Abstract
In this study we analyzed the structure and function of a truncated form of hemolysin A (HpmA265) from Proteus mirabilis using a series of functional and structural studies. Hemolysin A belongs to the two-partner secretion pathway. The two-partner secretion pathway has been identified as the most common protein secretion pathway among Gram-negative bacteria. Currently, the mechanism of action for the two-partner hemolysin members is not fully understood. In this study, hemolysis experiments revealed a unidirectional, cooperative, biphasic activity profile after full-length, inactive hemolysin A was seeded with truncated hemolysin A. We also solved the first x-ray structure of a TpsA hemolysin. The truncated hemolysin A formed a right-handed parallel β-helix with three adjoining segments of anti-parallel β-sheet. A CXXC disulfide bond, four buried solvent molecules, and a carboxyamide ladder were all located at the third complete β-helix coil. Replacement of the CXXC motif led to decreased activity and stability according to hemolysis and CD studies. Furthermore, the crystal structure revealed a sterically compatible, dry dimeric interface formed via anti-parallel β-sheet interactions between neighboring β-helix monomers. Laser scanning confocal microscopy further supported the unidirectional interconversion of full-length hemolysin A. From these results, a model has been proposed, where cooperative, β-strand interactions between HpmA265 and neighboring full-length hemolysin A molecules, facilitated in part by the highly conserved CXXC pattern, account for the template-assisted hemolysis.
Hemolysin A (HpmA)2 and B (HpmB) from Proteus mirabilis belong to the Type Vb or two-partner secretion pathway (1), the most widespread of the five porin-type protein translocating systems found within bacterial, fungal, plant, and animal kingdoms (2). Cell surface adhesions, iron-acquisition proteins, and cytolysins/hemolysins all use two-partner secretion pathways (3–5). The A-component of the two-partner secretion in P. mirabilis is a 166-kDa virulence factor capable of mammalian blood cell lysis upon secretion from the cell. This is accomplished by Sec-dependent transport to the periplasm followed by N-terminal proteolytic processing. Extracellular secretion occurs by transport through the B-component, HpmB, which is a 16-stranded β-barrel transmembrane channel (6). In addition to its role in efficient secretion, HpmB is also necessary for activation of the larger exoprotein A-component (HpmA) (7–10).
Studies on hemolytic TpsA members report that: 1) a truncated TpsA containing the N-terminal secretion cap (11) complements and restores hemolytic activity within a non-secreted/inactive pool of full-length TpsA (12), 2) the conserved cysteine residues within a CXXC motif are not required for secretion (12), and 3) the first asparagine within a NPNG hemagglutinin motif is required for efficient secretion (13). Other investigations demonstrate significant conformational change within TpsA members during B-component dependent secretion (8, 14–16).
Recent x-ray crystal structures for two TpsA adhesion orthologs, hemagglutinin from Bordetella pertussis (FHA) and high molecular weight protein from Haemophilus influenzae (HMW1) adopt a right-handed parallel β-helix similar to pectate lyase (11, 18, 19). The 301-residue N-terminal FHA fragment (Fha30) contains a 37 parallel stranded β-helix. Stabilization of type I β-turns at two highly conserved regions: 66NPNL and 105NPNG is proposed to play a large role in the ability of this N-terminal fragment to rapidly adopt β-helix architecture (11, 20). Despite 21% sequence identity, the 371-residue HMW1 structure (HMW1-PP) is a similar 47 parallel stranded β-helix. A hypothesis arose from these β-helix structures that suggests extracellular secretion through TpsB channels, and the progressive folding of TpsA members is energetically coupled. Full-length TpsA adhesion members have been proposed to have a filamentous appearance built from a right-handed β-helix fold (21). To date, there is little known about the full-length HpmA domain architecture. However, there are two filamentous hemagglutinin type domains. The N-terminal domain is positioned between residues 30 and 167, whereas the C-terminal domain lies between residues 1200 and 1264 and has been proposed to facilitate cellular aggregation.
In this work, we investigated the functional and structural role of truncated hemolysin A (HpmA265) during the template-assisted activation of hemolysis. A previous investigation with ShlA, a homologous TpsA member from Serratia marcescens, has shown similar complementation using a 255-amino acid fragment (12). Here, we demonstrate that HmpA265 can cooperatively cross seed an inactive pool of full-length hemolysin A (HpmA*) to form an exotoxin measured by our template-assisted hemolytic assay (TAHA). We also report that the CXXC motif provides structural stability and facilitates reversible re-folding. The structure reveals a right-handed β-helix, similar to those of FHA and HMW1. A number of conserved features found at the putative subunit interface suggest a mechanism by which activation of inactive HpmA* occurs.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Cells, and Culture Conditions
Escherichia coli strains used in this study were B834 (DE3), HMS174 (DE3) (EMD Biosciences), RAU126 (23), and XL-10 Gold (Stratagene) cells. All liquid E. coli cultures were grown in Luria-Bertani (LB) broth at 37 °C with rotary shaking. All chemicals were purchased from Fisher Scientific unless stated otherwise.
Construction of hpmA265
A 5′ segment of hpmA was amplified via PCR in the presence of pHpmA358, (+)hpmA265- sense and (−)hpmA265-antisense primers (Midland Certified Reagent Co.). The plasmid pHPMA358 harbors the first 358 N-terminal residues of HpmA. The (+)hpmA265-sense primer harbored an NdeI restriction site, a region encoding the ATG start site, and a 21-base sequence complementary to hpmA. The (−)hpmA265-antisense primer withholds a HindIII restriction site, a stop codon, base pairs coding for the 6 histidines, and a 19-base sequence complementary to hpmA. Following amplification, the PCR fragment, incorporating the N-terminal 265 amino acids of HpmA complete with an in-frame C-terminal six histidine sequence was cloned into the NdeI and HindIII sites of pET-24a(+) (EMD Biosciences). Potential positive clones were subjected to DNA sequencing (UW Biotechnology Center, Madison, WI). The resultant positive clone harboring hpmA265 within pET24a+ has been termed pLB265.
Site-directed Mutagenesis
Sets of oligonucleotide primers were designed to replace both cysteine 144 and cysteine 147 with either alanine or serine. The QuikChange Site-Directed Mutagenesis Kit was utilized according to published procedures (Stratagene). Positive clones were verified by DNA sequencing (University of Wisconsin Biotechnology Center, Madison, WI).
Protein Purification
All protein expression experiments were conducted in E. coli B834 (DE3) harboring hpmB within pACYC184 (23), as well as pLB265. The CXXC replacement constructs, termed either pDMA (double alanine mutant) or pDMS (double serine mutant) were used in replace of pLB265 during expression of the CXXC mutant proteins. Transformants were grown at 37 °C in the presence of 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. Cell cultures were induced overnight by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside after reaching A600 of 0.6–0.8. Cellular culture supernatant was collected via centrifugation at 12,000 × g for 30 min. The pH of the supernatant was adjusted to 7.6 using NaOH and then passed twice over 20 ml of Ni2+-nitrilotriacetic acid-agarose (Qiagen) entrapped in a 600-ml sintered glass funnel pre-equilibrated with Buffer A (50 mm NaH2PO4, 300 mm NaCl, pH 7.8). Contaminant proteins were removed with 40 column volumes of Buffer A, followed by 10 column volumes of Buffer B (50 mm NaH2PO4, 10 mm imidazole, 300 mm NaCl, pH 7.8). The resultant Ni2+-nitrilotriacetic acid-agarose was then transferred into a 2.8-cm × 13-cm chromatography column (Bio-Rad) and His-tagged HpmA265 was eluted via the addition of two column volumes of Buffer C (50 mm NaH2PO4, 400 mm imidazole, 300 mm NaCl, pH 7.8).
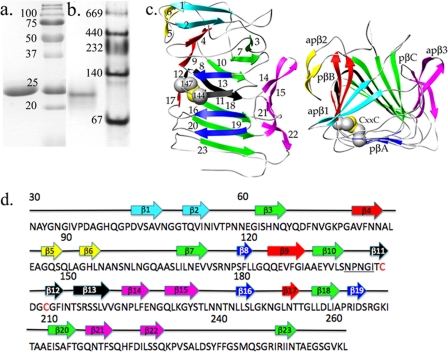
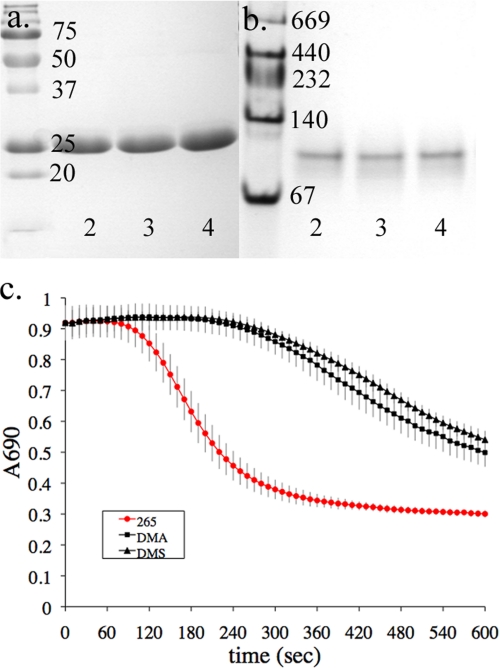
The protein eluant was adjusted to 10 mm EDTA and dialyzed exhaustively versus 10 mm Tris-HCl, pH 7.4, at 4 °C. After dialysis, HpmA265 was concentrated using a SpeedVac Plus SC110A (Savant) and re-dialyzed twice versus 4 liters of 10 mm Tris-HCl. Protein concentrations were determined via A280 using ϵ280 = 0.277 for HpmA265 (W. M. Keck Foundation Biotechnology Resource Laboratory). SDS-PAGE (15%) and native gel electrophoresis (4–15% Tris-HCl) were used to confirm purity (Fig. 1, a and b).
FIGURE 1.
Electrophoretic and structural analysis of HpmA265. a and b, denaturing and non-denaturing electrophoresis results, respectively, where lane 1 represents HpmA265 and lane 2 molecular weight markers. c, the β-helix structure of HpmA265 has been shown perpendicular and parallel to the helical axis. Each extended side of parallel β-sheet has been colored independently, where pβA is red, pβB is green, and pβC is blue. In addition, the three sections of anti-parallel β-sheet have been colored cyan for apβ1, yellow for apβ2, and magenta for apβ3. Cysteines 144 and 147 are positioned on the third complete coil and have been colored by atom type in spacefilling format, where gray is carbon and yellow is sulfur. d, various β-strands have been mapped onto the amino acid sequence of HpmA265. The highly conserved NPNG motif has been underlined, while the cysteine residues have been colored red.
HpmA* Preparation
Full-length inactive HpmA (HpmA*) expression was induced for 5 h via the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside to E. coli strain RAU126 grown to an A600 of 0.6–0.8 at 37 °C in the presence of 30 μg/ml kanamycin. Cellular pellets were collected in 50-ml conical tubes (Fisher) and stored at −80 °C. Crude HpmA* was prepared by ultrasonication (five 20-s intervals) after resuspension of the cellular pellet into 25 ml of 10 mm NaH2PO4 150 mm NaCl, pH 7.4 (PBS). This source of inactive full-length HpmA, produced in the absence of HpmB-dependent transport, was the source of HpmA* used throughout this study.
Template-assisted Hemolytic Assay
A TAHA was used to study the ability of HpmA265 to act as a template to fold and activate HpmA*. The TAHA was performed by mixing 50 μl of PBS, 10 μl of bovine serum albumin (10 mg/ml), 20 μl of 10% sheep red blood cells (ENOVA, White Bear Lake, MN), 10 μl of HpmA*, and 392 nm HpmA265 in a flat bottom 96-well microtiter plate (Fisher). After addition, the suspension was mixed by repeated pipetting, and hemolysis was monitored via decreasing optical density at 690 nm (24, 25).
HpmA** Reactivation
Preincubation experiments were conducted by mixing HpmA265 and HpmA* in volumes proportional to those used during the TAHA. After preincubation and inactivation of HpmA* → HpmA**, a series of components was added back to this mixture. Components added back to HpmA** were HpmA265 (392 nm), HpmA* (10 μl), HMS174 (DE3) (10 μl), and heated HpmA* (10 μl).
Endpoint Hemolysis Assay
To conduct the HpmA265 and HpmA* dose-response experiments, a series of quadruple end point analyses were conducted. Briefly, 200 μl of sheep red blood cells were combined with either 50 μl of HpmA* combined with increasing amounts of HpmA265 or 0.392 μm HpmA265 mixed with increasing amounts of HpmA*. Final volumes were adjusted to 500 μl with PBS. Reactions were stopped via centrifugation at 10,000 × g for 10 min, and hemoglobin release was measured at 540 nm. A control for complete lysis was determined by mixing 200 μl of red blood cells in 300 μl of PBS adjusted to 20% SDS (v/v) (24).
CD Spectroscopy
Secondary structure of these proteins were measured on a Jasco Spectropolarimeter 810 (JASCO Inc., Easton, MD) by following the far-UV-CD spectrum from 260 to 200 nm, taking measurements every 1 nm with a scanning speed of 10 nm/min. far-UV-CD spectra were also measured at 5 °C temperature intervals from 10 to 90 °C with an equilibration time of 15 min prior to scanning each protein. Protein concentrations were 13 μm.
Crystallization, Data Collection, and Processing
Crystals of HpmA265 were obtained at 16 °C using the hanging drop, vapor-diffusion method by mixing protein and mother liquor in a 1:1 ratio. Crystals were obtained from a protein concentration of 10 mg/ml in the presence of 0.8 m Na/KH2PO4 and 0.1 m HEPES, pH 7.5, and cryoprotected in the presence of xylitol. The diffraction data were collected in-house at 100 K on a Rigaku Microfocus 007 with R-Axis IV2+ detector, Varimax optics, and Cryostream 2000. The data were indexed, integrated, merged, and scaled using HKL2000 (26). Data collection statistics are reported in Table 1. One monomer was found within the asymmetric unit.
TABLE 1.
Crystallographic data
| Data collection | |
| Space group | P21212 |
| Unit cell dimensions | |
| a, b, c (Å) | 56.27, 119.53, 34.20 |
| α = β = γ (deg)a | 90 |
| Resolution range (Å) | 56.27-1.80 |
| Rmergeb (%), overall/highest bin | 9.2 (50.3) |
| I/σI, overall/highest bin | 13.9 (4.8) |
| Completeness (%), overall/highest bin | 100/100 |
| Redundancy, overall/highest bin | 17.38 (17.16) |
| No. unique reflections, overall/highest bin | 22,214 (2,167) |
| Refinement | |
| Resolution (Å)c | 56.27-1.80 |
| Protein atoms | 1,937 |
| Water molecules | 359 |
| Rworkd (%), overall/highest bin | 15.1 (18.9) |
| Rfree (%), overall/highest bin | 20.3 (29.8) |
| r.m.s.d.e bond length (Å) | 0.019 |
| r.m.s.d.e angle (deg) | 1.745 |
| Ramachandran statisticsf | |
| Most favored region (%) | 87.2 |
| Additionally allowed region (%) | 12.3 |
a Highest resolution bin 1.86-1.80 Å.
bRmerge = Σ〈I − 〈I〉/ΣI, where I equals the measured intensity and 〈I〉 equals the average intensity.
c Reflections during refinement were 22,192, and the intensity cutoffs were set at the default value of zero.
dR = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo is the observed structure factor and Fc is that calculated from the model. Rfree was generated by randomly selecting 5% of the reflections, while Rwork used all other reflections.
e r.m.s.d., root mean square deviations of bond lengths and angles from ideal geometry.
f PROCHECK (43) was used to determine ϕ, ψ angles.
Structure Determination and Refinement
Molecular replacement with MOLREP (version 9.2) was used, using the coordinates of Fha30 (1RWR) (11, 20) to obtain initial phases (27, 28). COOT and Refmac5 were used during repeated cycles of manual model building and refinement using reflections ranging from 56- to 1.8-Å resolution (29, 30). The final refinement and model statistics are reported in Table 1. All drawings were generated with Chimera (31). The atomic coordinates were submitted to the Protein Data bank with the identification code 3FY3.
Laser Scanning Confocal Microscopy
Red blood cells were washed three times in PBS and suspended to a final concentration of 10% (v/v). Red blood cell smears were prepared on a slide via fixing in −20 °C methanol for 30 s then washed three times in PBS. The fixed red blood cells were blocked for 10 min with 5% bovine serum albumin and subsequently washed three times more in PBS. Three separate slides preparations were then flooded with 500 μl of either HpmA* from RAU126 lysate, HpmA from a 10-min preincubation of HpmA265/RAU126 lysate, or HpmA** from an overnight preincubation of HpmA265/RAU126. Each slide was washed three times in PBS and then reacted with 100 μl of HpmA 1°-antibody for 1 h (32). Slides were washed three times with PBS and then subjected to 100 μl of Alexa Fluor 488 goat anti-mouse IgG antibody (Invitrogen), then again washed three additional times with PBS prior to laser scanning confocal microscopy. Images were collected using a Nikon CS1 confocal TE-1000 Plan Apo lens (60× and 1.4 numerical aperture).
RESULTS
HpmA265 Characterization
An HpmA fragment (HpmA265) containing the first 265 N-terminal amino acids was expressed, secreted, and purified to homogeneity (Fig. 1, a and b). Electrospray ionization mass spectrometry indicated 25,460 Da for HpmA265, essentially identical to the predicted 25,457 Da (UW Biotechnology Center, Madison, WI) from SDS-PAGE (Fig. 1a) (15, 16, 33, 34). The mass spectrum data suggested that HpmA265 was not covalently modified during TpsB-dependent activation and secretion as was previously reported (7). Additionally, native gel electrophoresis (Fig. 1b) and dynamic light scattering-size exclusion chromatography (W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University; data not shown) studies were performed to characterize the oligomeric state of HpmA265. Both methods indicated HpmA265 exists as an oligomeric species; dynamic light scattering-size exclusion chromatography results indicated a dimer of Mr 48,000, while the native-PAGE band was calculated to be 98,000 Da (Fig. 1b).
Three-dimensional Structure of HpmA265
The x-ray structure of HpmA265 harbored a 3-sided, 16-stranded, right-hand parallel β-helix, adjoining 2- and 4-stranded anti-parallel sheets (apβ1, apβ2, and apβ3) with final dimensions of 40 Å × 37 Å × 27 Å (Fig. 1c). Overall, the β-helix was built from three parallel sheets, pβA, pβB, and pβC (Fig. 1c). These sides derived from strands: 8, 11, 16, and 19 (pβA); 2, 4, 9, 12, and 17 (pβB); and 3, 7, 10, 13, 18, 20, and 23 (pβC) (Fig. 1, c and d). The three adjoining anti-parallel sheets lie at the top edge of pβB and off the side of pβC and were built from β-strands 1/2 (apβ1), 5/6 (apβ2), and 14/15:21/22 (apβ3) (Fig. 1, c and d). There were two main regions where aromatic stacking between Phe and Tyr residues occurred. First, there was a stack of aromatic groups positioned on the under side of apβ3 facing pβC. The underside of apβ3 donated Phe-164, Tyr-172, and Phe-221 from strands 14, 15, and 21, whereas Phe-225 laid within the turn connecting strands 21 and 22. The parallel β-helix donated Phe-215 from pβC to complete this aromatic stacking arrangement. The second aromatic stack was located within the inner core of the β-helix. Two of the three parallel sides donated aromatic groups into this area. Strands 3 and 10 of pβC donated Tyr-68 and Tyr-134, strands 4 and 12 of pβB donate Phe-80 and Phe-149, whereas the turn spanning strands 3 and 4 donated Phe-71 to complete this inner core aromatic stack.
Comparison of Fha30 and HMW1 to HpmA265
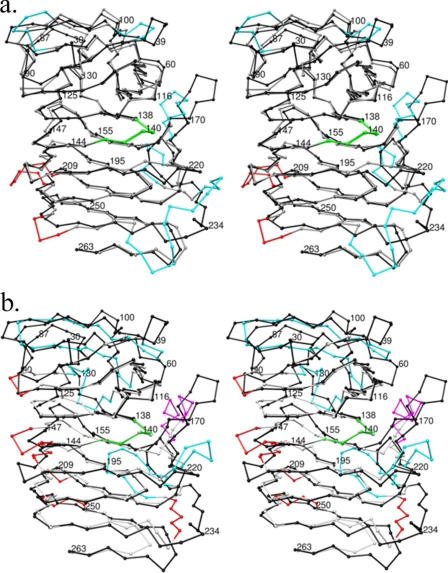
Fha30 from B. pertussis was the first reported x-ray structure for the TpsA family (11). This structure was followed most recently by the structure of H. influenzae HMW1-PP (18). The Fha30 and HMW1-PP shared 35.9 and 28.6% sequence identity with HpmA over 265 residues of the HpmA construct. Although there is only slight comparative percent amino acid identity between HpmA, Fha30, and HMW1-PP, there was significant structural similarity. Both truncated Cα coordinate files of Fha30 and HMW1-PP were superpositioned onto the HpmA265. The superposition of Cα coordinates translated to a root mean square deviation of 1.9 Å for 226 structurally related Fha30 Cα atoms and 2.7 Å for 195 structurally related HMW1-PP Cα atoms (35). As expected, the β-helix cores aligned well between all three Cα coordinate files. Fha30 was structurally more similar to HpmA265 than HMW1-PP, because both HpmA265 and Fha30 have their respective β-helices capped and flanked by sets of anti-parallel β-sheets (Fig. 2a). However, these anti-parallel segments still led to the largest structural differences between HpmA265 and Fha30. At the N-terminal end, the turn between β1 and β2 had a significant structural shift. This shift most likely allowed accommodation of a four-residue insertion within HpmA265 spanning Asp-35 to His-38. Additionally, the flanking anti-parallel strands β14/15 and β21/22 within apβ3 shifted quite dramatically between HpmA265 and Fh30 (Fig. 2a). A highly hydrophobic 6-residue (240YFFGSM) insertion probably led to this main-chain structural shift.
FIGURE 2.
The crystal structures of HpmA265, Fha30, and HMW1-PP show differences within the top and flanking anti-parallel β-sheet regions. a, the Cα coordinates have been superimposed between HpmA265 and Fha30. The trace for HpmA Cα atoms has been colored black in both images. Structural differences between HpmA265 and Fha30 have been colored cyan for the top and flanking regions apβ1, apβ2, and apβ3, while differences within β-arc regions have been colored red. The largest structural differences between HpmA265 and Fha30 lie at the flanking anti-parallel sheet apβ3. This shift accommodates a 12-residue insertion between β22 and β23 in HpmA265 and preserves the integrity of the β-helix core alignment. b, structural differences between HpmA265 and HMW1-PP have been colored cyan for anti-parallel regions, magenta for α-helix regions, and red for β-arcs. The large shift at the N-terminal end derives from a global superposition of three anti-parallel strands within HMW1-PP onto the four anti-parallel strands within HpmA265. This large difference stems from moving β2 of HMW1-PP between β1 and β2 of HpmA265. The N-terminal superposition also leaves β5 and β6 of HpmA265 without matching HMW1-PP strands. Additionally, HMW1-PP has an α-helix in place of two of the anti-parallel strands within apβ3. This creates another location of large structural difference. In both instances, the structural agreement between β-helix core Cα atoms is well maintained, especially within the NPNG motif (colored green).
Although the β-helix core was structurally conserved, there were major differences between HpmA265 and HMW1-PP at: 1) the capping and flanking anti-parallel regions and 2) some of the turns connecting parallel β-strands within the core. The N-terminal end HMW1-PP only has three anti-parallel strands, whereas HpmA265 has four. In addition, HpmA265 had an eight-residue insertion between Gln-67 and Gly-74. These differences created a global shift in the structural superposition within both apβ1 (Asp-42 to Asn-54) and apβ2 (Ala-64 to Leu-95) of HpmA265 (Fig. 2b). Additionally, HMW1-PP harbors a small α-helical insertion (Asn-176 to Arg-182) in place of apβ3 of HpmA265. The insertion of the α-helix within this region created a major structural difference between the areas flanking HpmA265 and HMW1-PP. Another set of structural differences existed within many of the turns connecting individual β-strands (Fig. 2b). Among all of the structural differences, the parallel β-helix was still well aligned within these regions: 1) Leu-108 to Phe-149 (β7-β12), 2) Leu-193 to Gly-205 (β18-β19), 3) Ile-212 to Asn-219 (β20-β21), and 4) Ile-250 to Asn-254 (β23). The conserved type I β-turn NPNG motif aligned well, and in fact it appeared to provide an anchoring point for the alignment between all three structures (Fig. 2b).
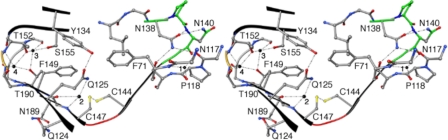
Stabilization of the Third Coil
We found characteristic interactions unlike any in the homologous Fha30 and HMW1-PP structures that appear to provide stability to the HpmA265 β-helix structure (Fig. 3). In this manuscript a distinction will be made between segments connecting anti-parallel and parallel β-strands. In the discussion to follow, a “coil” refers to one complete revolution of the parallel β-helix, while an “arc” refers to the segments adjoining these parallel strands. In anti-parallel sheets, the regions adjoining β-strands have been termed “turns.” The third coil implemented an N-bl β-arc, a buried disulfide bond, inner core hydrogen bonded solvent molecules, and a carboxyamide ladder. First, stabilization of the arc leading into coil three utilized the highly conserved Asn-138 (138NPNG141) to form hydrogen bonds to both Asn-40 within the N-bl β-arc and to Ser-119 and Asn-17 (Fig. 3). This network of hydrogen bonds appeared to be a major stabilizing force within the first arc of coil three. Four solvent molecules (1–4) were also observed within the HpmA265 β-helix core, and all were juxtapositioned within the third coil (Fig. 3). Solvent 1 formed hydrogen bonds to Phe-71-N, Phe-71-O, Pro-118-O, and Asn-17-O to further stabilize arc one of coil three.
FIGURE 3.
Stabilization of the β-helix third coil. The stereo-diagram illustrates a view down the β-helical axis where partial coils above and below the CXXC motif have been maintained. Each β-arc connecting each of the three sides (a–c) has been colored independently. The β-arc containing the NPNG motif has been colored green, the β-arc between the C144:C147 disulfide bond has been colored red, and the third β-arc has been colored gold. Solvent molecules (1–4) have been represented as black spheres. Atoms have been colored by type: carbon (gray), oxygen (red), nitrogen (blue), and sulfur (yellow). The third coil utilizes a highly conserved hemagglutinin pattern, a CXXC motif, four buried solvent molecules (1–4), and a carboxyamide ladder and thereby, provides unique stabilization architecture within the three respective β-arcs. The first arc implements Asn-38 and Asn-40 from the conserved hemagglutinin pattern (NPNG) and solvent molecule (1) for stabilization. The series of hydrogen bonds within this arc forms a stabilizing bridge between coils 1, 2, and 3. The disulfide bond shared between Cys-144 and Cys-147 within the CXXC motif stabilizes the second β-arc. Additionally, solvent 2 forms a trifurcated hydrogen bond with Gly-148-CO, Gly-148-N, and Gln-125-Nϵ2. Hydrogen bonding between solvents 3 and 4 and neighboring protein atoms stabilize the single residue β-arc three and interconnect coils 3 and 4. Additionally, the carboxyamide ladder shared between Gln-124 of β9 (coil 2) and Asn-89 of β17 (coil 4) adds further stability to β12 (coil 3).
The second β-arc located between cysteines 144 and 147 established a new genre of two-residue bl β-arc, where a disulfide bond is implemented in place of the hydrogen-bonded asparagine residue as found within previously described two-residue N-bl arcs (36). Asp-145 and Gly-146, the two residues lying between C144 and C147 had ϕ, ψ dihedral angles of −159°, 90° (β-sheet region) and 80°, 31° (left-handed α-helix region), respectively. Additionally, a Phe residue (Phe-149) was found within the trailing parallel β-strand as reported for N-bl arcs (36). Solvent 2 has also been recruited to this β-arc, and it forms hydrogen bonds with main-chain atoms Gly-148-N and Gly-148-O. Solvent 2 also forms a hydrogen bond to Gln-125-Nϵ2 within the β-helix core, and this effectively stabilized β12 and tethered coils three and two together (Fig. 3). Additionally, the hydrogen bond between Gln-125-Nϵ2 and Tyr-134-OH within the β-helix core further stabilized the arc completing the third full coil. These residues appear unique to TpsA hemolysin members (Fig. 3).
The third coil within the HpmA265 structure also contained a single residue β-arc, involving asparagine 151, which lies between Ile-150 and Thr-152. A highly organized hydrogen bond network between solvent and protein atoms stabilized this single residue arc (Fig. 3). Solvent 3 formed hydrogen bonds to Ser-155-Oγ and Thr-152-Oγ1 of coil three. Solvent 3 also formed interactions with Thr-190-Oγ1 and Leu-193-O of the fourth coil. Solvent 4 formed hydrogen bonds to Thr-152-N, Asn-51-N, Thr-152-Oγ1, and Thr-190-Oγ1. In essence, the four solvent molecules (1–4) formed a series of hydrogen-bonded interactions that stabilized coils two through four.
Finally, stabilization of the third coil was assisted via a cross-coil carboxyamide ladder. This type of interaction has been documented and was referred to as Asn or Gln ladders in other β-helix structures (11, 18, 19). The ladder was established via a hydrogen bond between Gln-124 from β9 of the second coil and Asn-89 from β17 within coil four. So, this hydrogen bond effectively stretched over β12 of coil three, and thereby locked coils two and four together. To accommodate the carboxyamide ladder, a glycine residue (Gly-148) appears to be required within β12 just after the 144CXXC147 disulfide bond. Thus, the lack of side-chain atoms at position 148 likely assisted third coil stability and subsequent β-helix folding.
Template-assisted Hemolytic Assay
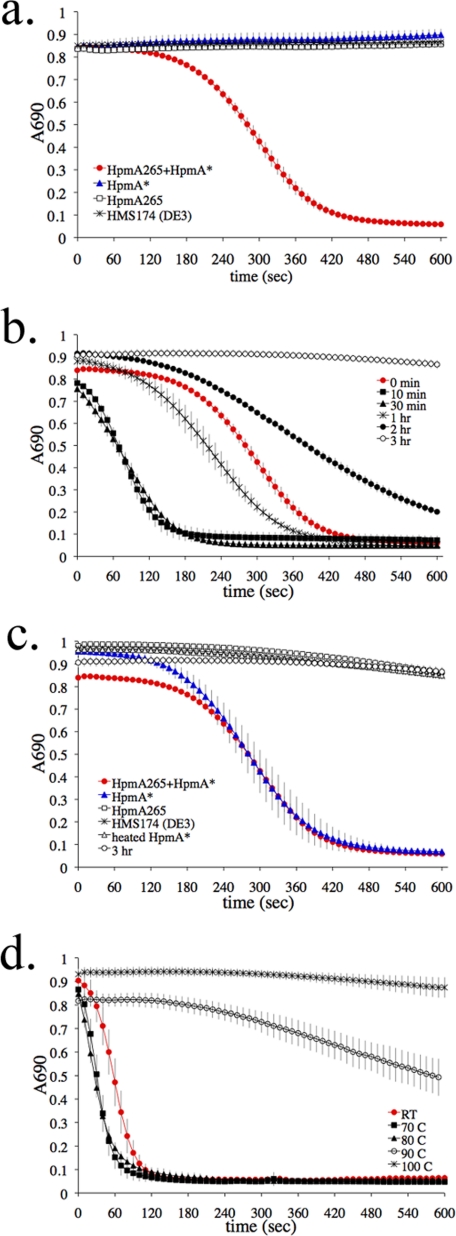
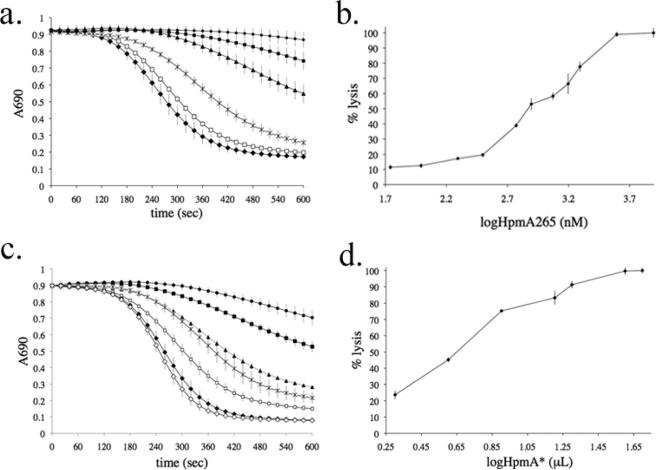
We used a microtiter plate hemolytic assay, which allowed high throughput and continuous kinetic analyses using minimal amounts of protein sample. The TAHA combined three critical components required for hemolysis: 1) HpmA265, a purified form of folded hemolysin A secreted in an HpmB-dependent fashion (Fig. 1a) that lacked the C-terminal pore-forming domain, 2) non-secreted full-length HpmA*, complete with the pore-forming domain obtained from an E. coli strain RAU126 (23), and 3) sheep red blood cells. By using these assays we found HpmA265 acted as a template to activate HpmA* (Fig. 4a, red circles) to form HpmA. We hypothesize HpmA265 aided in proper folding of the full-length exotoxin, and thereby activated hemolytic activity. In controls, HpmA265 (open squares) was unable to induce hemolysis and HpmA* (blue triangles) induced little hemolysis (Fig. 4a). Furthermore, lysate prepared from E. coli strain HMS 174 (DE3), the parent E. coli strain for RAU126 lacking HpmA* (X), was unable to complement HpmA265 and activate hemolysis. A note of clarification needs to be made regarding HpmA*, which unaided displayed hemolytic activity during long incubation times in the presence of red blood cells. For every hour of incubation between HpmA* and red blood cells the optical density at 690 nm dropped 0.1 (data not shown). Therefore, while HpmA* alone has low level activity, the rate of hemolysis was substantially increased after the addition of HpmA265.
FIGURE 4.
Template-assisted hemolysis. a, template-assisted hemolysis is induced by mixing HpmA265 in the presence of HpmA* and sheep red blood cells (●). The control samples were set up by mixing sheep red blood cells with (1) HpmA* (▲), (2) HpmA265 (□), or (3) HMS174 (DE3) lysate (×) with phosphate-buffered saline within a microtiter plate. b, hemolysis was monitored after preincubation of HpmA265 and HpmA* for the following time periods (10 min, 30 min, 1 h, 2 h, and 3 h). Long preincubation resulted in inactivation. c, add back experiments were conducted in the presence of HpmA**, the 3-h inactivated HpmA265/HpmA* (○) mixture shown in panel b. d, the temperature stability of HpmA265 was monitored prior to mixing with HpmA* and sheep red blood cells. Error bars represent standard deviations where n = 8.
From initial TAHA results (Fig. 4a, red circles), a biphasic activity profile was evident where a slow phase gave way to a much faster rate of hemolysis. The early slow phase was shortened by preincubating HpmA265 and HpmA* for 10–60 min before adding any sheep red blood cells (Fig. 4b, black squares, black triangles, and X). Longer periods of preincubation produced a newly discovered species, HpmA** (Fig. 4b, closed circles and open circles), that was hemolytically less active and resistant to HpmA265 reactivation (data not shown). To verify these losses in hemolytic activity were due to a HpmA265/HpmA* association longer than 60 min, HpmA265 and HpmA* were re-analyzed without preincubation and shown to establish the original activity profile.
A series of components were added to the 3-h preincubation samples containing the HpmA** species then retested by TAHA (Fig. 4c). Components added back were as follows: 1) HpmA265 (open squares), 2) HpmA* (blue triangles), 3) HMS174 (DE3) lysate (X), and 4) HpmA* heated to 60 °C (open triangles). Only additions of unheated HpmA* to the HpmA** sample were able to re-activate hemolysis (Fig. 4c). Intriguingly, little difference in the hemolysis rate was now found when adding either HpmA* or HpmA* preincubated with HpmA265. We observed in the past that HpmA* became inactivated in the absence of HpmA265 over a few days while being stored at 4 °C. Thus, progressive HpmA* inactivation was accelerated by HpmA265 during preincubation in the absence of red blood cells. We do not yet know if the HpmA** is structurally the same as non-HpmA265-inactivated HpmA*. To explore the stability of the β-helix fold indirectly via TAHA measurements, HpmA265 was treated to a variety of temperatures before co-incubation with HpmA*. Interestingly, only high temperatures above 90 °C seemed to have an effect (Fig. 4d).
To further establish that an interaction occurs between HpmA265 and HpmA*, we conducted a dose-response hemolytic analysis using the TAHA (Fig. 5a) and an end point assay (Fig. 5b) in the presence of a fixed amount of HpmA* and increasing amounts of HpmA265. First, although increasing amounts of HpmA265 within the TAHA sped hemolysis, we were unable to completely remove the slow portion of the previously observed biphasic activity. Second, the end-point hemolysis assay demonstrated a cooperative profile (Fig. 5b). In comparison, increasing amounts of HpmA* also failed to eliminate the slow portion of the biphasic hemolytic activity (Fig. 5c). However, the HpmA* dose-response curve does not display the cooperative behavior as reported for HpmA265 (Fig. 5d). Thus, HpmA265 acts in a cooperative manner to interconvert HpmA* → HpmA. However, the conformation of HpmA* is unable to cooperatively self-propagate the production of HpmA.
FIGURE 5.
HpmA265 and HpmA* dose-response curves. a and b, the dose-response curves contained amounts HpmA265 concentrations as follows: a, 7.85E1 (●), 1.57E2 (■), 3.14E2 (▲), 7.85E2 (*), 1.57E3 (□), 1.96E3 nm (♦) or b, 5.5E1 to 7.85E3 nm and fixed amounts of red blood cells and HpmA*. a, the HpmA265 dose-response was monitored using the TAHA. Increasing amounts of HpmA265 shorten, but do not eliminate the initial slow phase of hemolysis. b, the HpmA265 endpoint hemolysis assay was used to demonstrate a cooperative interaction between HpmA265 and HpmA*. c and d, represent HpmA* dose-response curves where HpmA* volumes were 2 (●), 4 (■), 8 (▲), 16 (*), 20 (□), 40 (♦), and 50 μl (◇) in the presence of a 3.92E2 nm HpmA265. c, similar to the HmpmA265 response, increasing amounts HpmA* shorten, but do not eliminate the slow phase. d, the HpmA* dose-response is non-cooperative. PBS was used to correct for all volume differences. Error bars represent standard deviations where n = 4.
Functional and Structural Role of the CXXC Motif
Using CD as a function of temperature, we then confirmed the exceptionally high thermal stability observed for HpmA265 (Fig. 4d).
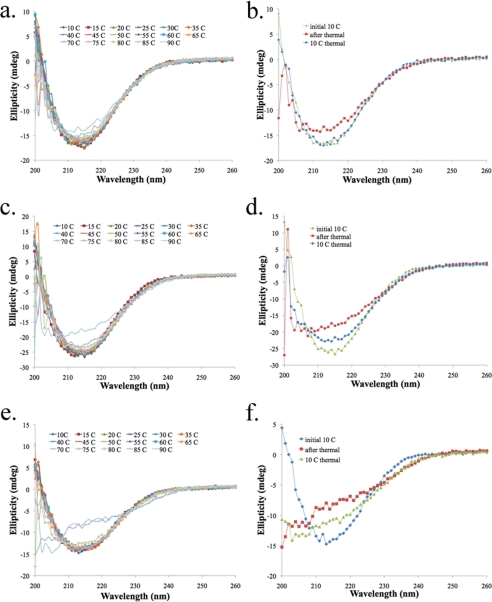
Double replacement mutants were constructed where both cysteine 144 and 147 were replaced with either alanine or serine residues. The resultant CXXC mutants have been identified as double alanine replacement (DMA) and double serine replacement (DMS), respectively. DMA and DMS were purified (Fig. 6a) and analyzed for hemolysis (Fig. 6c). Both CXXC mutants were also analyzed via native gel electrophoresis (Fig. 6b) and dynamic light scattering-size exclusion chromatography (data not shown). From these results replacement of the two cysteine residues did not alter the dimeric nature of HpmA265. Moreover, alteration of the two cysteines residues does not affect secretion of HpmA265 as has been previously determined within other TpsA hemolysins (12). However, alteration of the CXXC motif does slow the corresponding hemolytic activity (Fig. 6c). After the TAHA results were collected, HpmA265, DMA, and DMS were analyzed for secondary structure differences using far-UV-CD. A series of scans were conducted from 10 to 90 °C for each protein (Fig. 7). The stability of the HpmA265 β-helix is evident from the maintenance of secondary structure through the final scan at 90 °C (Fig. 7a). Additionally, HpmA265 was able to refold reversibly after reincubation 10 °C (Fig. 7b, 10 °C thermal). In comparison, DMA and DMS have less stability than HpmA265. The double alanine replacement, DMA, started to lose secondary structure at 90 °C (Fig. 7c) and was unable to completely refold (Fig. 7d, 10 °C thermal). The double serine replacement, DMS, was even more destabilized as it started to lose significant secondary structure at 85 °C and was unable to refold into the structure it had prior to thermal denaturation (Fig. 7f, 10 °C thermal). These results indicate that the CXXC motif stabilized the β-helix structure and afforded the ability to refold after thermal denaturation.
FIGURE 6.
Electrophoretic mobility and template-assisted activity of the CXXC mutants. Panels a and b reflect (a) denaturing and (b) native gel electrophoresis results for HpmA265 (lanes 2), DMA (lanes 3), and DMS (lanes 4). c, TAHA results for DMA and DMS in comparison to HpmA265. Error bars represent standard deviations where n = 4.
FIGURE 7.
Comparative far-UV-CD results for HpmA265, DMA, and DMS. a, c, and e represent far-UV-CD results during thermal scans ranging from 10 to 90 °C for HpmA265, DMA, and DMS, respectively. b, d, and f represent an initial baseline scan at 10 °C (initial 10 °C scan), a scan after thermal denaturation at 90 °C (after thermal), and a scan after cooling the protein back to 10 °C (10 °C thermal) for HpmA265, DMA, and DMS, respectively.
Laser Scanning Confocal Microscopy
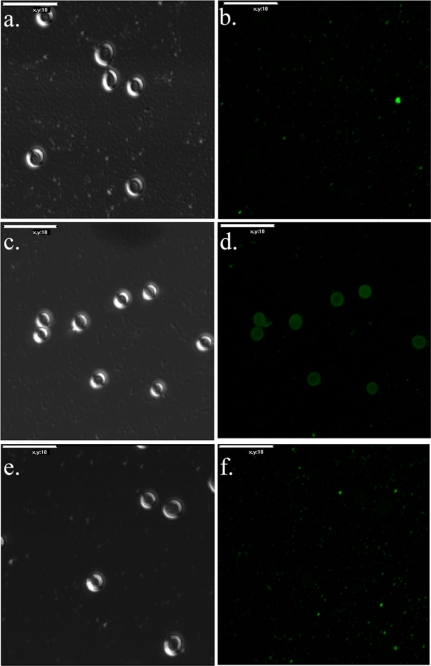
Based upon the TAHA results three forms of full-length HpmA have now been identified: 1) HpmA*, 2) HpmA, and 3) HpmA**. The accelerated formation of two of these forms, HpmA and HpmA**, was shown to be HpmA265-dependent. Fixed red blood cells were reacted with either HpmA*, HpmA, or HpmA** from either an E. coli RAU126 crude lysate or a RAU126/HpmA265-mixed lysate. The hemolytically active hemolysin, HpmA, bound to the surface of red blood cells to a much greater extent (Fig. 8d) then either HpmA* (Fig. 8b) or HpmA** (Fig. 8f). Both HMS174 (DE3) lysate, which is the E. coli parent strain for the source of HpmA*, and HpmA265, were also tested, and neither provided any fluorescence on the red blood cell surface (data not shown). These results supported a structural alteration of HpmA* → HpmA → HpmA** via HpmA265.
FIGURE 8.
HpmA265 induced alterations of HpmA* lead to differences in red blood cell binding. Fixed red blood cells were incubated in the presence of HpmA*, HpmA, and HpmA**. Both differential interference contrast (DIC) and fluorescence images have been provided. a and b, DIC and fluorescence images for HpmA*-reacted cells. c and d, DIC and fluorescence images for HpmA reacted cells, where d noted a significant increase in fluorescence of the red blood cells. e and f, DIC and fluorescence images for HpmA**-reacted cells. Based upon these results, the HpmA265 template-assisted activation was attributed to enhanced red blood cell binding capacity.
DISCUSSION
The monomeric β-helix architecture has been proposed to be a very common passenger domain for Type V secretory proteins. This structural topology is evolutionarily conserved as 97% of proteins secreted from pathogenic Gram-negative bacteria are predicted to form parallel β-helices. The final step of Type V protein secretion is an energy-independent C → N-terminal threading of the passenger domain through the outer membrane, effected in part by the C-terminal domain, along with or followed by C → N-terminal folding. The C-terminal rungs of the β-helix have been proposed to act as a template during membrane secretion, and these rungs could provide the requisite interactions needed to circumvent energy dependence during β-helix translocation (37). Similar to the current model for Type V protein secretion, our results provide evidence that the HpmA β-helix architecture functioned as a template. However, we further extend the current model to include a two-stage process. During stage 1, monomeric HpmA265 interacts cooperatively within multiple regions of HpmA* creating a dry, dimeric interface. At stage 2 these cooperative interactions eventually induce a conformational change leading to increased red blood cell binding capacity and subsequent hemolysis.
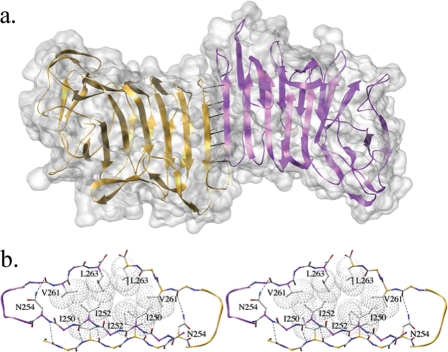
First, we know that HpmA265 facilitates the HpmA* → HpmA conversion. Previous results with ShlA, a closely related homolog of HpmA, have suggested both monomers and dimers were hemolytic. In these studies, gel filtration in the presence of 6 m urea detected monomers, dimers, tetramers, and even larger oligomers. Furthermore, the fraction representing ShlA monomers had a higher specific activity than the dimer fraction (14). From these results, monomeric ShlA was proposed to insert into the red blood cell membrane and then assemble into oligomers with increased pore size. From our own results, we know that an oligomeric form of HpmA265 exists in solution. Therefore, we used the unit cell to analyze for putative HpmA265 dimer interfaces. Based upon unit cell packing, HpmA265 forms three crystallographic interfaces. Two interacting surfaces involve very few protein-protein interactions and are filled with a number of bound waters (wet dimers). The third dimer forms a completely desolvated dry interface. The dry dimer forms the most stable of the three interfaces and main-chain hydrogen bonding between exposed edge β-strands of neighboring monomers facilitates dimerization. In this fashion, the β-helix fold propagates an anti-parallel β-sheet arrangement leading to a fibril appearance (Fig. 9a). Beyond main-chain hydrogen bonding interactions, this dimer stacks a series of inner core hydrophobic residues, including Met-245, Ile-250, Ile-252, and Val-261 to create a dry, sterically compatible protein interface of 712 Å2 (Fig. 9b). We suggest that this unit cell species is the dimer observed within the dynamic light scattering-size exclusion chromatography experiments conducted with purified HpmA265, because unprotected β edge strands, bordered on only one side by another β-strand, are prone to forming dimeric interfaces (38).
FIGURE 9.
HpmA265 crystallographic dry dimer interface leading to a filamentous appearance. a, the HpmA265 dry dimer forms a filamentous structure. The solid lines represent hydrogen bonds shared between β23 strands of both subunits. b, the dry dimer interface has been displayed where β23 from each monomer has been colored independently. The anti-parallel interstrand hydrogen-bonding network creates dry, sterically compatible dimer interface. Dashed lines represent hydrogen bonds, while van der Waal's surfaces have been provided for various hydrophobic side chains.
The presence of the CXXC pattern within all known TpsA hemolysin members has been confusing, because alteration of these residues does not affect TpsB-dependent secretion. CXXC motifs in other proteins have been shown to destabilize their structure and provide redox-dependent signaling and catalytic function (39). Based upon the far-UV-CD results, we propose an energetic argument to support the high conservation of cysteines 144 and 147 within the TpsA hemolysin family. From earlier investigations, there was found to be a large entropic barrier associated with the assembly of the β-helix, which was associated with a characteristic lag phase during folding (40). From crude estimates, the transition-state complex leading to successful β-helix formation was three strands or coils, and if a four-stranded coil were formed, additional helical coils would add rapidly to extend the β-helix (41).
Additionally, the hydrogen bond strength was shown to increase in a non-linear fashion within the first four coils, and this was surmised to reflect cooperativity among adjacent β-sheets (42). The far-UV-CD with HpmA25, DMA, and DMS suggests that the disulfide shared between Cys-144 and Cys-147 may have an intertwined dual role within the TpsA hemolysin family. First, replacement of the CXXC motif thermally destabilizes the β-helix structure so that loss of secondary structure occurs at lower temperatures (Fig. 7, a, c, and e). In fact DMA and DMS behave quite differently. The double alanine replacements at positions 144 and 147 are better accommodated than the isosteric serine residues. From the HpmA265 structure this makes sense structurally and chemically. The β-arc spanning cysteine 144 through cysteine 147 has a fairly compact arrangement, and thus, the smaller alanine side chain would fit better into this region and, thereby, maintain β-helix stability. Also, most of the internal environment nearest this arc is hydrophobic. Thus, the polar serine residue would lead to decreased stability and inability to refold after thermal denaturation (Fig. 7, e and f). This is not unexpected, because the polar serine would destabilize the hydrophobic internal core of the β-helix. Therefore, the disulfide appears to thwart complete unfolding and thereby, preserves the requisite first three helical coils leading to rapid refolding of the β-helix (Fig. 7, b, d, and f).
Thus, the entropic barrier of refolding HpmA265 is overcome by maintaining the CXXC disulfide bond within the second β-arc of coil three. Furthermore, all four buried solvent molecules (1–4) are located within the third coil and thereby, provide increased structural integrity to this region (Fig. 3). In fact, we propose these buried solvents to be an intrinsic part of the TpsA hemolysin β-helix fold. The CXXC disulfide bond, along with four solvent molecules, may prevent complete unfolding of the β-helix, and thereby maintain the HpmA multivalent activation surface required for the template-assisted activity we observed.
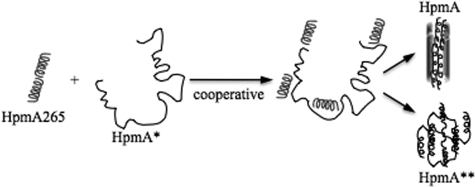
From our results, we propose that the dry, dimeric interface acts as the template to cooperatively assist the HpmA* → HpmA conversion. First, HpmA265, stabilized within the third coil by a CXXC disulfide bond, hydrogen-bonded solvent molecules, and a cross-coil carboxyamide ladder, would cooperatively implement parallel β-strand interactions at multiple rungs with neighboring HpmA* during conformational change into HpmA (Fig. 10). Evidence for the β-helix structure with multiple rungs has been suggested as a model for the full-length structure of the adhesion TpsA member, Fha (21). Using in register exposed β-strands like we propose for TpsA activation is also analogous to the protein tau filament growth theory. In this theory, unstructured protein Tau forms a series of on edge intermolecular β-strand hydrogen bonds with an exposed tau filament (17). Zipper-like interactions between exposed tau filament β-strands and the new protein layer facilitate growth. Thus, at increasing concentrations, monomeric HpmA265 binds multiple HpmA* rungs through a dry interface, and thereby, facilitates a cooperative and unidirectional conformational change of HpmA* → HpmA. Prolonged incubation of HpmA* and HpmA265 in the absence of red blood cells irreversibly establishes a newly discovered and hemolytically incompetent species termed, HpmA**. Our model also has HpmA* acting non-cooperatively (Fig. 10) and analogously to ShlA, where the assembly of oligomers and subsequent pore formation was preceded by the insertion of ShlA* monomers into the membrane (14). We suggest that the insertion of ShlA* monomers into the membrane, as previously reported, would effectively elevate the local ShlA* concentration and thereby, mimic our cooperative response reported for HpmA. Ultimately, the activation of hemolysis stems from the ability of HpmA265 to cooperatively enhance the red blood cell binding capacity of full-length hemolysin A (Fig. 8d). Similarly, the conversion of ShlA* → ShlA was proposed to be conformationally dependent based upon gel-filtration chromatography, protease sensitivity, CD, intrinsic fluorescence, and electrophoretic mobility (16).
FIGURE 10.
Model for HpmA265 template-assisted hemolysis. HpmA265 acts cooperatively via intermolecular β-strand interactions to bind multiple positions within HpmA*. These interactions 1) conformationally change HpmA* → HpmA resulting in productive red blood cell binding and subsequent hemolysis or 2) in the absence of red blood cells, interconvert HpmA* → HpmA → HpmA**, where HpmA** is unable to bind red blood cells and promote hemolysis. The time required for HpmA265 to bind and change HpmA* → HpmA would be attributed to the lag during hemolysis. Once HpmA has been established, in the presence or absence of red blood cells, hemolysis ensues rapidly.
Finally, from our results we suggest that the evolutionary pressure to recruit and maintain the right-handed, parallel β-helix architecture complete with the CXXC disulfide bond resides from both its structural stability and its ability to interact cooperatively with mis-folded neighboring molecules. These combined attributes would preserve functional TpsA virulence factors, like adhesions and hemolysins for numerous Gram-negative bacteria. Additional studies are needed to determine 1) the HpmA265 interface responsible for HpmA* activation, 2) the stoichiometry of HpmA265/HpmA* interactions, and 3) the hemolytically competent oligomeric state of HpmA. In conclusion, this work establishes a new model for red blood cell binding and hemolysis for the TpsA hemolysin family.
Acknowledgment
We thank Scott Cooper (University of Wisconsin-La Crosse, Dept. of Biology) for many helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AI057437 (to T. W.). This work was also supported by National Science Foundation Grant MCB0744754 (to T. W.), by the University of Wisconsin (UW)-La Crosse Faculty Research Program (to T. W.), and by a UW-La Crosse, College of Science and Health Dean's Summer Fellowship (to G. W.) supported the work. A National Science Foundation Grant DBI619289 (to Jennifer Miskowski, UW-La Crosse, Dept. of Biology) supported the laser scanning confocal microscopy studies.
The atomic coordinates and structure factors (code 3FY3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HpmA
- hemolysin A
- HpmB
- hemolysin B
- FHA
- hemagglutinin from B. pertussis
- HMW1
- high molecular weight protein from H. influenzae
- Fha30
- 301-residue N-terminal FHA fragment
- TAHA
- template-assisted hemolytic assay
- PBS
- phosphate-buffered saline
- DMA
- double alanine mutant
- DMS
- double serine mutant
- DIC
- differential interference contrast.
REFERENCES
- 1.Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen M. R., Peabody C. R., Partovi S. M., Zhai Y., Tseng Y. H., Saier M. H. (2002) Biochim. Biophys. Acta 1562, 6–31 [DOI] [PubMed] [Google Scholar]
- 3.Hertle R., Hilger M., Weingardt-Kocher S., Walev I. (1999) Infect. Immun. 67, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertle R. (2002) J. Membr. Biol. 189, 1–14 [DOI] [PubMed] [Google Scholar]
- 5.Schiebel E., Braun V. (1989) Mol. Microbiol. 3, 445–453 [DOI] [PubMed] [Google Scholar]
- 6.Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., Villeret V. (2007) Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 7.Hertle R. (2005) Curr. Protein Pept. Sci. 6, 313–325 [DOI] [PubMed] [Google Scholar]
- 8.Hertle R. (2000) Curr. Protein Pept. Sci. 1, 75–89 [DOI] [PubMed] [Google Scholar]
- 9.Jacob-Dubuisson F., Locht C., Antoine R. (2001) Mol. Microbiol. 40, 306–313 [DOI] [PubMed] [Google Scholar]
- 10.Thanassi D. G., Stathopoulos C., Karkal A., Li H. (2005) Mol. Membr. Biol. 22, 63–72 [DOI] [PubMed] [Google Scholar]
- 11.Clantin B., Hodak H., Willery E., Locht C., Jacob-Dubuisson F., Villeret V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6194–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönherr R., Tsolis R., Focareta T., Braun V. (1993) Mol. Microbiol. 9, 1229–1237 [DOI] [PubMed] [Google Scholar]
- 13.Hertle R., Süssmuth R., Braun V., Jung G. (2000) J. Chromatogr. B Biomed. Sci. Appl. 737, 13–23 [DOI] [PubMed] [Google Scholar]
- 14.Schönherr R., Hilger M., Broer S., Benz R., Braun V. (1994) Eur. J. Biochem. 223, 655–663 [DOI] [PubMed] [Google Scholar]
- 15.Hertle R., Brutsche S., Groeger W., Hobbie S., Koch W., Könninger U., Braun V. (1997) Mol. Microbiol. 26, 853–865 [DOI] [PubMed] [Google Scholar]
- 16.Walker G., Hertle R., Braun V. (2004) Infect. Immun. 72, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margittai M., Langen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeo H. J., Yokoyama T., Walkiewicz K., Kim Y., Grass S., Geme J. W., 3rd (2007) J. Biol. Chem. 282, 31076–31084 [DOI] [PubMed] [Google Scholar]
- 19.Mayans O., Scott M., Connerton I., Gravesen T., Benen J., Visser J., Pickersgill R., Jenkins J. (1997) Structure 5, 677–689 [DOI] [PubMed] [Google Scholar]
- 20.Wilson D. R., Finlay B. B. (1997) Protein Eng. 10, 519–529 [DOI] [PubMed] [Google Scholar]
- 21.Kajava A. V., Cheng N., Cleaver R., Kessel M., Simon M. N., Willery E., Jacob-Dubuisson F., Locht C., Steven A. C. (2001) Mol. Microbiol. 42, 279–292 [DOI] [PubMed] [Google Scholar]
- 22.Deleted in proof [Google Scholar]
- 23.Uphoff T. S., Welch R. A. (1990) J. Bacteriol. 172, 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernheimer A. W. (1988) Methods Enzymol. 165, 213–217 [DOI] [PubMed] [Google Scholar]
- 25.Young J. D., Leong L. G., DiNome M. A., Cohn Z. A. (1986) Anal. Biochem. 154, 649–654 [DOI] [PubMed] [Google Scholar]
- 26.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 28.Vagin A. A., Isupov M. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Steiner R. A., Lebedev A. A., Murshudov G. N. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 2114–2124 [DOI] [PubMed] [Google Scholar]
- 31.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 32.Swihart K. G., Welch R. A. (1990) Infect. Immun. 58, 1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F. L., Braun V. (2000) Int. J. Med. Microbiol. 290, 529–538 [DOI] [PubMed] [Google Scholar]
- 34.Braun V., Ondraczek R., Hobbie S. (1993) Zentralbl. Bakteriol. 278, 306–315 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Skolnick J. (2005) Nucleic Acids Res. 33, 2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennetin J., Jullian B., Steven A. C., Kajava A. V. (2006) J. Mol. Biol. 358, 1094–1105 [DOI] [PubMed] [Google Scholar]
- 37.Junker M., Schuster C. C., McDonnell A. V., Sorg K. A., Finn M. C., Berger B., Clark P. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siepen J. A., Radford S. E., Westhead D. R. (2003) Protein Sci. 12, 2348–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wouters M. A., George R. A., Haworth N. L. (2007) Curr. Protein Pept. Sci. 8, 484–495 [DOI] [PubMed] [Google Scholar]
- 40.Cellmer T., Douma R., Huebner A., Prausnitz J., Blanch H. (2007) Biophys. Chem. 125, 350–359 [DOI] [PubMed] [Google Scholar]
- 41.Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., Eisenberg D. (2005) Nature 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsemekhman K., Goldschmidt L., Eisenberg D., Baker D. (2007) Protein Sci. 16, 761–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]