Abstract
Allene oxides are reactive epoxides biosynthesized from fatty acid hydroperoxides by specialized cytochrome P450s or by catalase-related hemoproteins. Here we cloned, expressed, and characterized a gene encoding a lipoxygenase-catalase/peroxidase fusion protein from Acaryochloris marina. We identified novel allene oxide synthase (AOS) activity and a by-product that provides evidence of the reaction mechanism. The fatty acids 18.4ω3 and 18.3ω3 are oxygenated to the 12R-hydroperoxide by the lipoxygenase domain and converted to the corresponding 12R,13-epoxy allene oxide by the catalase-related domain. Linoleic acid is oxygenated to its 9R-hydroperoxide and then, surprisingly, converted ∼70% to an epoxyalcohol identified spectroscopically and by chemical synthesis as 9R,10S-epoxy-13S-hydroxyoctadeca-11E-enoic acid and only ∼30% to the 9R,10-epoxy allene oxide. Experiments using oxygen-18-labeled 9R-hydroperoxide substrate and enzyme incubations conducted in H218O indicated that ∼72% of the oxygen in the epoxyalcohol 13S-hydroxyl arises from water, a finding that points to an ionic intermediate (epoxy allylic carbocation) during catalysis. AOS and epoxyalcohol synthase activities are mechanistically related, with a reacting intermediate undergoing a net hydrogen abstraction or hydroxylation, respectively. The existence of epoxy allylic carbocations in fatty acid transformations is widely implicated although for AOS reactions, without direct experimental support. Our findings place together in strong association the reactions of allene oxide synthesis and an ionic reaction intermediate in the AOS-catalyzed transformation.
A diverse spectrum of signaling molecules is biosynthesized in nature from polyunsaturated fatty acids, their peroxides, and further transformations of the fatty acid peroxides. The peroxides are formed by two classes of dioxygenase enzyme. The hemoprotein dioxygenases include prostaglandin H synthase (cyclooxygenase) in animals (1), α-dioxygenase in plants (2), and several linoleate dioxygenases in fungi (3–5). The non-heme iron lipoxygenases are even more widespread, being almost ubiquitous among organisms that contain polyunsaturated fatty acids (6–8). Although further biosynthetic transformation is sometimes accomplished by an additional catalytic activity of the initiating dioxygenase (e.g. leukotriene A4 synthase (9) or aldehyde-synthesizing hydroperoxide cleaving activity (10) among the LOX2 enzymes), commonly another distinct enzyme is used to rearrange or otherwise modify the reactive fatty acid peroxide intermediate. Two hemoprotein types are found that have become specialized for this biosynthetic role: cytochrome P450s and catalase-related enzymes.
The fatty acid peroxide-metabolizing P450s are by far the better known and include CYP5 (thromboxane synthase) and CYP8A (prostacyclin synthase) in animals (11), and the entire family of CYP74 in plants (12). The individual CYP74 enzymes include allene oxide synthase (AOS), one of which catalyzes a key step in cyclopentenone synthesis in the jasmonate pathway, hydroperoxide lyase, divinyl ether synthase, and epoxyalcohol synthase (12, 13). The catalase-related enzymes are distinctive in that they have been found naturally as a fusion protein with the LOX enzyme that forms their hydroperoxide substrate (14). The known activities include AOS in Plexaura homomalla and other marine corals (with a different specificity for fatty acid hydroperoxide compared with the plant P450 AOS) (15, 16), and the unique bicyclobutane synthase and other allylic epoxide synthase activities of the enzyme in the cyanobacterium Anabaena PCC-7120 (17, 18). Currently we are trying to understand the scope of the reactions catalyzed by this catalase-related family of enzymes. Our objectives are to help understand the structure-function relationships in the reactions with peroxides, to provide new insights on the mechanism of these hemoprotein-catalyzed transformations, as well as, by reflection, to give a different perspective on the parent hemoprotein, the hydrogen peroxide-metabolizing true catalase.
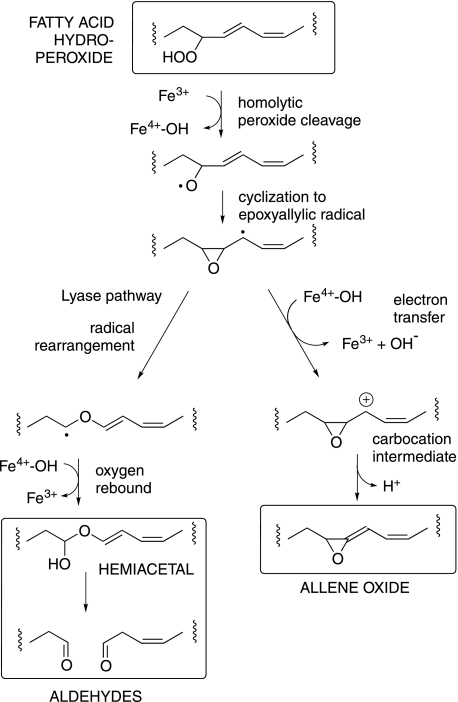
The underlying chemistry of the fatty acid hydroperoxide transformations by the specialized P450 and catalase-related hemoproteins can be written as purely free radical in character, or ionic, or with facets of both (19–22). Reaction is generally considered to be initiated by homolytic cleavage of the peroxide O–O bond (see Fig. 1). While subsequent reactions can be construed as following a radical pathway to product (e.g. in the hydroperoxide lyase reaction (19)), others are considered to involve an electron transfer step, giving a carbocation intermediate in the penultimate steps to product (22) (Fig. 1). The underlying grounds for these mechanisms are more a subject of debate and of comparison to other chemistry than of defining evidence on the reactions in question. The results reported here add a measure of experimental support for ionic events in the peroxide transformation by a catalase-related hemoprotein.
FIGURE 1.
Hemoprotein-catalyzed transformation of fatty acid hydroperoxides via diverging routes. A radical pathway can lead to aldehydes (left side) and an ionic pathway via a putative carbocation intermediate to allene oxides (right side).
Recently genome sequencing was completed on the cyanobacterium Acaryochloris marina (23). A. marina is a focus of attention owing to its harboring a light-harvesting complex containing the unusual chlorophyll d, considered a bridge in the evolutionary development of photosynthetic mechanisms (24, 25). BLAST searches of the A. marina genome reveal three individual LOX sequences and additionally, the presence of DNA encoding a putative fusion protein of lipoxygenase and catalase-related hemoprotein, the topic of this report. We find an unexpected by-product in the reaction with one particular fatty acid (C18.2ω6); the partial incorporation of 18O from water in a newly formed hydroxyl group has implications related to the mechanism of hydroperoxide transformation.
EXPERIMENTAL PROCEDURES
Materials
Fatty acids were purchased from NuChek Prep Inc. (Elysian, MN). Standards of racemic HPODEs, HODEs, HPOTrEs, HOTrEs, HPETEs, and HETEs were prepared by vitamin E-controlled autoxidation (26). Chiral hydroperoxides were prepared enzymatically using the A. marina LOX domain for conversion of linoleic acid to 9R-HPODE, α-linolenic acid to 12R-HPOTE, and arachidonic acid to 11R-HPETE. Labeled water H218O, nominally with ≥95% oxygen-18, was purchased 25 years ago from the Monsanto Research Corp. (Miamisburg, OH).
Cloning, Expression, and Purification of the Full-length Protein of A. marina
Genomic DNA from A. marina MBIC11017 was a kind gift from Dr. Robert E. Blankenship of Washington University in St. Louis. cDNA encoding the full-length fusion protein, designated as a hypothetical protein (ABW27596 of 805 amino acids) was cloned by PCR. Forward and reverse primers in PCR reaction contained NdeI and XhoI restriction sites, and an N-terminal (His)6 tag was engineered for the full-length construct. The sequence of the forward primer was 5′-TCCATATGCATCACCATCACCATCACGATAGTCGTGATCCGCGC-3′ and that of the reverse primer was 5′-GACTCGAGTTAGATATTGGTGCTCATCATAAG-3′. The correct PCR product was subsequently cut with NdeI and XhoI restriction enzymes and inserted into the same sites of the expression vector pET17b. DNA sequencing confirmed the identity to the published sequence in GenBankTM (accession number CP000828.1, gene AM1–2589 and protein ABW27596) and at CyanoBase. A cDNA encoding only the N-terminal (catalase-related) domain was similarly prepared using the upstream primer 5′-TCCATATGGATAGTCGTGATCCGCGC-3′ together with the downstream primer 5′-GACTCGAGTTAGTGATGGTGATGCACTAGGCTGGAGCTAGGAGT-3′, including a His4 tag. Expression of the N-terminal and full-length domains in Escherichia coli BL21(DE3) cells (Novagen) was accomplished with methods described previously (27). The LOX domain required a lower temperature for expression, 14 °C instead of 28 °C, and use of 2× YT (16 g Tryptone, 10 g yeast extract, and 5 g NaCl in a final volume of 1 L, adjusted to pH 7.0 using NaOH) medium, to be described in detail elsewhere. Purification of the His-tagged full-length protein and N-terminal domain followed the protocol of Imai et al. (28) and Boutaud et al. (27) with some modifications, as described recently (29). The frozen pellet from a 50-ml bacterial culture was resuspended in 10 ml of BugBuster® protein extraction reagent (Novagen) by sonication and homogenized using glass Dounce homogenizer (Wheaton). The 16,000 × g supernatant was loaded onto a nickel-nitrilotriacetic acid column (0.5-ml bed volume, Qiagen) equilibrated with 50 mm potassium phosphate buffer, pH 7.2, 500 mm NaCl at 0.5 ml/min. The column was then washed with the equilibration buffer, and the nonspecific bound proteins were eluted with 50 mm potassium phosphate buffer, pH 7.2, 1 m NaCl, 70 mm glycine. A final wash was performed by 50 mm potassium phosphate buffer, pH 7.2, 500 mm NaCl, 20 mm imidazole. The His tag protein was then eluted with 50 mm potassium phosphate buffer, pH 7.2, 500 mm NaCl, 250 mm imidazole. The full-length fusion protein was dark green in color. Fractions containing the most protein as judged by SDS-PAGE were pooled and dialyzed overnight against 50 mm Tris buffer, pH 7.5, 500 mm NaCl, 20% glycerol, aliquoted and frozen at −80 °C for later use.
Incubation, Extraction, and HPLC Product Analysis
Small scale incubations were performed at room temperature in 1 ml of 50 mm Tris, pH 7.5, 150 mm NaCl using the fatty acids C18.4ω3, C18.3ω3, C20.4ω6, or C18.2ω6 (10–30 μg) added in 5 μl of ethanol. Incubations with fatty acid hydroperoxides were conducted in a 1-ml quartz cuvette, and the rate of reaction was monitored by repetitive scanning from 350 to 200 nm using a Lambda-35 spectrophotometer (PerkinElmer Life Sciences). Larger scale incubations were conducted in 50 ml of oxygenated buffer with an appropriate amount of A. marina full-length enzyme and 10 mg of fatty acid substrate. Reaction was complete within 5 min, whereupon the samples were acidified to pH 6 with 1 n HCl and extracted using a 1-ml Oasis HLB cartridge (Waters) or 1-g size for the larger incubations.
Products were analyzed initially by RP-HPLC using a C18 Waters Symmetry 5-μm column (0.46 × 25 cm) at a flow rate of 1 ml/min with methanol/water/acetic acid (70/30/0.01, by volume). Peaks were monitored using an Agilent 1100 diode array detector. To detect radiolabeled products, a Packard A-100 Flo-One Radiomatic liquid scintillation detector was connected to the Agilent 1100 diode array detector. For larger scale samples, a semi-preparative 5-μm Beckman ODS column (10 mm × 25 cm) was used for the separation of incubation products; the major peaks were collected and extracted from the solvent with dichloromethane, taken to dryness under a stream of nitrogen, and stored in methanol at −20 °C. The major products collected from RP-HPLC were further purified as the methyl esters by SP-HPLC using a Beckman 5-μm Ultrasphere silica column (25 × 0.46 cm, or 25 × 1 cm for milligram quantities of product) with a solvent system of hexane/isopropanol (100:1.5, v/v).
Synthesis of Allene Oxide from C18.3ω3
Reaction of the A. marina enzyme with 12R-HPOTrE was conducted at 0 °C with the substrate initially in ice-cold hexane (10 ml and 100 μm) and layered over the A. marina enzyme (0.8 nmol) in 200 μl of phosphate buffer, pH 7. The reaction was initiated by vigorous vortex mixing of the two phases, which was continued for 2 min; then the test tube was placed back on ice. The hexane phase was scanned from 200–350 nm in UV using a PerkinElmer Lambda-35 spectrophotometer. The combined hexane phases were evaporated to a small volume (∼2 ml) by using a vigorous stream of nitrogen. The sample was then treated with ethanol (20 μl) and ethereal diazomethane for 30 s at 0 °C and then rapidly evaporated to dryness and stored in hexane at −80 °C until further analysis. The sample was purified as described previously for an allene oxide derived from 13S-HPOTrE using a 4.5- × 0.46-cm silica column operated at −15 °C using a solvent system of hexane/diethyl ether (100:1, v/v) run at a flow rate of 3 ml/min (30).
Liquid Chromatography-Mass Spectrometry
LC-MS was performed using an LTQ ion-trap instrument (ThermoFinnigan) monitoring negative ions. A Waters Symmetry C18 column (2.1 × 150 mm) was used and eluted with CH3CN/H2O/HAc (55/45/0.01, v/v) at 0.2 ml/min. In some cases, samples were run on a Waters Acquity HSS T3 1.8 μm ultra-pressure liquid chromatography column (2.1 × 100 mm) eluted with a programmed gradient from solvent A (methanol/water/ammonium acetate of 5/95/10 mm, v/v/concentration) to B (methanol/water/ammonium acetate = 95/5/10 mm, v/v/concentration) at 0.3 ml/min. Isocratic runs were also performed with the solvent system of CH3CN/H2O/NH4Ac (35/65/10 mm, v/v/concentration) at a flow rate of 0.2 ml/min using the same column.
Derivatization and GC-MS Analysis
Methyl esters were prepared using ethereal diazomethane in methanol. Catalytic hydrogenations were performed in 100 μl of ethanol using ∼1 mg of palladium and bubbling with hydrogen for 2 min at room temperature. The hydrogenated products were recovered by the addition of water and extraction with ethyl acetate. Methoxime derivatives were prepared by treatment with 5 μl of methoxylamine hydrochloride in pyridine (10 mg/ml) overnight at room temperature. Trimethylsilyl ether were prepared using bis(trimethylsilyl)trifluoracetamide (10 μl) at room temperature for 2 h or overnight. Subsequently, the reagents were evaporated under a stream of nitrogen, and the samples were dissolved in hexane for GC-MS. Analysis of the methyl ester trimethylsilyl derivatives or the further methoxime derivatives of the products were carried out in the positive ion electron impact mode (70 eV) using a ThermoFinnigan DSQ mass spectrometer. The initial temperature was set for 150 °C, held for 1 min, and then increased to 300 °C at 20 °C/min increment and held at 300 °C for 3 min.
NMR Analysis
1H NMR and 1H,1H-COSY NMR spectra were recorded on a Bruker DXR 500- or 600-MHz spectrometer at 298 K. Analysis of the allene oxide methyl ester from C18.3ω3 was conducted at 253 K in d-hexane (with ppm values normalized to the hexane peak at 1.30). Other ppm values are reported relative to residual non-deuterated solvent (δ = 7.16 ppm for C6H6).
18O2 Oxygenation in the Synthesis of Labeled 9R-HPODE
The 18O2 labeled 9R-HPODE was prepared using Anabaena 9R-LOX (31) reacted with C18.2ω6 in pH 7.5 Tris buffer under an atmosphere of 18O2. The 18O2 oxygenation product, labeled 9R-HPODE, was purified and quantified by UV spectroscopy (ϵ = 23,000 (32)). The 18O content was measured by negative ion electrospray LC-MS as shown under “Results.”
Incubation Reaction in 18O-labeled Water (H218O)
The incubation buffer for H218O experiments was prepared by addition of 150 μl H218O (≥95% oxygen-18) to 50 μl of 200 mm Tris, 600 mm NaCl, pH 7.5, to generate a buffer containing 50 mm Tris, 150 mm NaCl, pH 7.5. The incubation reactions were conducted in this 50 mm Tris buffer containing the 72% H218O or using 18O2-labeled 9R-HPODE in regular Tris buffer. Control reactions were performed in parallel using 9R-HPODE in regular Tris buffer.
Analysis of the 13-Hydroxyl Stereoconfiguration in the Linoleic Acid-derived Epoxyalcohol Product of the A. marina Enzyme
Authentic 13S-HODE or 13RS-HODE methyl esters were epoxidized using equimolar mCPBA in dichloromethane at 0 °C for 30 min. The epoxidation products were separated by RP-HPLC with methanol/water (85/15, by volume) at a flow rate of 1 ml/min. The peak co-chromatographing with the enzymatic epoxyalcohol product described under “Results” was collected and further separated by SP-HPLC using hexane/isopropanol (100/1, by volume) at 2 ml/min, again collecting the peak that co-chromatographed with the enzymatic product. The enantiomers of the epoxide originating from 13RS-HODE were well resolved using a Chiralpak AD column (25 × 0.46 cm) with a solvent of hexane/ethanol (100/5, by volume). Chromatograms from the chiral column are given in the supplemental materials. The first-eluting peak co-chromatographed with the enzymatic product and was confirmed to have an identical NMR spectrum.
Analysis of the Epoxide Stereoconfiguration in the Linoleic Acid-derived Epoxyalcohol Product of the A. marina Enzyme
This was achieved in five steps as follows: (i) Synthesis of 9,10-epoxy-12Z-octadecenoic acid (supplemental Scheme 1): Linoleic acid (50 mg) was epoxidized using equimolar mCPBA in dichloromethane on ice for 30 min. The sample was washed with 0.1 m NaHCO3 and water, dried, and stored in methanol. Although the two major products, the 12,13- and 9,10-cis-epoxides separate on RP-HPLC (eluting in that order; e.g. Ref. 33), SP-HPLC using the solvent system hexane/isopropanol/glacial acetic acid (100/1/0.1, by volume) gave the same order of elution and a more satisfactory separation (34). (ii) Isolation of the cis-9,10-epoxide enantiomers: the chemically synthesized 9,10-epoxy-12Z-octadecenoic acid enantiomers were resolved using a Chiralpak AD column with a solvent system of hexane/methanol/glacial acetic acid (100/2/0.05, by volume) giving retention times of 24.7 and 30.3 min. (The peaks eluted earlier when ≥100-μg amounts were chromatographed and with tailing, thus limiting the amounts that could be purified per injection). A semi-preparative Chiralpak AD column (25 × 1 cm) was used to prepare milligram quantities. (iii) The absolute stereochemistries were assigned after chemical transformation to 9-HODE (methyl ester acetate derivative) following procedures reported for conversion of epoxyeicosatrienoic acids to HETEs with minor modifications (35). Each 9,10-epoxide enantiomer (4 mg) as the methyl ester was dissolved in tetrahydrofuran (500 μl), ice-cold acetic acid (1 ml) was added, saturated aqueous potassium iodide (200 μl) was then added to the mixture, and the reaction was stirred in the dark for 16 h at room temperature to prepare a mixture of the 9-hydroxy-10-iodo- and 9-iodo-10-hydroxy derivatives. Following evaporation to dryness and extraction three times with Et2O, the combined organic extracts were washed with H2O, 5% Na2SO3, 5%NaHCO3, and brine. The organic layer was dried under nitrogen, dissolved in dichloromethane (500 μl), pyridine and acetic anhydride (300 μl, 2:1, v/v) were added, and the mixture was allowed to react overnight at room temperature. The resulting iodo-acetates were taken to dryness, redissolved in Et2O, and washed with H2O, 5% NaHCO3, and brine. The samples were taken to dryness, redissolved in dry benzene (800 μl), and treated with 5 equivalents of 1,8-diazabicyclo[5,4,0]undec-7-ene (10 μl). After overnight reaction, the mixture of 9-HODE methyl ester acetates was washed with concentrated CuSO4 solution (to remove the 1,8-diazabicyclo[5,4,0]undec-7-ene), and the organic layer was dried under nitrogen, and redissolved in ethanol. The 9-HODE methyl ester acetate samples were purified by SP-HPLC with a solvent system of hexane/isopropanol (100/0.5, by volume). The chemically derived 9-HODE ester acetates were chromatographed on a Chiralpak AD column using a solvent system of hexane/methanol (100/1, by volume) run at 1 ml/min. Authentic standards prepared directly from 9R-HODE and 9RS-HODE were also run on the same system to assign the stereoconfiguration, with the 9S enantiomer eluting first at 4.4 min and the 9R at 5.0 min. In this way it was shown that of the two 9,10-epoxides resolved earlier on the chiral column, the first-eluting enantiomer was converted to 9S-HODE and thus was itself 9S,10R-epoxy-12Z-octadecenoic acid, and the second peak, which gave 9R-HODE, was 9R,10S-epoxy-12Z-octadecenoic acid. (iv) Each epoxide enantiomer (2 mg of the methyl ester) was reacted with singlet oxygen using methylene blue as sensitizer (1 ml of a 0.6 mg/15 ml methanol solution) and a strong source of visible light at 0 °C for 2 days. At this stage, ∼50% was converted to a more polar product on TLC. The samples were taken to dryness, redissolved in ethyl acetate (5 ml), and passed through the Bond-Elut silica cartridge (1 g) to remove the methylene blue. After esterification with diazomethane and treatment with a 1.5-fold excess of triphenylphosphine to reduce hydroperoxides, each sample was resolved into two major 9,10-epoxy-13-hydroxy diastereomers by SP-HPLC using a Beckman Ultrasphere silica column (25 × 1 cm) with a solvent system of hexane/isopropanol (100/2, by volume, 4 ml/min, retention times of 13.7 and 14.5 min). The first eluting diastereomer from each original chiral epoxide co-chromatographed with the natural product. These enantiomers were collected separately for further comparison with the enzymatic product and structural confirmation by NMR. (v) To distinguish between the two chemically synthesized enantiomeric 9,10-epoxy-13-hydroxy products that matched the natural product on SP-HPLC, they were compared with the enzymatic product by chiral column analysis using a Chiralpak AD column with a solvent system of hexane/ethanol (100/5, by volume) at flow rate of 1 ml/min. The two chemically synthesized enantiomers resolved at retention times of 14.5 and 16.5 min, and the first-eluting enantiomer, the one derived from the 9R,10S-epoxide, co-chromatographed with the enzymatic product.
Alkali Isomerization of the C18.3ω3-derived Cyclopentenone
The cyclopentenone ester of C18.3ω3 was treated with 0.1 m methanolic KOH solution for 30 min at room temperature. After extraction with dichloromethane (and re-esterification with diazomethane to ensure it was still 100% methyl ester), it was compared with an untreated sample on SP-HPLC and RP-HPLC with the solvent system of hexane/isopropanol (100/1.5, by volume) or methanol/H2O (85/15, by volume), respectively.
RESULTS
Cloning, Expression, and Purification of the Mini-catalase/Lipoxygenase Fusion Protein
BLAST searches using the catalase-related allene oxide synthase domain of Plexaura homomalla identified a good match (E value, 3e-30; 29% amino acid identity) on the main chromosome of A. marina MBIC11017, a hypothetical protein of 805 amino acids designated as ABW27596. The polypeptide consisted of an N-terminal catalase-related sequence and C-terminal lipoxygenase (and an alignment with related fusion proteins is given in supplemental Fig. S1). The full-length cDNA was cloned from A. marina genomic DNA with an N-terminal His tag encoded in the upstream PCR primer. Separate constructs were also prepared of the N-terminal catalase-related domain and the C-terminal lipoxygenase. After insertion into the expression vector pET17b the proteins were expressed in Escherichia coli BL21(DE3) and subsequently purified by nickel-nitrilotriacetic acid affinity chromatography. The full-length fusion protein was recovered with a yield of ∼110 mg of the green-colored protein per liter of bacterial culture. The UV-visible spectrum exhibited a typical hemoprotein absorbance with the main Soret band at 405 nm. SDS-PAGE showed a major band at the predicted molecular mass of 92 kDa with a purity of ∼90% (supplemental Fig. S2). Expression of the catalase-related domain by itself (amino acids 1–351) gave strong expression of the protein but with low (<10%) incorporation of heme (data not shown). Nearly all the experiments described below used the full-length protein, although we confirmed that the N-terminal construct exhibited the same activity in metabolism of fatty acid hydroperoxides.
Activity of the A. marina LOX and Hemoprotein Domains
In results to be reported in detail elsewhere, the LOX domain of the A. marina fusion protein was found to oxygenate stearidonic acid (C18.4ω3, the most prominent polyunsaturated fatty acid in A. marina) to its 12R-hydroperoxide, to similarly metabolize α-linolenic acid to 12R-HPOTrE, while the two ω6 fatty acids tested, linoleic acid and arachidonic acid, were converted to 9R-HPODE and 11R-HPETE, respectively. These hydroperoxides were not further converted by the LOX-only domain, whereas they were rapidly metabolized by the full-length protein containing the hemoprotein domain and similarly by the hemoprotein-only construct. Three of the hydroperoxide substrates were prepared, and their rates of metabolism by the full-length A. marina fusion protein were monitored by disappearance of the conjugated diene chromophore at 235 nm. The turnover rates calculated from the rate of decrease of absorbance at 235 nm were 669 ± 24 s−1 for 12R-HPOTrE, 562 ± 61 s−1 for 9R-HPODE, and 671 ± 32 s−1 (mean ± S.E.) for 11R-HPETE at 60 μm substrate concentrations, respectively.
Stable End-products from C18.3ω3 and C18.4ω3
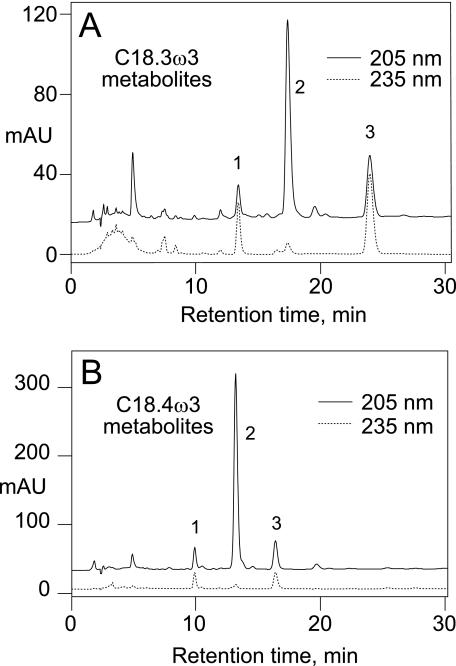
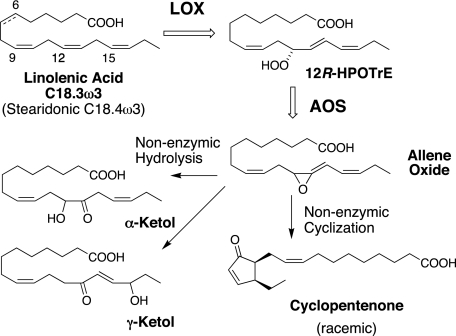
RP-HPLC analysis of the products from C18.3ω3 or C18.4ω3 revealed three prominent peaks with a characteristic profile in the UV (Fig. 2, peaks 1–3). In the case of C18.3ω3, we established that a similar profile is obtained starting with the fatty acid or purified 12R-HPOTrE. The first-eluting α-linolenic acid product, the minor of the three, was identified from its UV spectrum (an enone chromophore with λmax at 225 nm in RP-HPLC solvent), LC-MS (M-H ion at m/z 309 in the negative ion mode, corresponding to a molecular weight of 310), and by GC-MS of the methyl ester TMS ether and methyl ester TMS ether methoxime derivatives (with and without hydrogenation) as the γ-ketol, 13-keto-16-hydroxyoctadeca-9,14-dienoic acid (supplemental materials). The second and third products were identified using the same methods and also using 1H NMR. The second and most abundant product, which only displays end-absorbance in the UV (no distinct chromophore above 205 nm), was identified as the α-ketol 12-hydroxy-13-ketooctadeca-9,15-dienoic acid (supplemental Table 1). The last main product has a distinctive enone chromophore (λmax at 223 nm in RP-HPLC solvent) and was identified by GC-MS and NMR as the cyclopentenone formed by bridging the carbon chain between C12 and C16 (supplemental Table 2). The cyclopentenone has the cis arrangement of the two side chains and is racemic (supplemental Fig. S3). The corresponding three products from stearidonic acid were identified from their HPLC elution characteristics, UV spectra and by GC-MS as the direct analogues of the three α-linolenate products, i.e. γ-ketol (13-keto-16-hydroxy), the major α-ketol (12-hydroxy-13-keto), and cyclopentenone (supplemental materials).
FIGURE 2.
RP-HPLC analyses of products from the reaction of the A. marina fusion protein with C18.3ω3 (A) and C18.4ω3 (B). A Waters Symmetry C18 column (25 × 0.46 cm) was eluted with a solvent system of methanol/water/acetic acid in the proportions 70/30/0.01 (v/v), at a flow rate of 1 ml/min, and with UV detection at 205 and 235 nm. (For clarity, the 205 nm recording is offset from zero.) The peaks labeled 1, 2, and 3 in each chromatogram were identified as the corresponding γ-ketol, α-ketol, and cyclopentenone, respectively (see main text).
Isolation and Identification of a Novel Allene Oxide
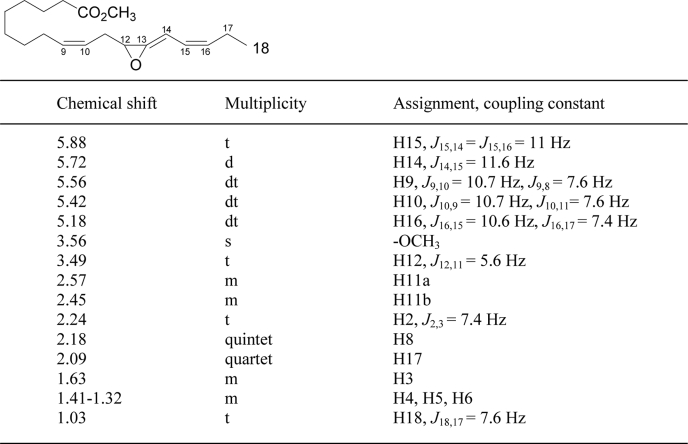
By precedent, the three stable end-products from C18.3ω3 (or C18.4ω3) are formed by hydrolysis or cyclization of an unstable allene epoxide (e.g. Ref. 36). Using methods we have developed for the biosynthesis, isolation, and HPLC of highly unstable epoxides, we prepared the intermediate by incubation and simultaneous extraction of 12R-HPOTrE with the full-length fusion protein at 0 °C and HPLC-purified the methyl ester derivative at subzero temperature (−15 °C, Fig. 3A). The allene oxide has a characteristic smooth and enone-like UV chromophore with λmax at 236.5 nm (Fig. 3B). The 1H NMR spectrum, recorded at −20 °C in d-hexane (Fig. 3C), resembles that of the allene oxide formed from 13S-HPOTrE in the jasmonate biosynthetic pathway (30). There are signals from five protons in the olefinic region, notably including a doublet at H14 coupling only to H15, with no proton at C13. H12 appears as a triplet at 3.49 ppm and the two H11 protons, with different chemical shifts due to the chiral center at C12, are located at 2.57 and 2.45 ppm. The NMR spectral data (Table 1) establish the structure as the novel allene oxide as 12,13-epoxyoctadeca-9Z,13,15Z-trienoic acid. The hemoprotein domain of the A. marina enzyme functions as an allene oxide synthase, and hydrolysis and cyclization of the epoxide account for formation of the three stable end-products formed from 12R-hydroperoxy-linolenic acid (Fig. 4).
FIGURE 3.
Isolation and characterization of the unstable allene oxide. A, SP-HPLC profile of the allene oxide methyl ester from 12R-hydroperoxy-linolenic acid chromatographed on a silica column (4.5 × 0.46 cm) with the solvent hexane/ether (100/1, v/v) at a flow rate of 3 ml/min. All procedures were performed at −15 °C. B, UV spectrum of the allene oxide showing the maximum absorbance at 236.5 nm. C, 1H NMR spectrum of the allene oxide methyl ester recorded at −20 °C in d-hexane.
TABLE 1.
1H NMR chemical shifts of the methyl ester of the novel allene oxide (12,13-epoxyoctadeca-9Z,13,15Z-trienoic acid)
The NMR spectrum was acquired at −20 °C in d-hexane solvent.
FIGURE 4.
Transformation of α-linolenic acid by the A. marina fusion protein. The same reactions occur with stearidonic acid (C18.4ω3), which has one additional cis double bond at Δ6 (shown as a dotted bond on the structure of α-linolenate). The allene oxide is unstable and rapidly undergoes non-enzymatic hydrolysis or cyclization.
A Distinctive Product Profile from C18.2ω6
Reaction of the full-length A. marina protein with two ω6 fatty acids was analyzed. Arachidonic acid and its oxygenation product 11R-HPETE were converted solely to the corresponding allene oxide, giving the typical profile on RP-HPLC of γ-ketol, α-ketol, and cyclopentenone (supplemental Fig. S4). Surprisingly, therefore, reaction of linoleic acid or its oxygenation product 9R-HPODE led to a distinctly different pattern of products on RP-HPLC. Although the expected stable end-products from an allene oxide were present, the chromatogram was dominated by a single new peak with strong absorbance at 205 nm in the UV (Fig. 5). LC-MS analysis indicated that the new product and the second-most prominent peak, the α-ketol formed by allene oxide hydrolysis, had the same molecular weight of 312. In the case of the secondary product, its structure was confirmed by 1H NMR and GC-MS as the α-ketol, 9-hydroxy-10-ketooctadec-12Z-enoic acid (supplemental Table 3). The corresponding γ-ketol and cyclopentenone were also identified by GC-MS as minor peaks on the RP-HPLC chromatogram (cf. Fig. 5, and data not shown).
FIGURE 5.
RP-HPLC analyses of products from the reaction of the A. marina fusion protein with C18.2ω6. RP-HPLC conditions are the same as in Fig. 2. The peaks labeled 1, 2, and 3 were identified as a linoleic acid-derived γ-ketol, α-ketol, and cyclopentenone, respectively. The major product, eluting at ∼17.5 min and labeled “new product” showed strong absorbance at 205 nm and was identified as an epoxyalcohol (see main text).
Identification of the Major Product from C18.2ω6
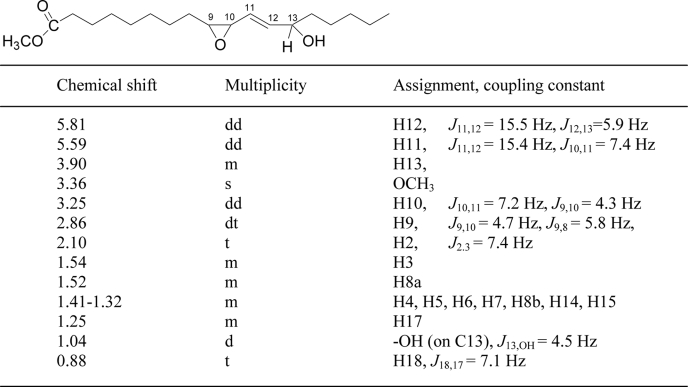
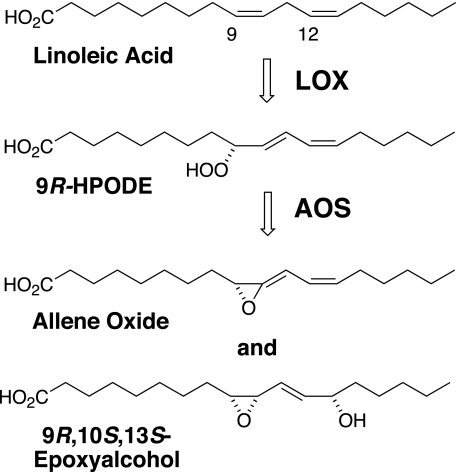
1H NMR analysis outlined a structure of cis-9,10-epoxy-13-hydroxy-octadeca-11E-enoic acid (Table 2). The complete stereochemistry of this epoxyalcohol was established by a series of chemical transformations to the natural product. The 13-hydroxyl configuration was determined via epoxidation of 13S-HODE and 13RS-HODE using mCPBA to a mixture of epoxyalcohols; by successive RP-, SP-, and chiral column-HPLC and final NMR analysis, it was shown that only a 13S-hydroxy-derived epoxyalcohol matched the enzymatic product (supplemental Fig. S5). To determine the epoxide stereochemistry, initially we epoxidized linoleic acid, isolated the 9,10-cis epoxide by SP-HPLC, and resolved the enantiomers by chiral column chromatography; the absolute stereochemistry of the enantiomeric 9R,10S- and 9S,10R-epoxyoctadeca-12Z-enoates was established by chemical transformation to 9-HODE (35), followed by chiral column analysis of the product (37). Then, the two 9,10-cis-epoxides of known configuration were oxidized with singlet oxygen, reduced to the corresponding hydroxy derivatives, and compared with the natural product on HPLC. Only an epoxyalcohol derived from the 9R,10S-epoxide matched the enzymatic product, and again its identity was confirmed by NMR. Thus, the complete stereochemistry of the main A. marina product from C18.2ω6 was proven to be 9R,10S-epoxy-13S-hydroxyoctadeca-11E-enoic acid (Fig. 6).
TABLE 2.
1H NMR chemical shifts of the methyl ester of the epoxyalcohol (9,10cis-epoxy-13-hydroxy-octadeca-11E-enoic acid)
The spectrum was acquired at room temperature in d6-benzene.
FIGURE 6.
Transformation of linoleic acid by the A. marina fusion protein.
Origin of the 13S-Hydroxyl in the Linoleate-derived Epoxyalcohol
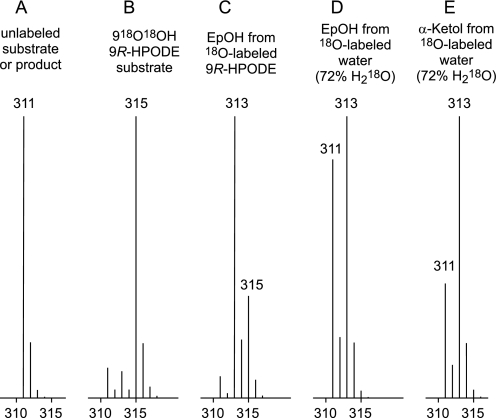
[18O2]9R-HPODE with 84% incorporation of 218O in the hydroperoxy group (Fig. 7, A and B) was synthesized using Anabaena 9R-LOX under an atmosphere of 18O2 and aliquots used as substrate for the A. marina enzyme. LC-MS analysis of the products indicated a marked loss of the 18O-labeled oxygens in transformation to the epoxyalcohol (Fig. 7C): on normalizing the substrate 218O content to 100%, only 27.5% of the epoxyalcohol molecules (27% and 28% in two separate experiments) retained both of the original hydroperoxide oxygens.
FIGURE 7.
Mass spectrometric analysis of oxygen-18 retention or incorporation in epoxyalcohols from 18O-labeled 9R-HPODE substrate or H218O. Partial mass spectral recordings of the prominent M-H ion of unlabeled 9R-HPODE, epoxyalcohol, or α-ketol (A), 18O-labeled 9R-HPODE substrate (B), epoxyalcohol (EpOH) from 18O-labeled 9R-HPODE substrate (C), epoxyalcohol from 18O-labeled water (72% H218O in the reaction buffer) (D), and α-ketol from 18O-labeled water (72% H218O in the reaction buffer (E).
The allene oxide hydrolysis product, the α-ketol, was analyzed in the same LC-MS runs, and as expected it showed complete loss of one of the original hydroperoxy oxygens (data not shown). Actually, the retention of label in the α-ketol was measured as less than one 18O label due to exchange of the oxygen in the 10-ketone during sample work-up, a phenomenon reported in the first account of 18O labeling of an α-ketol from a fatty acid hydroperoxide (38). The loss of label was particularly obvious when the α-ketol was pre-purified by HPLC prior to LC-MS analysis. We also confirmed that pre-purification had no effect on the label measured in the epoxyalcohol.
To account for the incorporation of the missing hydroperoxy oxygen in the epoxyalcohol, we also analyzed the products from incubations of unlabeled 9R-HPODE and A. marina enzyme in buffer containing 72% H218O. These experiments showed a measured 52% incorporation of one 18O from water in the epoxyalcohol (Fig. 7D), which, when normalized to 100% H218O, corresponds to 72% incorporation of 18O1. By comparison, the α-ketol, which is expected to incorporate 100% of one 18O from water in the 9-hydroxyl during hydrolysis of its allene oxide precursor (and this 9-hydroxy oxygen is non-exchangeable), gave a measured content of 66% (Fig. 7E), which when normalized to 100% H218O corresponds to 92% 18O1 incorporation.
Epoxyalcohol from the [18O2]9R-HPODE and H218O experiments was analyzed by GC-MS to locate the position of the 18O labels. Catalytic hydrogenation converts the 9,10-epoxy-13-hydroxy epoxyalcohol partly to the 9,13-dihydroxy hydrogenolysis product (cf. Ref 39), which gives prominent ions for the 9- and 13-hydroxyls when analyzed as the trimethylsilyl derivative. The results showed that the original hydroperoxy oxygen was fully retained at C9 and that oxygen from water (∼70%) is incorporated at C13 (data not shown).
Incubation pH and 18O Incorporation in the Epoxyalcohol
As considered more fully under “Discussion,” one explanation for the appearance of oxygen from water in the epoxyalcohol is exchange of the heme ferryl oxygen with the solvent. This process can be pH-dependent due to involvement of an ionizable distal heme histidine in the exchange (40–43). To examine the possibility of a pH dependence in the oxygen incorporations with the A. marina enzyme, in a first experiment we incubated [18O2]9R-HPODE with enzyme at pH 5.0, 7.5, and 9.5 while monitoring the transformation by UV spectroscopy (repetitive scanning at 200–300 nm to observe disappearance of the conjugated diene chromophore). In the three incubations the incorporation of two hydroperoxy oxygens in the epoxyalcohol product (normalized to 100% 218O content of the substrate) was 32, 27, and 29%, respectively, essentially showing no effect of pH. As the enzyme reaction worked well at the higher pH and because the pKa of the distal heme His reported for catalase and peroxidases is around pH 8.5–9.0 (40, 42), in a second experiment we extended the incubation range up to pH 11. At pH 11 the enzyme maintained a reaction rate of ∼21% compared with pH 7.5 (and ∼43% at pH 10). LC-MS analysis of the epoxyalcohol formed from [18O2]9R-HPODE showed similar 18O retentions at pH 7.5, 10, and 11 (31, 33, and 36%, respectively, again normalized to 100% 218O content of the substrate).
DISCUSSION
Characteristics of the A. marina Protein Sequence
The primary structure of the A. marina catalase-related domain has the closest overall homology to that of Anabaena PCC 7120 (∼30% amino acid identity) (see alignment in supplemental Fig. S1), although the two exhibit different catalytic activities (cf. Refs. 17, 18). The percent identity is relatively low (20–25%) to P. homomalla and other coral AOS even though their reactions are more closely related. At the position of the distal heme His-39, A. marina has the sequence RATH. This is similar to all the fatty acid hydroperoxide-metabolizing catalase-related enzymes characterized to date in having a Thr immediately before the distal His, and contrasts with the typical Val of true catalases (44). The Thr has been shown to support efficient catalytic activity while preventing any reaction with hydrogen peroxide (possibly by hydrogen bonding with an imidazole nitrogen of the His and consequently holding it in an unproductive orientation for catalatic reaction) (44). According to the alignment (supplemental Fig. S1), the A. marina enzyme substitutes Ser (Ser-120) for the distal heme Asn typical of catalases and the coral AOS. Around the proximal heme ligand, Tyr-334, A. marina exhibits the typical signature sequence of catalases (RxxxYxxxxxxR). This is the same in catalase and its structural relatives, except Anabaena, which based on alignments lacks the first Arg and has a His replacing the Tyr as the proximal heme ligand (17). Perhaps related to this, like the true catalases the A. marina and coral enzymes are dark green in color, whereas the Anabaena enzyme appears straw-colored or brownish.
Main Products of the A. Marina Fusion Protein
In transformations that probably mimic the natural pathway in A. marina, the LOX domain of the fusion protein oxygenates stearidonic acid (and similarly α-linolenic acid) with a unique specificity forming the 12R-hydroperoxide, and then the AOS activity of the catalase-related domain converts this to a novel 12R,13-epoxy allene oxide (Fig. 4). The relative abundance of C18.4ω3 in A. marina and its transformation to a single product by the AOS/LOX is supportive of the primary importance of this route of metabolism. By analogy to the jasmonate pathway in plants (8), the C18.4 allene oxide may be the metabolic precursor of a lipid signaling molecule in A. marina, or alternatively, by analogy to unstable mediators of mammalian systems (thromboxane A2, and prostacyclin) (45), it might also function in that role itself.
The jasmonate pathway in plants is associated with an allene oxide cyclase that is specific for the 13S-hydroperoxide of α-linolenic acid (46, 47). In marine invertebrates expressing a catalase-related AOS, it is inferred that the pathway leads to cyclopentenone-derived products (e.g. clavulones and punaglandins (48, 49)), although an enzymatic allene oxide cyclization has yet to be identified. The same applies to the allene oxide biosynthesis we describe here in A. marina. Just as the plant CYP74 AOS and the cAOS are unrelated hemoproteins (that specifically metabolize S and R configuration hydroperoxides, respectively), so the putative cyclase could also be quite distinct in structure and not identifiable based on protein homology. Lipidomic analyses on these cyanobacteria may establish the existence of allene oxide-derived products and help direct future enzymological studies.
A Mechanistically Significant By-product
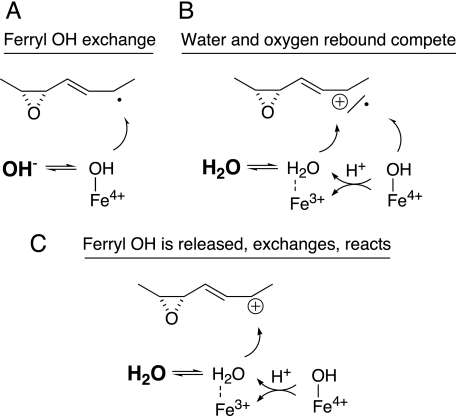
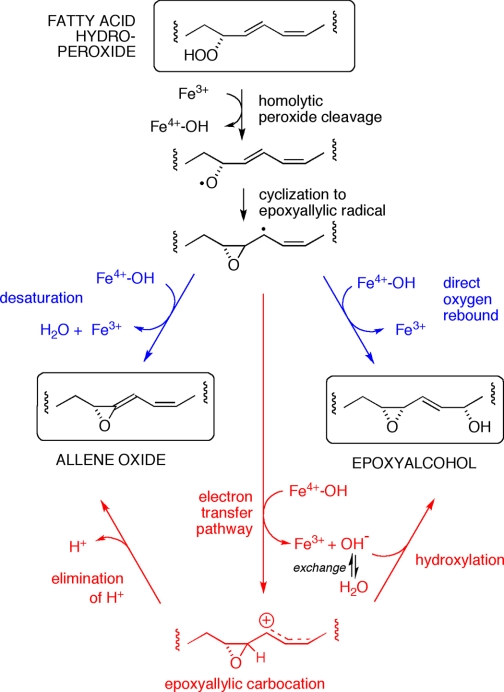
Considering that the two C18 ω3 fatty acids and arachidonic acid (and 11R-HPETE) are transformed cleanly to allene oxides, it was surprising to find that linoleic acid and its 9R-hydroperoxide yield the allene oxide as a relatively minor product. The major end-product from linoleic acid is the epoxyalcohol 9R,10S-epoxy-13S-hydroxyoctadec-11E-enoic acid. This single isomer is unusual among fatty acid hydroperoxide-derived epoxyalcohols in being a cis epoxide. Although the specific transformation of hydroperoxy fatty acids to cis-epoxy products has been described before (by plant peroxygenase enzymes (50) and by a hydroperoxide isomerase from the fungus Saprolegnia parasitica (51)), these mechanisms involve oxygen transfer and epoxidation of a cis double bond. Recently a P450 CYP74-catalyzed transformation of 13S-hydroperoxylinoleic acid to an 11-hydroxy-12,13-cis-epoxyalcohol was reported (13), which is more akin to our findings. In both cases the cis epoxide forms from the carbon bearing the hydroperoxide moiety. Unexpectedly, we found that the 13-hydroxyl in the 9R-HPODE-derived epoxyalcohol derives only partly (∼30%) from the distal oxygen of the hydroperoxide group; 70% arises by incorporation of oxygen from water. We consider it significant that this stereochemically pure epoxyalcohol is formed partly by incorporating oxygen from water as it provides strong evidence of an ionic intermediate in the reaction of enzyme with hydroperoxide. As to the mechanism of exchange, three potential explanations (illustrated diagrammatically in Fig. 8) are considered in the following discussion: (i) the heme ferryl oxygen exchanges with water prior to oxygen rebound, (ii) hydroxylation with water occurs in parallel to oxygen rebound, and (iii) the ferryl oxygen is released whereupon it mixes with water, which reacts to give the hydroxyl at C13.
FIGURE 8.
Three potential mechanisms explaining the appearance of oxygen from water in the 13-hydroxyl of the linoleic acid-derived epoxyalcohol. A, exchange of the ferryl heme oxygen with water prior to oxygen rebound. B, formation of the 13-hydroxyl via two routes, direct oxygen rebound of the ferryl oxygen, and reaction of water with a carbocation intermediate. C, water reacts with a carbocation intermediate, and the water derives partly from release of the ferryl oxygen, and partially from H2O in the enzyme active site.
Discussion of Ferryl Heme Oxygen Exchange
The first possibility is that solely oxygen from the heme accounts for the formation of the single specific epoxyalcohol and that this ferryl heme oxygen partly exchanges with water prior to rebound (Fig. 8A). Such an exchange contrasts with evidence on P450 transformations that the heme ferryl oxygen does not exchange (52). (There is an exceptional and not directly relevant P450-catalyzed exchange of the oxygen in the oxygen donor iodosobenzene with solvent, producing an activated ferryl heme with oxygen from water, but this reaction occurs in the absence of substrate (52–55).) To the best of our knowledge, the only reported example of ferryl oxygen exchange in P450 catalysis is the 8% incorporation of 18O from H218O in the cumene hydroperoxide-dependent hydroxylation of cyclohexane by rabbit P450LM (CYP2B4) (56); in the same study, no exchange was seen using NADPH and the reductase with molecular oxygen. Cumene hydroperoxide will oxidize the P450 heme to Compound I (Fe4+ = O), at which point the by-product cumyl alcohol has to get out of the way (if not out of the active site) to permit interaction of the cyclohexane substrate with the ferryl oxygen. And only after hydrogen abstraction from cyclohexane will the resulting Compound II be formed that is equivalent to the ferryl oxygen in AOS catalysis. So, with the added involvement of Compound I and the lack of immediate access of the cyclohexane, there is more opportunity for exchange of a ferryl oxygen in the cumene hydroperoxide-activated monooxygenase system.
Although not generally seen with P450s, oxygen exchange is a recognized phenomenon in some other hemoproteins, including horseradish peroxidase, cytochrome c peroxidase, lactoperoxidase, myeloperoxidase, and bovine liver catalase (40–42, 57, 58). As these enzymes are not oxygenases, the ferryl oxygen does not appear in the product, and therefore the exchange is observed spectroscopically, using resonance Raman. Compound II is generated chemically and is sufficiently stable to allow spectral recording over several minutes. It is in this long lasting species that exchange occurs. Definite rates of exchange are not given. Although it is usually assumed to be fast, in some cases it is implied to occur within a few minutes. The exchange is pH-sensitive and is linked to ionization of the distal heme His; it is curtailed at 1–2 pH units above the His pKa, reported at 8.5–8.8 for Compound II of horseradish peroxidase (40, 42). In our study, 18O incorporation into the epoxyalcohol was unaffected by pH within the range 5–11. Besides the issue of ferryl oxygen exchange, this is remarkable, because it suggests that the distal heme His (with a likely pKa within the pH range 5–9) is not ionizing during catalysis or that its ionization is impervious to external pH.
We can measure the turnover rate of the A. marina hemoprotein at ∼600 per second (readily observed as disappearance of the conjugated diene chromophore of the hydroperoxide substrate). This rate encompasses all phases of the substrate-enzyme interaction, namely, encountering and binding the substrate, catalysis, and product release. Consider also that the ferryl heme is not present until the peroxyl oxygens are cleaved, at which point it exists as a Compound II, poised for oxygen rebound or electron transfer, either of which returns the heme to the ferric state. In typical P450 reactions the oxygen rebound step can be very fast indeed, in the order of 109-1010 s−1 (59), although the delocalized fatty acid radical or carbocation intermediates in our system could tolerate longer lifetimes. (Nonetheless, contrast this evanescent existence with the long lasting Compound II used for spectral measurements of exchange.) We are uncertain as to how fast a ferryl oxygen could equilibrate with water in the active site, but if it occurred here it would need to be able to out-compete the rate of oxygen rebound, which seems unlikely.
Two Routes to the Linoleate Epoxyalcohol?
The next possibility is that the linoleate epoxyalcohol is formed both by oxygen rebound and by reaction with solvent, giving the same product (Fig. 8B). We would argue against this possibility on the grounds that the formation of the one specific epoxyalcohol, when there is the potential for multiple isomers, argues for strict enzymatic control. By this scheme the enzyme needs to accomplish solely 13S-hydroxylation both via radical and carbocation pathways, and it is difficult to accept that one single specific epoxyalcohol isomer is formed with the hydroxyl oxygen coming through two different mechanisms.
Exchange Occurs After Release of Ferryl Oxygen
The third explanation avoids the dilemma of two separate routes to the one product and can also account for the exchange. If the ferryl oxygen is released, returning the enzyme to the resting ferric state, the free OH− could be partly exchangeable with solvent and yet amenable to an enzyme-directed reaction with the fatty acid intermediate (Fig. 8C). By this mechanism, there is no doubt that the OH− reacts with an ionic species of intermediate. Reaction with this “free” OH− begs the question as to how the enzyme controls the stereochemistry of the hydroxylation. Presumably the ferric heme can bind a water molecule, and perhaps the active site His-39 and Ser-120 (the substitute in A. marina for the usual distal heme Asn) may also play a role.
Implications for the Mechanism of Allene Oxide Synthesis
As noted in the introduction, the mechanism of the specific fatty acid hydroperoxide transformations catalyzed by the CYP74 hemoproteins has been considered for many years and has been presented as proceeding via a radical or carbocation pathway, or by incorporating elements of both (11, 20, 60). The same issues pertain to the catalase-related enzymes, especially in relation to AOS activity, which is common to both classes of hemoprotein. The consensus is that reaction is initiated by a homolytic cleavage of the hydroperoxide (Fig. 9). Cyclization of the resulting alkoxyl radical gives an epoxyallylic carbon radical. At this stage, the required loss of a hydrogen atom (H·) to give the allene oxide raises a red flag. In most cases some type of proton-coupled electron transfer process is considered more feasible than an actual hydrogen abstraction (61). So it is proposed that completion of the allene oxide synthase reaction proceeds via transfer of an electron from the carbon, forming a carbocation intermediate, with loss of a proton yielding the final product (11). Part of the argument in support of this line of reasoning is based on feasibility as determined by theory and part by chemical precedent (21), yet with the current state of knowledge this leaves unresolved issues on both counts. Accordingly, we consider it significant that we have identified a specific product formed with a clearly demonstrable ionic component to the mechanism, namely the major epoxyalcohol product from linoleic acid.
FIGURE 9.
Radical and ionic pathways to allene oxide and epoxyalcohol. After homolytic hydroperoxide cleavage and formation of the epoxyallylic radical (top), the subsequent diverging transformations can be represented as radical reactions (blue), or as ionic via a carbocation intermediate (red). The incorporation of oxygen from water in the epoxyalcohol product supports the existence of the carbocation, the intermediate that best accounts for the chemistry of allene oxide synthesis (22).
Both the CYP74 AOS and the cAOS catalyze a dehydrase reaction on the fatty acid hydroperoxide, and therefore both classes of hemoprotein produce water as OH− and H+ in consecutive steps of the catalytic cycle. Also, note that when using their natural substrates, water does not intercept a reacting intermediate and an allene oxide is the sole product with both classes of hemoprotein. We had argued before that the appearance of epoxyalcohols in association with allene oxide synthesis by a purified CYP74 enzyme is evidence of a close parallel in the mechanism of their production (60). (A parallel can be made with conventional P450s, which typically hydroxylate their substrate yet a few can also catalyze desaturation (62–67).) The fatty acid epoxyalcohols were produced previously when flaxseed AOS (CYP74A1) was presented with unnatural hydroxy-hydroperoxy fatty acid substrates (60). It appeared that these unnatural substrates upset the enzymatic control and allowed “oxygen rebound” instead of the usual hydrogen atom abstraction. Indeed, we found complete retention of the hydroperoxide oxygens in these CYP74A1-derived epoxyalcohols. Although the association of allene oxide and epoxyalcohol synthesis remains valid, what the previous evidence does not provide is a link to any ionic reactions in the course of, or parallel with, allene oxide synthesis. Here the evidence is different in several ways. The substrate is a natural fatty acid, only one pure epoxyalcohol derivative is formed, it is solely a cis epoxide, and, most significantly, it arises largely via a final ionic pathway incorporating OH from water in the 13S-hydroxyl. As it can reasonably be assumed that oxygen from water reacted with an intermediate carbocation in the enzyme active site, the findings present an intimate link between AOS synthesis and an ionic reaction intermediate. We can at least conclude that it is likely that the ferryl heme releases the bound OH as a fatty acid carbocation intermediate is formed, and that the fatty acid carbocation is hydroxylated by OH− partially equilibrated with water molecule(s) in the active site. In addition, in relation to AOS activity per se, for the first time ionic events can be detected in parallel with synthesis of the allene epoxide.
Supplementary Material
Acknowledgments
We thank Alessandro Repetto for help with the NMR.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-074888.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Tables S1–S3, and Scheme 1.
- LOX
- lipoxygenase
- AOS
- allene oxide synthase
- cAOS
- catalase-related allene oxide synthase
- H(P)ODE
- hydro(pero)xyoctadecadienoic acid
- H(P)OTrE
- hydro(pero)xyoctadecatrienoic acid
- H(P)ETE
- hydro(pero)xyeicosatetraenoic acid
- mCPBA
- meta-chloroperoxybenzoic acid
- RP-HPLC
- reversed-phase high pressure liquid chromatography
- SP-HPLC
- straight-phase HPLC
- GC-MS
- gas chromatography mass spectrometry
- LC-MS
- liquid chromatography-mass spectrometry.
REFERENCES
- 1.Smith W. L., Song I. (2002) Prostaglandins Other Lipid Mediat. 68–69, 115–128 [DOI] [PubMed] [Google Scholar]
- 2.Hamberg M., Ponce de Leon I., Rodriguez M. J., Castresana C. (2005) Biochem. Biophys. Res. Commun. 338, 169–174 [DOI] [PubMed] [Google Scholar]
- 3.Garscha U., Oliw E. (2008) Biochem. Biophys. Res. Commun. 373, 579–583 [DOI] [PubMed] [Google Scholar]
- 4.Garscha U., Oliw E. H. (2009) J. Biol. Chem. 284, 13755–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodhun F., Göbel C., Hornung E., Feussner I. (2009) J. Biol. Chem. 284, 11792–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brash A. R. (1999) J. Biol. Chem. 274, 23679–23682 [DOI] [PubMed] [Google Scholar]
- 7.Oliw E. H. (2002) Prostaglandins Other Lipid Mediat. 68–69, 313–323 [DOI] [PubMed] [Google Scholar]
- 8.Liavonchanka A., Feussner I. (2006) J. Plant Physiol. 163, 348–357 [DOI] [PubMed] [Google Scholar]
- 9.Rådmark O., Werz O., Steinhilber D., Samuelsson B. (2007) Trends Biochem. Sci. 32, 332–341 [DOI] [PubMed] [Google Scholar]
- 10.Senger T., Wichard T., Kunze S., Göbel C., Lerchl I., Pohnert G., Feussner I. (2005) J. Biol. Chem. 280, 7588–7596 [DOI] [PubMed] [Google Scholar]
- 11.Ullrich V., Brugger R. (1994) Angew. Chem. Int. Ed. Engl. 33, 1911–1919 [Google Scholar]
- 12.Stumpe M., Feussner I. (2006) Phytochem. Rev. 5, 347–357 [Google Scholar]
- 13.Lee D. S., Nioche P., Hamberg M., Raman C. S. (2008) Nature 455, 363–368 [DOI] [PubMed] [Google Scholar]
- 14.Tijet N., Brash A. R. (2002) Prostaglandins Other Lipid Mediat. 68–69, 423–431 [DOI] [PubMed] [Google Scholar]
- 15.Koljak R., Boutaud O., Shieh B. H., Samel N., Brash A. R. (1997) Science 277, 1994–1996 [DOI] [PubMed] [Google Scholar]
- 16.Oldham M. L., Brash A. R., Newcomer M. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider C., Niisuke K., Boeglin W. E., Voehler M., Stec D. F., Porter N. A., Brash A. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18941–18945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niisuke K., Boeglin W. E., Murray J. J., Schneider C., Brash A. R. (2009) J. Lipid Res. 50, 1448–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grechkin A. N., Brühlmann F., Mukhtarova L. S., Gogolev Y. V., Hamberg M. (2006) Biochim. Biophys. Acta 1761, 1419–1428 [DOI] [PubMed] [Google Scholar]
- 20.Crombie L., Morgan D. O. (1991) J. Chem. Soc. Perkin Trans. 1, 581–587 [Google Scholar]
- 21.Gerwick W. H. (1996) Lipids 31, 1215–1231 [DOI] [PubMed] [Google Scholar]
- 22.Ullrich V. (2003) Arch. Biochem. Biophys. 409, 45–51 [DOI] [PubMed] [Google Scholar]
- 23.Swingley W. D., Chen M., Cheung P. C., Conrad A. L., Dejesa L. C., Hao J., Honchak B. M., Karbach L. E., Kurdoglu A., Lahiri S., Mastrian S. D., Miyashita H., Page L., Ramakrishna P., Satoh S., Sattley W. M., Shimada Y., Taylor H. L., Tomo T., Tsuchiya T., Wang Z. T., Raymond J., Mimuro M., Blankenship R. E., Touchman J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mimuro M., Tomo T., Tsuchiya T. (2008) Photosynth. Res. 97, 167–176 [DOI] [PubMed] [Google Scholar]
- 25.Ohashi S., Miyashita H., Okada N., Iemura T., Watanabe T., Kobayashi M. (2008) Photosynth. Res. 98, 141–149 [DOI] [PubMed] [Google Scholar]
- 26.Peers K. E., Coxon D. T. (1983) Chem. Phys. Lipids 32, 49–56 [Google Scholar]
- 27.Boutaud O., Brash A. R. (1999) J. Biol. Chem. 274, 33764–33770 [DOI] [PubMed] [Google Scholar]
- 28.Imai T., Globerman H., Gertner J. M., Kagawa N., Waterman M. R. (1993) J. Biol. Chem. 268, 19681–19689 [PubMed] [Google Scholar]
- 29.Gao B., Boeglin W. E., Brash A. R. (2008) Arch. Biochem. Biophys. 477, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brash A. R., Baertschi S. W., Ingram C. D., Harris T. M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3382–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y., Boeglin W. E., Schneider C., Brash A. R. (2008) J. Biol. Chem. 283, 5138–5147 [DOI] [PubMed] [Google Scholar]
- 32.Gibian M. J., Vandenberg P. (1987) Anal. Biochem. 163, 343–349 [DOI] [PubMed] [Google Scholar]
- 33.Bylund J., Ericsson J., Oliw E. H. (1998) Anal. Biochem. 265, 55–68 [DOI] [PubMed] [Google Scholar]
- 34.Laethem R. M., Balazy M., Koop D. R. (1996) Drug Metab. Dispos. 24, 664–668 [PubMed] [Google Scholar]
- 35.Falck J. R., Manna S., Jacobson H. R., Estabrook R. W., Chacos N., Capdevila J. (1984) J. Am. Chem. Soc. 106, 3334–3336 [Google Scholar]
- 36.Grechkin A. N., Kuramshin R. A., Safonova E. Y., Latypov S. K., Ilyasov A. V. (1991) Biochim. Biophys. Acta 1086, 317–325 [DOI] [PubMed] [Google Scholar]
- 37.Schneider C., Boeglin W. E., Brash A. R. (2000) Anal. Biochem. 287, 186–189 [DOI] [PubMed] [Google Scholar]
- 38.Veldink G. A., Vliegenthart J. F., Boldingh J. (1970) FEBS Lett. 7, 188–190 [DOI] [PubMed] [Google Scholar]
- 39.Antón R., Puig L., Esgleyes T., de Moragas J. M., Vila L. (1998) J. Invest. Dermatol. 110, 303–310 [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto S., Tatsuno Y., Kitagawa T. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 2417–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oertling W. A., Hoogland H., Babcock G. T., Wever R. (1988) Biochemistry 27, 5395–5400 [DOI] [PubMed] [Google Scholar]
- 42.Chuang W. J., Heldt J., Van Wart H. E. (1989) J. Biol. Chem. 264, 14209–14215 [PubMed] [Google Scholar]
- 43.Proshlyakov D. A., Paeng I. R., Paeng K. J., Kitagawa T. (1996) Biospectroscopy 2, 317–329 [Google Scholar]
- 44.Tosha T., Uchida T., Brash A. R., Kitagawa T. (2006) J. Biol. Chem. 281, 12610–12617 [DOI] [PubMed] [Google Scholar]
- 45.Funk C. D., FitzGerald G. A. (2007) J. Cardiovasc. Pharmacol. 50, 470–479 [DOI] [PubMed] [Google Scholar]
- 46.Ziegler J., Wasternack C., Hamberg M. (1999) Lipids 34, 1005–1015 [DOI] [PubMed] [Google Scholar]
- 47.Feussner I., Wasternack C. (2002) Annu. Rev. Plant Biol. 53, 275–297 [DOI] [PubMed] [Google Scholar]
- 48.Kikuchi H., Tsukitani Y., Iguchi K., Yamada Y. (1982) Tetrahedron Lett. 23, 5171–5174 [Google Scholar]
- 49.Baker B. J., Okuda R. K., Yu T. K., Scheuer P. J. (1985) J. Am. Chem. Soc. 107, 2976–2977 [Google Scholar]
- 50.Blée E., Wilcox A. L., Marnett L. J., Schuber F. (1993) J. Biol. Chem. 268, 1708–1715 [PubMed] [Google Scholar]
- 51.Hamberg M., Herman R. P., Jacobsson U. (1986) Biochim. Biophys. Acta 879, 410–418 [Google Scholar]
- 52.Ortiz de Montellano P. R. (ed) (1995) in Cytochrome P450: Structure, mechanism, and biochemistry, Second Ed., pp. 245–303Plenum Press, New York [Google Scholar]
- 53.Heimbrook D. C., Sligar S. G. (1981) Biochem. Biophys. Res. Commun. 99, 530–535 [DOI] [PubMed] [Google Scholar]
- 54.Macdonald T. L., Burka L. T., Wright S. T., Guengerich F. P. (1982) Biochem. Biophys. Res. Commun. 104, 620–625 [DOI] [PubMed] [Google Scholar]
- 55.Ortiz de Montellano P. R. (ed) (1986) in Cytochrome P450: Structure, mechanism, and biochemistry, pp. 217–271Plenum Press, New York [Google Scholar]
- 56.Nordblom G. D., White R. E., Coon M. J. (1976) Arch. Biochem. Biophys. 175, 524–533 [DOI] [PubMed] [Google Scholar]
- 57.Hashimoto S., Teraoka J., Inubushi T., Yonetani T., Kitagawa T. (1986) J. Biol. Chem. 261, 11110–11118 [PubMed] [Google Scholar]
- 58.Rector C. L., Stec D. F., Brash A. R., Porter N. A. (2007) Chem. Res. Toxicol. 20, 1582–1593 [DOI] [PubMed] [Google Scholar]
- 59.Groves J. T. (2006) J. Inorg. Biochem. 100, 434–447 [DOI] [PubMed] [Google Scholar]
- 60.Song W. C., Baertschi S. W., Boeglin W. E., Harris T. M., Brash A. R. (1993) J. Biol. Chem. 268, 6293–6298 [PubMed] [Google Scholar]
- 61.Mayer J. M. (2004) Annu. Rev. Phys. Chem. 55, 363–390 [DOI] [PubMed] [Google Scholar]
- 62.Nagata K., Liberato D. J., Gillette J. R., Sasame H. A. (1986) Drug Metab. Dispos. 14, 559–565 [PubMed] [Google Scholar]
- 63.Rettie A. E., Rettenmeier A. W., Howald W. N., Baillie T. A. (1987) Science 235, 890–893 [DOI] [PubMed] [Google Scholar]
- 64.Korzekwa K. R., Trager W. F., Nagata K., Parkinson A., Gillette J. R. (1990) Drug Metab. Dispos. 18, 974–979 [PubMed] [Google Scholar]
- 65.Wang R. W., Kari P. H., Lu A. Y., Thomas P. E., Guengerich F. P., Vyas K. P. (1991) Arch. Biochem. Biophys. 290, 355–361 [DOI] [PubMed] [Google Scholar]
- 66.Guengerich F. P., Kim D. H. (1991) Chem. Res. Toxicol. 4, 413–421 [DOI] [PubMed] [Google Scholar]
- 67.Rettie A. E., Sheffels P. R., Korzekwa K. R., Gonzalez F. J., Philpot R. M., Baillie T. A. (1995) Biochemistry 34, 7889–7895 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.