Abstract
Transcription factors Oct4 and Sox2 are key players in maintaining the pluripotent state of embryonic stem cells (ESCs). Small changes in their levels disrupt normal expression of their target genes. However, it remains elusive how protein levels of Oct4 and Sox2 and expression of their target genes are precisely controlled in ESCs. Here we identify PARP1, a DNA-binding protein with an NAD+-dependent enzymatic activity, as a cofactor of Oct4 and Sox2 to regulate expression of their target gene FGF4. We demonstrate for the first time that PARP1 binds the FGF4 enhancer to positively regulate FGF4 expression. Our data show that PARP1 interacts with and poly(ADP-ribosyl)ates Sox2 directly, which may be a step required for dissociation and degradation of inhibitory Sox2 proteins from the FGF4 enhancer. When PARP1 activity is inhibited or absent, poly(ADP-ribosyl)ation of Sox2 decreases and association of Sox2 with FGF4 enhancers increases, accompanied by an elevated level of Sox2 proteins and reduced expression of FGF4. Significantly, specific knockdown of Sox2 expression by RNA interference can considerably abrogate the inhibitory effect of the poly(ADP- ribose) polymerase inhibitor on FGF4 expression. Interestingly, PARP1 deficiency does not affect undifferentiated ESCs but compromises cell survival and/or growth when ESCs are induced into differentiation. Addition of FGF4 can partially rescue the phenotypes caused by PARP1 deficiency during ESC differentiation. Taken together, this study uncovers new mechanisms through which Sox2 protein levels and FGF4 expression are dynamically regulated during ESC differentiation and adds a new member to the family of proteins regulating the properties of ESCs.
Embryonic stem cells (ESCs),2 derived from the inner cell mass of the blastocyst-stage embryo, are pluripotent. They can differentiate into all cell types of an organism and self-renew indefinitely in vitro (1, 2). Intensive research over past decades has demonstrated that transcription factors Oct4 and Sox2 are key players in maintaining the pluripotent state of ESCs (3, 4). Recently, their central position in stem cell biology has been further highlighted by their critical role in the establishment of induced pluripotent stem cells (5–7). It is also clear that Oct4 and Sox2 cooperatively regulate their own expression as well as that of different sets of target genes, such as FGF4 (8), Nanog (9), and UTF1 (10). Remarkably, small changes in the levels of Oct4 and Sox2 disrupt normal expression of their target genes and alter cell fate determination in ESCs (11–15). Therefore, levels of Oct4 and Sox2 as well as of their target genes must be tightly controlled. However, to date, our knowledge of the molecular mechanisms controlling their expression is limited. Obviously, answers to these questions are not only fundamental to ESC maintenance and differentiation but also have important implications for efficient generation of induced pluripotent stem cells.
As is the case for most other transcription factors, Oct4 and Sox2 are regulated at both transcriptional and post-transcriptional levels. However, past emphasis has been mainly placed on their transcriptional regulation, whereas their post-transcriptional control has been little touched upon. Previous studies in our laboratory demonstrated that Oct4 could be ubiquitinated and sumoylated and that ubiquitination and sumoylation jointly maintain the protein level of Oct4 in a normal range in ESCs (16, 17). Recently, phosphorylation of Oct4 was also reported (18). As for Sox2, one study indicated that Sox2 sumoylation negatively regulates its transcriptional activity, although its role in control of the Sox2 protein level is not known (19). In addition to ubiquitination, sumoylation, and phosphorylation, a wide variety of post-translational modifications, such as glycosylation and poly(ADP-ribosyl)ation, exists, and enzymes mediating these modifications make a great contribution to modulation of transcription factors. One such enzyme is poly(ADP-ribose) polymerase-1 (PARP1), a 114-kDa, abundant nuclear DNA-binding protein that catalyzes the covalent attachment of poly(ADP-ribose) (PAR) from NAD+ to itself and other nuclear protein acceptors such as topoisomerase I and II, NF-κB, p53, and histones (20–25). In contrast to Oct4 and Sox2, which are specifically expressed in pluripotent stem cells, PARP1 is a constitutively expressed protein (26, 27). Although the best studied function of PARP1 is in the maintenance of genomic integrity (28), studies over the past decade have demonstrated its role in the regulation of gene expression (29–32). However, the underlying mechanism responsible for its functions in transcription regulation is not well defined, and its role in ESC proliferation and differentiation has not been explored.
Recent studies indicate that additional factors are involved in the regulation of target genes of Oct4 and Sox2 and that these factors function in a gene-specific manner. For example, Nakatake et al. (33) reported that Klf4 cooperates with Oct4 and Sox2 to activate Lefty1 expression. However, its presence is not required for expression of Oct4 and UTF1 under the same conditions (33). Moreover, Esrrb was found to interact with Oct4, positively regulating Nanog expression (34). Recruitment of cofactors provides an important mechanism by which transcriptional factors may regulate expression of their target genes precisely and differentially in a tissue-specific and developmentally stage-specific manner. In the search for more such regulatory factors mediating Oct4 and Sox2 functions, the FGF4 distal enhancer provides a good model. FGF4 is essential for survival of the postimplantation embryo (35) and plays important roles at multiple stages of development (36). It is expressed in the ICM of the blastocyst in vivo and in ESCs as well as embryonal carcinoma (EC) cells in vitro. However, its transcription is silent in the adult and down-regulated when ESCs are induced to differentiate (37, 38). Uniquely, FGF4 transcription is regulated by a powerful distal enhancer located in the 3′-untranslated region, containing the POU and the HMG cassettes (8). Previous studies have demonstrated that Oct4 and Sox2 bind both in vitro and in vivo to the POU motif and the HMG motif, respectively, to control the expression of FGF4 (39, 40). However, it is unclear whether other factors, together with Oct4 and Sox2, are implicated in FGF4 expression in undifferentiated ESCs and during their differentiation.
In this study, we identify PARP1 as a novel cofactor of Oct4 and Sox2 and as a regulator of FGF4 expression. It binds to the FGF4 enhancer together with Oct4 and Sox2 and is required for appropriate expression of FGF4 during ESC differentiation. Our data show that PARP1 interacts with Sox2 directly and modifies Sox2 by poly(ADP-ribosyl)ation, which may be a step required for dissociation of excessive Sox2 from the FGF4 enhancer. When PARP1 activity is absent or inhibited, Sox2 poly(ADP-ribosyl)ation is reduced, which is associated with an elevated Sox2 protein level and increased binding of Sox2 to the FGF4 enhancer. Moreover, specific knockdown of Sox2 expression by RNA interference can rescue reduction in FGF4 expression caused by inhibition of PARP activity. In addition, we find that FGF4 can partially rescue PAPR1 deficiency-associated phenotypes in differentiating ESCs. Therefore, the study establishes links among PARP activity, Sox2 poly(ADP-ribosyl)ation, and FGF4 expression, uncovering new mechanisms through which the Sox2 protein level and FGF4 transcription are dynamically regulated.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Rabbit polyclonal antibodies against GST, Oct4, and Sox2 were raised and affinity-purified in our laboratory using bacterially expressed GST, GST-Oct4N (16), and GST-Sox2C (114-amino acid residues at the C terminus of the Sox2 protein) fusion proteins, respectively. Antibodies against the FLAG (Sigma), tubulin (Sigma), PARP1 (Santa Cruz Biotechnology), and PAR (Alexis Biochemical) were used for immunoblotting. Anti-human FGF4-neutralizing antibody (R & D Systems) was reconstituted in 1× phosphate-buffered saline and used for neutralization of FGF4 activity secreted by ESCs. 3AB, FGF4, and NAD+ were purchased from Sigma, and PJ34 was obtained from Calbiochem.
Plasmids and Primers
The cDNA sequences corresponding to the full-length proteins of PARP1, Oct4, and Sox2 were amplified by reverse transcriptase-PCR using the RNA of mouse ESCs as templates, and they were cloned into pET-30a(+) (Novagen) and pGEX-4T-1 (Amersham Biosciences) vectors for expression in bacteria or pPyCAGIP vector (a kind gift of I. Chambers and A. Smith) for expression in mammalian cells. Probes and primers used in this study are listed in supplemental Table S1.
RNA Interference
To make small interfering RNA vectors for Sox2 (Sox2 RNAi), two independent 19-bp sequences (5′-GGTTGATATCGTTGGTAAT-3′ and 5′-CCCTGCAGTACAACTCCAT-3′) within the coding region of the murine Sox2 gene were selected and cloned into the pTER+ vectors (41). Small interfering RNA vector for EGFP (EGFP RNAi) was made by selecting a 19-bp sequence in the coding region of the EGFP gene (5′-GGCTACGTCCAGGAGCGCA-3′).
Cell Culture and Transient Transfection
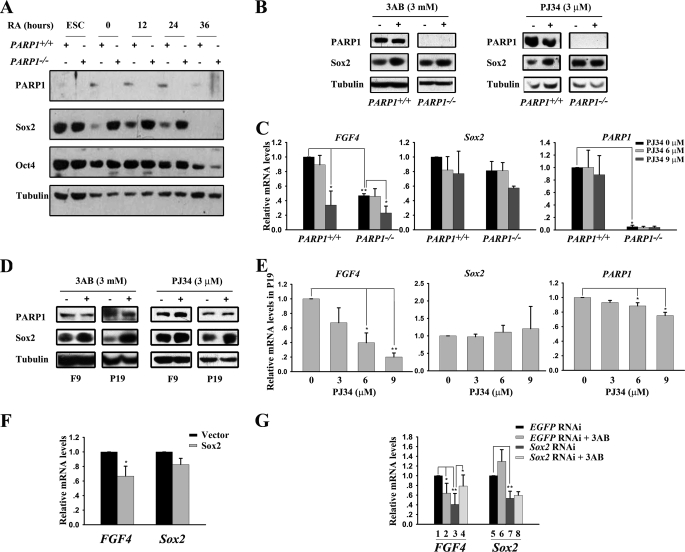
PARP1+/+ and PARP1−/− ESCs were grown in media consisting of Dulbecco's modified Eagle's media (Invitrogen) supplemented with 15% (v/v) fetal bovine serum (Hyclone), 2 mm l-glutamine (Invitrogen), 0.1 mm nonessential amino acids (Invitrogen), 1 mm sodium pyruvate (Invitrogen), 1000 units/ml leukemia inhibitory factor (LIF, Chemicon), 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 0.1 mm β-mercaptoethanol (Invitrogen). ESCs were maintained on a feeder layer. To induce differentiation by retinoic acid (RA, Fisher), ESCs were cultured on gelatin-coated dishes without a feeder layer at a density of 5 × 105 cells per 35-mm dish for 24 h. Subsequently, RA at a concentration of 1 μm was added to the culture medium containing LIF. For spontaneous differentiation at low cell density, ESCs were cultured in a gelatin-coated 6-well plate at a density of 3000 per well for 7 days.
P19 and F9 EC cells were maintained in the Dulbecco's modified Eagle's media/F-12 medium supplemented with 10% fetal bovine serum, 1.5% NaHCO3 (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin. Both ESCs and EC cells were transiently transfected with LipofectamineTM 2000 (Invitrogen) according to manufacturer's instructions. HEK 293 cells were cultured under standard conditions and transfected using the calcium phosphate method.
For Sox2 RNAi experiments, P19 cells were transfected with EGFP RNAi or Sox2 RNAi plasmids. Twelve hours after transfection, the cells were incubated in fresh medium and selected by Zeocin at 100 μg/ml for 12 h. Twenty four hours later, the transfected cells were trypsinized and replated in 24-well plates. After an additional 24 h, 3AB at a concentration of 6 mm was added. The cells were collected 48 h later.
Nuclear Extract Preparation and Protein Purification by Affinity Chromatography
Preparation of nuclear extract (NE), oligonucleotide probes, and protein purification by affinity chromatography were accomplished as described previously (42) with minor changes (details are provided in the supplementary material).
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays using formaldehyde cross-linking and specific antibodies were performed as described previously (17).
Luciferase Reporter Assays
ESCs were cultured in a gelatin-coated 24-well plate at a density of 1 × 105 per well. After 24 h, cells were cotransfected with 200 ng of the plasmid DNA construct (FGF4/pTAL) containing the enhancer fragment of the mouse FGF4 gene in the pTAL reporter gene or empty vector pTAL and 20 ng of pRL-TK (Promega) as an internal control to normalize the transfection efficiency. Forty eight hours after transfection, samples were collected and analyzed with the dual luciferase reporter assay system (Promega).
Coimmunoprecipitation (CoIP)
The NE of cells was prepared in CoIP buffer (10 mm Hepes, pH 7.6, 250 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA, 1 mm NaF, and 1 mm phenylmethylsulfonyl fluoride) and incubated with a specific antibody overnight at 4 °C, followed by incubation with protein A-Sepharose beads for 2 h. The samples were analyzed using Western blotting.
GST Pulldown
GST and His fusion proteins were expressed and purified according to the manufacturer's instructions from Amersham Biosciences and Novagen, respectively. GST pulldown experiments were performed as described previously (16).
Poly(ADP-ribosyl)ation Assay in Vivo and in Vitro
For the assay in vivo, cells were harvested and boiled in the lysis buffer (100 mm Tris-HCl, pH 7.5, 1% SDS) for 10 min to disassociate protein-protein interaction. The lysate was then diluted by 10-fold in the CoIP buffer and sonicated briefly, followed by centrifugation at 14,000 × g for 10 min. The supernatant was immunoprecipitated with specific antibodies overnight at 4 °C, followed by incubation with protein A-Sepharose beads for 2 h. In vitro poly(ADP-ribosyl)ation assays were performed as described previously (24, 27). Briefly, purified GST fusion proteins (1 μg) were incubated in the poly(ADP-ribosyl)ation reaction buffer (100 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol) with purified his-PARP1 fusion proteins (100 ng), 0.5 mm NAD+, and 1 μg of sonicated salmon sperm DNA for 40 min at 37 °C. Poly(ADP-ribosyl)ated proteins were detected by Western blot analysis using anti-PAR antibody.
RNA Extraction and Quantitative Real Time PCR (qPCR)
RNAs were extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNAs were synthesized with the ReverTra Ace® reverse transcriptase (Toyobo, Osaka, Japan). qPCR was performed using SYBR Green Master Mix on an ABI PRISM 7900 machine as described previously (17). The Ct value of each gene was normalized against that of the housekeeping gene GAPDH.
Statistical Analysis
Data were analyzed using Student's t test. Data are shown as the mean ± S.D. of at least three experiments. We considered the difference between comparisons to be significant when p < 0.05 for all the statistical analysis. All experiments were performed at least three times.
RESULTS
PARP1 Binds Specifically to the FGF4 Enhancer Both in Vitro and in Vivo
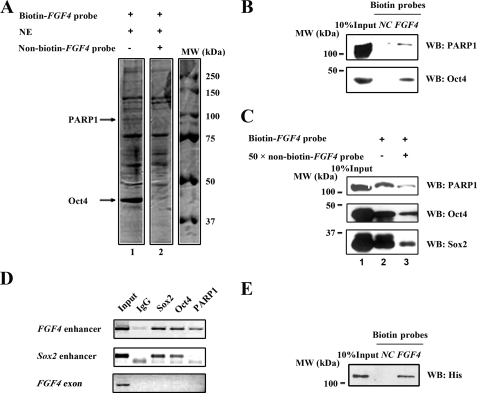
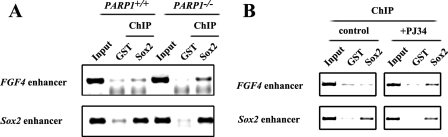
To identify proteins associated with control of FGF4 expression, in addition to its known regulators, Oct4 and Sox2, we conducted affinity chromatography with the NE of F9 mouse EC cells, using synthetic biotinylated oligonucleotides containing the 3′-untranslated region of FGF4 (biotin-FGF4 probe). F9 cells were chosen because of their relatively easy and low cost culture as compared with ESCs. The isolated proteins were visualized by Coomassie blue staining (Fig. 1A). Multiple protein bands were seen in the NE column with use of the biotin-FGF4 probe (Fig. 1A, lane 1). To exclude nonspecific binding proteins, only the protein bands, which vanished when a high concentration of nonbiotinylated FGF4 probe (non-biotin-FGF4 probe) was present (compare lanes 1 and 2), were excised and analyzed by mass spectrometry. Among these proteins, the peptides from the 45-kDa protein band matched with Oct4, supporting the validity of the approach used. Interestingly, the peptides derived from the 100-kDa protein band corresponded to PARP1. The implication of PARP1 in transcriptional regulation prompted us to investigate further. We repeated the DNA-protein binding assay described in Fig. 1A and examined the probe-binding proteins by Western blot analysis. Using the probe (2×mIUR-NF-κB) known not to bind PARP (43) as a negative control (NC probe), PARP1 was found to bind the FGF4 probe, in addition to Oct4 and Sox2 (Fig. 1, B and C). Moreover, the binding intensity was dramatically reduced for all three proteins when the non-biotin-FGF4 probe was included in the reaction mixture (Fig. 1C, lane 3), indicating the specificity of their binding to the FGF4 enhancer. Next, to examine whether PARP1 was associated with the FGF4 enhancer under physiological conditions, ChIP assays were conducted. We found that PARP1 was specifically recruited to the FGF4 enhancer but not to the Sox2 enhancer or the FGF4 exon, whereas, as reported previously (12, 39, 40), Oct4 and Sox2 bound to the enhancers of both FGF4 and Sox2 (Fig. 1D). In addition, we found that purified His-PARP1 was capable of binding the biotin-FGF4 probe directly in vitro (Fig. 1E). Thus, our data demonstrate for the first time that PARP1 is directly associated with the FGF4 enhancer, together with Oct4 and Sox2, both in vitro and in vivo.
FIGURE 1.
PARP1 binds specifically to the FGF4 enhancer in vitro and in vivo. A, purification of FGF4 enhancer-binding proteins by affinity chromatography using biotin-labeled oligonucleotide of the FGF4 enhancer containing Oct4 and Sox2 binding sequence (biotin-FGF4 probe). Purified proteins were subjected to SDS-PAGE and visualized with Coomassie Blue staining. B, Western blot (WB) analysis of affinity-purified DNA-binding proteins. NC is a negative control probe. C, PARP1 bound specifically to the FGF4 enhancer. F9 nuclear proteins were subjected to SDS-PAGE with the biotin-FGF4 probe in the presence or absence of 50-fold excess unlabeled FGF4 probe (non-biotin-FGF4 probe). D, ChIP assay was performed in F9 EC cells with antibodies indicated. An antibody against IgG was used as a negative control. E, PARP1 bound to the FGF4 enhancer directly. Bacterially expressed fusion protein of his-PARP1 was purified and subjected to SDS-PAGE with the indicated biotin-labeled probes.
PARP1 Is Required for FGF4 Expression and Recombinant FGF4 Can Functionally Rescue Phenotypes of PARP1-deficient Cells
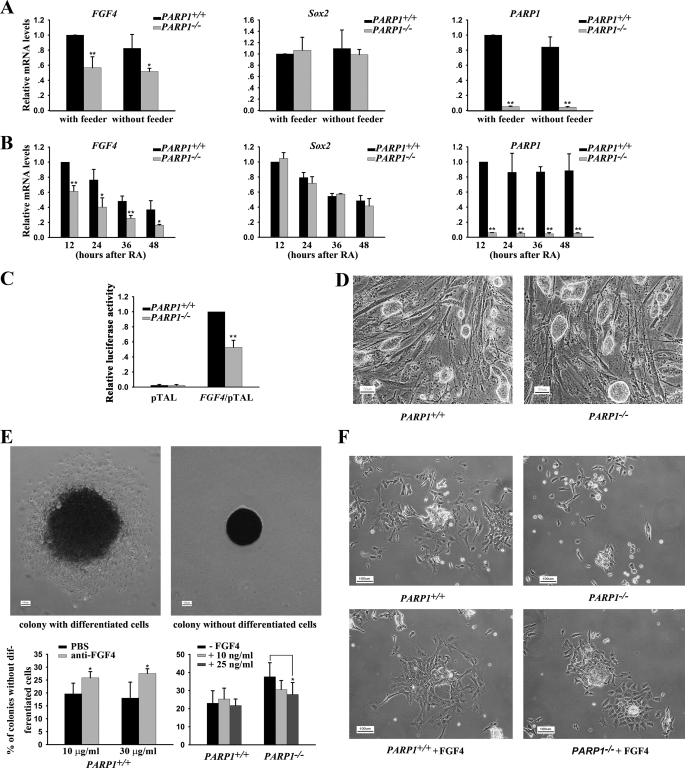
To learn whether PARP1 recruited to the FGF4 enhancer has any function, we compared mRNA levels of FGF4 in PARP1+/+ and PARP1−/− ESCs cultured with or without feeder cells for 24 h. As shown in Fig. 2A, the level of FGF4 transcripts was significantly lower in PARP1 knock-out cells than in wild type cells under both conditions. Furthermore, the mRNA level of FGF4 decreased in both wild type and PARP1-deficient ESCs during ESC differentiation induced by RA (Fig. 2B), consistent with a previous report (38). Significantly, FGF4 expression was lower in PARP1−/− cells than in PARP1+/+ cells at all time points tested (Fig. 2B). In contrast, there was no detectable difference in the mRNA level of Sox2 between wild type and PARP1 knock-out cells (Fig. 2, A and B). The results indicate that PARP1 may play a positive role in maintaining FGF4 expression in ESCs and during their differentiation. To verify that the effect of PARP1 on FGF4 expression is mediated through the FGF4 enhancer, we transfected the FGF4/pTAL luciferase reporter containing the FGF4 enhancer sequence into PARP1+/+ and PARP1−/− ESCs cultured without feeder cells, and we found that the luciferase activity was significantly lower in PARP1 knock-out cells than in wild type cells (Fig. 2C). This result indicates that PARP1 may regulate FGF4 expression, at least partially, through the FGF4 enhancer.
FIGURE 2.
PARP1 is implicated in FGF4 expression. A and B, qPCR analysis of gene expression in undifferentiated and differentiating PARP1+/+ and PARP1−/− ESCs cultured without a feeder layer and treated with 1 μm RA. *, p < 0.05; **, p < 0.01, n = 3. C, activity of FGF4/pTAL luciferase activity in PARP1+/+ and PARP1−/− ESCs cultured without a feeder layer was compared. **, p < 0.01, n = 3. D, morphology of PARP1+/+ and PARP1−/− ESCs. PARP1+/+ and PARP1−/− ESCs were cultured for several passages on feeder layers. The scale bar is 50 μm. E, ESCs were cultured at a low cell density for 7 days without a feeder layer on gelatin-coated surfaces in the presence or absence of an anti-human FGF4-neutralizing antibody or FGF4. Culture media of cells were refed with fresh media every day. Prior to photography, the cells were stained for alkaline phosphatase expression, and colonies were subclassed into colonies with or without differentiated cells on their periphery. The scale bar is 100 μm. *, p < 0.05, n = 3. F, morphology of differentiated cells derived from PARP1+/+ and PARP1−/− ESCs. ESCs were cultured on gelatin-coated surfaces in the medium containing LIF and induced to differentiate with RA (1 μm) for 3 days in the presence or absence of FGF4. The scale bar is 100 μm. The representative results of three independent experiments are shown.
We further reasoned that the phenotypes of PARP1-deficient cells should resemble those of FGF4−/− ESCs to a certain extent if PARP1 is an important regulator of FGF4 expression. It is known that undifferentiated FGF4−/− ESCs proliferate normally in vitro. However, the growth and/or survival of RA-induced differentiated cells is severely compromised in the absence of FGF4 (44). Similarly, we found that PARP1−/− ESCs grew normally as wild type ESCs did when they were cultured under undifferentiated conditions (Fig. 2D). We next induced differentiation of PARP1−/− ESCs by two culture conditions as follows: spontaneous differentiation at low cell density and RA-induced differentiation, respectively. In both cases, the cells were cultured in the presence of LIF but in the absence of a feeder layer. At low cell density, the majority of colonies formed by wild type ESCs contained differentiated alkaline phosphatase-negative cells surrounding central undifferentiated alkaline phosphatase-positive cells (Fig. 2E, upper left panel). A minority of colonies was small and did not have differentiated cells peripherally (Fig. 2E, upper right panel). Addition of the anti-human FGF4-neutralizing antibody significantly increased the number of small colonies in wild type ESCs (Fig. 2E, bottom left panel). In contrast, PARP1−/− ESCs produced more small colonies without differentiated cells than wild type ESCs did. Importantly, addition of recombinant FGF4 significantly decreased the number of small colonies in PARP1−/− ESCs in a dosage-dependent manner (Fig. 2E, bottom right panel). Next, more dead floating cells, fewer surviving cells, and slower cell growth rates were observed in differentiating PARP1−/− cells than in differentiating PARP1+/+ cells, when RA was used to induce ESCs to differentiate. However, the differences in cell growth and/or survival between wild type and PARP1−/− ESCs after RA treatment were diminished when FGF4 was included in the culture media (Fig. 2F and data not shown). Intriguingly, the above described phenotypes seen in both low density and RA-treated culture of PARP1−/− ESCs were reported, to a great extent, for FGF4−/− ESCs (44). Therefore, it is reasonable to propose that a low level of FGF4 expression is at least partially responsible for the phenotypes observed in PARP1 deficiency and that PARP1 plays a positive role in FGF4 expression during ESC differentiation.
PARP1 Physically Interacts with Sox2
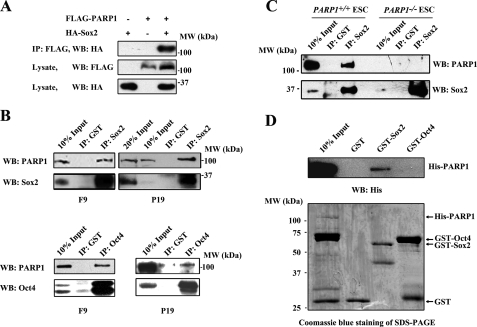
The next question involves the manner in which PARP1 positively participates in the regulation of FGF4 expression. Because it has been well documented that Oct4 and Sox2 are key factors binding to the FGF4 enhancer and modulating its expression (39, 45), we suspected that PARP1 might associate with Oct4, Sox2, or both. To test this hypothesis, CoIP experiments were conducted in HEK 293 cells expressing FLAG-PARP1, together with HA-Sox2 or HA-Oct4. Western blot analysis indicated that FLAG-PARP1 associated with HA-Sox2 (Fig. 3A). However, we were not able to detect an interaction between HA-Oct4 and FLAG-PARP1 (data not shown). Surprisingly, we detected the interaction not only between endogenous PARP1 and Sox2, but also between PARP1 and Oct4, when CoIP experiments were performed in F9 or P19 EC cells (Fig. 3B). We also conducted such experiments in wild type ESCs and found that Sox2 interacted with PARP1 as it did in EC cells. As expected, such interaction was not observed in PARP1 knock-out ESCs, arguing for a specific association between endogenous Sox2 and PARP1 in vivo (Fig. 3C). The reason for our inability to detect an association between PARP1 and Oct4 in HEK 293 cells could be explained by the possibility that PARP1 may interact with Oct4 indirectly in F9/P19 EC cells and that the factor(s) mediating PARP1 and Oct4 association in EC cells are not present in HEK 293 cells. We therefore further examined whether PARP1 directly interacts with Sox2 or Oct4 using an in vitro GST pulldown assay. The results show that His-PARP1 could be pulled down by GST-Sox2, but not by GST-Oct4 or GST alone, indicating that Sox2, but not Oct4, directly interacts with PARP1 (Fig. 3D). Thus, it appears that PARP1 controls FGF4 expression through its direct association with Sox2 and post-translational modification of Sox2.
FIGURE 3.
Association of PARP1 with Sox2 or Oct4. A, CoIP of FLAG-PARP1 and HA-Sox2 in HEK 293 cells, which were transiently cotransfected with FLAG-PARP1 and HA-Sox2 or vector. B, association of endogenous PARP1 with Sox2 or Oct4 in F9 and P19 cells. The NE of F9 or P19 cells was subjected to CoIP with anti-Sox2 antibody or anti-Oct4 antibody. Anti-GST antibody was used as a negative control. C, association of endogenous PARP1 with Sox2 in PARP1+/+ and PARP1−/− ESCs. D, Sox2 but not Oct4 interacts directly with PARP1 in vitro. A GST-pulldown assay was performed with purified GST-Sox2, GST-Oct4, and His-PARP1 fusion proteins. The representative results of three independent experiments are shown. IP, immunoprecipitation; WB, Western blot.
Poly(ADP-ribosyl)ation of Sox2 by PARP1
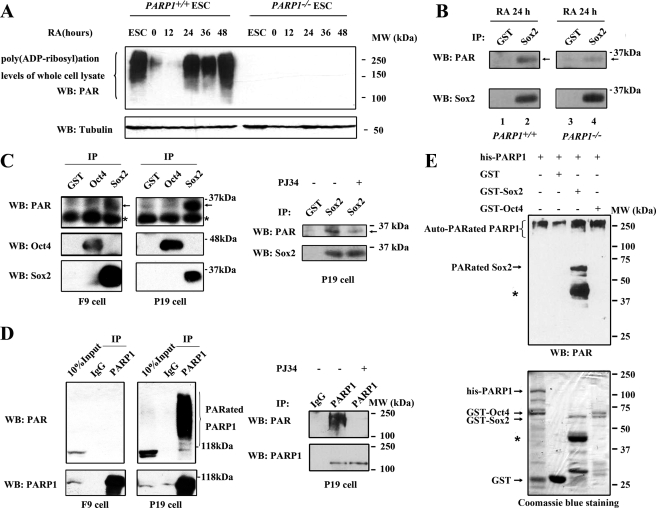
Before examining whether PARP1 could modify Sox2 post-translationally, we assessed the PARP enzymatic activity in ESCs and differentiating ESCs cultured without feeder cells for 24 h and further induced by RA treatment. Assessment was accomplished using Western blot analysis of the total PAR level. As shown in Fig. 4A, PARP enzymatic activity existed in PARP1+/+ cells but was not detectable in PARP1−/− cells. There was a drastic drop of the activity at the onset of ESC differentiation induced by withdrawal of feeder cells. However, the activity recovered after addition of RA and relatively high levels of the PARP activity were detected in PARP1+/+ cells treated with RA for 24–48 h (Fig. 4A). We termed the culture condition the “differentiation condition” (feeder-free and RA treatment for 24–48 h). Subsequently, we found that Sox2 was modified by PAR in differentiating wild type ESCs (Fig. 4B, lane 2). The modification was substantially weak in PARP1 knock-out cells (Fig. 4B, lane 4). The residual modification could be catalyzed by other members of the PARP family (46). As in differentiating ESCs, PAR-modified Sox2, but not PAR-modified Oct4, existed in F9 and P19 EC cells (Fig. 4C, left). Both cell lines are considered to be in a differentiated state as compared with ESCs, with P19 cells being further differentiated than F9 cells, and present a good in vitro model of an early differentiation stage of pluripotent embryonic cells (47–49). Remarkably, a stronger signal for PAR-modified Sox2 was detected in P19 cells (Fig. 4C, left) than in F9 cells, consistent with our finding that a higher PARP activity is present in P19 cells as compared with that in F9 cells (Fig. 4D, left). Furthermore, we tested whether the PARP inhibitors (3AB and PJ34) could block the Sox2 and PARP1 PAR modification detected in P19 EC cells. As expected, inhibition of PARP enzymatic activity by PJ34 (Fig. 4, C, right, and D, right) or 3AB (data not shown) decreased the poly(ADP-ribosyl)ation level of Sox2 and PARP1 evidently. Finally, an in vitro poly(ADP-ribosyl)ation assay with purified GST fusion proteins and His-PARP1 provided biochemical evidence that Sox2 is a direct substrate of PARP1 (Fig. 4E). Collectively, our studies clearly demonstrate that PARP1 can efficiently modify Sox2 both in vivo and in vitro.
FIGURE 4.
Sox2 is poly(ADP-ribosyl)ated in vivo and in vitro. A, poly(ADP-ribosyl)ation levels of PARP1+/+ and PARP1−/− cell extracts were evaluated using anti-PAR antibody. PARP1+/+ and PARP1−/− ESCs were cultured without a feeder layer for 24 h (time 0) and then induced to differentiate with 1 μm RA for different lengths of time. B, poly(ADP-ribosyl)ation status of endogenous Sox2 in PARP1+/+ and PARP1−/− differentiating cells. Immunoprecipitation (IP) experiments were performed with the indicated antibodies. The precipitated proteins were analyzed with anti-PAR antibody (top panel). The same blot was probed sequentially with anti-Sox2 antibody (bottom panel). Arrows indicate poly(ADP-ribosyl)ated Sox2. C, Sox2 but not Oct4 is poly(ADP-ribosyl)ated in F9 and P19 EC cells. The experiments were performed as in B. Arrows indicate poly(ADP-ribosyl)ated Sox2. For experiments using the PARP inhibitor, P19 cells were treated with 3 μm of PJ34 for 48 h to inhibit the PARP enzyme activity before harvesting. Asterisks indicate nonspecific bands. D, poly(ADP-ribosyl)ation levels of endogenous PARP1 in F9 and P19 EC cells. Lysates from F9 and P19 cells were immunoprecipitated with anti-PARP1 antibody. The precipitated proteins were analyzed with anti-PAR antibody (top panel). The same blot was probed sequentially with anti-PARP1 antibody (bottom panels). For experiments using the PARP inhibitor, P19 cells were treated with 3 μm of PJ34 for 48 h to inhibit the PARP enzyme activity before harvesting. PARated, poly(ADP-ribosyl)ated. E, in vitro poly(ADP-ribosyl)ation of Sox2 by PARP1. In vitro poly(ADP-ribosyl)ation reactions were performed using indicated sets of purified fusion proteins. Proteins were separated on 12% SDS-PAGE followed by Western blot (WB) analysis. One μg of the purified fusion proteins was resolved on 12% SDS-PAGE and stained with Coomassie Blue (bottom). Asterisks indicate the degraded GST-Sox2 protein occurred during purification. The representative results of three independent experiments are shown.
PARP1 Modulates Sox2 Protein Levels to Control FGF4 Expression
To investigate the functional consequences of Sox2 modification by PAR, we compared the protein levels of Oct4 and Sox2 in PARP1+/+ and PARP1−/− ESCs under both undifferentiated and differentiating conditions. Western blot analysis showed that the Sox2 protein level in differentiating PARP1−/− cells was markedly higher than that in differentiating PARP1+/+ cells at each time point after RA treatment. In contrast, a similar Oct4 protein level was found between these two cell types (Fig. 5A). However, there was no distinctive difference in the Sox2 protein level between the two types of undifferentiated ESCs. The obviously higher steady-state level of Sox2 proteins in differentiating PARP1−/− cells implies an important role for PARP1 in the regulation of the Sox2 protein level during differentiation. Next, we examined whether the PARP activity is involved in regulation of the Sox2 protein level. We found that inhibition of the PARP enzymatic activity by either 3AB or PJ34 elevated the Sox2 protein level in PARP1+/+ cells cultured under differentiation conditions, but the inhibition only influenced the Sox2 protein level slightly in PARP1−/− cells (Fig. 5B). We also determined the expression of FGF4 in PARP1+/+ and PARP1−/− cells treated with PJ34 or 3AB at different concentrations, and we found that both inhibitors caused a decrease in the mRNA level of FGF4 in differentiating PARP1+/+ cells at a higher dosage but caused less of a decrease in PARP1−/− cells (Fig. 5C and supplemental Fig. 1A). These observations suggest that the effect of inhibitors on the Sox2 protein level and FGF4 expression is mainly mediated via their inhibition of PARP1 activity. The inhibitory role of PJ34 on FGF4 expression in PARP1−/− cells at 9 μm could be due to the activity of other members of the PARP family. To further characterize the effect of the PARP activity on the Sox2 protein level and FGF4 expression, similar experiments were performed with F9/P19 EC cell lines. As observed in differentiating ESCs, inhibition of the PARP enzymatic activity by either 3AB or PJ34 elevated the Sox2 protein level in both F9 and P19 cells, with a stronger effect in P19 cells (Fig. 5D). Interestingly, PJ34 or 3AB produced a dosage-dependent inhibitory effect on FGF4 expression in P19 cells (Fig. 5E and supplemental Fig. 1B) but affected FGF4 expression only slightly in F9 cells (supplemental Fig. 1B and Fig. 2). On the other hand, PJ34 had little effect on Sox2 and PARP1 expression in P19 cells, suggesting a specific role for PARP activity in regulation of FGF4 transcription. Based upon the above results, we came to the conclusion that PARP1 is implicated in the maintenance of an appropriate level of Sox2 proteins and FGF4 expression in an enzymatic activity-dependent manner.
FIGURE 5.
PARP1 modulates the Sox2 protein level to control FGF4 expression. A, Sox2 protein level is markedly elevated in differentiating PARP1−/− ESCs. ESCs were cultured under the differentiation condition for different lengths of time, as indicated, and the levels of various proteins were evaluated by Western blot analysis. B, ESCs were cultured under the differentiation condition and treated with the PARP inhibitor 3AB or PJ34 for 48 h. The Sox2 protein level was determined by Western blot analysis. C, ESCs were cultured under the differentiation condition and treated with PJ34 at different concentrations for 48 h. Gene expression levels were determined by qPCR. *, p < 0.05; **, p < 0.01, n = 3. D, inhibition of PARP enzymatic activity by 3AB or PJ34 for 48 h increases the Sox2 protein level in F9 and P19 cells as determined by Western blot analysis. E, qPCR analysis of gene expression in P19 EC cells treated with PJ34 at different concentrations for 48 h. *, p < 0.05; **, p < 0.01, n = 3. F, expression of FGF4 and Sox2 in P19 cells is inhibited by transient overexpression of Sox2. Transcript levels of FGF4 and Sox2 were measured using qPCR. *, p < 0.05, n = 3. G, Sox2 RNAi blocks the inhibitory effect of the PARP inhibitor on FGF4 expression in P19 cells. P19 cells expressing EGFP or Sox2 RNAi were treated with 3AB at 6 mm for 48 h. Data were obtained from qPCR analysis. *, p < 0.05; **, p < 0.01, n = 4.
To provide a clear answer to the question of whether the increased Sox2 protein level is responsible for the reduction in FGF4 expression, we transiently transfected P19 EC cells with a Sox2 expressing construct and examined transcript levels of FGF4 and Sox2 (Fig. 5F). P19 cells were used here because they have a constitutively high PARP activity (Fig. 4D). As expected and consistent with previous reports (11, 14), overexpression of Sox2 reduced mRNA levels of FGF4 and also of Sox2 (Fig. 5F). Overexpression of Sox2 in P19 cells was verified by Western blot analysis (data not shown). Thus, our results further confirmed the inhibitory effect of excessive Sox2 on expression of FGF4 and of itself. We then examined whether knockdown of Sox2 could rescue the reduced expression of FGF4 observed in PARP activity-inhibited P19 cells. As shown in Fig. 5G, 3AB treatment (column 2) significantly reduced FGF4 expression. Notably, this inhibitory effect of 3AB on FGF4 expression was considerably abrogated when Sox2 expression was knocked down by Sox2-specific RNAi (Fig. 5G, column 4), supporting the notion that the elevated Sox2 protein level is a major cause for reduced FGF4 expression in the case of low or absent PARP activity. Furthermore, an evidently low level of FGF4 mRNA observed in cells expressing Sox2 RNAi alone (Fig. 5G, column 3) highlights the fact that an appropriate expression level of Sox2 is essential for FGF4 expression. The efficiency of Sox2 RNAi was confirmed by a simultaneous measurement of the Sox2 mRNA level (Fig. 5G, columns 5–8). These findings clearly indicate that the inhibitory effect of 3AB on FGF4 expression is secondary to an elevated Sox2 protein level and that an abnormal level of Sox2 would disrupt normal expression of FGF4.
Putative Mechanism for PARP1 in Control of FGF4 Expression
As a transcription factor, Sox2 exerts its function primarily via binding to regulatory sequences of its target genes. Therefore, to understand how PARP1-mediated Sox2 post-translational modification is involved in regulation of FGF4 expression, we compared the Sox2 association with the FGF4 enhancer in differentiating PARP1+/+ and PARP1−/− cells. Data from ChIP assays revealed that more FGF4 enhancers were associated with Sox2 in differentiating PARP1−/− cells than in differentiating PARP1+/+ cells (Fig. 6A, top row), whereas there was little difference between these two cell types in Sox2 association with its own enhancer (Fig. 6A, bottom row). Furthermore, Sox2 recruitment to the FGF4 enhancer was detectable when P19 cells were treated with PJ34 for 48 h but not in untreated control cells (Fig. 6B, top row), although the recruitment to its own enhancer was detected in both control and PJ34-treated P19 cells (Fig. 6B, bottom row). A similar result was obtained when P19 cells were treated with 3AB (data not shown). These findings indicate that more FGF4 enhancers, but not Sox2 enhancers, were associated with Sox2 proteins when Sox2 proteins are not modified by PAR, leading to inhibition of FGF4 expression. In other words, PARP1 may modify Sox2 and promote its dissociation from FGF4 enhancers, relieving its inhibition of FGF4 expression in normally differentiating cells.
FIGURE 6.
A putative mechanism for PARP1 in control of FGF4 expression. A, ChIP analysis of association of Sox2 with the FGF4 or Sox2 enhancer in PARP1+/+ and PARP1−/− ESCs cultured under the differentiation condition. B, association of Sox2 with the FGF4 enhancer in P19 cells was determined by ChIP assay in the presence or absence of PJ34 (3 μm). The representative results of four independent experiments are shown.
DISCUSSION
Despite intensive studies, little is known about how other transcriptional coregulators act in concert with Oct4 and Sox2 to precisely control FGF4 expression during development. Here we bring a new member, PARP1, into the FGF4 expression regulation network. Our study demonstrates that PARP1 binds to FGF4 enhancer directly and maintains a normal level of FGF4 through poly(ADP-ribosyl)ating Sox2. Moreover, we find that differentiating PARP1−/− ESCs display some phenotypes, similar to those seen in FGF4-deficient ESCs (44), such as fewer surviving cells (Fig. 2, E and F). Functional rescue of the phenotypes by recombinant FGF4 further supports the functional link between PARP1 and FGF4 expression (Fig. 2, E and F). Nevertheless, the phenotypes caused by their deletion in animal models are quite different (35, 50). This could be explained by functional redundancy among members of the PARP family during development. The fact that single null mutant PARP1 or PARP2 mice are viable but double null mutant PARP1 and PARP2 embryos die early in development at the onset of gastrulation (28) clearly demonstrates the essential role of poly(ADP-ribosyl)ation and redundant functions of PARP1 and PARP2 during early embryonic development. Our observation that a small inhibitory effect of PARP inhibitors on FGF4 expression (Fig. 5C and supplemental Fig. 1A) and a weak signal of Sox2 poly(ADP-ribosyl)ation (Fig. 4B) exist in PARP1 knock-out cells provides further evidence to support the existence of functional redundancy among members of this family, whereas PARP1 is the major enzyme responsible for poly(ADP-ribosyl)ation of cellular proteins (46). In addition, our results reveal that PARP1 activity is dynamically regulated by ESC differentiation processes, instead of DNA breaks. This is in agreement with previous reports showing that PARP1 can be activated by signals such as increased activity of calmodulin kinase II and ERK2 independently of DNA damage (51, 52). It is interesting to know why PARP activities declined initially after withdrawal of feeder cells but is reactivated during further differentiation induced by RA treatment. Taken together, the establishment of physical and functional links among ESC differentiation, PARP1 activity, and FGF4 expression provides new evidence for an important role of PARP1 in regulating gene expression and early development.
PARP1 regulates gene expression through diverse mechanisms that are likely to vary in an activator-specific and gene-specific manner (53). In some cases, PARP1 enzymatic activity is not required for its coregulatory activity, e.g. with NF-κB (30, 54), whereas in others it is required, e.g. with HES1 and NFAT (51, 55). In many of the latter cases, DNA-binding factors or other components of the coregulatory complex are targets for PARP1-dependent poly(ADP-ribosyl)ation (51, 55, 56). For example, PARP1 was reported to poly(ADP-ribosyl)ate several components of a TLA (transducin-like enhancer of split) corepressor complex, resulting in dismissal of the complex from the MASH1 promoter and neuronal differentiation (51). In this study, we found that PARP1 is recruited to the FGF4 enhancer together with Oct4 and Sox2, and poly(ADP-ribosyl)ates Sox2 directly. It is possible that PARP1-mediated addition of an anionic polymer on the Sox2 protein causes dismissal of Sox2 from the FGF4 enhancer, because of repulsion among anionic molecules as proposed previously by other researchers (46, 51, 57). Furthermore, we found that PARP1 deficiency or inhibition of the PARP activity specifically affects FGF4 but little, if any, Sox2 transcription (Fig. 2, A and B, and Fig. 5, C and E). It is probably explained by our observations that binding of Sox2 to its own enhancer was not affected by the absent or relatively low PARP1 activity (Fig. 6, A and B) and that PARP1 was not recruited to the Sox2 enhancer (Fig. 1D). However, expression of Sox2 itself and its other target genes, to which PARP1 is not recruited, could be reduced when the Sox2 protein level is dramatically elevated, such as in the case of overexpression of Sox2 (Fig. 5F) (11). On the other hand, Sox2 is also essential for expression of its target genes (33, 58, 59) (Fig. 5G). Thus, a dynamic and fine regulation is required for normal functions of Sox2. In addition, the mechanism by which PARP1 regulates FGF4 expression in undifferentiated ESCs may be different from that in differentiating ESCs as PARP1 deficiency did not affect the Sox2 protein level (Fig. 5A) but reduced FGF4 expression in undifferentiated ESCs (Fig. 2A). The manner by which PARP1 regulates FGF4 expression in undifferentiated ESCs needs further investigation.
In summary, we have identified PARP1 as a coregulator of Oct4 and Sox2 in FGF4 expression and demonstrated that poly(ADP-ribosyl)ation of Sox2 plays an important and direct role in maintaining an appropriate level of Sox2 proteins as well as FGF4 expression during differentiation of ESCs. Our work provides the first evidence for the importance of post-translational modifications in control of Sox2 protein levels and opens up a new way to address the critical issue of how the Sox2 expression level is precisely and dynamically controlled during development. Nevertheless, it remains unanswered as to how the accumulated Sox2 at the FGF4 enhancer inhibits FGF4 expression. It is possible that excessive Sox2 disrupts the equilibrium among factors functioning in FGF4 expression, such as enhanced recruitment of transcription repressors. Identification of other proteins potentially implicated in FGF4 expression will help us to understand why a precise level of Sox2 protein is critical for its function.
Supplementary Material
Acknowledgments
We thank Dr. Zhaoqi Wang for providing PARP1+/+ and PARP1−/− ESCs and Dr. Naihe Jing for providing P19 EC cells. We also thank Chunliang Li and Junjie Gu for preparing mouse embryonic fibroblast feeder cells.
This work was supported by National Natural Science Foundation of China Grant 30730051, Shanghai Science and Technology Developmental Foundation Grant 08JC1413100, National High Technology Research and Development Program of China Grants 2006CB943901 and 2007CB947904, and Shanghai Leading Academic Discipline Project S30201.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1 and 2, and Table S1.
- ESC
- embryonic stem cell
- EC
- embryonic carcinoma
- LIF
- leukemia inhibitory factor
- NE
- nuclear extract
- RA
- retinoic acid
- ChIP
- chromatin immunoprecipitation
- CoIP
- coimmunoprecipitation
- qPCR
- quantitative PCR
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- EGFP
- enhanced green fluorescent protein
- GST
- glutathione S-transferase
- RNAi
- RNA interference
- 3AB
- 3-aminobenzamide.
REFERENCES
- 1.Brook F. A., Gardner R. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5709–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers I., Smith A. (2004) Oncogene 23, 7150–7160 [DOI] [PubMed] [Google Scholar]
- 3.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 5.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 7.Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., Zaehres H., Schöler H. R. (2009) Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- 8.Yuan H., Corbi N., Basilico C., Dailey L. (1995) Genes Dev. 9, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 9.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 10.Nishimoto M., Fukushima A., Okuda A., Muramatsu M. (1999) Mol. Cell. Biol. 19, 5453–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boer B., Kopp J., Mallanna S., Desler M., Chakravarthy H., Wilder P. J., Bernadt C., Rizzino A. (2007) Nucleic Acids Res. 35, 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong H., Hohenstein K. A., Donovan P. J. (2008) Stem Cells 26, 1931–1938 [DOI] [PubMed] [Google Scholar]
- 14.Kopp J. L., Ormsbee B. D., Desler M., Rizzino A. (2008) Stem Cells 26, 903–911 [DOI] [PubMed] [Google Scholar]
- 15.Niwa H., Miyazaki J., Smith A. G. (2000) Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 16.Xu H. M., Liao B., Zhang Q. J., Wang B. B., Li H., Zhong X. M., Sheng H. Z., Zhao Y. X., Zhao Y. M., Jin Y. (2004) J. Biol. Chem. 279, 23495–23503 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z., Liao B., Xu M., Jin Y. (2007) FASEB J. 21, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 18.Saxe J. P., Tomilin A., Schöler H. R., Plath K., Huang J. (2009) PLoS ONE 4, e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuruzoe S., Ishihara K., Uchimura Y., Watanabe S., Sekita Y., Aoto T., Saitoh H., Yuasa Y., Niwa H., Kawasuji M., Baba H., Nakao M. (2006) Biochem. Biophys. Res. Commun. 351, 920–926 [DOI] [PubMed] [Google Scholar]
- 20.Kasid U. N., Halligan B., Liu L. F., Dritschilo A., Smulson M. (1989) J. Biol. Chem. 264, 18687–18692 [PubMed] [Google Scholar]
- 21.Scovassi A. I., Mariani C., Negroni M., Negri C., Bertazzoni U. (1993) Exp. Cell Res. 206, 177–181 [DOI] [PubMed] [Google Scholar]
- 22.Kameoka M., Ota K., Tetsuka T., Tanaka Y., Itaya A., Okamoto T., Yoshihara K. (2000) Biochem. J. 346, 641–649 [PMC free article] [PubMed] [Google Scholar]
- 23.Smulson M. E., Simbulan-Rosenthal C. M., Boulares A. H., Yakovlev A., Stoica B., Iyer S., Luo R., Haddad B., Wang Z. Q., Pang T., Jung M., Dritschilo A., Rosenthal D. S. (2000) Adv. Enzyme Regul. 40, 183–215 [DOI] [PubMed] [Google Scholar]
- 24.Huang J. Y., Chen W. H., Chang Y. L., Wang H. T., Chuang W. T., Lee S. C. (2006) Nucleic Acids Res. 34, 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M. Y., Zhang T., Kraus W. L. (2005) Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 26.Kanai M., Tong W. M., Wang Z. Q., Miwa M. (2007) Biochem. Biophys. Res. Commun. 359, 426–430 [DOI] [PubMed] [Google Scholar]
- 27.Kanai M., Hanashiro K., Kim S. H., Hanai S., Boulares A. H., Miwa M., Fukasawa K. (2007) Nat. Cell Biol. 9, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 28.Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J. C., Dierich A., LeMeur M., Sabatier L., Chambon P., de Murcia G. (2003) EMBO J. 22, 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemberger M., Nozaki T., Winterhager E., Yamamoto H., Nakagama H., Kamada N., Suzuki H., Ohta T., Ohki M., Masutani M., Cross J. C. (2003) Dev. Biol. 257, 371–381 [DOI] [PubMed] [Google Scholar]
- 30.Kraus W. L., Lis J. T. (2003) Cell 113, 677–683 [DOI] [PubMed] [Google Scholar]
- 31.Masutani M., Nozaki T., Watanabe M., Ochiya T., Hasegawa F., Nakagama H., Suzuki H., Sugimura T. (2001) Mutat. Res. 477, 111–117 [DOI] [PubMed] [Google Scholar]
- 32.Wacker D. A., Frizzell K. M., Zhang T., Kraus W. L. (2007) Subcell. Biochem. 41, 45–69 [DOI] [PubMed] [Google Scholar]
- 33.Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Mol. Cell. Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Berg D. L., Zhang W., Yates A., Engelen E., Takacs K., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2008) Mol. Cell. Biol. 28, 5986–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman B., Poueymirou W., Papaioannou V. E., DeChiara T. M., Goldfarb M. (1995) Science 267, 246–249 [DOI] [PubMed] [Google Scholar]
- 36.Niswander L., Martin G. R. (1992) Development 114, 755–768 [DOI] [PubMed] [Google Scholar]
- 37.Basilico C., Moscatelli D. (1992) Adv. Cancer Res. 59, 115–165 [DOI] [PubMed] [Google Scholar]
- 38.Schoorlemmer J., Kruijer W. (1991) Mech. Dev. 36, 75–86 [DOI] [PubMed] [Google Scholar]
- 39.Ambrosetti D. C., Basilico C., Dailey L. (1997) Mol. Cell. Biol. 17, 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luster T. A., Rizzino A. (2003) Gene 323, 163–172 [DOI] [PubMed] [Google Scholar]
- 41.van de Wetering M., Oving I., Muncan V., Pon Fong M. T., Brantjes H., van Leenen D., Holstege F. C., Brummelkamp T. R., Agami R., Clevers H. (2003) EMBO Rep. 4, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhar S. K., Lynn B. C., Daosukho C., St Clair D. K. (2004) J. Biol. Chem. 279, 28209–28219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amiri K. I., Ha H. C., Smulson M. E., Richmond A. (2006) Oncogene 25, 7714–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilder P. J., Kelly D., Brigman K., Peterson C. L., Nowling T., Gao Q. S., McComb R. D., Capecchi M. R., Rizzino A. (1997) Dev. Biol. 192, 614–629 [DOI] [PubMed] [Google Scholar]
- 45.Ambrosetti D. C., Schöler H. R., Dailey L., Basilico C. (2000) J. Biol. Chem. 275, 23387–23397 [DOI] [PubMed] [Google Scholar]
- 46.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 47.Yeom Y. I., Fuhrmann G., Ovitt C. E., Brehm A., Ohbo K., Gross M., Hübner K., Schöler H. R. (1996) Development 122, 881–894 [DOI] [PubMed] [Google Scholar]
- 48.Boer B., Bernadt C. T., Desler M., Wilder P. J., Kopp J. L., Rizzino A. (2006) J. Cell. Physiol. 208, 97–108 [DOI] [PubMed] [Google Scholar]
- 49.Johnson L. R., Lamb K. A., Gao Q., Nowling T. K., Rizzino A. (1998) Mol. Reprod. Dev. 50, 377–386 [DOI] [PubMed] [Google Scholar]
- 50.Shall S., de Murcia G. (2000) Mutat. Res. 460, 1–15 [DOI] [PubMed] [Google Scholar]
- 51.Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. (2004) Cell 119, 815–829 [DOI] [PubMed] [Google Scholar]
- 52.Cohen-Armon M., Visochek L., Rozensal D., Kalal A., Geistrikh I., Klein R., Bendetz-Nezer S., Yao Z., Seger R. (2007) Mol. Cell 25, 297–308 [DOI] [PubMed] [Google Scholar]
- 53.Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassa P. O., Hottiger M. O. (2002) Cell. Mol. Life Sci. 59, 1534–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olabisi O. A., Soto-Nieves N., Nieves E., Yang T. T., Yang X., Yu R. Y., Suk H. Y., Macian F., Chow C. W. (2008) Mol. Cell. Biol. 28, 2860–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaniolo K., Desnoyers S., Leclerc S., Guérin S. L. (2007) BMC Mol. Biol. 8, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferro A. M., Olivera B. M. (1982) J. Biol. Chem. 257, 7808–7813 [PubMed] [Google Scholar]
- 58.Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.