order oligomers and fibrillar structures. Hence, the region surrounding helix II of PrP is a likely source of interaction with the initial phases of the transformation to a β-rich conformation. Indeed, this region of PrP has been identified to be the most amyloidogenic within PrP (81).
Presently, there is uncertainty over the precise cause of prion-mediated neurodegeneration (82), but there is growing evidence that prion protein neurotoxicity may be mediated through toxic oligomers, with smaller intermediate oligomeric species being biologically more active than larger amyloid fibrils (82). In common with native PrPSc, oligomeric β-PrP is a potent inhibitor of the 26 S proteasome, an integral part of the ubiquitin-proteasome system (46), which protects the cell against potentially toxic effects of protein aggregation. In contrast, amyloid fibrils generated from recombinant protein, and from other amyloidogenic proteins, do not elicit such a response. Together with recent data suggesting that prion disease is associated in vivo with dysfunction of the ubiquitin-proteasome system (46), and the observation that antibodies raised against β-PrP are capable of recognizing native PrPSc, but not PrPC (42–44), this suggests that β-PrP is likely to be of biological significance in prion disease. Here, we have established that the oligomeric species that make up the majority of β-PrP share a common β-sheet structure, suggesting that the arrangements of PrP chains within the oligomers is common throughout, and it is the macromolecular organization that is the predominant difference between the components of the oligomeric mixture. Hence, further dissection of the nature of the oligomeric species within β-PrP should lead to a greater understanding of the molecular basis of prion toxicity.
Acknowledgment
We are grateful to Ray Young for assistance in preparation of figures for this manuscript.
This work was supported by the Medical Research Council.
- PrP
- prion protein
- AUC
- analytical ultracentrifugation
- CD
- circular dichroism
- DTT
- 1,4-dithiothreitol
- PrPC
- cellular PrP isoform
- PrPSc
- pathogenic (scrapie) PrP isoform
- PrP91–231
- human prion protein (residues 91–231)
- HSQC
- heteronuclear single quantum coherence
- TSP
- sodium 3-trimethylsilyl-2,2,3,3-(2H4)-propionate.
REFERENCES
- 1.Sipe J. D., Cohen A. S. (2000) J. Struct. Biol. 130, 88–98 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collinge J. (2001) Annu. Rev. Neurosci. 24, 519–550 [DOI] [PubMed] [Google Scholar]
- 4.McKinley M. P., Bolton D. C., Prusiner S. B. (1983) Cell 35, 57–62 [DOI] [PubMed] [Google Scholar]
- 5.Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. (1996) Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 6.Hosszu L. L., Jackson G. S., Trevitt C. R., Jones S., Batchelor M., Bhelt D., Prodromidou K., Clarke A. R., Waltho J. P., Collinge J. (2004) J. Biol. Chem. 279, 28515–28521 [DOI] [PubMed] [Google Scholar]
- 7.Jackson G. S., Murray I., Hosszu L. L., Gibbs N., Waltho J. P., Clarke A. R., Collinge J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98, 8531–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells M. A., Jelinska C., Hosszu L. L., Craven C. J., Clarke A. R., Collinge J., Waltho J. P., Jackson G. S. (2006) Biochem. J. 400, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells M. A., Jackson G. S., Jones S., Hosszu L. L., Craven C. J., Clarke A. R., Collinge J., Waltho J. P. (2006) Biochem. J. 399, 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter E. D., Chattopadhyay M., Millhauser G. L. (2006) Biochemistry 45, 13083–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klewpatinond M., Davies P., Bowen S., Brown D. R., Viles J. H. (2008) J. Biol. Chem. 283, 1870–1881 [DOI] [PubMed] [Google Scholar]
- 12.Davies P., Brown D. R. (2008) Biochem. J. 410, 237–244 [DOI] [PubMed] [Google Scholar]
- 13.Hosszu L. L., Baxter N. J., Jackson G. S., Power A., Clarke A. R., Waltho J. P., Craven C. J., Collinge J. (1999) Nat. Struct. Biol. 6, 740–743 [DOI] [PubMed] [Google Scholar]
- 14.Hosszu L. L., Wells M. A., Jackson G. S., Jones S, Batchelor M., Clarke A. R., Craven C. J., Waltho J. P., Collinge J. (2005) Biochemistry 44, 16649–16657 [DOI] [PubMed] [Google Scholar]
- 15.Griffith J. S. (1967) Nature 215, 1043–1044 [DOI] [PubMed] [Google Scholar]
- 16.Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 17.Collinge J., Clarke A. R. (2007) Science 318, 930–936 [DOI] [PubMed] [Google Scholar]
- 18.Bessen R. A., Kocisko D. A., Raymond G. J., Nandan S., Lansbury P. T., Caughey B. (1995) Nature 375, 698–700 [DOI] [PubMed] [Google Scholar]
- 19.Saborio G. P., Permanne B., Soto C. (2001) Nature 411, 810–813 [DOI] [PubMed] [Google Scholar]
- 20.Supattapone S. (2004) J. Mol. Med. 82, 348–356 [DOI] [PubMed] [Google Scholar]
- 21.Hornemann S., Glockshuber R. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 6010–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson G. S., Hosszu L. L., Power A., Hill A. F., Kenney J., Saibil H., Craven C. J., Waltho J. P., Clarke A. R., Collinge J. (1999) Science 283, 1935–1937 [DOI] [PubMed] [Google Scholar]
- 23.Swietnicki W., Morillas M., Chen S. G., Gambetti P., Surewicz W. K. (2000) Biochemistry 39, 424–431 [DOI] [PubMed] [Google Scholar]
- 24.Nandi P. K., Leclerc E., Nicole J. C., Takahashi M. (2002) J. Mol. Biol. 322, 153–161 [DOI] [PubMed] [Google Scholar]
- 25.Lee S., Eisenberg D. (2003) Nat. Struct. Biol. 10, 725–730 [DOI] [PubMed] [Google Scholar]
- 26.Kuwata K., Li H., Yamada H., Legname G., Prusiner S. B., Akasaka K., James T. L. (2002) Biochemistry 41, 12277–12283 [DOI] [PubMed] [Google Scholar]
- 27.Kachel N., Kremer W., Zahn R., Kalbitzer H. R. (2006) BMC Struct. Biol. 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. (2002) J. Biol. Chem. 277, 21140–21148 [DOI] [PubMed] [Google Scholar]
- 29.Baskakov I. V., Legname G., Gryczynski Z., Prusiner S. B. (2004) Protein Sci. 13, 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Sullivan D. B., Jones C. E., Abdelraheim S. R., Thompsett A. R., Brazier M. W., Toms H., Brown D. R., Viles J. H. (2007) Biochem. J. 401, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luhrs T., Zahn R., Wuthrich K. (2006) J. Mol. Biol. 31, 833–841 [DOI] [PubMed] [Google Scholar]
- 32.Gerber R., Tahiri-Alaoui A., Hore P. J., James W. (2007) J. Biol. Chem. 282, 6300–6307 [DOI] [PubMed] [Google Scholar]
- 33.Gerber R., Tahiri-Alaoui A., Hore P. J., James W. (2008) Protein Sci. 17, 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2004) Science 305, 673–676 [DOI] [PubMed] [Google Scholar]
- 35.Sasaki K., Gaikwad J., Hashiguchi S., Kubota T., Sugimura K., Kremer W., Kalbitzer H. R., Akasaka K. (2008) Prion 2, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turk E., Teplow D. B., Hood L. E., Prusiner S. B. (1988) Eur. J. Biochem. 176, 21–30 [DOI] [PubMed] [Google Scholar]
- 37.Brown P., Liberski P. P., Wolff A., Gajdusek D. C. (1990) Proc. Natl. Acad. Sci. U. S. A. 87, 7240–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann L. M., Caughey B. (1998) Neuroreport 9, 2457–2461 [DOI] [PubMed] [Google Scholar]
- 39.Knaus K. J., Morillas M., Swietnicki W., Malone M., Surewicz W. K., Yee V. C. (2001) Nat. Struct. Biol. 8, 770–774 [DOI] [PubMed] [Google Scholar]
- 40.Tattum M. H., Cohen-Krausz S., Thumanu K., Wharton C. W., Khalili-Shirazi A., Jackson G. S., Orlova E. V., Collinge J., Clarke A. R., Saibil H. R. (2006) J. Mol. Biol. 357, 975–985 [DOI] [PubMed] [Google Scholar]
- 41.Uversky V. N., Fink A. L. (2004) Biochim. Biophys. Acta 1698, 131–153 [DOI] [PubMed] [Google Scholar]
- 42.Khalili-Shirazi A., Quaratino S., Londei M., Summers L., Tayebi M., Clarke A. R., Hawke S. H., Jackson G. S., Collinge J. (2005) J. Immunol. 174, 3256–3263 [DOI] [PubMed] [Google Scholar]
- 43.Khalili-Shirazi A., Kaisar M., Mallinson G., Jones S., Bhelt D., Fraser C., Clarke A. R., Hawke S. H., Jackson G. S., Collinge J. (2007) Biochim. Biophys. Acta 1774, 1438–1450 [DOI] [PubMed] [Google Scholar]
- 44.Paquet S., Langevin C., Chapuis J., Jackson G. S., Laude H., Vilette D. (2007) J. Gen. Virol. 88, 706–713 [DOI] [PubMed] [Google Scholar]
- 45.Maiti N. R., Surewicz W. K. (2001) J. Biol. Chem. 276, 2427–2431 [DOI] [PubMed] [Google Scholar]
- 46.Kristiansen M., Deriziotis P., Dimcheff D. E., Jackson G. S., Ovaa H., Naumann H., Clarke A. R., van Leeuwen F. W., Menéndez-Benito V., Dantuma N. P., Portis J. L., Collinge J., Tabrizi S. J. (2007) Mol. Cell 26, 175–188 [DOI] [PubMed] [Google Scholar]
- 47.Redfield C., Schulman B. A., Milhollen M. A., Kim P. S., Dobson C. M. (1999) Nat. Struct. Biol. 6, 948–952 [DOI] [PubMed] [Google Scholar]
- 48.Schulman B. A., Kim P. S., Dobson C. M., Redfield C. (1997) Nat. Struct. Biol. 4, 630–634 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Alexandrescu A. T., Shortle D. (1995) Philos. Trans. R. Soc. Lond. B Biol. Sci. 348, 27–34 [DOI] [PubMed] [Google Scholar]
- 50.Redfield C. (2004) Methods Mol. Biol. 278, 233–254 [DOI] [PubMed] [Google Scholar]
- 51.Bax A., Ikura M. (1991) J. Biomol. NMR 1, 99–104 [DOI] [PubMed] [Google Scholar]
- 52.Wittekind M., Mueller L. (1993) J. Magn. Reson. Ser. B 101, 201–205 [Google Scholar]
- 53.Muhandiram D. R., Kay L. E. (1994) J. Magn. Reson. Ser. B 103, 203–216 [Google Scholar]
- 54.Kay L. E., Xu G. Y., Yamazaki T. (1994) J. Magn. Reson. Ser. A 109, 129–133 [Google Scholar]
- 55.Reed M. A., Hounslow A. M., Sze K. H., Barsukov I. G., Hosszu L. L., Clarke A. R., Craven C. J., Waltho J. P. (2003) J. Mol. Biol. 330, 1189–1201 [DOI] [PubMed] [Google Scholar]
- 56.Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S. J., Sørensen O. W., Griesinger C. (1994) J. Biomol. NMR 4, 301–306 [DOI] [PubMed] [Google Scholar]
- 57.Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson G. S., Hill A. F., Joseph C., Hosszu L., Power A., Waltho J. P., Clarke A. R., Collinge J. (1999) Biochim. Biophys. Acta 1431, 1–13 [DOI] [PubMed] [Google Scholar]
- 59.Zahn R., von Schroetter C., Wüthrich K. (1997) FEBS Lett. 417, 400–404 [DOI] [PubMed] [Google Scholar]
- 60.McParland V. J., Kalverda A. P., Homans S. W., Radford S. E. (2002) Nat. Struct. Biol. 9, 326–331 [DOI] [PubMed] [Google Scholar]
- 61.Yao J., Dyson H. J., Wright P. E. (1997) FEBS Lett. 419, 285–289 [DOI] [PubMed] [Google Scholar]
- 62.Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 63.Homans S. W. (1989) A Dictionary of Concepts in NMR, Oxford University Press, Oxford [Google Scholar]
- 64.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 65.Wishart D. S., Nip A. M. (1998) Biochem. Cell Biol. 76, 153–163 [DOI] [PubMed] [Google Scholar]
- 66.Fitzmaurice T. J., Burke D. F., Hopkins L., Yang S., Yu S., Sy M. S., Thackray A. M., Bujdoso R. (2008) Biochem. J. 409, 367–375 [DOI] [PubMed] [Google Scholar]
- 67.Ziegler J., Sticht H., Marx U. C., Müller W., Rösch P., Schwarzinger S. (2003) J. Biol. Chem. 278, 50175–50181 [DOI] [PubMed] [Google Scholar]
- 68.Cavagnero S., Nishimura C., Schwarzinger S., Dyson H. J., Wright P. E. (2001) Biochemistry 40, 14459–14467 [DOI] [PubMed] [Google Scholar]
- 69.Schwarzinger S., Mohana-Borges R., Kroon G. J., Dyson H. J., Wright P. E. (2008) Protein Sci. 17, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki T., Kurokawa T., Hashimoto H., Sugiyama M. (2002) Biochem. Biophys. Res. Commun. 294, 912–917 [DOI] [PubMed] [Google Scholar]
- 71.Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 72.Muramoto T., Scott M., Cohen F. E., Prusiner S. B. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 15457–15462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown D. R., Schmidt B., Kretzschmar H. A. (1996) Nature 380, 345–347 [DOI] [PubMed] [Google Scholar]
- 74.Wilkins D. K., Grimshaw S. B., Receveur V., Dobson C. M., Jones J. A., Smith L. J. (1999) Biochemistry 38, 16424–16431 [DOI] [PubMed] [Google Scholar]
- 75.Zandomeneghi G., Krebs M. R., McCammon M. G., Fändrich M. (2004) Protein Sci. 13, 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zahn R., Liu A. Z., Lührs T., Riek R., von Schroetter C., López García F., Billeter M., Calzolai L., Wider G., Wüthrich K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cobb N. J., Sonnichsen F. D., McHaourab H., Surewicz W. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 18946–18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu K., Cho H. S., Hoyt D. W., Nguyen T. N., Olds P., Kelly J. W., Wemmer D. E. (2000) J. Mol. Biol. 303, 555–565 [DOI] [PubMed] [Google Scholar]
- 79.Hou L., Shao H., Zhang Y., Li H., Menon N. K., Neuhaus E. B., Brewer J. M., Byeon I. J., Ray D. G., Vitek M. P., Iwashita T., Makula R. A., Przybyla A. B., Zagorski M. G. (2004) J. Am. Chem. Soc. 126, 1992–2005 [DOI] [PubMed] [Google Scholar]
- 80.Calloni G., Lendel C., Campioni S., Giannini S., Gliozzi A., Relini A., Vendruscolo M., Dobson C. M., Salvatella X., Chiti F. (2008) J. Am. Chem. Soc. 130, 13040–13050 [DOI] [PubMed] [Google Scholar]
- 81.Tartaglia G. G., Pawar A. P., Campioni S., Dobson C. M., Chiti F., Vendruscolo M. (2008) J. Mol. Biol. 380, 425–436 [DOI] [PubMed] [Google Scholar]
- 82.Caughey B., Lansbury P. T., Jr. (2003) Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 83.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 84.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]