Abstract
The hyperthermophile Aquifex aeolicus belongs to the deepest branch in the bacterial genealogy. Although it has long been recognized that this unique Gram-negative bacterium carries genes for different steps of lipopolysaccharide (LPS) formation, data on the LPS itself or detailed knowledge of the LPS pathway beyond the first committed steps of lipid A and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) synthesis are still lacking. We now report the functional characterization of the thermostable Kdo transferase WaaA from A. aeolicus and provide evidence that the enzyme is monofunctional. Compositional analysis and mass spectrometry of purified A. aeolicus LPS, showing the incorporation of a single Kdo residue as an integral component of the LPS, implicated a monofunctional Kdo transferase in LPS biosynthesis of A. aeolicus. Further, heterologous expression of the A. aeolicus waaA gene in a newly constructed Escherichia coli ΔwaaA suppressor strain resulted in synthesis of lipid IVA precursors substituted with one Kdo sugar. When highly purified WaaA of A. aeolicus was subjected to in vitro assays using mass spectrometry for detection of the reaction products, the enzyme was found to catalyze the transfer of only a single Kdo residue from CMP-Kdo to differently modified lipid A acceptors. The Kdo transferase was capable of utilizing a broad spectrum of acceptor substrates, whereas surface plasmon resonance studies indicated a high selectivity for the donor substrate.
Lipopolysaccharide (LPS)7 is the major constituent of the outer leaflet of the outer membrane (OM) of virtually all Gram-negative bacteria. LPS is a unique amphiphilic molecule composed of a hydrophilic heteropolysaccharide and a covalently bound lipid moiety, lipid A, which anchors the molecule in the OM. The polysaccharide component of many wild-type bacteria can be subdivided into a highly variable O-specific polysaccharide and a structurally less heterogeneous outer and inner core oligosaccharide (1). The 8-carbon sugar 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) links the lipid A to the carbohydrate domain of LPS and is the only conserved structural element found in all inner core regions investigated to date (2).
The ubiquitous nature of Kdo within LPS structures and its essential role in maintaining OM integrity and viability of the majority of Gram-negative bacteria has prompted detailed studies into its biosynthesis. The Kdo pathway is initiated by the enzyme d-arabinose-5-phosphate isomerase, which catalyzes the interconversion of d-ribulose 5-phosphate and d-arabinose 5-phosphate (3). The Kdo-8-phosphate synthase KdsA subsequently condenses d-arabinose 5-phosphate with phosphoenolpyruvate to form Kdo 8-phosphate (4), followed by hydrolysis of Kdo 8-phosphate to Kdo and inorganic phosphate by the Kdo-8-phosphate phosphatase KdsC (5) and activation of Kdo to CMP-Kdo by the CMP-Kdo synthetase KdsB, before finally Kdo is transferred from CMP-Kdo to the lipid A moiety by the glycosyltransferase WaaA (6). In Escherichia coli, the Kdo-dependent late acyltransferases LpxL and LpxM subsequently transfer the fatty acids laurate and myristate, respectively, to Kdo2-lipid IVA to generate the characteristic acyloxyacyl units of hexaacylated Kdo2-lipid A (7).
It has long been recognized that Kdo transferases are unusual glycosyltransferases. WaaA is bifunctional in bacteria with LPS that contains an α-(2→4)-linked Kdo disaccharide in the inner core region, such as E. coli (6), Klebsiella pneumoniae (8), Legionella pneumophila (9), Acinetobacter baumannii, and Acinetobacter haemolyticus (10). In E. coli, CMP-Kdo is utilized for the transfer of Kdo to the tetraacylated lipid A precursor lipid IVA, resulting in an α-(2→6)-linkage between the distal glucosamine (GlcN) of the lipid A backbone and the first Kdo residue and an α-(2→4)-linkage between a second Kdo residue and the first one. Thus, WaaA is capable of catalyzing the formation of two different glycosidic bonds, tolerating acceptor molecules with varying extents of acylation but strictly depending on the 4′-phosphate group of the tetraacyldisaccharide 1,4′-bisphosphate intermediate (6). In chlamydiae, however, which express an LPS composed of a Kdo trisaccharide with an unusual α-(2→8)-linkage between the second and a third Kdo residue (11), the Kdo transferases were shown to display at least trifunctional activity (12). The LPS of Chlamydophila psittaci consists of up to four Kdo residues of the structure α-Kdo-(2→4)-[α-Kdo-(2→8)]-α-Kdo-(2→4)-α-Kdo (13), and heterologous expression of the waaA gene in E. coli was found to be sufficient for synthesis of the complete chlamydial Kdo structure (12). Finally, the Kdo transferases of Haemophilus influenzae and Bordetella pertussis were shown to be monofunctional (14, 15), consistent with the presence of a single phosphorylated Kdo residue in their respective LPS (16, 17).
On the basis of phylogenetic analyses of 16 S ribosomal RNA sequences, members of the family Aquificaceae with growth-temperature maxima near 95 °C are thought to represent the deepest branching species of the kingdom Bacteria (18). The cells are Gram-negative with a rather complex cell envelope of a surface protein layer, murein, and an OM (19). Previous studies provided the first direct evidence for the presence of smooth form LPS in Aquifex pyrophilus (20). Furthermore, KdsA and the UDP-(3-O-(R-3-hydroxymyristoyl))-N-acetylglucosamine deacetylase (LpxC) of A. aeolicus, a close relative of A. pyrophilus, have been characterized in detail, and it was demonstrated that these enzymes catalyze the first committed steps in Kdo and lipid A formation, respectively (21–23). Moreover, a number of genes presumably encoding different steps of LPS biosynthesis have been identified on the A. aeolicus genome, including putative kdsB and waaA orthologues for Kdo activation and subsequent incorporation of the sugar into LPS (24). However, the number of Kdo residues transferred by WaaA of A. aeolicus remains unknown. We herein provide evidence that the A. aeolicus enzyme is a strictly monofunctional Kdo transferase through the characterization of its enzymatic activity and the chemical analysis of the native A. aeolicus LPS.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
All strains and plasmids used in the present study are described in Table 1. Standard cultures of E. coli and Salmonella enterica sv. Typhimurium strains were routinely grown aerobically with shaking (250 rpm) at 37 °C in LB medium (10 g/liter Bacto-tryptone, 5 g/liter yeast extract) supplemented with 10 g/liter NaCl to avoid colanic acid production in E. coli Kdo pathway mutants (KPM designation) (25), whereas BL21-SI212 and BL21-SI216 were propagated under non-inducing conditions in LBON (LB without NaCl). When necessary, kanamycin (30 μg/ml), chloramphenicol (30 μg/ml), ampicillin (100 μg/ml), or carbenicillin (50 μg/ml) was added to the media.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or Ref. |
|---|---|---|
| E. coli strains | ||
| BW30270 | E. coli K-12 MG1655 rph+fnr+ | CGSC 7925 |
| KPM22 | BW30270 ΔgutQ ΔkdsD yhjD400 | Ref. 28 |
| KPM50 | KPM22 kdsD+ | This study |
| KPM53 | KPM50 ΔwaaC | This study |
| KPM56 | KPM53 ΔwaaA | This study |
| KPM122 | KPM56 with pUM210 carrying the waaA gene of C. psittaci 6BC | This study |
| KPM123 | KPM56 with pUM211 carrying the waaA gene of H. influenzae I69 | This study |
| KPM124 | KPM56 with pUM212 carrying the waaA gene of E. coli K-12 | This study |
| KPM125 | KPM56 with pUM216 carrying the waaA gene of A. aeolicus | This study |
| XL1-Blue | E. coli K-12 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (TetR)] | Stratagene |
| BL21-CodonPlus(DE3)-RIL | E. coli B F−ompT hsdS(rB– mB–) dcm+ TetRgal λ(DE3) endA Hte [argU ileY leuW CmR] | Stratagene |
| BL21-RIL01 | BL21-CodonPlus(DE3)-RIL with pT7-kdsB | This study |
| BL21-SI | F–ompT lon hsdSB(rB– mB–) gal dcm endA1 proUp ::T7 RNAP ::malQ-lacZ (TetS) | Invitrogen |
| BL21-SI212 | BL21-SI with pUM212 | This study |
| BL21-SI216 | BL21-SI with pUM216 | This study |
| S. enterica sv. Typhimurium strains | ||
| SL3770 | rfa+; smooth form LPS | SGSC 225 |
| SA1627 | Δ(rfb-his); Ra chemotype of LPS | SGSC LPS mutant kit |
| SL3749 | rfaL446; Ra chemotype of LPS | SGSC 228 |
| SL733 | rfaK953; Rb1 chemotype of LPS | SGSC 389 |
| SL3750 | rfaJ417; Rb2 chemotype of LPS | SGSC 229 |
| SL3748 | rfaI432; Rb3 chemotype of LPS | SGSC 227 |
| SL3769 | rfaG471; Rd1 chemotype of LPS | SGSC 231 |
| SL1102 | rfaE543; Re chemotype of LPS | SGSC 258 |
| Corynebacterium glutamicum R163/pJKB72 | Restriction-deficient mutant of C. glutamicum AS019 harboring the expression plasmid pJKB72 with kdsB of E. coli K-12 | Ref. 30 |
| Plasmids | ||
| pT7-7 | AmpR; T7 expression vector | Ref. 79 |
| pKD46 | AmpR; λ Red recombinase expression plasmid | Ref. 27 |
| pKD4 | AmpR, KanR; template plasmid for kanamycin resistance cassette | Ref. 27 |
| pCP20 | AmpR, CmR; FLP recombinase expression plasmid | Ref. 27 |
| pT7-kdsB | pT7-7 carrying the kdsB gene of A. aeolicus | This study |
| pUM140 | Cm+; pHSG398 carrying the waaA gene of C. psittaci 6BC | Ref. 29 |
| pCB23 | KanR; E. coli/C. glutamicum shuttle plasmid carrying the waaA gene of H. influenzae I69 | Ref. 14 |
| pET-16b | AmpR; T7 expression vector for N-terminal His-tag fusions | Novagen |
| pUM210 | pET-16b carrying the waaA gene of C. psittaci 6BC | This study |
| pUM211 | pET-16b carrying the waaA gene of H. influezae I69 | This study |
| pUM212 | pET-16b carrying the waaA gene of E. coli K-12 | This study |
| pUM216 | pET-16b carrying the waaA gene of A. aeolicus | This study |
DNA Methods
Standard recombinant DNA methods were used for nucleic acid preparation and analysis (26). Primer sequences are listed in supplemental Table S1. To both restore gene function and construct gene deletions, the phage λ Red recombinase procedure was used essentially as described (27), except plasmids pKD46 and pCP20 were cured at 37 °C to accommodate the temperature sensitive phenotype of lipid IVA-expressing KPM22 derivatives (28). For generation of a kdsD insertion in KPM22, primers ECOyrbG and ECOkdsC were used to amplify the kdsD gene of E. coli K-12 wild-type strain BW30270, followed by transformation of KPM22/pKD46 with the resulting PCR product and selection for restored LPS biosynthesis on McConkey agar plates to yield strain KPM50. The ΔwaaC ΔwaaA strain KPM56 was obtained by successive deletion of the waaC gene of KPM50 and waaA of the resulting ΔwaaC strain KPM53, for which the primer pairs ECOwaaCH1/ECOwaaCH and ECOwaaAH1/ECOwaaAH2 with pKD4 as template were used to construct the insert cassettes targeting waaC and waaA, respectively.
To clone kdsB (aq_692) of A. aeolicus into pT7-7 and waaA genes of different origin into pET-16b, genomic DNAs were used as templates to amplify the kdsB gene with primer pair AAE5NdeIkdsB/AAE3BamHIkdsB and the waaA genes of A. aeolicus (aq_326) and E. coli K-12 with primers AAE5NdeIwaaA/AAE3BamHIwaaA and ECO5NdeIwaaA/ECO3BamHIwaaA, respectively, whereas the plasmids pCB23 (14) and pUM140 (29) served as templates for amplification of the waaA genes of H. influenzae I69 (primers HIN5NdeIwaaA/HIN3BamHIwaaA) and C. psittaci 6BC (primers CPS5NdeIwaaA/CPS3BamHIwaaA), respectively. The purified amplification products were digested with NdeI and BamHI and ligated into the NdeI and BamHI sites of the expression vectors. The ligation mixtures were used to transform E. coli XL1-Blue cells. Plasmid DNA isolated from several of the respective transformants and identified by restriction analysis to contain the PCR products was subjected to DNA sequencing to confirm the sequences of the desired genes. Plasmids pUM210 with waaACPS, pUM211 (waaAHIN), pUM212 (waaAECO), and pUM216 (waaAAAE) were subsequently used for transformation of KPM56 to obtain the strains KPM122, KPM123, KPM124, and KPM125, respectively. For overexpression of the histidine-tagged Kdo transferases of A. aeolicus and E. coli, plasmids pUM216 and pUM212 were introduced into BL21-SI to yield BL21-SI216 and BL21-SI212, respectively. Strain BL21-RIL01 was generated by transformation of BL21-CodonPlus(DE3)-RIL cells with pT7-kdsB to overexpress KdsB of A. aeolicus.
Overexpression and Purification of KdsB from A. aeolicus
To induce expression of KdsBAAE in BL21-RIL01 cells, isopropyl-β-d-thiogalactoside was added to a final concentration of 1 mm when the culture reached the mid-exponential growth phase at an A600 of 0.6–0.8. The cells were harvested 3 h post-induction by centrifugation (9000 × g, 4 °C, 20 min), washed with phosphate-buffered saline, and resuspended in buffer A (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm 2-mercaptoethanol) at 0.1 g of wet biomass/ml. The suspension was incubated with lysozyme (0.2 mg/ml), DNase I (0.1 mg/ml), and RNase A (0.1 mg/ml) at 20 °C for 30 min with continuous stirring and placed on ice for 15 min, and phenylmethylsulfonyl fluoride was added to a final concentration of 1 mm. The cells were broken by passage three times through a French pressure cell at 20,000 p.s.i., followed by centrifugation of the homogenate to remove cellular debris (27,000 × g, 4 °C, 30 min). The supernatant was heated to 85 °C in a boiling water bath and incubated at 85 °C for an additional 15 min with continuous swirling. The suspension was allowed to cool to 22 °C and placed on ice for 15 min, and the precipitated protein was removed by centrifugation (27,000 × g, 4 °C, 30 min). The supernatant was loaded onto a DEAE-Sepharose Fast Flow column (GE Healthcare) pre-equilibrated with buffer A. The column was eluted with a linear gradient from 0 to 0.2 m NaCl in the same buffer. Fractions containing KdsBAAE were pooled, and solid (NH4)2SO4 was added to a final concentration of 20% (w/v). The sample was subsequently applied to a phenyl-Sepharose CL-4B column (GE Healthcare) pre-equilibrated with 20% (NH4)2SO4 (w/v) in buffer A. A reverse gradient from 20% (w/v) to 0% (NH4)2SO4 in buffer A was used to elute KdsBAAE. The fractions containing KdsBAAE were combined, dialyzed against buffer A, and concentrated by chromatography on a HiTrap Q-Sepharose HP (5 ml) column (GE Healthcare) using a linear gradient from 0 to 0.5 m NaCl in buffer A. For final KdsBAAE polishing, the pooled fractions were directly loaded onto a size exclusion HiLoad 26/100 Superdex 200 pg column (GE Healthcare) and eluted with buffer B (20 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 5 mm MgCl2, 2 mm 2-mercaptoethanol). The final preparation was concentrated using Centricon Plus-20 concentrators (Amicon), dialyzed against buffer B, and stored at 4 °C.
Overexpression and Purification of KdsB from E. coli
The synthetase KdsBECO has been previously obtained by immobilized metal affinity chromatography, using Corynebacterium glutamicum R163/pJKB72 as the expression host (30). Here we have upgraded the published procedure to purify KdsBECO to apparent homogeneity. The C. glutamicum R163/pJKB72 culture was grown to an A600 of 0.6–0.8, followed by expression of KdsBECO in the presence of 0.2 mm isopropyl-β-d-thiogalactoside for an additional 16 h. The cells were harvested by centrifugation (9000 × g, 4 °C, 20 min), washed with phosphate-buffered saline, resuspended at 0.3 g of wet biomass/ml of buffer C (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 5 mm 2-mercaptoethanol) supplemented with 10 mm imidazole, and incubated with continuous stirring at 37 °C for 45 min with lysozyme (1 mg/ml), lipase (1 mg/ml), DNase I (0.1 mg/ml), and RNase A (0.1 mg/ml). Then phenylmethylsulfonyl fluoride was added to a final concentration of 1 mm before the chilled cells were disrupted by passage three times through a French pressure cell at 20,000 p.s.i. After removal of cellular debris by centrifugation (27,000 × g, 4 °C, 30 min), the supernatant was loaded onto a nickel-Sepharose HP column (GE Healthcare) pre-equilibrated with 10 mm imidazole in buffer C. KdsBECO was eluted with 250 mm imidazole in buffer C, following an extensive washing step using 80 mm imidazole in the same buffer. The fractions containing KdsBECO were dialyzed against buffer A and subjected to ion exchange chromatography on HiTrap Q-Sepharose HP and gel filtration on HiLoad 26/100 Superdex 200 pg as described above for KdsBAAE purification. The final concentrated protein sample was dialyzed against buffer B and stored at −80 °C.
Overexpression and Purification of WaaA from A. aeolicus
The conditions used to express soluble WaaAAAE in BL21-SI216 were adapted from a previously published protocol for recovery of active heterologous protein from E. coli cells grown at decreased temperature and acidic pH values (31). Strain BL21-SI216 was grown to the mid-exponential growth phase (A600 of 0.6–0.8) at 20 °C with vigorous shaking (250 rpm) in a medium containing 10 g/liter peptone and 20 g/liter yeast extract. At this point, the medium was supplemented with NaCl (120 mm), glucose (1%), glycerol (0.4%), and potassium phosphate (100 mm), pH 4.5, to induce WaaA expression along with acidification of the medium. The cells were allowed to grow at 20 °C for an additional 18 h before they were sedimented (9000 × g, 4 °C, 20 min), washed with phosphate-buffered saline, and resuspended at 0.3 g of wet biomass/ml of buffer D (50 mm Tris-HCl, pH 8.7, 10% glycerol, 0.1% Triton X-100, 5 mm 2-mercaptoethanol). After incubation of the suspension with lysozyme (0.2 mg/ml) at 20 °C for 30 min and continuous stirring, the cells were chilled on ice and supplemented with DNase I (0.1 mg/ml), RNase A (0.1 mg/ml), and Complete protease inhibitor mixture (EDTA-free) according to the recommendations of the manufacturer (Roche Applied Science) and disintegrated by passage four times through a French pressure cell at 20,000 p.s.i. The cellular debris was removed by centrifugation (27,000 × g, 4 °C, 30 min), and the supernatant was loaded onto a DEAE-Sepharose Fast Flow column pre-equilibrated with buffer D. Under these conditions, WaaAAAE was eluted in the flow-through, whereas impurities found to interact with WaaAAAE in subsequent purification steps remained bound on the column. The eluate was mixed with an equal volume of a solution containing 2 m NaCl, 10% glycerol, 0.1% Triton X-100, 5 mm 2-mercaptoethanol, and 20 mm imidazole, followed by incubation of the mixture on ice for 2 h with stirring, centrifugation (27,000 × g, 4 °C, 30 min), and filtration through a 0.22-μm membrane. The supernatant was subjected to immobilized metal affinity chromatography on a nickel-Sepharose HP column pre-equilibrated with buffer E (25 mm Tris-HCl, pH 8.7, 10% glycerol, 0.1% Triton X-100, 5 mm 2-mercaptoethanol) containing 1 m NaCl and 10 mm imidazole. The column was extensively washed applying a stepwise gradient of 20, 80, and 100 mm imidazole before WaaAAAE was eluted with 500 mm imidazole in buffer E supplemented with 1 m NaCl. The fractions containing WaaAAAE were combined, dialyzed against 0.1 m NaCl in buffer E, heated to 80 °C in a boiling water bath, and incubated at 80 °C for an additional 20 min with continuous swirling. The solution was chilled on ice for 15 min, followed by sedimentation of the denatured protein (27,000 × g, 4 °C, 30 min) and affinity chromatography of the cleared supernatant on a HiTrap Heparin-Sepharose HP (5 ml) column (GE Healthcare) pre-equilibrated with the above buffer. The column was then washed with 0.5 m NaCl in buffer E, and WaaAAAE was eluted with 0.9 m NaCl in the same buffer. Finally, the WaaAAAE pool was diluted with buffer E to decrease the NaCl concentration to 0.1 m and loaded once more onto a HiTrap heparin-Sepharose HP (1 ml) column. The washing and elution steps were performed essentially as described above, except buffer E was used with 50 mm octyl-β-d-glucopyranoside instead of 0.1% Triton X-100. The combined WaaAAAE peak fractions were dialyzed against 25 mm Tris-HCl, pH 8.7, 0.1 m NaCl, 10% glycerol, 50 mm octyl-β-d-glucopyranoside, and 5 mm 2-mercaptoethanol and stored at 4 °C.
Overexpression and Purification of WaaA from E. coli
The conditions used for overexpression of the Kdo transferase from E. coli in BL21-SI212 cells as well as purification of the enzyme were essentially the same as described above for WaaAAAE, except buffers D and E contained 1% Triton X-100, heating of the WaaAECO solution at 80 °C was omitted, 1.5 m NaCl was necessary to elute the protein from the HiTrap Heparin-Sepharose HP (5 ml) column, and final exchange of the detergents was not performed due to a strong propensity of the protein to aggregate in octyl-β-d-glucopyranoside-containing buffers.
SDS-PAGE and Immunoblotting
Protein samples were denatured at 95 °C for 2 min in sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.7 m 2-mercaptoethanol, 0.002% bromphenol blue), resolved with 10% SDS-PAGE (32), and stained with Coomassie Brilliant Blue R-250. For immunoblot analysis, protein samples were electrotransferred to polyvinylidene difluoride membranes (Millipore) as described (33). The blots were incubated with penta-His antibody and processed in accordance with the protocols of the manufacturer (Qiagen).
KdsB Activity Assay
The activities of the CMP-Kdo synthetases from A. aeolicus and E. coli were monitored by colorimetric measurement of CMP-Kdo formation essentially as reported previously (34), except KdsBAAE was assayed in 100 mm glycine-NaOH buffer, pH 9.0, at the assayed temperature, at 50 °C for 30 s.
In Vitro Analysis of WaaA Activity
The in vitro reactions were performed under optimized conditions in a total volume of 100 μl containing 100 mm HEPES, pH 7.5, at the assayed temperature, 10% glycerol, 10 mm MgCl2, 2 mm Kdo, 5 mm CTP, 3.2 mm Triton X-100, 17 milliunits of either KdsBAAE or KdsBECO, 75 μm either WaaAAAE or WaaAECO, and a synthetic lipid A acceptor. The tetraacyl-1,4′-bisphosphate lipid A precursor 406 used at a concentration of 107 μm, the tetraacyl-1-monophosphate precursor 405 (113 μm), the hexaacyl-1,4′-bisphosphate lipid A compound 506 (83 μm), the 1-carboxymethyl (CM) derivative CM-506 of hexaacyl lipid A (84 μm), and the tetraacyl-1,4′-CM compound Bis-CM-406 (110 μm) have been synthesized as described (35–39). Tetraacylated lipid A of A. pyrophilus was applied at a concentration of 91 μm. To assay WaaAAAE, the reactions were routinely incubated in thin walled PCR tubes at 60 °C for 1 h using the GeneAmp PCR system 9700 thermocycler (PerkinElmer Life Sciences), whereas the WaaAECO reactions were incubated at 37 °C. Triton X-100 in the reactions was depleted by subsequent treatment of the mixtures with Calbiosorb Adsorbent, as recommended by the supplier (Calbiochem), followed by extensive dialysis against H2O, lyophilization, and analysis of the reaction products by electrospray-ionization Fourier-transformed ion cyclotron mass spectrometry (ESI FT-ICR MS).
Thermostability Measurements
Purified enzyme was either separately preincubated at 70, 80, 90, or 99 °C for 1 h or constantly maintained at 80 °C for 16 h prior to in vitro analysis of WaaA activity using the standard assay.
LPS Isolation and Analysis
The LPS profiles of proteinase K-digested whole cell lysates were investigated on silver-stained 13% SDS-polyacrylamide gels according to the method of Hitchcock and Brown (40). The LPS of E. coli strains was extracted from the dried biomass of stationary phase 2-liter cultures using either the original phenol/chloroform/light petroleum (PCP) procedure (41) or a modified PCP protocol for strains lacking the K-12 core oligosaccharide (42). For isolation and purification of A. aeolicus LPS, the dried biomass was extracted using hot phenol/water supplemented with 2% sodium N-lauroylsarcosine (w/v), as described previously (20). Oligosaccharides were obtained by hydrolysis of the LPS in 0.1 m HCl at 100 °C for 90 min, followed by removal of lipid A using ultracentrifugation (105,000 × g, 4 °C, 4 h). The carbohydrate portion (supernatant) was purified by gel permeation chromatography on a Biogel P4 column using a pyridine/acetic acid/water (4 ml/10 ml/986 ml) mixture as eluent. Combined GLC/MS analyses were performed to identify the sugar components of LPS and the oligosaccharide fraction either after weak (0.5 m HCl in methanol, 85 °C, 45 min, followed by peracetylation) or strong methanolysis (2 m HCl in methanol, 85 °C, 20 h, followed by peracetylation) or after hydrolysis with 4 m trifluoroacetic acid (100 °C, 4 h) and reduction with NaBH4 or NaBD4 to confirm the presence of GalA (43). Neutral sugars were analyzed quantitatively by GLC following hydrolysis of the samples in 2 m trifluoroacetic acid at 120 °C for 2 h (43). The content of Kdo was determined photometrically using the thiobarbituric acid assay as published (44). The preparation of O-deacylated lipid A from A. pyrophilus has been described previously (20). The A. aeolicus VF5 biomass was purchased from Harald Huber (Institute of Microbiology, University of Regensburg, Germany).
Electrospray-Ionization Fourier-transformed Ion Cyclotron Mass Spectrometry
Mass spectra were recorded in the negative ion mode using an APEX II and a hybrid Apex Qe Instrument (Bruker Daltonics) equipped with a 7-Tesla actively shielded magnet. Details on sample preparation and MS characteristics of LPS have been published (45). The mass spectra were charge-deconvoluted, and the mass numbers given refer to the monoisotopic peaks of the neutral molecules.
Surface Plasmon Resonance Binding Studies
Surface plasmon resonance (SPR) experiments were conducted in a BIAcore 3000 instrument (Biacore AB). A CM5 sensor chip, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), N-hydroxysuccinimide, 1 m ethanolamine-HCl, pH 8.5, and 10 mm sodium acetate, pH 4.0, were purchased from Biacore. The Bistris propane running buffer consisted of a 50 mm Bistris propane solution in double-distilled water containing 150 mm NaCl and 10 mm MgCl2. The pH was adjusted to 7.4 at 25 °C with a solution of 17.5% HCl. All buffers were filtered through a 0.22-μm membrane and degassed prior to use. All experiments were carried out at 25 °C.
The Kdo transferase of A. aeolicus was covalently immobilized to the carboxyl-methylated dextran surface of the CM5 sensor chip according to the amine coupling protocol of the manufacturer (Biacore AB). The surface was activated at a flow rate of 5 μl/min with a 7-min pulse of N-hydroxysuccinimide/EDC, followed by WaaAAAE immobilization (20 μm in 10 mm sodium acetate, pH 4.0) and subsequent neutralization of unreacted N-hydroxysuccinimide-esters with a 7-min pulse of 1 m ethanolamine-HCl, pH 8.5. A total of 7406 resonance units (RU) of the Kdo transferase was immobilized to one flow cell of the sensor chip, whereas 8954 RU of neutravidin (17 μm in 10 mm sodium acetate buffer, pH 4.0) was immobilized by the same procedure to a reference flow cell to mimic the environment of the WaaAAAE-containing cell.
To investigate the interaction of WaaAAAE with CMP, a concentrated solution of CMP (200 mm in Bistris propane running buffer) was serially diluted, yielding concentrations in the range between 39 μm and 20 mm. Samples of 10 μl were injected over the sensor chip at a flow rate of 10 μl/min. CMP dissociated very fast from the surface so that further regeneration steps were not necessary. A 10-μl pulse of buffer was injected between ligand injections. The titration was repeated under the same experimental conditions to ensure reproducibility. The resulting sensorgrams were processed and evaluated with BIAevaluation software version 3.2 (Biacore AB) and the Scrubber software package (BioLogic Software), following a procedure based on the double referencing method (46). The response from the neutravidin-coated flow cell was first subtracted from the response in the flow cell containing WaaAAAE. Second, the averaged response of three buffer injections was subtracted to obtain the final processed sensorgrams. The equilibrium dissociation constant was determined by steady state analysis of the sensorgrams, using non-linear square curve fitting to the equation, [AB]eq = ABmax(1/(1 + KD/[A])), with A and AB being the analyte (CMP) and the analyte-ligand complex (CMP-WaaAAAE), respectively, ABmax the maximum surface binding capacity (RU), and KD the equilibrium dissociation constant (m). Further, SPR binding studies were performed to investigate the interaction of AMP, TMP, UMP, or GMP with WaaAAAE, employing the same methodology as described for evaluation of the affinity between CMP and the Kdo transferase. Finally, SPR was used to examine the binding of Kdo to WaaAAAE, for which a concentrated solution of Kdo (200 mm) in Bistris propane running buffer was serially diluted to obtain concentrations ranging from 625 μm to 20 mm and processed as described above.
RESULTS
Characterization of the LPS from A. aeolicus
Although the presence of KdsA (21), LpxC (23), and presumably other LPS-related functions (24) indicates the ability of A. aeolicus to synthesize LPS and incorporate Kdo into LPS, data on the LPS itself, including the number of Kdo residues to predict WaaA functionality, is still lacking. Analysis by SDS-PAGE revealed a ladder-like appearance of the LPS preparation, suggesting the synthesis of smooth form LPS in A. aeolicus (supplemental Fig. S1, lane 7). In contrast to the LPS of A. pyrophilus with a decidedly non-modal O-specific polysaccharide banding (supplemental Fig. S1, lane 8), the LPS of A. aeolicus showed the characteristic pattern of modal chain length distribution of the supposed O-polysaccharide, with a significant LPS fraction apparently being not ligated with O-specific polysaccharide. Electrospray-ionization Fourier-transformed ion cyclotron mass spectrometry (ESI FT-ICR MS) of the lipid A identified a molecular mass of 1917.33 u (data not shown), consistent with the previously published pentaacylated structure of A. pyrophilus lipid A (20). In contrast to A. pyrophilus (20), investigation of the native LPS from A. aeolicus by MS revealed a remarkably homogeneous pattern of the unligated LPS population with three prominent molecular peaks (supplemental Fig. S2A). Assignment of the masses suggested a chemical composition of lipid A, one Kdo, two heptose, one hexose, and three hexosuronic acid residues for peak M1 at 3211.67 u (calculated mass 3211.614 u), whereas peak M2 at 4161.03 u corresponded to a species containing in addition one hexose, one N-acetyl hexosamine, and four deoxyhexose moieties (calculated mass 4160.978 u). The peak M3 at 4323.10 u apparently originated from a molecule with another hexose attached (calculated mass 4323.030 u) (supplemental Table S2). Under conditions of unspecific fragmentation in the collision cell, which leads to cleavage of the labile linkage between lipid A and Kdo of the carbohydrate portion of the LPS (45), the presence of all three unligated oligosaccharide fractions in the A. aeolicus LPS sample was confirmed. The spectra displayed peaks at 1917.33 u, 1294.31 u (M4), 2243.70 u (M5), and 2405.75 u (M6) (supplemental Fig. S2B) that were in excellent agreement with the calculated masses of free lipid A and the oligosaccharide components lacking lipid A (Δm = 1917.3 u), respectively (supplemental Table S2). Compositional analysis of the LPS and the oligosaccharide fraction isolated after hydrolysis of the LPS identified Rha, Man, Glc, Gal, GalA, Hep, and Kdo (supplemental Table S3), confirming that the polysaccharide chains of the LPS of A. aeolicus and A. pyrophilus (20) were of a quite different nature. Noteworthy, thiobarbituric acid-reactive material could be liberated only upon strong hydrolysis of the LPS and the oligosaccharide fraction but not using mild acid treatment, indicating the lack of acid-labile side chain Kdo substitutions (44). Thus, the application of harsh hydrolysis conditions enabled the determination of an average molar ratio of the common inner core constituents Kdo and Hep of 1.0:1.8 for the LPS and 1.0:1.7 for the LPS-derived oligosaccharide, with Kdo being probably a constituent of the main core oligosaccharide chain. Taken together, the results suggested that the LPS of A. aeolicus contains a single Kdo residue, and therefore, it was assumed that WaaA of A. aeolicus is a monofunctional Kdo transferase.
Expression of WaaA from A. aeolicus in E. coli
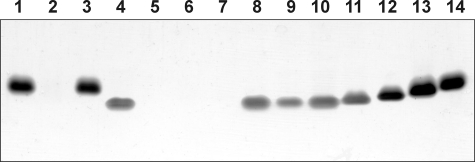
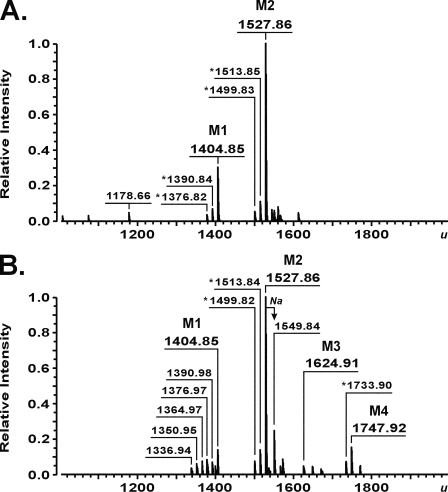
Preliminary in vitro experiments revealed that, although WaaA of A. aeolicus is thermophilic (see below), it is still capable of catalyzing Kdo transfer at a temperature of 37 °C (data not shown). In order to investigate the in vivo activity of the A. aeolicus enzyme as well as for characterization of other heterologous Kdo transferases in E. coli, the strain KPM56, a kdsD+ ΔwaaC ΔwaaA derivative of KPM22, was constructed. Strain KPM22, defective in the d-arabinose-5-phosphate isomerases KdsD and GutQ, is a nonconditional E. coli K-12 Kdo pathway null mutant that lacks Kdo and is viable despite predominantly elaborating the lipid A precursor lipid IVA (28). The lethal ΔKdo phenotype of KPM22 was shown recently to be suppressed by an Arg to Cys substitution at position 134 of the inner membrane protein YhjD, a gain-of-function mutation that converts the normally essential Kdo pathway genes into nonessential genes (28, 42). Thus, the presence of the yhjD400 suppressor allele in KPM22 enabled the derivation of the target KPM56 in three steps. First, the Kdo pathway of KPM22 was reconstituted by insertion of the kdsD wild-type gene, which yielded strain KPM50 with restored K-12 LPS biosynthesis (Fig. 1, lane 3). Second, to facilitate subsequent analyses of the LPS from strains expressing heterologous Kdo transferases, the LPS pathway following attachment of the Kdo residues to lipid A was interrupted in KPM50 by deleting the waaC gene for heptosyltransferase I, which resulted in strain KPM53 expressing LPS of the Re chemotype (Kdo2-lipid A) (Fig. 1, lane 4). Finally, KPM53 served as the host for the waaA knock-out to yield strain KPM56, which was indeed viable and capable of maintaining the normally lethal ΔwaaA mutation. Like KPM22 (Fig. 1, lane 2) (28), silver-stained SDS-polyacrylamide gels detected no LPS bands for KPM56 (Fig. 1, lane 5). Analysis of the KPM56 LPS by ESI FT-ICR MS, however, revealed two prominent LPS-related peaks with molecular masses of 1404.85 and 1527.86 u, consistent with the structures of the tetraacyl-1,4′-bisphosphate LPS precursor lipid IVA (calculated mass 1404.854 u) and lipid IVA modified with a phosphoethanolamine (P-EtN) group (calculated mass 1527.863 u), respectively (Fig. 2A). Although SDS-PAGE analysis of the LPS isolated from control strains KPM124 and KPM122 readily demonstrated complementation of the ΔwaaA mutation by Kdo transferases from E. coli and C. psittaci (Fig. 1, lanes 8 and 9), no LPS bands could be observed for KPM125 and the KPM123 control strain carrying the Kdo transferases of A. aeolicus and H. influenzae (Fig. 1, lanes 6 and 7), respectively. However, when the LPS samples of A. aeolicus and H. influenzae were examined using MS, we could not only confirm the monofunctional activity of WaaA from H. influenzae by detecting a nearly exclusive peak of Kdo-lipid IVA in KPM123 (1624.91 u) (supplemental Fig. S3A and Table S4) but also could detect peaks at 1624.91 and 1747.92 u in the KPM125 sample, which indicated the presence of only a single Kdo residue attached to lipid IVA and P-EtN-modified lipid IVA, respectively (Fig. 2B). Although substantial amounts of the lipid IVA precursors remained nonglycosylated, the results suggested that the Kdo transferase of A. aeolicus is monofunctional in E. coli. Strikingly, a common feature of KPM123 and KPM125 was the lack of the secondary lauroyl and myristoyl chains on Kdo-lipid IVA derivatives. To verify that their parental strain KPM56 still contained functional copies of the late acyltransferases LpxL and LpxM, we analyzed the LPS of KPM124 by ESI FT-ICR MS. As shown in supplemental Fig. S3B and Table S4, complementation of the chromosomal waaA deletion with the homologous, bifunctional Kdo transferase resulted in expression of mainly hexaacylated lipid A species, each carrying the secondary acyl chains and two Kdo residues. Although MS revealed a rather heterogeneous mixture of LPS molecules with acyl chains of various lengths differing in mass each by 14.02 u, the results indicated the capability of LpxL and LpxM to form the characteristic acyloxyacyl units of lipid A in KPM56-derived strains, provided that lipid IVA was glycosylated with two Kdo residues. Thus, our data indicating lipid IVA precursors remaining underacylated with only a single Kdo residue attached corroborate previous in vitro (47, 48) and in vivo (28) findings of an extremely high specificity of LpxL and LpxM for the Kdo2-lipid IVA substrate.
FIGURE 1.
SDS-PAGE analysis of proteinase K-digested whole cell lysates of KPM22-derived E. coli and S. enterica sv. Typhimurium reference strains. LPS samples were subjected to electrophoresis on a 13% polyacrylamide gel followed by silver nitrate staining. Lane 1, wild-type E. coli strain BW30270; lane 2, KPM22; lane 3, KPM50; lane 4, KPM53; lane 5, KPM56; lane 6, KPM125; lane 7, KPM123; lane 8, KPM124; lane 9, KPM122; lane 10, S. enterica sv. Typhimurium SL1102 (Re); lane 11, S. enterica sv. Typhimurium SL3769 (Rd1); lane 12, S. enterica sv. Typhimurium SL3748 (Rb3); lane 13, S. enterica sv. Typhimurium SL3750 (Rb2); lane 14, S. enterica sv. Typhimurium SL3749 (Ra).
FIGURE 2.
Charge deconvoluted ESI FT-ICR mass spectra in negative ion mode of LPS molecules isolated from E. coli strains KPM56 (A) and KPM125 (B). M1, lipid IVA (1404.85 u); M2, lipid IVA modified with a P-EtN group (1527.86 u); M3, Kdo-lipid IVA (1624.91 u); M4, Kdo-lipid IVA substituted with a P-EtN group (1747.92 u). Peaks presumably representing molecules with variations in acyl chain length (Δm = 14.02 u) are labeled with an asterisk. The molecule corresponding to triacylated lipid A (1178.66 u) is probably an artifact produced during LPS isolation, since it is not consistent with a known pathway intermediate.
Overexpression and Purification of WaaA from A. aeolicus
Our initial attempts to express the Kdo transferase of A. aeolicus at high levels in T7-promoter based expression hosts were unsuccessful due to sequestration of the protein within insoluble inclusion bodies. Soluble and catalytically active WaaAAAE could only be obtained in sufficient quantities when WaaAAAE expression was performed in BL21-SI216, which allowed tightly regulated protein production at moderate levels. The formation of inclusion bodies was nearly avoided when the growth rate of the induced cells was reduced under suboptimal conditions in acidic medium at 20 °C. Under these conditions, the maximal ratio of recombinant to cellular protein was achieved by expression of the Kdo transferase for a minimum of 18 h.
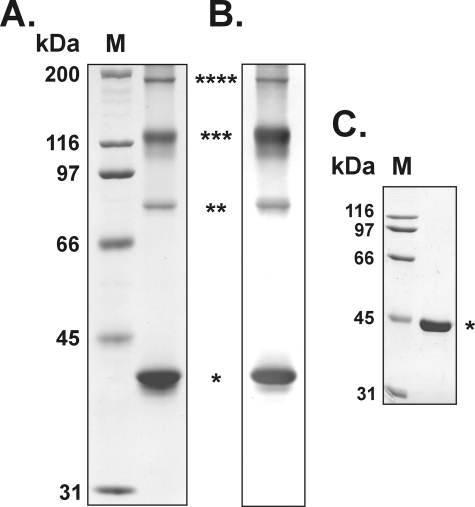
Purification to apparent homogeneity of recombinant WaaAAAE was accomplished in three chromatographic steps using anion exchange chromatography on DEAE-Sepharose, immobilized metal affinity chromatography on nickel-Sepharose, and affinity chromatography on heparin-Sepharose. Like the Kdo transferase of E. coli (6), WaaA of A. aeolicus displayed reversible affinity for the heparin resin, and therefore, it was used for purification of the enzyme. Taking advantage of the enzyme's tolerance to high temperatures (see below), residual host cell protein could be easily removed by heat treatment of the samples. The Kdo transferase of A. aeolicus as analyzed by SDS-PAGE and verified by immunoblotting using an anti-histidine tag antibody appeared as four bands with apparent relative molecular masses of about 42, 83, 122, and 185 kDa (Fig. 3, A and B). The apparent relative molecular mass of the 42 kDa band was in good agreement with the calculated mass of the subunit of 40.7 kDa fused to a 2.5-kDa N-terminal histidine tag, whereas the other three bands migrated with apparent relative molecular masses about 2, 3, and 4 times the mass of the subunit, suggesting the capability of WaaAAAE to form remarkably stable oligomers that were refractory to the normally denaturing conditions used for SDS-PAGE.
FIGURE 3.
SDS-PAGE and immunoblot analysis of recombinant Kdo transferases from A. aeolicus and E. coli. The proteins were resolved using 10% polyacrylamide gels and stained with Coomassie Blue. Molecular mass protein markers (kDa) were run in lanes M. The asterisks indicate the positions of the supposed WaaA subunits, dimers, trimers, and tetramers. A, SDS-PAGE of purified WaaAAAE (15 μg); B, immunoblot of WaaAAAE (5 μg) probed with anti-histidine tag and alkaline phosphatase-conjugated goat anti-mouse IgG (H + L) antibodies and developed in the presence of nitro blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate substrate; C, SDS-PAGE of purified WaaAECO (5 μg).
To go further into the question of reliability of the herein developed methods for WaaAAAE overexpression and purification, as well as to address the question of an equivalent control for subsequent in vitro assays, we attempted to express the Kdo transferase of E. coli in BL21-SI212 cells and purify the enzyme essentially as outlined for the Kdo transferase of A. aeolicus. Using identical conditions for expression and minor modifications of the basic purification scheme (for details, see “Experimental Procedures”), we could purify the Kdo transferase of E. coli to apparent homogeneity as well. The subunit as shown by SDS-PAGE under denaturing conditions migrated as a single band with an apparent relative molecular mass of about 45 kDa (Fig. 3C), which was marginally different from the calculated mass of 49.8 kDa but in accord with the relative mobility previously observed for WaaAECO in SDS-PAGE (6).
In Vitro Analysis of WaaAAAE Activity
To ascertain whether the Kdo transferase of A. aeolicus was monofunctional, the WaaA-catalyzed transfer of Kdo from CMP-Kdo to synthetic lipid acceptors was assayed in vitro at 60 °C, using the herein developed novel method for detection of WaaA reaction products by MS (for details, see “Experimental Procedures”). Since CMP-Kdo is known to be highly unstable (49), a CMP-Kdo-generating system consisting of CTP, Kdo, Mg2+, and CMP-Kdo synthetase is required to determine WaaA activity in vitro (6). For this purpose and like the Kdo transferases of A. aeolicus and E. coli, we have purified the homologous CMP-Kdo synthetases to apparent homogeneity as described under “Experimental Procedures” (data not shown).
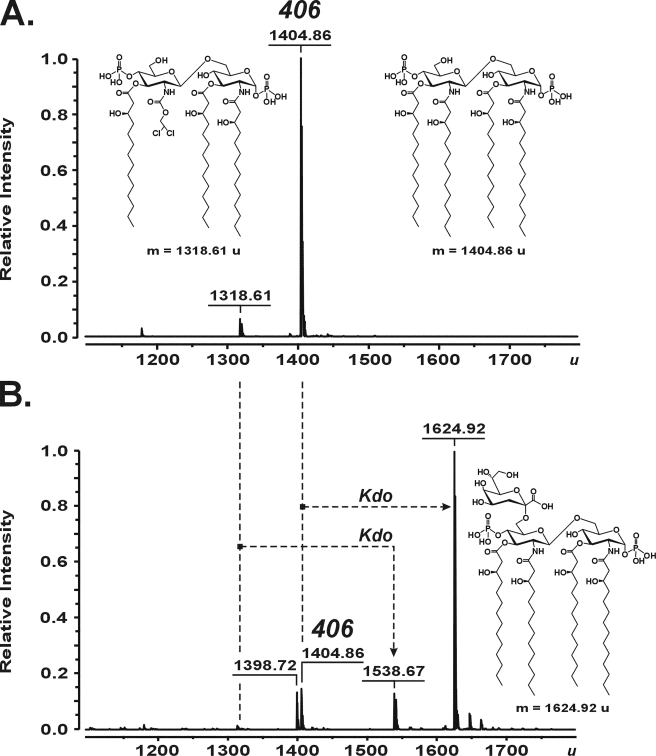
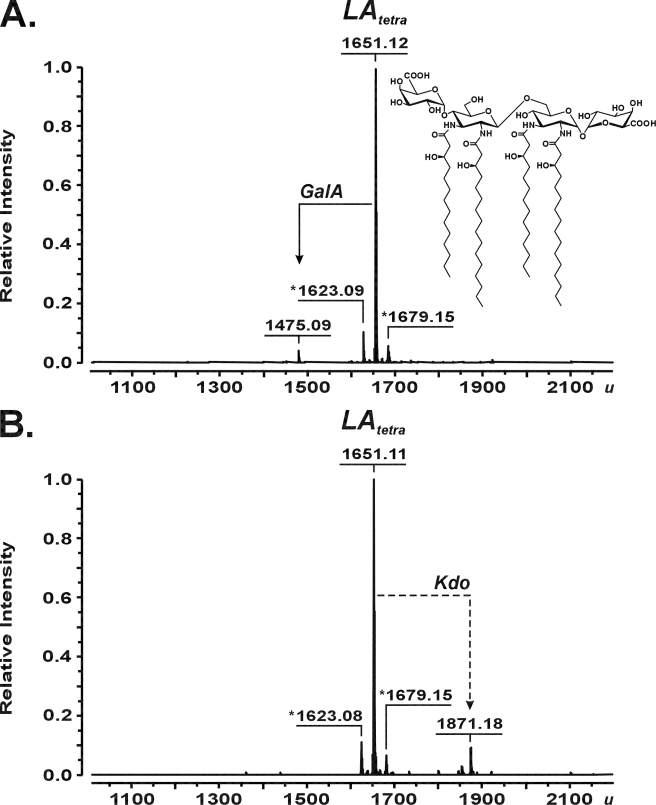
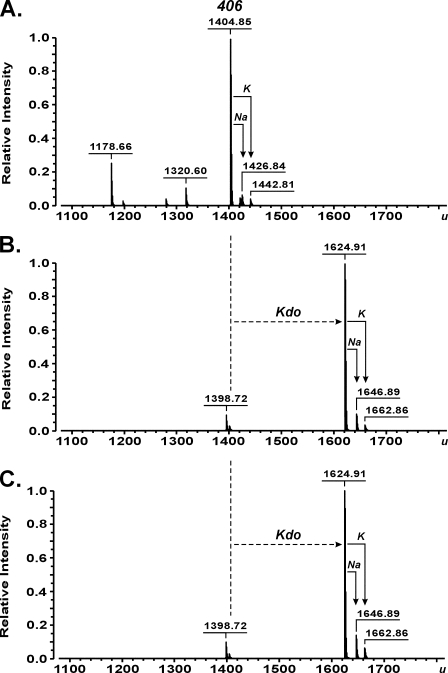
As demonstrated by ESI FT-ICR MS, the Kdo transferase of A. aeolicus was capable of converting the synthetic tetraacyl-1,4′-bisphosphate lipid A precursor 406 with a molecular mass of 1404.86 u (Fig. 4A) into a molecule with a molecular mass of 1624.92 u, consistent with the structure of 406 being substituted with a single Kdo residue (calculated mass 1624.912 u) (Fig. 4B). Further, a minor batch-dependent triacylated byproduct of 406 synthesis with a mass of 1318.61 u (Fig. 4A) was converted into a product with a mass of 1538.67 u (Fig. 4B), indicating the attachment of only a single Kdo residue to the artificial acceptor. No products were obtained in reactions when either WaaAAAE or KdsBAAE was omitted (not shown). For the positive control experiments, WaaA of E. coli was assayed under identical conditions, except the incubation temperature was adjusted to 37 °C. As shown in supplemental Fig. S4, incubation of compound 406 with the bifunctional E. coli enzyme indeed resulted in formation of a peak at 1844.99 u, corresponding to Kdo2-406 with a calculated mass of 1844.971 u. In order to investigate the specificity of WaaAAAE for lipid acceptors, we intentionally have chosen a compound 506 sample containing a mixture of penta-, tetra-, and triacylated degradation products as a result of gradual loss of the ester-linked acyl groups upon long term storage of the synthetic hexaacyl-1,4′-bisphosphate lipid A molecule in solution. Thus, ESI FT-ICR MS of the lipid mixture identified a prominent peak of compound 506 (1797.22 u) together with peaks of 506 lacking either the primary 14:0(3-OH) at position 3 of the reducing GlcN I (1571.02 u), the 14:0[3-O(14:0)] attached to position 3′ of the non-reducing GlcN II (1360.82 u), or even both units (1134.63 u) (supplemental Figs. S5A and S6 showing the structures of the acceptors). When the mixture was incubated with the Kdo transferase of A. aeolicus, the mass of each lipid increased by 220.07 u (supplemental Fig. S5B), which indicated the glycosylation of each differently acylated lipid molecule with a single Kdo residue. Furthermore, to address the question of whether a decrease in the negative net charge at the 1- and 4′-positions of the GlcN disaccharide backbone of acceptor molecules affected the transfer of Kdo, the tetraacyl-1-monophosphate precursor 405, lacking the phosphate group at position 4′, as well as carboxymethyl derivatives of compounds 506 (CM-506) and 406 (Bis-CM-406) were subjected to WaaAAAE in vitro reactions. Under standard conditions, the enzyme converted not all but still significant amounts of compound 405 (1324.89 u) into Kdo-405 (1544.94 u) (supplemental Fig. S7), suggesting that the 4′-phosphate group of the GlcN disaccharide backbone of lipid A is not an absolute requirement for the addition of Kdo. Compound CM-506, which contained a minor fraction of tetraacylated compound CM-406, and compound Bis-CM-406 turned out to be good acceptors for Kdo. Regardless of whether the 1-phosphate of the hexaacylated compound 506 or the tetraacylated compound 406 was substituted for CM or whether the phosphate groups at positions 1 and 4′ of the tetraacylated precursor 406 were each replaced by a CM moiety, a single Kdo residue was transferred to each of the artificial lipid A analogs by WaaAAAE (supplemental Figs. S8 and S9). In contrast, when O-deacylated lipid A of A. pyrophilus lacking the ester-bound octadecanoic acid (1651.12 u) was incubated with WaaAAAE, only a marginal fraction of the acceptor was glycosylated with a single Kdo (1871.18 u) (Fig. 5), indicating that tetraacylated lipid A with GalA residues in place of the phosphate groups at positions 1 and 4′ is a poor substrate for the Kdo transferase. Collectively, the results showed that WaaA of A. aeolicus is a monofunctional Kdo transferase also in vitro.
FIGURE 4.
Charge deconvoluted ESI FT-ICR mass spectra in negative ion mode, demonstrating the conversion of the tetraacyl-1,4′-bisphosphate lipid A precursor 406 (inset structure; 1404.86 u) (A) into Kdo-406 (inset structure; 1624.92 u) (B) by the Kdo transferase of A. aeolicus. The reaction mixtures containing 100 mm HEPES, pH 7.5, 10% glycerol, 10 mm MgCl2, 2 mm Kdo, 5 mm CTP, 3.2 mm Triton X-100, and 107 μm compound 406 were incubated at 60 °C for 1 h with no enzymes (A), and in the presence of 17 milliunits of purified KdsBAAE and 75 μm purified WaaAAAE (B). A triacylated byproduct of 406 synthesis (inset structure; 1318.61 u) (A) served as an acceptor for the WaaAAAE-catalyzed transfer of a single Kdo residue to yield the molecule at 1538.67 u (B). The peak at 1398.72 u (B) is probably a triacylated degradation product of Kdo-406 produced during sample preparation.
FIGURE 5.
Charge-deconvoluted ESI FT-ICR mass spectra in negative ion mode showing the O-deacylated lipid A sample from A. pyrophilus (LAtetrainset structure; 1651.12 u (20)). The lipid A molecule (91 μm) was used as an acceptor in reactions containing 100 mm HEPES, pH 7.5, 10% glycerol, 10 mm MgCl2, 2 mm Kdo, 5 mm CTP, and 3.2 mm Triton X-100. The mixtures were incubated at 60 °C for 1 h with no enzymes (A) and in the presence of 17 milliunits of purified KdsBAAE and 75 μm purified WaaAAAE (B). Peaks presumably representing molecules with variations in acyl chain length are labeled with an asterisk.
Studies of the Interaction between WaaAAAE and Donor Substrates
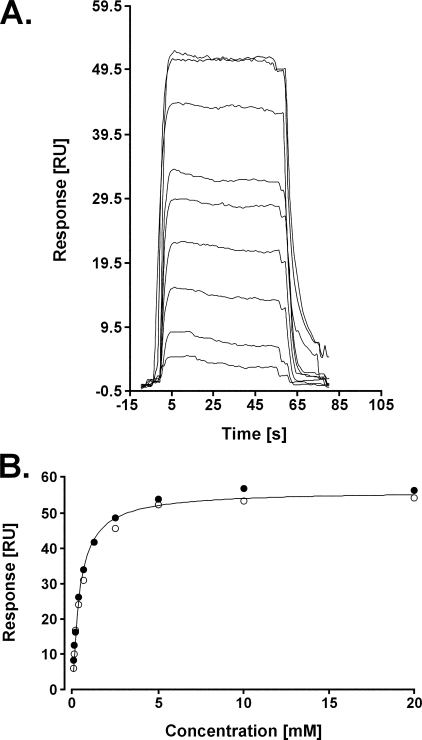
Due to the high instability of CMP-Kdo (49), interaction of CMP and Kdo with the Kdo transferase of A. aeolicus was investigated separately using SPR technology (50). WaaAAAE (ligand) was immobilized to the surface of a CM5 sensor chip, and different concentrations of CMP or Kdo (analytes) were injected to the surface, as described under “Experimental Procedures.” Binding of the analyte to the ligand was monitored as an increase of the response (RU) versus time (sensorgram). When CMP was injected to the surface with WaaAAAE, an increase of the response was detected, which suggested binding of CMP to the Kdo transferase. The resulting sensorgrams were characteristic of a binding event with fast on and off rates, and thus, only steady state analysis of the sensorgrams was possible (Fig. 6A). The corresponding binding isotherm was fitted to a 1:1 model, and a KD of 401 ± 25 μm was determined (Fig. 6B). When Kdo was delivered to the surface coated with WaaAAAE, we could not measure any increase of the response, indicating no binding of Kdo to the Kdo transferase under the experimental conditions used (data not shown). Injection of AMP, TMP, UMP, and GMP detected much lower signals compared with the response caused by binding of CMP to WaaAAAE, and plot of the Req versus NMP concentration resulted in all cases in linear graphs characteristic of nonspecific electrostatic interactions between the ligand and the analyte (supplemental Fig. S10). Taken together, the data indicated that CMP but not Kdo alone and other NMPs are specifically recognized by the Kdo transferase of A. aeolicus.
FIGURE 6.
Sensorgrams of the binding between CMP and the Kdo transferase of A. aeolicus immobilized to a CM5 sensor chip (A) and steady-state analysis of the interaction between CMP and WaaAAAE (B). Req (RU) from two data sets were plotted versus the concentration of CMP (mm). Fitting of the isotherm to a 1:1 Langmuir model resulted in a KD of 401 ± 25 μm.
The Effect of High Temperature on WaaAAAE Activity
The hyperthermophilic origin of WaaAAAE prompted us to examine the effect of temperature on the WaaAAAE-catalyzed conversion of compound 406 into Kdo-406 as a function of time. Pretreatment of the enzyme at 70 and 80 °C for 1 h prior to the determination of residual activity at 60 °C did not affect the ability of WaaAAAE to perform transfer of a single Kdo residue to the lipid A acceptor, whereas preincubation of the enzyme at 90 and 99 °C for 1 h resulted in gradual loss of its activity (supplemental Fig. S11). Remarkably, we could not observe any detrimental effect on Kdo glycosylation of compound 406 after long term storage of the Kdo transferase at 80 °C for 16 h (Fig. 7). Thus, the extraordinary thermostability demonstrated by the recombinant Kdo transferase of A. aeolicus allowed us to conclude that the enzyme was produced and isolated in its native state.
FIGURE 7.
Charge deconvoluted ESI FT-ICR mass spectra in negative ion mode demonstrating the WaaAAAE-catalyzed conversion of the tetraacyl-1,4′-bisphosphate lipid A precursor 406 (1404.85 u) (A) into Kdo-406 (1624.91 u) without (B) and with treatment of the Kdo transferase at 80 °C for 16 h (C) prior to the determination of enzymatic activity. The activity was assayed in reaction mixtures containing 100 mm HEPES, pH 7.5, 10% glycerol, 10 mm MgCl2, 2 mm Kdo, 5 mm CTP, 3.2 mm Triton X-100, and 107 μm compound 406. The reactions were incubated at 60 °C for 1 h with no enzymes (A) and in the presence of 17 milliunits of purified KdsBAAE and 75 μm either untreated (B) or pretreated WaaAAAE (C). The peak at 1320.60 u is consistent with a batch-dependent byproduct of 406 synthesis (A). The peaks at 1178.66 u (A) and 1398.72 u (B and C) are probably triacylated degradation products of compound 406 and Kdo-406 produced during sample preparation, respectively.
DISCUSSION
This study describes the characterization of WaaA from A. aeolicus and represents the first report on a Kdo transferase from an extreme hyperthermophile. The results presented herein strongly support the conclusion that WaaAAAE belongs to the group of monofunctional Kdo transferases of LPS biosynthesis.
A. aeolicus Synthesizes LPS Containing a Single Kdo Residue
Except for Helicobacter pylori and Francisella novicida, the LPSs of which possess a single Kdo in the inner core region while expressing bifunctional Kdo transferases (51, 52), the number of Kdo residues in the core oligosaccharide of LPS normally predicts the functionality of WaaA. Our data on the initial characterization of the A. aeolicus LPS indicate that the molecule contains a single Kdo residue and, therefore, implicates a monofunctional Kdo transferase in LPS biosynthesis of A. aeolicus. In fact, we provide direct evidence for the existence of LPS in A. aeolicus, thus confirming the results of previous investigations, which predicted the presence of LPS in A. aeolicus on the basis of genome analysis (24), biochemical and structural characterization of KdsA (21, 22) and LpxC (23, 53), or detection of LPS and a unique lipid A structure in A. pyrophilus, a close relative of A. aeolicus (20). Since lipid A is considered to be a valuable chemotaxonomic and phylogenetic marker (54), the close relation of A. aeolicus and A. pyrophilus is further supported by our ESI FT-ICR MS experiments that suggest that the structure of A. aeolicus lipid A is identical to that of A. pyrophilus (20). However, we observed a striking differences between the LPS from A. aeolicus and A. pyrophilus by SDS-PAGE and ESI FT-ICR MS as well as a different sugar composition in the LPS of A. aeolicus, indicating significant strain-specific variations in structure and biosynthesis of the LPS polysaccharide chain within the genus Aquifex.
A. aeolicus waaA Encodes a Monofunctional Kdo Transferase That Can Complement a ΔwaaA Mutation in E. coli
In vivo examination of Kdo transferases of different origin has usually been performed in E. coli to gain insight into the structure of the resulting LPS. In consideration of the fact that viability of wild-type E. coli cells is normally dependent upon the presence of a functional waaA copy (55), several strategies have been developed to reduce or even avoid LPS complexity stemming from co-expression of the heterologous and the host-specific Kdo transferase, such as cloning of Kdo transferases in an E. coli Re mutant (56, 57), plasmid-encoded expression of WaaA in an E. coli waaA null mutant by displacement under nonpermissive conditions of a functional waaAECO copy provided in trans (9, 15, 55), or homologous recombination of WaaA expression cassettes into the waaA locus of a recBC sbcBC E. coli strain (12, 14). Here we have constructed the ΔwaaA suppressor strain KPM56 as a novel host for expression and in vivo analysis of exclusively plasmid-borne Kdo transferases. Although our results show that WaaA from A. aeolicus is monofunctional in KPM125, the enzyme apparently does not reach its full catalytic activity in the mesophilic E. coli host. Our in vitro data demonstrating conclusively the conversion of chemically synthesized compound 406 into Kdo-406 do not give us reason to assume that its natural counterpart lipid IVA represents a poor substrate of WaaAAAE but suggest a diminished capability of the thermophilic enzyme to glycosylate the lipid A precursor at a suboptimal temperature of 37 °C. Although thermophilic enzymes usually develop optimal activity at high temperatures close to the optimal growth temperatures of their hosts, they are thought to adopt structures of high rigidity at mesophilic temperatures below 40 °C, which in turn may cause a decrease in catalytic efficiency (58, 59). The successful expression of WaaA from H. influenzae in KPM123, leading to predominant formation of Kdo-lipid IVA, indicates that KPM56 is a suitable host also for characterization of monofunctional Kdo transferases. This is emphasized especially in view of the fact that investigation of monofunctional Kdo transferases has been met with restrictions under conditions of customarily used E. coli ΔwaaA complementation systems, such as the need to co-express the Kdo kinase KdkA from H. influenzae in order to compensate for the lethal phenotype associated with the substitution of waaAECO for waaA of H. influenzae (14). In view of Kdo-lipid IVA being a very poor substrate of LpxL in vitro (60) and, as we show now, also in vivo, with LpxM apparently being unable to transfer a myristate chain to Kdo-lipid IVA in the absence of laurate, the underacylated and underglycosylated precursor is expected to accumulate in large toxic amounts in the inner membrane due to the high selectivity of the ATP binding cassette transporter MsbA for hexaacylated LPS/lipid A substrates (61, 62). E. coli KPM123, however, is viable despite predominantly elaborating Kdo-lipid IVA. Therefore, we propose that the YhjDR134C suppressor protein, which has been suggested to participate in transmembrane transport of lipid IVA in E. coli KPM22 (42), can also transit Kdo-lipid IVA molecules across the inner membrane of KPM22-derived strains, restoring viability by supplementing the insufficient amounts of Kdo-lipid IVA being translocated by MsbA alone.
WaaAAAE Is a Thermostable Kdo Transferase
Kdo transferases have been described to share a low degree of similarity at the primary amino acid sequence level (10, 29). Comparison of the deduced amino acid sequence of 353 residues of the A. aeolicus waaA gene with the predicted sequences of other Kdo transferase genes identified a total of 81 amino acid residues within the Aquifex enzyme that are either identical or similar to amino acids being conserved across Kdo transferases of mesophilic origin (supplemental Fig. S12). Unlike its mesophilic E. coli orthologue, WaaA of A. aeolicus, as herein demonstrated by SDS-PAGE, is able to form highly stable oligomers of up to quadruple the mass of its subunit, suggesting strong intersubunit interactions that resist dissociation when exposed to denaturants, such as SDS. Moreover, we have shown that WaaA of A. aeolicus is a thermostable Kdo transferase, the stability of which falls into a range of the thermostability exhibited by other proteins isolated from hyperthermophilic bacteria (63). An increasing body of evidence shows that thermophilic enzymes are generally more robust than their mesophilic counterparts, including a higher resistance to chemical denaturation (59, 64, 65). Although it has become widely accepted that stabilization of thermophilic enzymes is not a result of a single type of reaction but rather occurs cumulatively at all stages of structure formation, oligomerization via stabilizing forces, such as hydrophobic interactions, hydrogen bond formation, and/or surface ion pair networks, is thought to be a major mechanism of stabilizing the proteins (58, 59). Together with the detection of WaaAAAE dimers and trimers using analytical gel filtration chromatography on a calibrated Superdex 200 pg column under native conditions (data not shown), the results give reason to assume that the thermophilic Kdo transferase features the intrinsic propensity to adopt extremely stable dimeric, trimeric, or even tetrameric forms in solution, which in turn could contribute to stabilization of the protein.
WaaAAAE Exhibits Relaxed Specificity for Acceptor Substrates
Our in vitro data provide further evidence that the Kdo transferase of A. aeolicus is indeed monofunctional. The results allow us to conclude that the enzyme shares some significant properties with its mesophilic orthologues. WaaA of A. aeolicus resembles Kdo transferases of E. coli (6), A. baumannii, A. haemolyticus (10), or L. pneumophila (9) in its ability to utilize a fairly broad spectrum of acceptor substrates for glycosylation. Under our standard reaction conditions to assay Kdo transferase activity in vitro, WaaAAAE was neither selective for a defined acylation pattern of the synthetic acceptor molecules tested nor dependent upon the β-(1→6)-linked 2,3-diamino-2,3-dideoxy-d-glucopyranose disaccharide backbone of Aquifex lipid A. The latter conclusion is supported by the finding that the presence of a β-(1→6)-linked GlcN disaccharide backbone in all chemically synthesized lipid A derivatives investigated in the present study did not detrimentally affect WaaAAAE activity. Phosphorylation of the 4′-OH group of the lipid A precursor has been suggested to be an absolute prerequisite for the activity of distal LPS enzymes, including Kdo transferases (6, 9). To the best of our knowledge, WaaA of A. aeolicus as shown by the partial conversion of the tetraacyl-1-monophosphate precursor 405 into Kdo-405 is the first Kdo transferase that does not strictly depend on a negatively charged phosphate group at position 4′ of the lipid A intermediate. Previous studies with CM derivatives of compound 506 indicated that the function of some mesophilic Kdo transferases is already abolished when the 4′-phosphate group of the acceptor is substituted for the less negatively charged CM moiety (66). The activity of WaaAAAE was not impaired by using either CM-506 or Bis-CM-406 as acceptor substrate, suggesting that the thermophilic Kdo transferase can tolerate a decrease in the negative net charge not only at position 4′ but also at positions 4′ and 1 of the lipid A carbohydrate backbone. Although we currently cannot respond to the question of whether the acceptor substrate promiscuity of WaaAAAE plays a role in biosynthesis of the A. aeolicus LPS in vivo, our in vitro results are consistent with the proposal for the existence of common although still unknown WaaA recognition motifs within the acceptor molecules (9). Although the enzyme apparently did not display any preference for most of the differently substituted lipid A acceptors tested, we cannot rule out differences in the rate of Kdo incorporation. Although ESI FT-ICR MS coupled to WaaA in vitro reactions currently does not allow the determination of kinetic values, it could be applied here for the first time to determine the simultaneous and direct monitoring of multiple WaaA substrate and product species with a high degree of specificity. Thus, MS may represent a valuable alternative to customarily used techniques that rely on multistep synthesis and purification of radioactively labeled WaaA substrates (9, 10, 14, 55, 67) and/or immunological detection of lipid A acceptors or Kdo epitopes of the glycosylated reaction products (9, 10).
GalA Substitution of Lipid A Inhibits Kdo Transfer
The ability of WaaAAAE to transfer a single Kdo residue to various lipid A acceptors in vitro is virtually abolished when authentic tetraacylated Aquifex lipid A is used as a substrate. The most straightforward explanation for this finding is that the presence of two GalA residues replacing the phosphate groups at positions 4′ and 1 of the 2,3-diamino-2,3-dideoxy-d-glucopyranose disaccharide backbone prevents the molecule from efficient glycosylation, suggesting that the acceptor binding site of WaaAAAE cannot accommodate more than a disaccharide unit of the acceptor substrate. This in turn argues for a Kdo transfer to a phosphorylated lipid A intermediate prior to GalA attachment in vivo, an assumption that is indirectly supported by the existence of an LpxK orthologue for incorporation of a phosphate moiety at O-4′ of the lipid A backbone in A. aeolicus (24). Thus, we suspect that lipid A processing, leading to mature Kdo-glycosylated lipid A in A. aeolicus, is similar to that of Rhizobium leguminosarum, F. tularensis, or H. pylori, the lipid A of which has been shown to lack one or both phosphate groups (68–70). In these bacteria, the biosynthesis of lipid A diverges from that of E. coli and other Gram-negative bacteria after the first seven enzymes of the constitutive E. coli lipid A pathway have generated the intermediate Kdo2-lipid IVA in the cytoplasmic compartment (1, 7). The lipid A molecules are subsequently completed at the periplasmic face of the inner membrane, which includes the removal of the 1- and/or 4′-phosphate groups by LpxE and/or LpxF as a prerequisite for additional species-specific lipid A modifications (71–75). Except for the UDP-2,3-diacylglucosamine hydrolase LpxH, all of the other six orthologues constituting the conserved Kdo-lipid IVA pathway have been discovered in A. aeolicus, namely LpxA, LpxC, LpxD, LpxB, LpxK, and WaaA (24). However, studies to shed light on the entire process of lipid A formation in A. aeolicus are hindered by the fact that no significant similarities to genes for possibly distantly related lipid A-modifying enzymes have so far been identified in the genome of the hyperthermophilic bacterium.
WaaAAAE Is Highly Specific for the Donor Substrate
Like all mesophilic Kdo transferases, WaaAAAE displayed an absolute dependence upon the presence of CTP, Kdo, and KdsBAAE (data not shown). Two lines of evidence support the conclusion that CMP-Kdo, although extremely labile at moderate temperatures (6, 49), is the donor for Kdo transfer in the hyperthermophilic A. aeolicus. First, at pH 7.5 and a temperature of 80 °C, KdsB of A. aeolicus was found to be highly specific for CTP and Kdo, with ATP and GTP being unable to replace CTP. UTP could act as a substitute for CTP, albeit to a much lesser extent.8 Second, the Kdo transferase of A. aeolicus as herein monitored in real time by SPR clearly displayed affinity for CMP but not for AMP, TMP, UMP, or GMP. Although the equilibrium dissociation constant of the reaction indicated that the strength of CMP binding to WaaAAAE is moderate under the conditions examined, the results suggest that utilization of CMP-Kdo for Kdo transfer in A. aeolicus is not exclusively defined by the specificity of the CMP-Kdo synthetase for CTP and Kdo but also by a high specificity of the Kdo transferase itself, at least for the nucleotide moiety of the donor substrate. However, our SPR measurements failed to detect any affinity of WaaAAAE for Kdo. It has been shown for other glycosyltransferases that the nucleotide and sugar moieties of the donor substrate play different roles in the interaction between the enzyme and the activated nucleotide sugar, with the nucleotide moiety being crucial to recognition of the substrate and the sugar being instead involved in determining the specificity of the enzyme (76). This supports the idea that CMP may guide CMP-Kdo to the donor substrate binding site of the Kdo transferase. The lack of binding of Kdo to the enzyme currently favors the hypothesis that WaaAAAE is strictly selective for the anomeric configuration of Kdo. Taking into account that Kdo in solution exists predominantly in the α-pyranose form, with a minor fraction of about 2% being β-pyranosidic (77), as well as that the latter Kdo anomer was demonstrated to be the preferred KdsB substrate for synthesis of CMP-Kdo containing Kdo in the β-configuration (78), we believe that Kdo must be β- but not α-configured to bind to the Kdo transferase of A. aeolicus.
In summary, we have characterized the Kdo transferase from the hyperthermophilic bacterium A. aeolicus and shown that the enzyme adds a single Kdo residue to lipid A. The work presented here suggests that the Kdo transferase of the phylogenetically ancient A. aeolicus is presumably part of the conserved constitutive Kdo-lipid IVA pathway before the phosphorylated lipid A moiety is eventually processed to yield the phosphateless lipid A of Aquifex LPS.
Supplementary Material
Acknowledgments
We thank Brigitte Kunz and Hermann Moll (Research Center Borstel) for technical assistance and Thomas Peters (Institute of Chemistry, University of Lübeck, Germany) for help with SPR experiments. Strain C. glutamicum R163/pJKB72 was kindly provided by Sabine Gronow (German Resource Center for Biological Material, Braunschweig, Germany), plasmid pCB23 by Werner Brabetz (Biotype AG, Dresden, Germany), and genomic DNA of A. aeolicus by Karl Otto Stetter (Department of Microbiology, University of Regensburg, Germany).
This work was supported by Deutsche Forschungsgemeinschaft Grant MA 1408/2-1 (to U. M.), Grant ME 2741/1-1 (to J. R. M. and R. H.), and Grant Li-448/4-1 (to B. L.). The BIAcore 3000 instrument was acquired with the support of Deutsche Forschungsgemeinschaft SFB470.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S12.
H. Schmidt and U. Mamat, unpublished results.
- LPS
- lipopolysaccharide
- u
- unified atomic mass unit
- OM
- outer membrane
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- ESI
- electrospray-ionization
- FT-ICR
- Fourier-transformed ion cyclotron
- MS
- mass spectrometry
- CM
- 1-carboxymethyl
- GLC/MS
- gas liquid chromatography
- SPR
- surface plasmon resonance
- RU
- resonance units
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- P-EtN
- phosphoethanolamine.
REFERENCES
- 1.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holst O. (2007) FEMS Microbiol. Lett. 271, 3–11 [DOI] [PubMed] [Google Scholar]
- 3.Meredith T. C., Woodard R. W. (2003) J. Biol. Chem. 278, 32771–32777 [DOI] [PubMed] [Google Scholar]
- 4.Radaev S., Dastidar P., Patel M., Woodard R. W., Gatti D. L. (2000) J. Biol. Chem. 275, 9476–9484 [DOI] [PubMed] [Google Scholar]
- 5.Wu J., Woodard R. W. (2003) J. Biol. Chem. 278, 18117–18123 [DOI] [PubMed] [Google Scholar]
- 6.Belunis C. J., Raetz C. R. (1992) J. Biol. Chem. 267, 9988–9997 [PubMed] [Google Scholar]
- 7.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noah C., Brabetz W., Gronow S., Brade H. (2001) J. Endotoxin Res. 7, 25–33 [PubMed] [Google Scholar]
- 9.Brabetz W., Schirmer C. E., Brade H. (2000) J. Bacteriol. 182, 4654–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode C. E., Brabetz W., Brade H. (1998) Eur. J. Biochem. 254, 404–412 [DOI] [PubMed] [Google Scholar]
- 11.Heine H., Müller-Loennies S., Brade L., Lindner B., Brade H. (2003) Eur. J. Biochem. 270, 440–450 [DOI] [PubMed] [Google Scholar]
- 12.Brabetz W., Lindner B., Brade H. (2000) Eur. J. Biochem. 267, 5458–5465 [DOI] [PubMed] [Google Scholar]
- 13.Rund S., Lindner B., Brade H., Holst O. (2000) Eur. J. Biochem. 267, 5717–5726 [DOI] [PubMed] [Google Scholar]
- 14.Brabetz W., Müller-Loennies S., Brade H. (2000) J. Biol. Chem. 275, 34954–34962 [DOI] [PubMed] [Google Scholar]
- 15.Isobe T., White K. A., Allen A. G., Peacock M., Raetz C. R., Maskell D. J. (1999) J. Bacteriol. 181, 2648–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helander I. M., Lindner B., Brade H., Altmann K., Lindberg A. A., Rietschel E. T., Zähringer U. (1988) Eur. J. Biochem. 177, 483–492 [DOI] [PubMed] [Google Scholar]
- 17.Le Dur A., Caroff M., Chaby R., Szabó L. (1978) Eur. J. Biochem. 84, 579–589 [DOI] [PubMed] [Google Scholar]
- 18.Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. (1992) Syst. Appl. Microbiol. 15, 352–356 [DOI] [PubMed] [Google Scholar]
- 19.Huber R., Wilharm T., Huber D., Trincone A., Burggraf S., König H., Rachel R., Rockinger I., Fricke H., Stetter K. O. (1992) Syst. Appl. Microbiol. 15, 340–351 [Google Scholar]
- 20.Plötz B. M., Lindner B., Stetter K. O., Holst O. (2000) J. Biol. Chem. 275, 11222–11228 [DOI] [PubMed] [Google Scholar]
- 21.Duewel H. S., Sheflyan G. Y., Woodard R. W. (1999) Biochem. Biophys. Res. Commun. 263, 346–351 [DOI] [PubMed] [Google Scholar]
- 22.Duewel H. S., Radaev S., Wang J., Woodard R. W., Gatti D. L. (2001) J. Biol. Chem. 276, 8393–8402 [DOI] [PubMed] [Google Scholar]
- 23.Whittington D. A., Rusche K. M., Shin H., Fierke C. A., Christianson D. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8146–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deckert G., Warren P. V., Gaasterland T., Young W. G., Lenox A. L., Graham D. E., Overbeek R., Snead M. A., Keller M., Aujay M., Huber R., Feldman R. A., Short J. M., Olsen G. J., Swanson R. V. (1998) Nature 392, 353–358 [DOI] [PubMed] [Google Scholar]
- 25.Meredith T. C., Mamat U., Kaczynski Z., Lindner B., Holst O., Woodard R. W. (2007) J. Biol. Chem. 282, 7790–7798 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. (2006) ACS Chem. Biol. 1, 33–42 [DOI] [PubMed] [Google Scholar]
- 29.Mamat U., Baumann M., Schmidt G., Brade H. (1993) Mol. Microbiol. 10, 935–941 [DOI] [PubMed] [Google Scholar]
- 30.Gronow S., Brabetz W., Brade H. (2000) Eur. J. Biochem. 267, 6602–6611 [DOI] [PubMed] [Google Scholar]
- 31.Kopetzki E., Schumacher G., Buckel P. (1989) Mol. Gen. Genet. 216, 149–155 [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 33.Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray P. H., Benedict C. D. (1982) Methods Enzymol. 83, 535–540 [DOI] [PubMed] [Google Scholar]
- 35.Fukase K., Oikawa M., Suda Y., Liu W. C., Fukase Y., Shintaku T., Sekljic H., Yoshizaki H., Kusumoto S. (1999) J. Endotoxin Res. 5, 46–51 [Google Scholar]
- 36.Imoto M., Yoshimura H., Yamamoto M., Shimamoto T., Kusumoto S., Shiba T. (1987) Bull. Chem. Soc. Jpn. 60, 2197–2204 [Google Scholar]
- 37.Kataoka M., Hashimoto M., Suda Y., Kusumoto S., Fukase K. (2006) Heterocycles 69, 395–415 [Google Scholar]
- 38.Kusumoto S., Fukase K. (2006) Chem. Rec. 6, 333–343 [DOI] [PubMed] [Google Scholar]
- 39.Liu W. C., Oikawa M., Fukase K., Suda Y., Kusumoto S. (1999) Bull. Chem. Soc. Jpn. 72, 1377–1385 [Google Scholar]
- 40.Hitchcock P. J., Brown T. M. (1983) J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galanos C., Lüderitz O., Westphal O. (1969) Eur. J. Biochem. 9, 245–249 [DOI] [PubMed] [Google Scholar]
- 42.Mamat U., Meredith T. C., Aggarwal P., Kühl A., Kirchhoff P., Lindner B., Hanuszkiewicz A., Sun J., Holst O., Woodard R. W. (2008) Mol. Microbiol. 67, 633–648 [DOI] [PubMed] [Google Scholar]
- 43.Kamerling J. P., Gerwig G. J. (2007) in Comprehensive Glycoscience: From Chemistry to Systems Biology, Vol. 2 ( Kamerling J. P., Boons G. H., Lee Y. C., Suzuki A., Taniguchi N., Voragen A. G. J., eds) Elsevier Ltd., Oxford [Google Scholar]
- 44.Brade H., Galanos C., Lüderitz O. (1983) Eur. J. Biochem. 131, 195–200 [DOI] [PubMed] [Google Scholar]
- 45.Kondakova A., Lindner B. (2005) Eur. J. Mass Spectrom. 11, 535–546 [DOI] [PubMed] [Google Scholar]
- 46.Myszka D. G. (1999) J. Mol. Recognit. 12, 279–284 [DOI] [PubMed] [Google Scholar]
- 47.Brozek K. A., Raetz C. R. (1990) J. Biol. Chem. 265, 15410–15417 [PubMed] [Google Scholar]
- 48.Six D. A., Carty S. M., Guan Z., Raetz C. R. (2008) Biochemistry 47, 8623–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C. H., Murray B. W., Ollmann I. R., Wong C. H. (1997) Biochemistry 36, 780–785 [DOI] [PubMed] [Google Scholar]
- 50.Karlsson R. (2004) J. Mol. Recognit. 17, 151–161 [DOI] [PubMed] [Google Scholar]
- 51.Raetz C. R., Guan Z., Ingram B. O., Six D. A., Song F., Wang X., Zhao J. (2009) J. Lipid Res. 50, S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stead C., Tran A., Ferguson D., Jr., McGrath S., Cotter R., Trent S. (2005) J. Bacteriol. 187, 3374–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClerren A. L., Zhou P., Guan Z., Raetz C. R., Rudolph J. (2005) Biochemistry 44, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 54.Zähringer U., Lindner B., Rietschel E. T. (1994) Adv. Carbohydr. Chem. Biochem. 50, 211–276 [PubMed] [Google Scholar]
- 55.Belunis C. J., Clementz T., Carty S. M., Raetz C. R. (1995) J. Biol. Chem. 270, 27646–27652 [DOI] [PubMed] [Google Scholar]
- 56.Holst O., Broer W., Thomas-Oates J. E., Mamat U., Brade H. (1993) Eur. J. Biochem. 214, 703–710 [DOI] [PubMed] [Google Scholar]
- 57.Holst O., Bock K., Brade L., Brade H. (1995) Eur. J. Biochem. 229, 194–200 [PubMed] [Google Scholar]
- 58.Sterner R., Liebl W. (2001) Crit. Rev. Biochem. Mol. Biol. 36, 39–106 [DOI] [PubMed] [Google Scholar]
- 59.Vieille C., Zeikus G. J. (2001) Microbiol. Mol. Biol. Rev. 65, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clementz T., Bednarski J. J., Raetz C. R. (1996) J. Biol. Chem. 271, 12095–12102 [DOI] [PubMed] [Google Scholar]
- 61.Doerrler W. T., Raetz C. R. (2002) J. Biol. Chem. 277, 36697–36705 [DOI] [PubMed] [Google Scholar]
- 62.Zhou Z., White K. A., Polissi A., Georgopoulos C., Raetz C. R. (1998) J. Biol. Chem. 273, 12466–12475 [DOI] [PubMed] [Google Scholar]
- 63.Adams M. W. (1993) Annu. Rev. Microbiol. 47, 627–658 [DOI] [PubMed] [Google Scholar]
- 64.Cacciapuoti G., Fusco S., Caiazzo N., Zappia V., Porcelli M. (1999) Protein Expr. Purif. 16, 125–135 [DOI] [PubMed] [Google Scholar]
- 65.Gentile F., Amodeo P., Febbraio F., Picaro F., Motta A., Formisano S., Nucci R. (2002) J. Biol. Chem. 277, 44050–44060 [DOI] [PubMed] [Google Scholar]
- 66.Seydel U., Schromm A. B., Brade L., Gronow S., Andrä J., Müller M., Koch M. H., Fukase K., Kataoka M., Hashimoto M., Kusumoto S., Brandenburg K. (2005) FEBS J. 272, 327–340 [DOI] [PubMed] [Google Scholar]
- 67.Belunis C. J., Mdluli K. E., Raetz C. R., Nano F. E. (1992) J. Biol. Chem. 267, 18702–18707 [PubMed] [Google Scholar]
- 68.Bhat U. R., Forsberg L. S., Carlson R. W. (1994) J. Biol. Chem. 269, 14402–14410 [PubMed] [Google Scholar]
- 69.Moran A. P., Lindner B., Walsh E. J. (1997) J. Bacteriol. 179, 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinogradov E., Perry M. B., Conlan J. W. (2002) Eur. J. Biochem. 269, 6112–6118 [DOI] [PubMed] [Google Scholar]
- 71.Karbarz M. J., Six D. A., Raetz C. R. (2009) J. Biol. Chem. 284, 414–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price N. P., Kelly T. M., Raetz C. R., Carlson R. W. (1994) J. Bacteriol. 176, 4646–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price N. P., Jeyaretnam B., Carlson R. W., Kadrmas J. L., Raetz C. R., Brozek K. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7352–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran A. X., Karbarz M. J., Wang X., Raetz C. R., McGrath S. C., Cotter R. J., Trent M. S. (2004) J. Biol. Chem. 279, 55780–55791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X., Karbarz M. J., McGrath S. C., Cotter R. J., Raetz C. R. (2004) J. Biol. Chem. 279, 49470–49478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blume A., Angulo J., Biet T., Peters H., Benie A. J., Palcic M., Peters T. (2006) J. Biol. Chem. 281, 32728–32740 [DOI] [PubMed] [Google Scholar]
- 77.Brade H., Zähringer U., Rietschel E. T., Christian R., Schulz G., Unger F. M. (1984) Carbohydr. Res. 134, 157–166 [Google Scholar]
- 78.Kohlbrenner W. E., Fesik S. W. (1985) J. Biol. Chem. 260, 14695–14700 [PubMed] [Google Scholar]
- 79.Tabor S., Richardson C. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.