Abstract
Epidermal growth factor (EGF)-like modules are defined in part by six cysteines joined by disulfides in a 1–3, 2–4, and 5–6 pattern. Thrombospondin-1 (TSP-1) is a multimodular glycoprotein with three EGF-like modules, E1, E2, and E3, arranged in tandem. These modules likely propagate conformational changes between surrounding C-terminal and N-terminal elements of TSP-1 and interact with other extracellular molecules. E1, E2, and their homologs in other TSPs are unique among EGF-like modules in having two residues rather than one between Cys-4 and Cys-5. In addition, E2 has a calcium-binding site and an unusually long loop between Cys-5 and Cys-6. The structure of E1, E2, or E3 expressed alone changed little upon heating as monitored by far-UV CD, whereas more marked changes occurred in E12, E23, and E123 tandem constructs. The individual modules denatured in differential scanning calorimetry experiments only at >85 °C. E12, E23, or E123 tandem constructs, however, had a transition in the range of 44–70 °C. The temperature of the transition was higher when calcium was present and higher with E123 than with E12 or E23. Isothermal titration calorimetry demonstrated KD values of binding of calcium to E2, E12, E23, or E123 at 25 °C of 11.5, 2.9, 2.2, or 0.3 μm, respectively. Monoclonal antibodies HB8432 and C6.7, which recognize epitopes in E2, bound to E12, E23, or E123 with greater affinity than to E2 alone. These results indicate that interactions among the modules of E123 influence the tertiary structure and calcium binding of E2.
Thrombospondins (TSPs)2 are multimodule, calcium-binding extracellular glycoproteins with various functions (1). TSP-1, which was the first TSP to be discovered and remains the best characterized, and TSP-2 are trimers. Each subunit is composed of an N-terminal module, oligomerization domain, von Willebrand factor type C module, three properdin or TSP type 1 modules, and the C-terminal signature domain that includes three EGF-like modules (E123), 13 aspartate-rich calcium-binding repeats of the wire module, and a lectin-like module (2–4). The five mammalian TSPs fall into two groups, trimeric (TSP-1 and TSP-2) and pentameric (TSP-3, TSP-4, and TSP-5) (1). All have a signature domain, with the major difference being the presence of four rather than three EGF-like modules in the signature domain of pentameric TSPs.
EGF-like modules exist in more than 300 human extracellular proteins and play important roles in biological processes such as blood clotting and cell-cell signaling (5–7). The modules are 30–50 residues long and characterized by six cysteine residues that form three disulfide bonds in the order 1–3, 2–4, and 5–6 (Fig. 1) (6, 7). The backbone structure of the EGF-like modules consists of two submodules, referred to as the major (N-terminal) and minor (C-terminal) submodules (6, 8, 9).
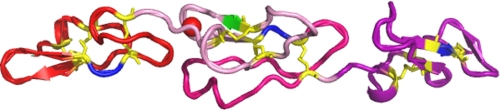
FIGURE 1.
Model of the structure of E123. The model is built based on the crystal structure of EGF modules in the TSP-2 signature domain (Protein Data Bank code 1YO8) using SYBYL 7.0. E1 is shown in red, E2 in pink, and E3 in purple. The cysteines are colored yellow; the backbones of the residues between the fourth and fifth Cys are in blue; Glu-609 recognized by HB8432 and C6.7 is shown in green; and the long loop in E2 between the fifth and sixth Cys is hot pink. Ca2+ bound to the binding site on E2 near the interface between E1 and E2 is depicted as a red ball.
The crystal structure of the three EGF-like modules of TSP-2 has been solved as part of the TSP-2 signature domain in 2 mm calcium (Ca2+) (Fig. 1) (4). All have the 1–3, 2–4, and 5–6 disulfide pattern. There is one Ca2+-binding site in the second EGF-like module (E2), located near the interface between the first and second EGF-like modules (E1 and E2) (Fig. 1). There is only one residue between the fourth and fifth cysteines in most EGF-like modules (6). However, E1 and E2 of TSP-1 and TSP-2 and three of the four EGF-like modules (E1, E2, and E2′) of pentameric TSPs have two residues between the fourth and fifth Cys. This difference is potentially important because the N-terminal major submodule of the repeat containing the 1–3 and 2–4 disulfides and the C-terminal submodule with the 5–6 disulfide have the potential to undergo hinge-like motions around the residues between the fourth and fifth Cys (6, 8, 9). Having two rather than one residue between these two Cys increases the potential flexibility. In addition, E2 modules in all five TSPs contain an unusually long loop of 23 residues between the fifth and sixth Cys (Fig. 1). In the TSP-2 signature domain structure, residues from the long loop interact with repeat 12N of the wire module (4). E3, which has one residue between the fourth and fifth Cys, interacts with the wire and the lectin-like module (3, 4). A common polymorphism (N700S) in wire repeat 1C of human TSP-1 influences the stability of the EGF-like modules (10). This finding suggests that the interactions between the EGF-like modules and more C-terminal elements of the signature domain allow conformational changes in the more C-terminal elements to be propagated N-terminally.
The EGF-like modules (E123) of TSP-1 denature in differential scanning calorimetry (DSC) with a melting temperature of ∼68 °C in 2 mm Ca2+ (10), although most EGF-like modules are stable to heating (7). We have investigated this transition in detail to learn its origins and the influence of Ca2+. The results indicate interactions among the modules of E123 that enhance Ca2+ binding and influence the tertiary structure of E2.
MATERIALS AND METHODS
Cloning of Truncation Constructs into the pAcGP67.coco Transfer Vector
E123 was generated previously (10). The other recombinant constructs encoding the sequences of EGF-like modules of TSP-1 were expressed in the baculovirus system. We used a modification of the pAcGP67 transfer vector, pAcGP67.coco (p.coco), in which cloning sites are flanked by 5′-DNA encoding a signal sequence and 3′-DNA encoding a polyhistidine tag (11). DNA encoding the recombinant proteins was generated by PCR amplification using full-length human TSP-1 cDNA as template. The forward primer contained an XmaI site, and the reverse primer contained an XbaI site. The PCR products were inserted into XmaI and XbaI sites of p.coco. Correct sequences of PCR-amplified DNA were verified by DNA sequencing. The inserted sequences were as follows: E1, DGCLSNPCFAGVKCTSYPDGSWKCGACPPGYSGNGIQCT (with introduced ADPG and LELVPRGSAAGHHHHHH N- and C-terminal tails by cloning); E2, DVDECKEVPDACFNHNGEHRCENTDPGYNCLPCPPRFTGSQPFGQGVEHATANKQVCK (with ADPG and GLELVPRGSAAGHHHHHH N- and C-terminal tails); and E3, PRNPCTDGTHDCNKNAKCNYLGHYSDPMYRCECKPGYAGNGIICGE (with ADPG and ALELVPRGSAAGHHHHHH N- and C-terminal tails). More information on the constructs is given in Table 1.
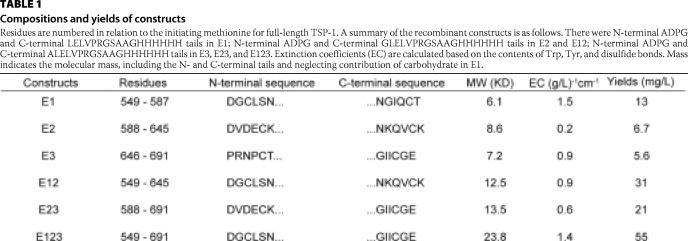
TABLE 1.
Compositions and yields of constructs
Residues are numbered in relation to the initiating methionine for full-length TSP-1. A summary of the recombinant constructs is as follows. There were N-terminal ADPG and C-terminal LELVPRGSAAGHHHHHH tails in E1; N-terminal ADPG and C-terminal GLELVPRGSAAGHHHHHH tails in E2 and E12; N-terminal ADPG and C-terminal ALELVPRGSAAGHHHHHH tails in E3, E23, and E123. Extinction coefficients (EC) are calculated based on the contents of Trp, Tyr, and disulfide bonds. Mass indicates the molecular mass, including the N- and C-terminal tails and neglecting contribution of carbohydrate in E1.
Expression and Purification of the Recombinant Proteins
Recombinant infectious viruses were generated as described (11). We used passage three of the virus (>100 plaque-forming units/ml) to infect High-Five insect cells in SF-900 II serum-free medium at 27 °C. After 60–65 h of incubation, ∼400 ml of conditioned medium was harvested and dialyzed into 10 mm MOPS, 300 mm NaCl, 2 mm CaCl2, pH 7.5 (MOPS buffer). Dialyzed medium was incubated with 3 ml of nickel-nitrilotriacetic acid resin at 4 °C overnight in the presence of 5 mm imidazole. The resin was collected by centrifugation and placed in a column. The column was washed with 45 ml of MOPS buffer with 15 mm imidazole. The protein was eluted in buffer containing 300 mm imidazole and then dialyzed into 10 mm MOPS, 150 mm NaCl, 2 mm CaCl2, pH 7.5. Proteins were purified further with a 1-ml HisTrap column eluted with a gradient of imidazole on a fast protein liquid chromatograph (GE Healthcare). Protein was stored at −80 °C in aliquots and thawed in a 25 °C water bath prior to use. Protein concentration was routinely estimated by A280 nm using extinction coefficients (Table 1) predicted by amino acid content (12). These calculations were validated by amino acid analysis, which agreed to within 8%.
Far-UV CD
Far-UV CD studies were done on a model 202 SF CD spectrophotometer (Aviv Biomedical). The samples were dialyzed against 10 mm Tris, 150 mm NaCl, 2 mm CaCl2, pH 7.5 (Ca2+-TBS), and placed in a 0.1-cm quartz cuvette holding a sample volume of 300 μl. Data were accumulated for 5 s at wavelengths from 200 to 260 nm at temperatures between 25 and 70 °C to define the denaturation profiles of the proteins. The temperature was raised at 10 °C/min, and the wavelength scans were started 5 min after the set temperature was reached. Protein concentrations were between 8 and 15 μm. Far-UV CD experiments were repeated three times.
DSC
DSC was done with a Microcal VP differential scanning calorimeter from 20–85 °C at a rate of 60 °C/h. Samples were cooled to 20 °C and heated again to assess the reversibility of the denaturation process. Prior to analysis, the proteins were treated with EDTA to remove bound Ca2+ and dialyzed in 10 mm MOPS, 150 mm NaCl without Ca2+ or with 0.2 or 2 mm CaCl2, pH 7.5. Dialyses were for 48 h with four changes of a >100-fold volume of buffer. The concentrations of the proteins were >1 mg/ml. The DSC data were analyzed and fitted with Origin 7.0 (OriginLab, Northampton, MA). The reference buffer data were subtracted from the sample data; the data were converted from cal/degree to cal/mol/degree, and the base lines were created with the software progress base-line function. ΔH values were obtained from the integral below the peak above the base line. DSC experiments were repeated twice.
Isothermal Titration Calorimetry (ITC)
A Microcal VP isothermal titration calorimeter was used. Samples were mixed with EDTA and then dialyzed with four changes against 10 mm MOPS, 150 mm NaCl, pH 7.5. Similar results were obtained when Ca2+ was removed with a Chelex resin (13). The samples of 30–70 μm E2, E12, E23, or E123 were added into the 1.42-ml sample cell. The buffer used for dialysis plus 3–5 mm CaCl2 were added to the injection syringe (rotating at 307 rpm) at 25 °C. 1 μl was injected in the first injection with a duration of 2 s, and for the others, 5 μl was injected into the sample cell over 10 s, with delay between injections of 240 s. The reference buffer titration data were subtracted from the sample data. To derive the heat associated with each injection, the area under each heat burst curve was determined by integration, and the data were then fitted to a one-site binding model (Origin 7.0). The first two injections of E2, E12, and E23 and the first injection of E123 data were deleted before the fitting process. ITC experiments were repeated at least three times.
Competition ELISA
ELISA plates (Costar 3590) were coated at 4 °C overnight with platelet TSP-1 diluted to a final concentration of 0.75 μg/ml in Ca2+-TBS and then washed one time with Ca2+-TBS plus 0.05% Tween 20 (Ca2+-TBST) and blocked with 5% bovine serum albumin in Ca2+-TBST for 1 h (14). Recombinant EGF-like modules were diluted in Ca2+-TBST plus 0.1% bovine serum albumin so that when combined with the diluted antibody HB8432 or C6.7 (14, 15), the final concentration of the EGF-like module construct was 3.0, 1.0, 0.3, 0.1, 0.03, 0.01, 0.003, or 0.001 μm, and the final concentration of the antibodies was 0.5 μg/ml. These mixtures were incubated for 1 h before addition to the wells coated with TSP-1 and incubated for another 2 h. The plates were then washed four times with Ca2+-TBST. Alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was diluted 1:5000 and added to the wells. The ELISA experiments were repeated at least three times.
RESULTS
The recombinant proteins were expressed using a baculovirus expression system and purified from conditioned medium with nickel chelate chromatography. The average yields of the recombinant proteins from 1 liter of conditioned medium varied from 5.6 mg for E3 to 55 mg for E123 (Table 1). Proteins had >95% purity by SDS-PAGE (data not shown).
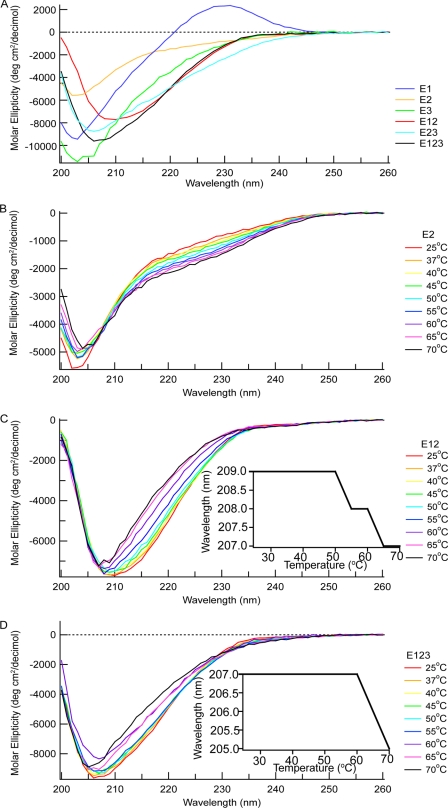
EGF modules typically have a far-UV CD spectrum with a minimum at ∼200 nm that is stable upon heating (5, 9, 16, 17). The spectra of E1, E2, E3, E12, E23, and E123 each had a negative band with slightly different minima at 25 °C as follows: 203 nm for E1, E2, and E3; 209 nm for E12; 205 nm for E23; and 207 nm for E123 (Fig. 2A). In addition, E1 had a positive peak at 230 nm. The positive maximum that was prominent only in the spectra of E1 likely arises from Trp-570, as described in the previous far-UV CD results of EGF-like modules with tryptophan (9, 17, 18). The shapes of the negative bands are compatible with the lack of α-helix or β-sheet in E1, E2, and E3 in the TSP-2 signature domain structure (4, 19). When the proteins were heated from 25 to 70 °C, the spectrum of E2 (Fig. 2B) or E1 or E3 (not shown) changed little. For E12, E23, and E123, the minima of the negative bands shifted to 203 nm (Fig. 2, C and D, and data not shown). The shift occurred at about 55 °C for E12 and E23 and 65 °C for E123 (Fig. 2, C and D, insets).
FIGURE 2.
Far UV CD of E1, E2, E3, E12, E23, and E123. A, spectra of the six proteins at 25 °C. B–D, spectral changes of E2 (B), E12 (C), and E123 (D) upon heating. Insets in C and D are changes in wavelengths of minima of E12 and E123 as a function of temperature. The buffer with 10 mm Tris, 150 mm NaCl, 2 mm CaCl2, pH 7.5, was used for far-UV CD experiments.
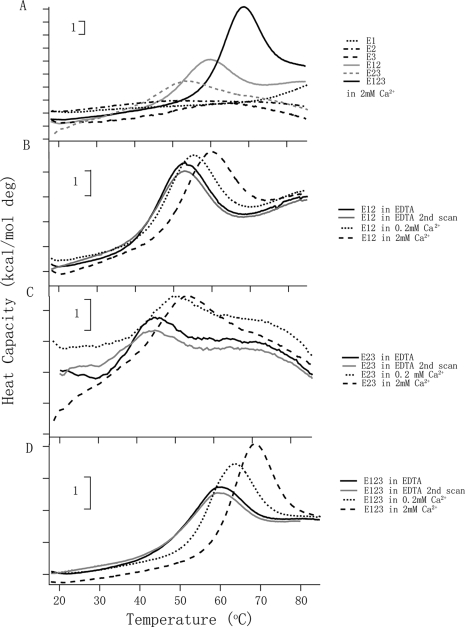
The far-UV CD experiments indicated that the individual EGF-like modules are stable to high temperature, but the tandem modules adopt a structure that melts at lower temperatures. The melting temperature for E123 matched the 68 °C transition found in DSC of the signature domain of TSP-2 in 2 mm Ca2+ and attributed to E123 (10). Therefore, DSC was performed on all six constructs. For E1, E2, and E3, there was no obvious melting temperature (Tm) up to 85 °C (Fig. 3A). In contrast, peaks with maxima at 58.9, 52.2, and 67.6 °C were found for E12, E23, and E123, respectively, in 2 mm Ca2+ (Fig. 3, B–D). Denaturations were reversible. The melting curves of E12 and E123 fitted a model with a single transition when analyzed by Origin 7.0. The melting temperatures and energies involved in the denaturation processes were different when the Ca2+ concentration was changed (Fig. 3, B–D, and Table 2). The melting temperatures of E12 shifted from 52.2 °C in EDTA to 54.9 °C in 0.2 mm Ca2+ and reached 58.9 °C in 2 mm Ca2+. The ΔH values were ∼50 kcal/mol. The Tm of E23 shifted from 44.3 °C in EDTA to 49.4 °C in 0.2 mm Ca2+ and to 52.2 °C in 2 mm Ca2+. The data for E23 and the quality of base lines did not allow curve fitting or estimations of ΔH values. The Tm of E123 changed from 59.0 °C in EDTA to 62.6 °C in 0.2 mm Ca2+ and to 67.6 °C in 2 mm Ca2+. The ΔH increased from 33 kcal/mol in EDTA to 44 kcal/mol in 0.2 mm Ca2+ and 63 kcal/mol in 2 mm Ca2+.
FIGURE 3.
DSC of E1, E2, E3, E12, E23, and E123. A, melting curves of E1, E2, E3, E12, E23, and E123 in 2 mm Ca2+. B, E12 is in different Ca2+ concentrations; the protein in EDTA is shown as solid lines, in 0.2 mm Ca2+ as dotted lines, and in 2 mm Ca2+ as broken lines. C, E23 in the three conditions of Ca2+ concentrations. D, E123 in the three conditions of Ca2+ concentrations. B–D, the repeat scan of the sample in EDTA is shown in gray to illustrate reversibility of the transition. Similar reversibility was found for the other samples. More analyses of the experimental data are in Table 2. The vertical bars indicate 1 kcal/mol degree.
TABLE 2.
Denaturation of E12, E23, and E123 as assessed by DSC
Numbers are means of two independent experiments. NC means not calculated.
| Protein | Tm | ΔH |
|---|---|---|
| °C | kcal/mol | |
| E12 in EDTA | 52.2 | 51 |
| E12 in 0.2 mm Ca2+ | 54.9 | 50 |
| E12 in 2 mm Ca2+ | 58.9 | 50 |
| E23 in EDTA | 44.3 | NC |
| E23 in 0.2 mm Ca2+ | 49.4 | NC |
| E23 in 2 mm Ca2+ | 52.2 | NC |
| E123 in EDTA | 59.0 | 33 |
| E123 in 0.2 mm Ca2+ | 62.6 | 44 |
| E123 in 2 mm Ca2+ | 67.6 | 63 |
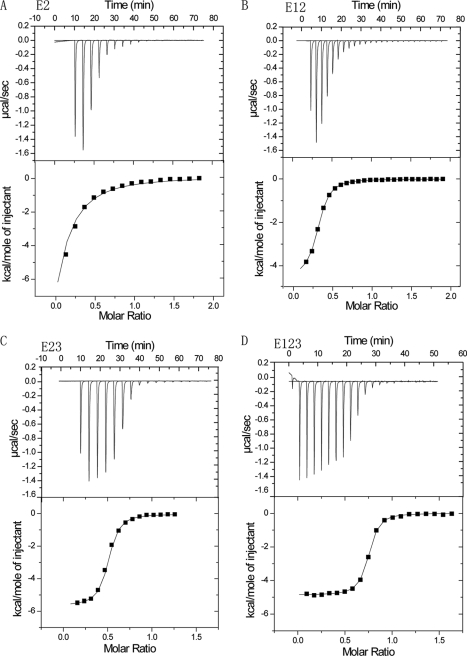
The DSC results indicate that E12, E23, and E123, but not any of the individual modules, adopt structures that are stabilized by Ca2+ and denature in a single reversible transition upon moderate heating. In the TSP-2 signature domain crystal, there is one Ca2+-binding site in E2 near the interface with E1 (4). We reasoned that binding of this site may be linked to the interaction of E2 with adjacent modules, and we therefore tested kinetics and energetics of Ca2+ binding to the site in recombinant E2, E12, E23, and E123 at 25 °C using ITC. Binding was tighter in the presence of E1 or E3 or of both E1 and E3, with KD values of 11.5 μm for E2, 2.9 μm for E12, 2.2 μm for E23, and 0.3 μm for E123 (Fig. 4 and Table 3). In addition, the data indicated that 1 mol of E123 bound 0.7 mol of Ca2+, whereas E2, E12, and E23 bound significantly less Ca2+, 0.2, 0.4, and 0.5 mol respectively. Finally, the tracings of E2, E12, and E23 were strange with less heat flow with the first 5-μl injections. Thermodynamically, the negative ΔH and positive ΔS indicated that the Ca2+ binding is driven by both enthalpy and entropy in all cases (Table 3).
FIGURE 4.
ITC of Ca2+ binding. CaCl2, 3–5 mm, was added repeatedly at 25 °C to sample cells containing E2 (A), E12 (B), E23 (C), or E123 (D). Top, calorimetric trace for titration; bottom, plot of the net heat change as a function of the molar ratio of Ca2+ to protein. The fitted binding sites were <1, ranging from 0.2 for E2 to 0.7 for E123. More analyses of these data are in Table 3.
TABLE 3.
Binding of calcium to E2-containing constructs as measured by ITC
| Protein | KD | −ΔH | TΔS |
|---|---|---|---|
| μm | kcal/mol | kcal/mol | |
| E2 | 11.5 ± 0.6 | 3.8 ± 0.2 | 2.9 |
| E12 | 2.9 ± 0.4 | 4.5 ± 0.0 | 3.0 |
| E23 | 2.2 ± 0.2 | 4.8 ± 0.2 | 2.8 |
| E123 | 0.3 ± 0.0 | 4.9 ± 0.1 | 4.0 |
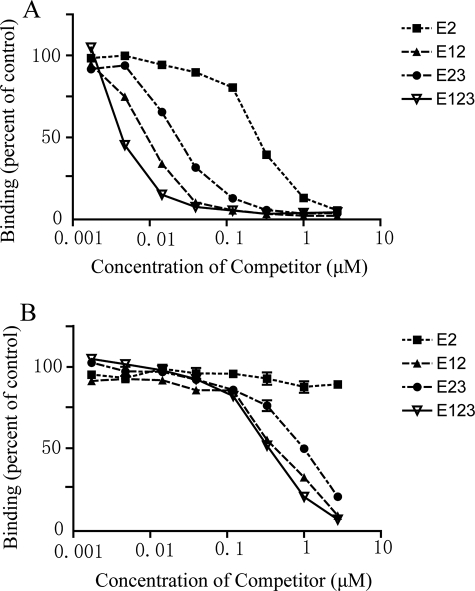
Differences in DSC and calcium binding of the four E2-containing constructs are strong evidence for differences in conformation of E2. This conclusion was tested further with two anti-TSP-1 monoclonal antibodies, HB8432 and C6.7, which recognize epitopes that include Glu-609 of E2 (14) (Fig. 1). HB4832 recognizes this epitope better when the complete signature domain is present, whereas C6.7 recognizes E12 and the complete signature domain equally well as assessed by a competitive ELISA (15). In competition ELISAs, recognition of E2 by HB8432 was 30–100-fold less than recognitions of E12, E23, or E123 (Fig. 5A). The EC50 values of the bindings were 120, 24, 30, and 2.8 μm for E2, E12, E23, and E123, respectively. C6.7 recognized E12, E23, and E123 equally well but failed to recognize E2 (Fig. 5B). Similar results were found in the presence of Ca2+ (Fig. 5) and in EDTA (data not shown).
FIGURE 5.
Competition ELISAs by soluble proteins. Competition by soluble E2, E12, E23, or E123 for binding of antibody HB8432 (A) and C6.7 (B) to substrate-bound intact TSP-1. ELISAs were performed in the presence of 2.0 mm Ca2+.
DISCUSSION
The EGF-like modules of group A TSPs are unusual in two respects as described in the Introduction and as follows: the presence of two residues between Cys-4 and Cys-5 of E1 and E2; and the intimate interactions among E2, E3, and the wire and lectin-like modules. Individual EGF-like modules are heat-stable in that the modules do not denature until the temperature is higher than 90 °C (7). Our results with far-UV CD and DSC of E1, E2, and E3 alone agree with this generalization; the far-UV CD of these recombinant EGF-like modules changed little as the temperature was raised, and no transition was evident in DSC up to 85 °C.
Far UV CD and DSC analysis revealed that, in contrast to the single modules, the tandem E12, E23, and E123 constructs had transitions with Tm values in the range of 44–68 °C. The transition seen with the tandem modules likely arises from perturbation of a conformation in E2 that is induced by interactions with the adjacent modules. Competition ELISA showed that HB4832 and C6.7 recognize epitopes on E2 that are exposed preferentially in constructs that include E1 or E3 or both E1 and E3. The effects of surrounding modules of the antigenicity of E2 were found in the presence and absence of Ca2+. There are two possible reasons for the epitope recognition differences. The first is that E2 is less well structured in the absence of the adjacent EGF-like modules, so that the binding site is exposed differently. The second possibility is that there are two or more distinct populations of E2 conformers, and the presence of adjacent modules influences the fraction of E2 that is in a given conformation. Based on the crystal structure of the TSP-2 signature domain (4) and residues that are identical in TSP-1 and TSP-2, one would expect that Arg-623 in the long loop between the fifth and sixth Cys of E2 forms a salt bridge with Asp-671 of E3, and 632QGVE of the long loop of E2 interacts with 577PGY of E1 through side chain interactions. Therefore, the long loop is positioned to interact with both E1 and E3 and may be the structural element of E2 that is most influenced by module-module interactions.
Approximately 35% of EGF-like modules in the data base are predicted to bind Ca2+ (8). Based on the structures of a number of Ca2+-binding EGF-like modules (20), the Ca2+-binding site on E2 in the TSP-2 signature domain crystal (4), and sequence alignments of the EGF-like modules of TSP-1 and TSP-2, one may predict the Ca2+ coordination pattern of E2 of TSP-1 as involving the side chains of Asp-588, Asn-610, and Glu-591 and main chain carbonyl groups of Val-589, Thr-611, and Gly-614. ITC of Ca2+ binding of the recombinant TSP-1 EGF-like modules E2, E12, E23, and E123 demonstrated that, when compared with E2 alone, the affinity increases 10-fold when E1 or E3 is adjacent to E2 and 100-fold in E123 and that there was also greater fractional occupancy when the adjacent modules were present. These findings are compatible with the scenario in which there are two or more distinct populations of E2 conformers, and the presence of adjacent modules influences the fraction of E2 that is capable of binding Ca2+. We cannot explain why E2, E12, and E23 required one injection of Ca2+ to facilitate Ca2+ binding, whereas E123 did not. β-Hydroxylation of the Asp/Asn in the sequence CX(D/N)XXXX(Y/F)XC between the third and fourth Cys is important for Ca2+ binding to EGF-like modules (21). The post-translational modification is often partial and requires the native conformation (22, 23). It is possible that the recombinant EGF-like modules adopt different conformations that influence Asn-610 β-hydroxylation and the ability to bind Ca2+.
DSC demonstrated that binding of Ca2+ stabilizes the conformation induced in E2 by the presence of E1 and/or E3. Most likely, Ca2+ stabilizes the interactions at the interfaces between E1 and E2 and between E2 and E3 and thereby enhances the effects of the interfaces on the conformation of E2 at higher temperatures, even though the antibody probes indicated that at 25 °C the conformation of E2 is not dependent on the occupation of the Ca2+.
In addition to the Ca2+-binding site in E2, the TSP-1 signature domain is predicted to have 26 binding sites in the wire and 4 binding sites in the lectin-like module (4, 10, 24). Because a transition attributable to the EGF-like modules is found in DSC of the complete signature domains of TSP-1 and TSP-2 (10) and HB4832 and C6.7 recognize E2 in the TSP-1 signature domain (14), it seems reasonable to assume that the Ca2+-binding site in E2 is similar in the tandem construct and signature domain. The sub-micromolar binding of Ca2+ to E123 is considerably tighter than the ∼100 μm binding of Ca2+ to the wire (25). The affinity for binding of Ca2+ to the lectin-like module is not known.
The presence of two rather than one residue between the fourth and fifth cysteines in E1 and E2 is predicted to increase the flexibility within these modules by hinge-like movement around the residues, as described in the Introduction. Such movement may be impeded in E2 compared with E1 because the long loop between the fifth and sixth cysteines interacts extensively with the N-terminal submodule. Interactions of the long loop in E2 with E1 and E3 may impede such flexibility further. We speculate that the interaction of E1 with E2 may influence the flexibility of E1 but has no evidence for such an hypothesis. In particular, intrinsic fluorescence and near UV CD, which would be expected to be influenced by changes in the environment of Trp-570 in E1, were identical in E1 and E12 (data not shown).
Pentameric TSPs have a fourth EGF-like module (E2′) that is interposed between E2 and E3. E2′ is predicted to have a Ca2+-binding site and has the same unusual cysteine spacing pattern present in E1 and E2 of TSP-1. The difference in effects of E123 and E122′3 groupings on the conformation of E2 and dynamics of the tandem modules may be profound. The signature domain of TSP-4 precipitates when subjected to DSC, whereas the signature domain of TSP-2 is stable (26). In contrast to the reversible denaturation of E123 of TSP-1 (Fig. 3D), recombinant E122′3 of TSP-4 subjected to DSC precipitates at ∼60 °C (data not shown). E2 is predicted to interact with the Ca2+-binding face of E2′ rather than E3 with the potential for cooperative binding of Ca2+ to the sites in E2 and E2′. In addition, there are three rather than two sites of potential intra-module flexibility in the E122′3 grouping. Further studies of the interactions among the EGF-like modules of the pentameric TSPs are needed, e.g. looking for the type of allosterism demonstrated for the four tandem EGF-like module of protein S, in which binding of Ca2+ the most C-terminal module changes the conformation of the most N-terminal module (13).
Several observations indicate that changes in the signature domain cause conformational changes in more N-terminal parts of TSP-1. Binding of monoclonal antibodies and heparin to the N-terminal modules is influenced by the presence or absence of Ca2+ in the signature domain (27). Furthermore, activation of latent transforming growth factor-β by the thrombospondin repeat 1 is blocked by antibody mAb133, which reacts with the signature domain (14, 28). The conformational changes of the wire and lectin-like module likely are transmitted to the EGF-like modules through the extensive interactions of E3 with the wire and lectin-like modules and the interaction between the repeat 12N of the wire and E2 (4). The present results indicate that the information may be transmitted to E1 and perhaps further N-terminally through the interactions among the EGF-like modules.
Acknowledgments
We thank C. Britt Carlson and Darrel McCaslin for helpful discussions. CD, DSC, and ITC data were obtained at the Biophysics Instrumentation Facility at the University of Wisconsin, which was established with support from National Institutes of Health Grant BIR-9512577 and National Science Foundation Grant S10 RR13790.
This work was supported, in whole or in part, by National Institutes of Health Grant ROIHL054462.
- TSP
- thrombospondin
- DSC
- differential scanning calorimetry
- ITC
- isothermal titration calorimetry
- EGF
- epidermal growth factor
- ELISA
- enzyme-linked immunosorbent assay
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Lawler J. (2000) Curr. Opin. Cell Biol. 12, 634–640 [DOI] [PubMed] [Google Scholar]
- 2.Adams J. C., Lawler J. (2004) Int. J. Biochem. Cell Biol. 36, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson C. B., Lawler J., Mosher D. F. (2008) Cell. Mol. Life Sci. 65, 672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson C. B., Bernstein D. A., Annis D. S., Misenheimer T. M., Hannah B. L., Mosher D. F., Keck J. L. (2005) Nat. Struct. Mol. Biol. 12, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanuttin F., Guarnaccia C., Pintar A., Pongor S. (2004) Eur. J. Biochem. 271, 4229–4240 [DOI] [PubMed] [Google Scholar]
- 6.Wouters M. A., Rigoutsos I., Chu C. K., Feng L. L., Sparrow D. B., Dunwoodie S. L. (2005) Protein Sci. 14, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell I. D., Bork P. (1993) Curr. Opin. Struct. Biol. 3, 385–392 [Google Scholar]
- 8.Drakenberg T., Ghasriani H., Thulin E., Thämlitz A. M., Muranyi A., Annila A., Stenflo J. (2005) Biochemistry 44, 8782–8789 [DOI] [PubMed] [Google Scholar]
- 9.Kohda D., Inagaki F. (1992) Biochemistry 31, 11928–11939 [DOI] [PubMed] [Google Scholar]
- 10.Carlson C. B., Liu Y., Keck J. L., Mosher D. F. (2008) J. Biol. Chem. 283, 20069–20076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosher D. F., Huwiler K. G., Misenheimer T. M., Annis D. S. (2002) Methods Cell Biol. 69, 69–81 [DOI] [PubMed] [Google Scholar]
- 12.Mach H., Middaugh C. R., Lewis R. V. (1992) Anal. Biochem. 200, 74–80 [DOI] [PubMed] [Google Scholar]
- 13.Persson K. E., Stenflo J., Linse S., Stenberg Y., Preston R. J., Lane D. A., Rezende S. M. (2006) Biochemistry 45, 10682–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annis D. S., Murphy-Ullrich J. E., Mosher D. F. (2006) J. Thromb. Haemost. 4, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annis D. S., Gunderson K. A., Mosher D. F. (2007) J. Biol. Chem. 282, 27067–27075 [DOI] [PubMed] [Google Scholar]
- 16.Holladay L. A., Savage C. R., Jr., Cohen S., Puett D. (1976) Biochemistry 15, 2624–2633 [DOI] [PubMed] [Google Scholar]
- 17.Kitamura M., Hojo H., Nakahara Y., Ishimizu T., Hase S. (2004) Glycoconj. J. 21, 197–203 [DOI] [PubMed] [Google Scholar]
- 18.Prestrelski S. J., Arakawa T., Wu C. S., O'Neal K. D., Westcott K. R., Narhi L. O. (1992) J. Biol. Chem. 267, 319–322 [PubMed] [Google Scholar]
- 19.Narhi L. O., Arakawa T., McGinley M. D., Rohde M. F., Westcott K. R. (1992) Int. J. Pept. Protein Res. 39, 182–187 [DOI] [PubMed] [Google Scholar]
- 20.Boswell E. J., Kurniawan N. D., Downing A. K. (2004) in Handbook of Metalloproteins (Messerschmidt A. ed) Vol. 3, pp. 553–570, Wiley, New York [Google Scholar]
- 21.Stenflo J., Stenberg Y., Muranyi A. (2000) Biochim. Biophys. Acta 1477, 51–63 [DOI] [PubMed] [Google Scholar]
- 22.Fernlund P., Stenflo J. (1983) J. Biol. Chem. 258, 12509–12512 [PubMed] [Google Scholar]
- 23.Stenflo J., Ohlin A. K., Owen W. G., Schneider W. J. (1988) J. Biol. Chem. 263, 21–24 [PubMed] [Google Scholar]
- 24.Kvansakul M., Adams J. C., Hohenester E. (2004) EMBO J. 23, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannah B. L., Misenheimer T. M., Pranghofer M. M., Mosher D. F. (2004) J. Biol. Chem. 279, 51915–51922 [DOI] [PubMed] [Google Scholar]
- 26.Misenheimer T. M., Mosher D. F. (2005) J. Biol. Chem. 280, 41229–41235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calzada M. J., Kuznetsova S. A., Sipes J. M., Rodrigues R. G., Cashel J. A., Annis D. S., Mosher D. F., Roberts D. D. (2008) Matrix Biol. 27, 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz-Cherry S., Murphy-Ullrich J. E. (1993) J. Cell Biol. 122, 923–932 [DOI] [PMC free article] [PubMed] [Google Scholar]