Abstract
The 43-kDa TAR DNA-binding protein (TDP-43) is known to be a major component of the ubiquitinated inclusions characteristic of amyotrophic lateral sclerosis and frontotemporal lobar degeneration with ubiquitin-positive inclusions. Although TDP-43 is a nuclear protein, it disappears from the nucleus of affected neurons and glial cells, implicating TDP-43 loss of function in the pathogenesis of neurodegeneration. Here we show that the knockdown of TDP-43 in differentiated Neuro-2a cells inhibited neurite outgrowth and induced cell death. In knockdown cells, the Rho family members RhoA, Rac1, and Cdc42 GTPases were inactivated, and membrane localization of these molecules was reduced. In addition, TDP-43 depletion significantly suppressed protein geranylgeranylation, a key regulating factor of Rho family activity and intracellular localization. In contrast, overexpression of TDP-43 mitigated the cellular damage caused by pharmacological inhibition of geranylgeranylation. Furthermore administration of geranylgeranyl pyrophosphate partially restored cell viability and neurite outgrowth in TDP-43 knockdown cells. In summary, our data suggest that TDP-43 plays a key role in the maintenance of neuronal cell morphology and survival possibly through protein geranylgeranylation of Rho family GTPases.
The 43-kDa TAR DNA-binding protein (TDP-43)2 has recently been identified as a major component of the ubiquitinated inclusions characteristic of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin-positive inclusions (1, 2). Subsequently several point mutations located in the glycine-rich domain of TDP-43 have been identified as the disease-causing mutations of familial and sporadic ALS (3–7). TDP-43 has been shown to be a fundamental component of ubiquitin-positive neuronal cytoplasmic and intranuclear inclusions as well as that of neuronal dystrophic neurites in the affected neurons or glial cells in these neurodegenerative diseases. TDP-43 is known to regulate gene transcription, exon splicing, and exon inclusion through interactions with RNA, heterogeneous nuclear ribonucleoproteins, and nuclear bodies (8–12). Recently it has been reported that TDP-43 stabilizes human low molecular weight neurofilament mRNA through direct interaction with the 3′-untranslated region (13) and that it regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression (14). However, the physiological function of TDP-43 in the central nervous system has not been fully elucidated, and it remains unclear how this protein is implicated in the pathogenesis of neurodegeneration.
The Rho family of GTPases are members of the Ras superfamily and are known for regulating actin cytoskeletal dynamics (15–18). RhoA, Rac1, and Cdc42, the most studied proteins of this family, also modulate functions such as cell movement, motility, transcription, cell growth, and cell survival (18). In neurons, RhoA, Rac1, and Cdc42 have been shown to regulate neurite outgrowth (19–21).
Although TDP-43 is localized in the nucleus of unaffected neurons, nuclear staining of this protein is significantly reduced in neurons bearing ubiquitin inclusions (1, 2, 22), suggesting that loss of TDP-43 function may play a role in neurodegeneration. In this study, we used small interfering RNA (siRNA) to investigate the effect of TDP-43 loss of function on cell death and neurite outgrowth and elucidated a novel relation between TDP-43 and the activities of RhoA, Rac1, and Cdc42.
EXPERIMENTAL PROCEDURES
siRNA Oligonucleotides and Construction of Expression Vectors
The oligonucleotide siRNA duplex was synthesized by Takara Bio (Shiga, Japan). The siRNA sequences were as follows: scrambled (control) siRNA-set1, 5′-GAAUCAGAUGCACAUGAGUTT-3′; -set2, 5′-ACGGCCUAAUCUAACAGACTT-3′; TDP-43 siRNA-set1, 5′-GAACGAUGAACCCAUUGAATT-3′; -set2, 5′-CCAAUGCUGAACCUAAGCATT-3′. Unless otherwise mentioned, set 1 siRNA was used for TDP-43 knockdown throughout the experiments.
The pEGFP-Rac1 construct was produced as described elsewhere (23, 24). Mouse TDP-43 (GenBankTM accession number NM_145556) cDNA was amplified by PCR from mouse brain cDNA using the following primers: 5′-GTGCTTCCTCCTTGTGCTTC-3′ and 5′-CCACACTGAACAAACCAATCTG-3′. The PCR product was cloned into the pCR-BluntII-TOPO vector (Invitrogen), and the entire coding region of mouse TDP-43 was inserted in-frame into either the KpnI and XbaI sites of the pcDNA3.1/V5His vector (Invitrogen) or the KpnI and BamHI sites of the pDsRed-Monomer-Hyg-N1 vector (Clontech). An siRNA-resistant form of the TDP-43 gene was generated by changing the targeted sequence of the siRNA to 5′-GAATGACGAGCCAATTGAA-3′ (mutated nucleotides are underlined) using the KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan).
Cell Culture and Transfection
Neuro-2a cells (American Type Culture Collection, Manassas, VA), a line derived from mouse neuroblastoma, were maintained as described previously (25). The transfection of siRNA into Neuro-2a cells was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. For the transfection of the intended plasmid and siRNA, cells were co-transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. To differentiate the Neuro-2a cells, the medium was changed to Dulbecco's modified Eagle's medium containing 2% fetal calf serum and 20 μm retinoic acid, and cells were cultured for 24 h. For the interventional studies, the cells were incubated for 24 h with differentiation medium containing Clostridium difficile toxin B (Sigma-Aldrich) or for 12 h with medium containing GGTI-298 (Calbiochem). Geranylgeranyl pyrophosphate (GGPP) (Sigma-Aldrich) was added to the differentiation medium at the indicated concentration 24 h after siRNA transfection, and cells were incubated for an additional 24 h.
Quantitative Assessment of Neurite Outgrowth
Neuro-2a cells cultured in 6-well dishes were photographed using an Olympus IX71 inverted phase-contrast microscope (Olympus, Tokyo, Japan). The length of the longest neurite was measured with Image Gauge version 4.22 software (Fujifilm, Tokyo, Japan). Averages of the lengths of over 100 transfected cells were analyzed. To confirm the efficacy of siRNA transfection, BLOCK-iTTM Alexa Fluor® Red Fluorescent Control (Invitrogen) was co-transfected with TDP-43 siRNA or control RNA. The efficacy of plasmid transfection was ensured using DsRed (Clontech). To assess neurite outgrowth in TDP-43 knockdown cells, we performed a time course analysis starting at 24 h after the siRNA transfection when the differentiation medium was changed.
Cell Viability and Apoptosis Analysis
The 3-(4,5-dimethylthiazol-2-yl)-5-(3-caboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)-based cell proliferation assay was carried out on the differentiated Neuro-2a cells 48 h post-transfection using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer's instructions. Absorbance at 490 nm was measured in a multiple plate reader (PowerscanHT, Dainippon Pharmaceutical). The assays were carried out in 6 wells for each condition. To assess cell apoptosis, differentiated Neuro-2a cells were stained with Alexa Fluor 488-conjugated Annexin V and with propidium iodide (PI) using the Vybrant Apoptosis Assay kit (Invitrogen) according to the manufacturer's instructions. TUNEL assays were performed using the APO-DIRECTTM kit (BD Biosciences). The extent of staining in 10,000 cells was monitored by FACSCaliburTM and CellQuest version 3.1.3 acquisition and analysis software (BD Biosciences) immediately after the staining.
Caspase-3/7 Assay
Neuro-2a cells were grown on black 96-well plates. The caspase-3/7 activity of differentiated Neuro-2a cells was analyzed using the Apo-ONE homogeneous caspase-3/7 assay (Promega) after 48 h of transfection or intended treatment according to the manufacturer's instructions. Fluorescence (485/528 nm) was measured in the multiple plate reader, and the assay was carried out in 6 wells for each condition.
Quantitative Real Time PCR
mRNA levels were determined by real time PCR as described before (26). Briefly total RNA from Neuro-2a cells was reverse transcribed into first strand cDNA, real time PCR was performed, and the product was detected by the iCycler system (Bio-Rad). For an internal standard control, the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was simultaneously quantified. The following primers were used: 5′-CCGCATGTCAGCCAAATACAAG-3′ and 5′-ACCAGAATTGGCTCCAACAACAG-3′ for TDP-43 and 5′-TGTGTCCGTCGTGGATCTGA-3′ and 5′-TTGCTGTTGAAGTCGCAGGAG-3′ for GAPDH.
Western Blot Analysis
Western blot analyses were performed as described before (27). Briefly Neuro-2a cells were lysed in Cellytic lysis buffer (Sigma-Aldrich) containing a protease inhibitor mixture (Roche Applied Science) 48 h after transfection. For subcellular fractionation, we used the ProteoExtact Subcellular Proteome Extraction kit (Calbiochem) according to the manufacturer's instructions. Cell lysates were separated by SDS-PAGE (5–20% gradient gel) and analyzed by Western blotting with ECL Plus detection reagents (GE Healthcare). Primary antibodies used were as follows: anti-TDP-43 rabbit polyclonal antibody (1:1000; ProteinTech, Chicago, IL), anti-Rac1 mouse monoclonal antibody (1:1000; Millipore, Temecula, CA), anti-RhoA mouse monoclonal antibody (1:1000; Cytoskeleton, Denver, CO), anti-Cdc42 rabbit polyclonal antibody (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-myosin phosphatase targeting subunit 1 (Thr-853) rabbit polyclonal antibody (1:500; Millipore), anti-myosin phosphatase targeting subunit 1 rabbit polyclonal antibody (1:1000; Santa Cruz Biotechnology), anti-H-Ras rabbit polyclonal antibody (1:1000; Santa Cruz Biotechnology), anti-Rab5 rabbit polyclonal antibody (1:1000; Santa Cruz Biotechnology), anti-GAPDH mouse monoclonal antibody (1:1000; BD Pharmingen) as a cytosol marker, anti-integrin β1 rabbit polyclonal antibody (1:1000; Santa Cruz Biotechnology) as a membrane marker, anti-green fluorescent protein mouse monoclonal antibody (1:2000; Millipore), anti-Rho GDP dissociation inhibitor rabbit polyclonal antibody (1:1000; Millipore), anti-geranylgeranyltransferase-1β mouse polyclonal antibody (1:1000; Abnova, Taipei, Taiwan), and anti-geranylgeranyl pyrophosphate synthase rabbit polyclonal antibody (1:1000; Abgent, San Diego, CA).
Rho Family GTPase Activation Analysis
To measure RhoA and Rac1/Cdc42 GTPase activities, we used the Rho binding domain (RBD) of the Rho effector protein with Rhotekin and Cdc42/Rac interactive binding (CRIB) region of the Cdc42/Rac effector protein with p21 activated kinase 1 (PAK), respectively. Pulldown assays were performed in the presence of glutathione S-transferase-tagged Rhotekin-RBD and PAK-CRIB protein-agarose beads (Cytoskeleton) according to the manufacturer's instructions.
Fluorescent Images of Neuro-2a Cells
Neuro-2a cells, which were transfected with EGFP-Rac1, DsRed-TDP-43, and siRNA (TDP-43 siRNA or control RNA), were fixed by 4% paraformaldehyde with 0.1% Triton X-100 for extraction of cytosol components. After washing, samples were mounted with VECTASHIELD mounting medium (Vector Laboratories, Inc., Burlingame, CA) and then photographed with a Zeiss Axio Imager M1 (Carl Zeiss AG).
Assay of Protein Geranylgeranylation
Neuro-2a cells were transfected with siRNA on 10-cm2 dishes. Twenty-four hours after the transfection, 20 μm lovastatin (Sigma) was added to the culture medium. Twenty-four hours after the addition of lovastatin, the cells were labeled by adding fresh culture medium containing 6.25 μCi/ml [14C]mevalonolactone (50–62 mCi/mmol) (GE Healthcare) and 20 μm lovastatin. Nineteen hours after labeling, the cells were harvested, and Rac1 was immunoprecipitated by incubating with 4 mg of anti-RhoA mouse monoclonal antibody (Santa Cruz Biotechnology) or anti-Rac1 mouse monoclonal antibody (Millipore) for 24 h followed by adding 20 μl of protein G-Sepharose (GE Healthcare). Immunoprecipitated proteins were separated by electrophoresis on a polyacrylamide-SDS gel. The 14C-labeled gels were fixed and soaked in Amplify Fluorographic Reagent (GE Healthcare) for 30 min. The gels were dried, and labeled proteins were visualized on a Typhoon 9410 Workstation (GE Healthcare) after exposure to a Storage Phosphor Screen (GE Healthcare) for 72 h. We validated this experiment using 20 μm GGTI-298.
Statistical Analysis
Statistical differences (not including neurite length data) were analyzed by analysis of variance and Bonferroni post hoc analyses for three or more group comparisons and the unpaired Student's t test for two-group comparisons. Neurite length differences were analyzed using the Mann-Whitney U test (SPSS version 15.0, SPSS Inc., Chicago, IL). Two-tailed p < 0.05 was regarded as statistically significant.
RESULTS
TDP-43 Depletion Induces Cell Death and Inhibits Neurite Outgrowth
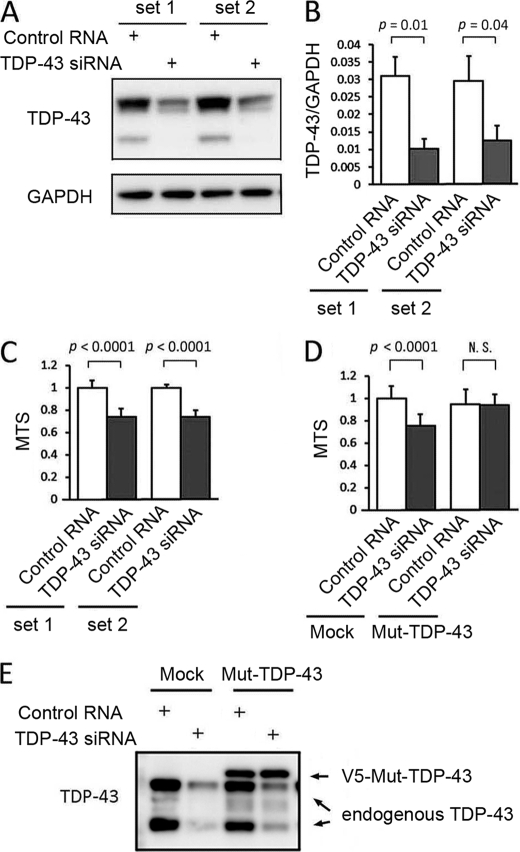
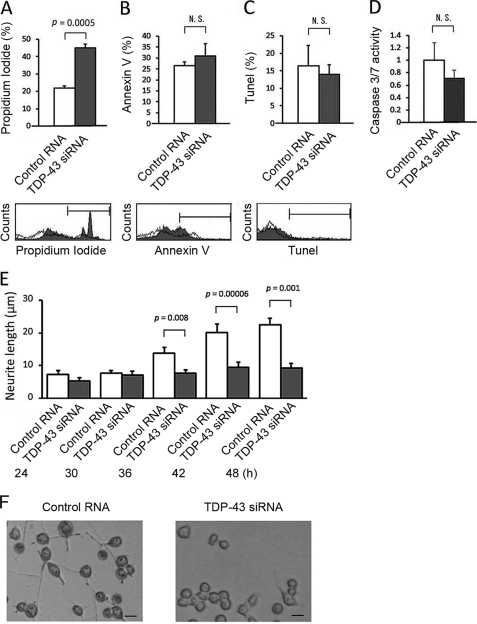
To analyze the effect of TDP-43 depletion, two sets of siRNA oligonucleotides were transfected into Neuro-2a cells. The efficiency of TDP-43 siRNA transfection was confirmed by Western blotting and quantitative real time RT-PCR (Fig. 1, A and B). To assess cell viability, we carried out an MTS-based cell proliferation assay in differentiated Neuro-2a cells after 48 h of siRNA transfection. The viability of knockdown cells was significantly decreased by both sets of siRNA compared with each control (Fig. 1C). To exclude the possibility of an off-target effect, a plasmid carrying an siRNA-resistant form of TDP-43 was co-transfected together with set 1 siRNA. As a result, the siRNA-resistant form of TDP-43 prevented the reduction of cell viability (Fig. 1D). The apoptotic process was quantified with Annexin V/PI staining and TUNEL labeling. The knockdown of TDP-43 significantly increased the number of PI-positive cells (Fig. 2A). However, the number of cells positive for Annexin V or TUNEL was not altered (Fig. 2, B and C). In the caspase-3/7 assay, there was no significant difference between knockdown and control cells (Fig. 2D). To clarify the effects of TDP-43 depletion on cellular morphology, we performed a time course experiment measuring the average length of neurites. Neurites of control cells extended over the course of 48 h, but neurite outgrowth was significantly inhibited in TDP-43-depleted cells (Fig. 2E).
FIGURE 1.
Effects of endogenous TDP-43 depletion on cell viability. A, anti-TDP-43 Western blot of Neuro-2a cells transfected with siRNAs. B, the mRNA expression levels of TDP-43 measured by real time RT-PCR. Data are shown as the ratio of the mRNA level of TDP-43 to that of GAPDH. C, the viability of Neuro-2a cells quantified by the MTS-based cell proliferation assays. TDP-43 depletion significantly reduced cell viability. D, a rescue experiment using the MTS assay. Mock plasmid and an siRNA-resistant form of TDP-43 (Mut-TDP-43) were co-transfected into Neuro-2a cells together with TDP-43 siRNA or control RNA. Mut-TDP-43 prevented the reduction of cell viability. E, Western blot of the cells transfected with mock plasmid or Mut-TDP-43. Error bars indicate S.D. N.S., not significant.
FIGURE 2.
Apoptosis analyses and quantitative assessment of neurite outgrowth in TDP-43-depleted Neuro-2a cells. A, B, and C, the number of the cells positive for PI, Annexin V, and TUNEL staining. We counted 10,000 cells using flow cytometry. Knockdown of TDP-43 significantly increased the number of cells stained with PI but not that with stained with Annexin V or TUNEL. D, caspase-3/7 activity of Neuro-2a cells transfected with TDP-43 siRNA or control RNA. E, time course analysis of neurite outgrowth in Neuro-2a cells transfected with TDP-43 siRNA or control RNA. Averages of the longest neurite lengths of over 100 transfected cells were analyzed 24 h after the transfection of siRNA. Neurites of control cells extended over the course of 48 h, but in TDP-43-depleted cells, neurite outgrowth was inhibited. Error bars indicate S.D. (A–D) or S.E. (E). F, phase-contrast images of Neuro-2a cells 48 h after the transfection of control RNA or TDP-43 siRNA. Scale bar, 10 μm. N.S., not significant.
Knockdown of TDP-43 Reduces Rho Family GTPase Activity
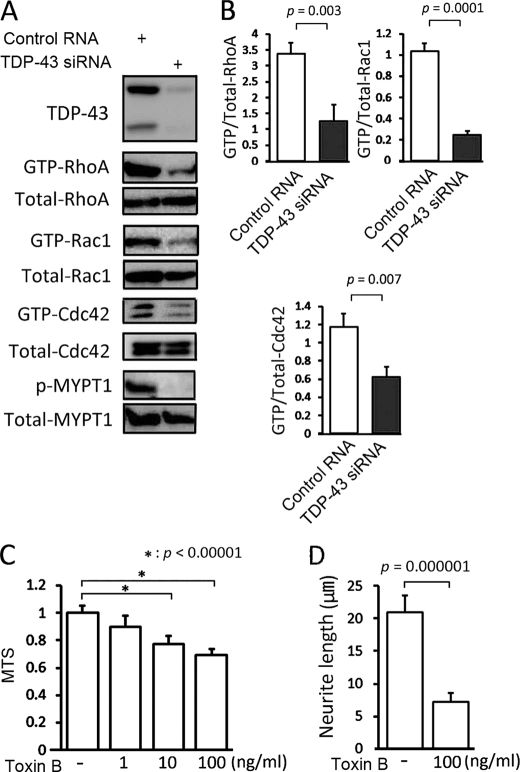
To elucidate the pathophysiology of neuronal cell death and the morphological alteration caused by TDP-43 depletion, we focused on the Rho family GTPases, which are potent regulators of neurite outgrowth and cell survival. First RhoA, Rac1, and Cdc42 GTPase activities were investigated by pulldown assays. In TDP-43-depleted cells, the activities of RhoA, Rac1, and Cdc42 GTPase were all decreased compared with the controls (Fig. 3, A and B). Inactivation of RhoA GTPase was also confirmed by a significant reduction in the level of the Thr-853 phosphorylation of myosin phosphatase targeting subunit 1, which is exclusively phosphorylated by Rho kinase (28) (Fig. 3A). To verify that inactivation of Rho family GTPases leads to neuronal damage, we tested the effects of C. difficile toxin B, a pan-inhibitor of Rho family proteins, on differentiated Neuro-2a cells. As a result, this toxin reduced cell viability and inhibited neurite outgrowth (Fig. 3, C and D).
FIGURE 3.
Activities of Rho family GTPases in TDP-43-depleted Neuro-2a cells. A, pulldown assays of RhoA, Rac1, and Cdc42 and Western blot using anti-phospho-myosin phosphatase targeting subunit 1 (p-MYPT1) in Neuro-2a cells transfected with TDP-43 siRNA or control RNA. TDP-43 knockdown significantly reduced Rho family GTPase activity. B, densitometric quantitations of three independent experiments. C, the viabilities of Neuro-2a cells incubated with the indicated concentrations of C. difficile toxin B. D, quantitation of neurite outgrowth in Neuro-2a cells incubated with 100 ng/ml toxin B compared with control. Error bars indicate S.D. (B and C) or S.E. (D).
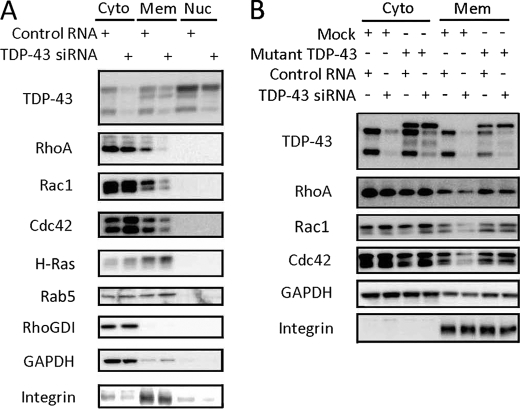
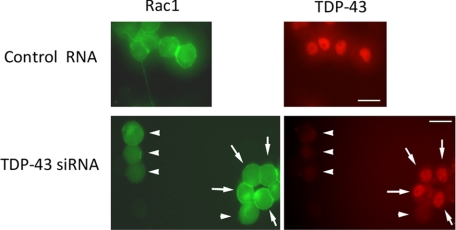
Small G proteins, including Rho and Ras family members, must be able to localize at the cell membrane to exert their biological functions (29). We thus investigated the intracellular distribution of Rho family proteins to elucidate the mechanism by which TDP-43 regulates their GTPase activity. Western blots showed that the amounts of RhoA, Rac1, and Cdc42, but not H-Ras or Rab5, in the membrane fractions were decreased in the TDP-43 knockdown cells (Fig. 4A). The subcellular fraction of Rho GDP dissociation inhibitor, another regulator of Rho activity, was not altered by TDP-43 knockdown (Fig. 4A). The siRNA-resistant form of TDP-43 rescued the reduction in the amount of membrane-bound Rho family GTPases (Fig. 4B). The fluorescent images also demonstrated that membrane-localized Rac1 was significantly reduced in TDP-43 knockdown cells in comparison with control cells (Fig. 5).
FIGURE 4.
Intracellular distribution of Rho family GTPases in TDP-43 knockdown Neuro-2a cells. A, Western blots of subcellular fractions (cytosol, membrane, and nucleus) of Neuro-2a cells transfected with TDP-43 siRNA or control RNA. The amount of Rho family members in the membrane fraction was significantly reduced in TDP-43 knockdown cells. This effect, however, was not observed with H-Ras or Rab5. Moreover the subcellular fraction of Rho GDP dissociation inhibitor (RhoGDI) was not altered by TDP-43 depletion. GAPDH and integrin β1 were used as a cytosol and membrane marker, respectively. Cyto, cytosol fraction; Mem, membrane fraction; Nuc, nuclear fraction. B, the effect of the siRNA-resistant form of TDP-43 (Mut-TDP-43) on the intracellular distribution of Rho family GTPases. Mut-TDP-43 prevented the reduction of membrane-bound Rho family members.
FIGURE 5.
Fluorescent images of Neuro-2a cells. Neuro-2a cells co-transfected with EGFP-Rac1, DsRed-TDP-43, and siRNA (TDP-43 siRNA or control RNA) were fixed by 4% paraformaldehyde with 0.1% Triton X-100. Membrane-localized Rac1 was significantly reduced in the TDP-43 depleted cells (arrowhead) compared with the cells that escaped knockdown of TDP-43 (arrow) and with the control RNA-transfected cells. Scale bar, 10 μm.
TDP-43 Regulates Rho GTPase Activity via Protein Geranylgeranylation
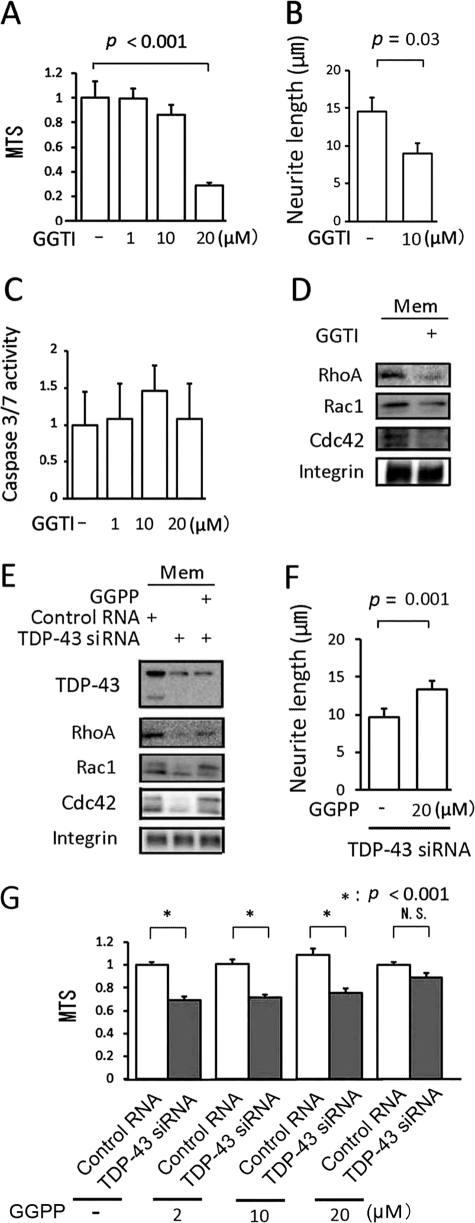
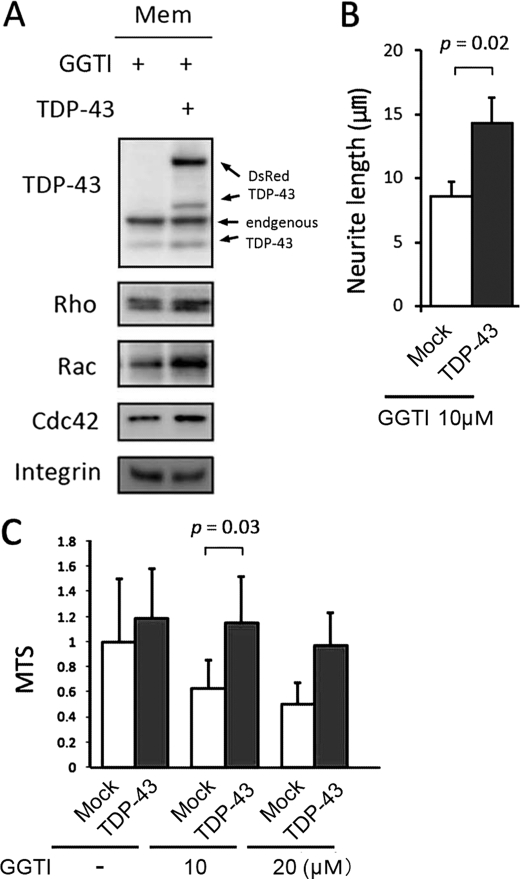
For membrane localization to occur, members of the Rho family of GTPases have to be catalyzed by transferring GGPP to the C-terminal motif (geranylgeranylation), whereas Ras family members, such as H-Ras, require farnesyl pyrophosphate. We thus tested two agents that modulate geranylgeranylation of Rho family molecules. GGTI-298, a specific inhibitor of geranylgeranylation, has been shown to increase neural cell death through the inactivation of Rho family GTPases (30, 31). In differentiated Neuro-2a cells, GGTI-298 inhibited neurite outgrowth and reduced cell viability in a dose-dependent manner without any evidence of caspase-3/7 activation (Fig. 6, A–C). GGTI-298 also decreased the amount of membrane-bound Rho GTPases (Fig. 6D). In contrast, GGPP, the final substrate of geranylgeranylation, prevented the reduction in the amounts of membrane-bound RhoA, Rac1, and Cdc42 caused by TDP-43 knockdown (Fig. 6E). In addition, GGPP restored viability and neurite outgrowth in TDP-43-depleted cells (Fig. 6, F and G). These findings indicate that impaired geranylgeranylation appears to be the molecular basis of TDP-43 depletion-induced cellular damage. We next examined whether TDP-43 regulates geranylgeranylation of Rho GTPases. Fig. 7, A–C, shows that TDP-43 increased the amount of membrane-bound Rho GTPases, augmented neurite outgrowth, and increased cell viability in GGTI-298-treated cells.
FIGURE 6.
The effect of modulation of geranylgeranylation on cell viability. A, the viability of Neuro-2a cells incubated with the indicated concentrations of GGTI-298 (GGTI). B, averages of neurite length in Neuro-2a cells incubated with 20 μm GGTI-298 compared with control. C, caspase-3/7 activity in Neuro-2a cells treated with GGTI-298. D, Western blot of the membrane fraction from Neuro-2a cells incubated with 20 μm GGTI-298 compared with control. E, Western blot of the membrane fraction from Neuro-2a cells incubated with 20 μm GGPP in the presence of TDP-43 siRNA. F, measurement of neurite length of Neuro-2a cells incubated with 20 μm GGPP. The cells were transfected with TDP-43 siRNA or control RNA. G, the viability of Neuro-2a cells incubated with the indicated concentrations of GGPP. Error bars indicate S.D. (A and C) or S.E. (B and F). N.S., not significant.
FIGURE 7.
Effect of TDP-43 in Neuro-2a cells on GGTI-298-induced cellular phenotype. A, Western blot of the membrane (Mem) fraction of 10 μm GGTI-298 (GGTI)-treated Neuro-2a cells transfected with TDP-43 or mock vector. B, neurite length of GGTI-298-treated Neuro-2a cells transfected with DsRed-TDP-43 or mock vector. C, the viability of Neuro-2a cells incubated with the indicated concentrations of GGTI-298. Error bars indicate S.E. (B) or S.D. (C).
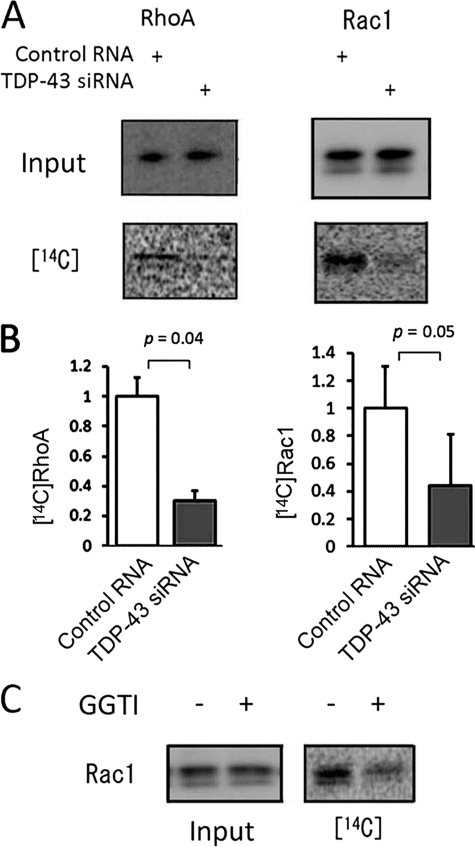
Taken together, these observations suggest that TDP-43 regulates the activities of Rho family members through protein geranylgeranylation. We thus investigated the effect of TDP-43 depletion on protein geranylgeranylation using [14C]mevalonic acid (MVA). Incorporation of [14C]MVA into RhoA or Rac1 was significantly decreased in the TDP-43-depleted Neuro-2a cells (Fig. 8, A and B), suggesting that the knockdown of TDP-43 inhibits geranylgeranylation of Rho family members. GGTI-298 reduced incorporation of [14C]MVA into Rac1, further confirming this conclusion (Fig. 8C).
FIGURE 8.
Protein prenylation of RhoA and Rac1 in Neuro-2a cells. A, Neuro-2a cells were metabolically labeled with [14C]MVA: left, Western blot with anti-RhoA or Rac1 antibody of cell lysates; right, incorporation of [14C]MVA into RhoA or Rac1. B, densitometric quantitation of four independent experiments. Error bars indicate S.D. C, incorporation of [14C]MVA into Rac1 in Neuro-2a cells incubated with or without GGTI-298 (GGTI).
Protein geranylgeranylation of the Rho family is regulated by specific enzymes: geranylgeranyltransferase-Iβ and geranylgeranyl pyrophosphate synthase-1, which is responsible for synthesis of GGPP. We therefore investigated the expression levels of these enzymes. However, the knockdown of TDP-43 did not alter the protein expression level of geranylgeranyltransferase-Iβ or geranylgeranyl pyrophosphate synthase-1 (data not shown).
DISCUSSION
TDP-43 as a Regulator of Rho Family GTPases
In the present study, we demonstrated that knockdown of TDP-43 inhibits neurite outgrowth and induces cell death in differentiated Neuro-2a cells, suggesting that loss of TDP-43 function plays a causative role in neurodegeneration. To elucidate the molecular mechanisms by which TDP-43 depletion causes neuronal cell damage, we examined the relationship between TDP-43 and Rho family GTPases. Neuronal morphology is determined in large part through the regulation of the cytoskeleton. One of the key regulators of the actin cytoskeleton is the Rho family of GTPases: RhoA, Rac1, and Cdc42 (15–17). Although each Rho family member has a distinct effect on cell morphology and plays an important role in the regulation of neuronal survival, we demonstrated that pan-inhibition of Rho family proteins by C. difficile toxin B suppressed neurite outgrowth and reduced cell viability as reported previously (19–21, 32, 33). We demonstrated here that the activities of RhoA, Rac1, and Cdc42 were all decreased in TDP-43-depleted cells, suggesting that knockdown of TDP-43 induces inhibition of neurite outgrowth and cell death through the inactivation of Rho family GTPases. There was no evidence of apoptosis mediated by TDP-43 knockdown in the model we used perhaps because of the biological characteristics of Neuro-2a cells in which the induction of apoptosis is known to be difficult (34, 35). In support of this view, we confirmed that GGTI-298 had no effect on apoptosis in Neuro-2a cells under the present conditions (Fig. 6C), although this reagent has been shown to efficiently induce apoptosis in non-neuronal cells (36, 37).
Inhibited Protein Geranylgeranylation of Rho Family Members in TDP-43-depleted Cells
How does TDP-43 regulate the activities of the Rho family? Various molecules have been shown to regulate the activity of Rho, Rac, and Cdc42, but few are capable of regulating all three GTPases concomitantly. To elucidate the molecular mechanism by which TDP-43 regulates Rho, Rac, and Cdc42, we directed our attention to the fact that membrane localization is the key regulatory factor common to these molecules. Small G proteins, including members of the Rho and Ras families, act as molecular switches cycling between an active, GTP-bound state and an inactive, GDP-bound state (18, 38, 39). Post-translational modification with a C-terminal prenyl moiety allows small G proteins to associate with the membrane where they can interact with and activate their effectors (29). Proteins that require prenylation include the farnesyl group, such as the Ras family, and the geranylgeranyl group, such as the Rho or the Rab family (29, 40). In the present study, we showed that knockdown of TDP-43 decreased membrane-bound RhoA, Rac1, and Cdc42 but did not affect the intracellular distribution of H-Ras or Rab5. TDP-43 knockdown also inhibited the incorporation of [14C]MVA, a tracer of the mevalonate pathway, into RhoA and Rac1 in differentiated Neuro-2a cells. In contrast, the expression level of Rho GDP dissociation inhibitor, which functions by extracting Rho family GTPases from membranes and solubilizing them in the cytosol (41, 42), was not significantly altered by TDP-43 knockdown. In addition, inhibition of geranylgeranylation by GGTI-298 reproduced the cytotoxic effects of TDP-43 depletion, whereas GGPP, the substrate of the geranylgeranylation pathway, restored cell viability and neurite outgrowth in TDP-43-depleted Neuro-2a cells. Furthermore in GGTI-298 treated cells, overexpression of TDP-43 restored the membrane localization of Rho GTPases as well as cell viability and neurite outgrowth. These findings suggest that TDP-43 depletion inactivates Rho family GTPases through inhibition of protein geranylgeranylation.
Protein geranylgeranylation of Rho family members is catalyzed by geranylgeranyltransferase-I using GGPP produced by geranylgeranyl pyrophosphate synthase-1 as the substrate (43). Geranylgeranyltransferase-I consists of two subunits, α and β, and the α subunit is also a component of protein farnesyltransferase (44). We thus assessed the mRNA and protein expression of geranylgeranyltransferase-Iβ and geranylgeranyl pyrophosphate synthase-1 in Neuro-2a cells. However, the expression levels of these enzymes were not altered by TDP-43 knockdown. These results imply that TDP-43 depletion decreases the activities of these enzymes.
Loss of TDP-43 Function in the Pathophysiology of TDP-43 Proteinopathies
Ubiquitinated cytoplasmic inclusions are a histopathological hallmark of ALS and frontotemporal lobar degeneration with ubiquitin-positive inclusions. Although the nature of these aggregates has not been fully elucidated, recent studies have identified TDP-43 as the major component of the ubiquitin-immunoreactive neuronal inclusions seen in ALS and frontotemporal lobar degeneration with ubiquitin-positive inclusions (1, 2). There has been a great deal of debate about whether loss or gain of function of TDP-43 causes neuronal dysfunction and eventual cell death. Although TDP-43 is a ubiquitously expressed, highly conserved nuclear protein, under the pathological conditions in ALS and frontotemporal lobar degeneration with ubiquitin-positive inclusions, TDP-43 completely disappears from the nuclei of the affected neurons (1, 2). These histopathological findings indicate that loss of nuclear TDP-43 may underlie neuronal degeneration, although it is also possible that neuronal inclusions possess cytotoxic properties. It has been reported that TDP-43 depletion leads to up-regulation of cyclin-dependent kinase 6 protein and transcript levels followed by misregulation of the cell cycle and apoptosis in cultured human epithelial cancer cells (14). In the present study, knockdown of TDP-43 inactivated Rho family GTPases and thereby induced cell death in differentiated Neuro-2a cells. Although this is not the model imitating the dislocation of TDP-43 from the nucleus to the cytoplasm, our findings suggest that loss of function of TDP-43 may induce neuronal degeneration probably through dysregulation of Rho family GTPases.
In summary, we have demonstrated that TDP-43 depletion inhibits neurite outgrowth and induces neuronal cell death. This phenomenon possibly results from a reduced membrane localization of Rho family GTPases due to the inhibition of protein geranylgeranylation.
This work was supported by a Center-of-Excellence grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan; grants from the Ministry of Health, Labor and Welfare of Japan; a grant from the Japan Intractable Diseases Research Foundation; and the Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology commissioned by the Ministry of Education, Culture, Sports, Science and Technology.
- TDP-43
- 43-kDa TAR DNA-binding protein
- ALS
- amyotrophic lateral sclerosis
- siRNA
- small interfering RNA
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-caboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- GGPP
- geranylgeranyl pyrophosphate
- MVA
- mevalonic acid
- PI
- propidium iodide
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 2.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 3.Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 5.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 6.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 9.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 10.Wang I. F., Reddy N. M., Shen C. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 12.Bose J. K., Wang I. F., Hung L., Tarn W. Y., Shen C. K. (2008) J. Biol. Chem. 283, 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strong M. J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. (2007) Mol. Cell. Neurosci. 35, 320–327 [DOI] [PubMed] [Google Scholar]
- 14.Ayala Y. M., Misteli T., Baralle F. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaibuchi K., Kuroda S., Amano M. (1999) Annu. Rev. Biochem. 68, 459–486 [DOI] [PubMed] [Google Scholar]
- 16.Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 17.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 18.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 19.Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 20.Bradke F., Dotti C. G. (1999) Science 283, 1931–1934 [DOI] [PubMed] [Google Scholar]
- 21.Ahmed I., Calle Y., Iwashita S., Nur-E-Kamal A. (2006) Mol. Cell. Biochem. 281, 17–25 [DOI] [PubMed] [Google Scholar]
- 22.Cairns N. J., Neumann M., Bigio E. H., Holm I. E., Troost D., Hatanpaa K. J., Foong C., White C. L., 3rd, Schneider J. A., Kretzschmar H. A., Carter D., Taylor-Reinwald L., Paulsmeyer K., Strider J., Gitcho M., Goate A. M., Morris J. C., Mishra M., Kwong L. K., Stieber A., Xu Y., Forman M. S., Trojanowski J. Q., Lee V. M., Mackenzie I. R. (2007) Am. J. Pathol. 171, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa M., Fukata M., Yamaga M., Itoh N., Kaibuchi K. (2001) J. Cell Sci. 114, 1829–1838 [DOI] [PubMed] [Google Scholar]
- 24.Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004) Dev. Cell 7, 871–883 [DOI] [PubMed] [Google Scholar]
- 25.Niwa J., Yamada S., Ishigaki S., Sone J., Takahashi M., Katsuno M., Tanaka F., Doyu M., Sobue G. (2007) J. Biol. Chem. 282, 28087–28095 [DOI] [PubMed] [Google Scholar]
- 26.Katsuno M., Adachi H., Minamiyama M., Waza M., Tokui K., Banno H., Suzuki K., Onoda Y., Tanaka F., Doyu M., Sobue G. (2006) J. Neurosci. 26, 12106–12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsuno M., Adachi H., Doyu M., Minamiyama M., Sang C., Kobayashi Y., Inukai A., Sobue G. (2003) Nat. Med. 9, 768–773 [DOI] [PubMed] [Google Scholar]
- 28.Feng J., Ito M., Ichikawa K., Isaka N., Nishikawa M., Hartshorne D. J., Nakano T. (1999) J. Biol. Chem. 274, 37385–37390 [DOI] [PubMed] [Google Scholar]
- 29.Adamson P., Marshall C. J., Hall A., Tilbrook P. A. (1992) J. Biol. Chem. 267, 20033–20038 [PubMed] [Google Scholar]
- 30.Tanaka T., Tatsuno I., Uchida D., Moroo I., Morio H., Nakamura S., Noguchi Y., Yasuda T., Kitagawa M., Saito Y., Hirai A. (2000) J. Neurosci. 20, 2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meske V., Albert F., Richter D., Schwarze J., Ohm T. G. (2003) Eur. J. Neurosci. 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 32.Linseman D. A., Laessig T., Meintzer M. K., McClure M., Barth H., Aktories K., Heidenreich K. A. (2001) J. Biol. Chem. 276, 39123–39131 [DOI] [PubMed] [Google Scholar]
- 33.Loucks F. A., Le S. S., Zimmermann A. K., Ryan K. R., Barth H., Aktories K., Linseman D. A. (2006) J. Neurochem. 97, 957–967 [DOI] [PubMed] [Google Scholar]
- 34.Sueyoshi N., Maehara T., Ito M. (2001) J. Lipid Res. 42, 1197–1202 [PubMed] [Google Scholar]
- 35.Saito M., Saito M., Cooper T. B., Vadasz C. (2005) Alcohol Clin. Exp. Res. 29, 1374–1383 [DOI] [PubMed] [Google Scholar]
- 36.Xia Z., Tan M. M., Wong W. W., Dimitroulakos J., Minden M. D., Penn L. Z. (2001) Leukemia 15, 1398–1407 [DOI] [PubMed] [Google Scholar]
- 37.Li X., Liu L., Tupper J. C., Bannerman D. D., Winn R. K., Sebti S. M., Hamilton A. D., Harlan J. M. (2002) J. Biol. Chem. 277, 15309–15316 [DOI] [PubMed] [Google Scholar]
- 38.Lamarche N., Hall A. (1994) Trends Genet 10, 436–440 [DOI] [PubMed] [Google Scholar]
- 39.Zhou K., Wang Y., Gorski J. L., Nomura N., Collard J., Bokoch G. M. (1998) J. Biol. Chem. 273, 16782–16786 [DOI] [PubMed] [Google Scholar]
- 40.Cox A. D., Der C. J. (1992) Curr. Opin. Cell Biol. 4, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 41.Fukumoto Y., Kaibuchi K., Hori Y., Fujioka H., Araki S., Ueda T., Kikuchi A., Takai Y. (1990) Oncogene 5, 1321–1328 [PubMed] [Google Scholar]
- 42.Dovas A., Couchman J. R. (2005) Biochem. J. 390, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ericsson J., Greene J. M., Carter K. C., Shell B. K., Duan D. R., Florence C., Edwards P. A. (1998) J. Lipid Res. 39, 1731–1739 [PubMed] [Google Scholar]
- 44.Moomaw J. F., Casey P. J. (1992) J. Biol. Chem. 267, 17438–17443 [PubMed] [Google Scholar]