Abstract
Noonan syndrome (NS) is an autosomal dominant congenital disorder characterized by multiple birth defects including heart defects and myeloproliferative disease (MPD). Approximately 50% of NS patients have germline gain-of-function mutations in PTPN11, which encodes the protein-tyrosine phosphatase, Shp2. We provide evidence that conditional ablation of Stat3 in hematopoietic cells and cardiac valvular tissues leads to myeloid progenitor hyperplasia and pulmonary stenosis due to the leaflet thickening, respectively. Consistently, STAT3 activation is significantly compromised in peripheral blood cells from NS patients bearing Shp2-activating mutations. Biochemical and functional analyses demonstrate that activated Shp2 is able to down-regulate Tyr(P)-Stat3 and that constitutively active Stat3 rescues activating mutant Shp2-induced granulocyte-macrophage colony-stimulating factor hypersensitivity in bone marrow cells. Collectively, our work demonstrates that Stat3 is an essential signaling component potentially contributing to the pathogenesis of NS and juvenile myelomonocytic leukemia caused by PTPN11 gain-of-function mutations.
Noonan syndrome (NS)4 is an autosomal dominant disorder characterized by unusual facial characteristics, short stature, heart defects, skeletal malformations, bleeding problems (1), and patients are also susceptible to myeloproliferative disorder, which occasionally progress to juvenile myelomonocytic leukemia (JMML) (2, 3). Gene sequencing studies demonstrated that about 50% of individuals with NS have germline mutations in the PTPN11 (1, 4, 5), which encodes the protein-tyrosine phosphatase, Shp2. Shp2 plays an important role in receptor-tyrosine kinase-mediated signaling pathways, especially in signaling through RAS-mitogen-activated protein kinase (MAPK) pathway (6), and is believed to be important to cell proliferation, differentiation, and migration (7). The contribution of Shp2 to NS was further confirmed using mutant mice bearing a common PTPN11 mutation, in which amino acid 61 was changed from aspartate to glycine (Shp2D61G). Shp2D61G mice largely recapitulate the NS phenotypes (8). In addition, ∼35% of children with JMML bear somatic activating PTPN11 mutations (9–11). These PTPN11 mutations found in NS and JMML patients are gain-of-function mutations that cause persistent activation of Shp2 phosphatase activity and up-regulate RAS-MAPK-mediated signaling (12, 13). In the past few years, several other genes coding for signaling transducers in the RAS/MAPK signaling pathways have also been found in a smaller fraction of NS patients, which include SOS1, KRAS, RAF1, and BRAF (1, 14–18). Interestingly, although common clinical features were found in NS patients with different genetic mutations, substantial phenotypic variations were observed. For instance, pulmonic stenosis is more commonly observed among individuals with mutations in the PTPN11 allele, whereas hypertrophic cardiomyopathy is more closely associated with RAF1 mutations (18, 19). These clinical observations suggested that multiple genetic factors contribute to the various phenotypes of NS and/or JMML.
Signal transducer and activator of transcription 3 (Stat3) belongs to a family of transcription factors that regulates a broad range of biological processes. In general, STAT activation is through the phosphorylation of a single tyrosine residue (Tyr705) that results in Src homology 2 domain-mediated dimerization, nucleus localization, DNA binding, and ultimately transcription activation or repression (20). Stat3 can be activated by receptor-tyrosine kinases, such as epidermal growth factor receptor (EGFR), cytokine receptor-associated Janus kinases or Src kinases. It plays a convergent role in integrating multiple signaling pathways. Ablation of Stat3 in mice gave rise to defects in embryonic pattern formation and early lethality (21). Stat3 has been generally viewed as a cellular survival factor or an oncogenic factor (21). However, the observations that activation of Stat3 is required for apoptotic action of interferon-β in primary murine pro-B cells (22), and that deletion of Stat3 in mammary gland epithelial cells led to the delay of involution due to the reduction of apoptosis (23) suggest that Stat3 plays a more complicated role in the regulation of cell survival and proliferation.
Previously, we generated mutant mice in which Stat3 was ablated specifically in the hematopoietic cell lineage using btie2-cre mice (Stat3f/f/btie2-cre) (24, 25). Interestingly, our initial characterization revealed that the Stat3f/f/btie2-cre mice demonstrated an increase in autonomous myeloid cell proliferation (24), similar to that observed in the Shp2D61G mutant mice (8), indicating that loss of Stat3 function contributes to phenotypic anomalies observed in NS. To follow up this initial observation and to further delineate the potential function of Stat3 in the pathogenesis of NS, we conducted a series of analyses on conditional Stat3 mutant mice, as well as examined the biochemical regulation of Stat3 by Shp2 phosphatase. Collectively, our data demonstrate that Stat3 is a direct target for Shp2 and is potentially an essential signaling component in PTPN11 mutation-associated NS and JMML.
MATERIALS AND METHODS
Mutant Mice and Histological Analysis
Stat3f/f/btie2-cre and Stat33f/f/nestin-cre mice were generated as previously described (24, 26). Embryos were harvested by cesarean section. Embryos and isolated hearts were fixed in 10% neutral buffered formalin, paraffin embedded and sectioned (6 μm), and stained with hematoxylin and eosin. To analyze phospho-Stat3 expression in the developing heart valve, we used a rabbit monoclonal antibody against phosphor-Stat3 (Tyr705) (Cell Signaling, D3A7) and a Vector staining system (Vector, PK-2200) according to the manufacturer's instructions.
NS and Healthy Control Blood Specimen Collection
Peripheral blood samples were collected in sodium heparin tubes. The Institutional Review Board at the Indiana University School of Medicine approved all protocols. Informed consent was obtained from donors or from the guardian of children less than 18 years of age in accordance with the Declaration of Helsinki.
Flow Cytometric Analyses
Single cell suspensions from BM or spleen were prepared as previously described (24). Aliquots of cells (1–2 × 106) were stained with fluorescence-conjugated antibodies specifically against Mac-1 (CD11b), Gr-1, or F480 (Pharmingen). For human peripheral blood tyrosine-phosphorylated STAT3 staining, 300 μl of human whole blood were incubated with IL-6 for 30 min, followed by treatment with fix/lysis and permeabilization buffer III (Pharmingen) according to manufacturer's instructions. Cells were then stained with phycoerythrin-conjugated antibody against Tyr(P)705-STAT3 (Pharmingen). Flow cytometry was performed using a FACScalibur (BD Biosciences), and data were analyzed using CELLQUEST software.
Retroviral Transduction
The retroviral vectors pMIEG3, pMIEG3-WT Shp2, pMIEG3-N308D, pMIEG3-E76K, and pMIEG3-hCD4 have been reported previously (27, 28). pMIEG3- Stat3CA-hCD4 was constructed by insertion of Stat3CA into the EcoRI site of pMIEG3-hCD4. Ecotropic retroviral supernatants were prepared using Eco-Phoenix packaging cells. Bone marrow low density mononuclear cells were isolated and transduced with pMIEG3, pMIEG3-WT Shp2, pMIEG3-N308D, pMIEG3-E76K, pMIEG3-hCD4, and pMIEG3-Stat3CA-hCD4, as previously described (27). Following transduction, cells were sorted for EGFP alone or EGFP and human CD4 antigen co-expression using fluorescence-activated cell sorting (FACS) to enrich for transduced cells (supplemental Fig. 6). The expression of Stat3 and Shp2 were further confirmed by Western blot analysis (supplemental Fig. 6).
BM Proliferation, Apoptosis, and Colony-forming Assay
BM single cell suspensions were prepared from Stat3f/f and Stat3f/f/ btie2-cre as previously described (24). For the BM cell proliferation assay, 7.5 × 104 cells of each genotype were plated per well in 96-well plates, subjected to growth factor and serum deprivation for 5 h, cultured with various cytokines for 16 h, pulsed with 1.0 μCi (0.037 MBq) of [3H]thymidine for 24 h, and harvested using an automated 96-well cell harvester (Brandel, Gaithersburg, MD) for counting thymidine incorporation (counts/min). To assay cell survival, cells were deprived of serum and growth factor for 5 h followed by treatment with granulocyte-macrophage colony stimulating factor (GM-CSF) at 0.1 and 1 ng/ml for 24 h and analyzed for apoptosis staining with Annexin-V (BD Biosciences). Colony assays were performed using MethoCult M3434 (Stem Cell Technology, Vancouver) in the absence or presence of increasing concentrations of GM-CSF (Peprotech, Rocky Hill, NJ), as previously described (27).
Western Blot and Electrophoretic Mobility Shift Assay (EMSA)
Western blots were performed using standard protocols. Phospho-Stat3 (Tyr(P)705) and phospho-p44/42 (Thr202/Tyr404) antibodies were from Cell Signaling Technologies (Beverly, MA). Anti-Stat3 (C-20) was from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-glyceraldehyde-3-phosphate dehydrogenase was from Biodesign International (Saco, ME). Stat3 EMSA was performed according the protocol previously described (29).
Shp2 Inhibitor IIB-08
The small molecule Shp2 inhibitor IIB-08 was identified from a small focused library of benzofuran salicylic acid derivatives. The design and synthesis of the library, and the identification and characterization of IIB-08 as a Shp2 inhibitor will be described elsewhere. IIB-08 exhibits an IC50 of 5.5 μm for Shp2 with little activity against a panel of mammalian PTPs, including Lyp, CD45, Cdc14, MKP3, and VHR.
In Vitro Phosphatase Assay
Raw 264.7 (mouse monocyte macrophage cell line) cells were treated with IL-6 (20 ng/ml) for 30 min before being subjected to lysis in a solution containing 10 mm Hepes, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 10 mm sodium fluoride, 1 mm Na3VO4. After centrifugation, cell extract was incubated with anti-Stat3 C-20 antibody (Santa Cruz Biotechnology) for Stat3 protein isolation. Stat3 immunoprecipitates were washed three times with 50 mm 3,3-dimethylglutarate buffer, pH 7.0. Two μg of recombinant constitutively active Shp2 protein (catalytic domain) was added to each sample and the reactions were incubated at 37 °C for the indicated time in 50 mm 3,3-dimethylglutarate buffer, pH 7.0. Reactions were quenched by adding SDS-loading buffer and boiling at 95 °C for 10 min. Samples were separated by SDS-PAGE and Stat3 dephosphorylation was determined by Western blotting analysis using anti-Tyr(P)705-Stat3 antibodies.
RESULTS
Ablation of Stat3 Contributes to Prominent Hypercellularity with a Mature Myeloid Predominance and Hypersensitivity to GM-CSF
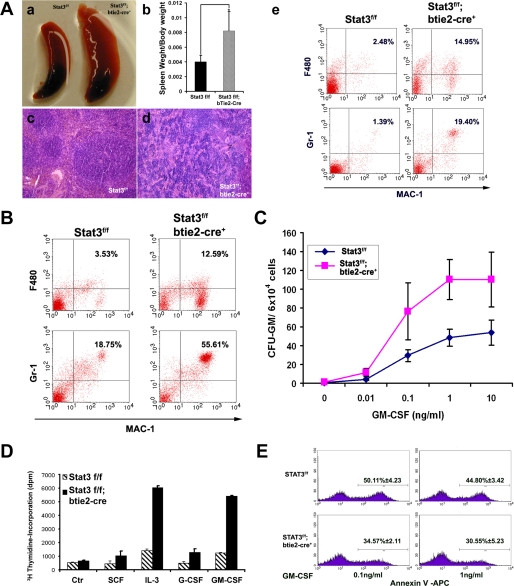
To further confirm and evaluate the role of Stat3 in myeloid cell function, we re-examined the hematopoietic compartment of Stat3f/f/btie2-cre mice. Stat3f/f/btie2-cre newborns were normal in size at birth, but became visibly smaller compared with littermates by 2 weeks of age, rapidly became sick, and died between 4 and 6 weeks of age. By gross anatomical analysis, we found that Stat3f/f/btie2-cre mice had dramatically enlarged spleens (Fig. 1A, a and b). Histology of these enlarged spleens revealed disruption of the white and red pulp architecture (Fig. 1A, c and d). Flow cytometric analysis of 3-week-old spleens demonstrated a significant expansion of both neutrophil (Mac-1+Gr-1+ cells, a 5.59-fold increase) and macrophage (Mac-1+F480+ cells, a 13.95-fold increase) in the Stat3f/f/btie2-cre spleens when compared with controls (Fig. 1A, e). Similarly, the frequency of Mac-1+F480+ and Mac-1+Gr-1+ cells in Stat3f/f/btie2-cre bone marrow was also significantly increased (Fig. 1B).
FIGURE 1.
Ablation of Stat3 in bone marrow cells gives rise to skewed myeloid cell differentiation and hypersensitivity to GM-CSF. A, morphological, histological, and flow cytometric analyses of spleens from Stat3f/f/btie2-cre and Stat3f/f littermate control mice. Panel a, splenomagaly was found in Stat3f/f/btie2-cre mice; panel b, comparison of spleen weight to body weight ratio of Stat3f/f and Stat3f/f/btie2-cre mice, p < 0.01 (Student's t test, 2 tails); panels c and d, disrupted spleen architecture in Stat3f/f/btie2-cre mice (d) when compared with Stat3f/f controls (c); panel e, both F480+Mac1+ and GR-1+Mac1+ populations were significantly increased in Stat3f/f/btie2-cre spleens compared with the Stat3f/f control spleens. B, bone marrow F480+Mac1+ and GR-1+Mac1+ populations were increased in the Stat3f/f/btie2-cre mice compared with Stat3f/f control mice. C, methylcellulose-based colony forming assays demonstrated increased bone marrow progenitor-derived colonies in response to increasing doses of GM-CSF from the Stat3f/f/btie2-cre mice compared with the Stat3f/f mice. D, increased proliferative activity of Stat3f/f/btie2-cre bone marrow cells in response to various cytokines. The final concentration of all cytokines, except IL-3 (100u/ml), was 30 ng/ml. E, decreased apoptosis in Stat3f/f/btie2-cre bone marrow cells when cultured in a low serum in the presence of 0.1 or 1 ng/ml of GM-CSF. Error bars represent S.D.
Peripheral blood progenitor cells from JMML patients and murine hematopoietic progenitors ectopically expressing gain-of-function Shp2 mutants demonstrate increased sensitivity to GM-CSF (8, 27, 30–32). To test this in hematopoietic progenitors lacking Stat3, we performed methylcellulose colony assays with increasing concentrations of GM-CSF (0–10 ng/ml). Similar to JMML patients or gain-of-function Shp2-transduced murine hematopoietic cells (27, 31), Stat3f/f/btie2-cre bone marrow low density mononuclear cells had significantly increased sensitivity to GM-CSF at concentrations as low as 0.1 ng/ml for colony-forming units granulocyte-macrophage formation (Fig. 1C). Because Stat3f/f/btie2-cre mutants showed a dramatic increase in myeloid progenitor cell population, we further tested the responses of Stat3f/f/btie2-cre and control bone marrow cells to a panel of cytokines (SCF, IL-3, G-CSF, and GM-CSF) in vitro. [3H]Thymidine incorporation assays demonstrated that Stat3f/f/btie2-cre mutant cells had A significantly higher labeling index in response to growth factor stimulation, especially to GM-CSF and IL-3 (Fig. 1D). In the presence of low concentrations of GM-CSF (0.1 and 1 ng/ml), Stat3f/f/btie2-cre mutant cells also demonstrated a reduction in apoptosis when compared with control cells (Fig. 1E). These data strongly suggest that the hematopoietic phenotypic is similar between NS and Stat3f/f/btie2-cre mutants.
Mutant Shp2 Negatively Regulates Stat3 Activation in Response to GM-CSF
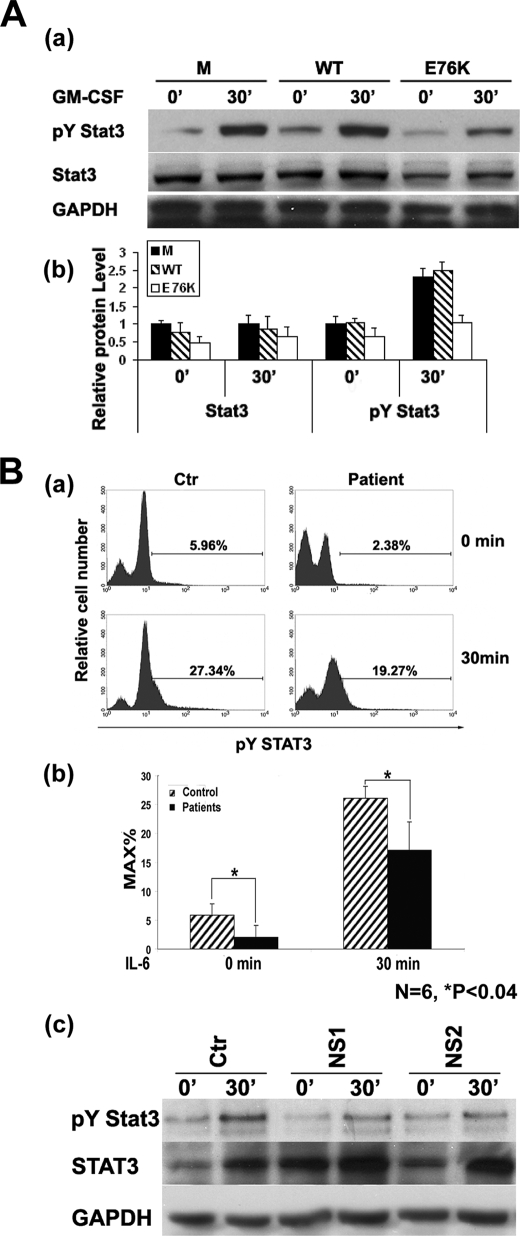
Previous studies have shown that ablation of Shp2 enhances Stat3 activation (33–35). To test whether Shp2 negatively regulates Stat3 activation in hematopoietic progenitors, we retrovirally transduced low density mononuclear cells with empty vector (MIEG3), WT Shp2, or Shp2E76K, one of the most common activating PTPN11 mutations observed in JMML (9, 10, 13, 36), sorted for transduced enhanced green fluorescent protein (EGFP)-expressing cells, and cultured into macrophage progenitors as previously described (27). Cells were serum-deprived, stimulated with GM-CSF, and analyzed for activation of Stat3 (Tyr(P)705) and expression of total Stat3. At baseline and upon GM-CSF stimulation, Shp2E76K-expressing macrophage progenitors expressed significantly lower total Stat3 as well as activated Stat3 when compared with MIEG3- or WT Shp2-transduced cells (Fig. 2A). This inhibition of Stat3 expression and function in Shp2E76K-transduced cells suggests that Shp2 plays a critical negative regulatory role in Stat3-mediated signaling in hematopoietic cells.
FIGURE 2.
Shp2 gain-of-function mutants negatively regulate Stat3 activation in murine bone marrow cells and peripheral blood nucleated cells from individuals with NS. A, panel a, Western blot analyses examining GM-CSF-stimulated (20 ng/ml) Stat3 activation in the mouse bone marrow-derived macrophage progenitors transduced with MIEG3 (empty vector), WT Shp2, or Shp2E76K. Panel b, density quantification of protein band in A, panel a; B, panel a, representative flow cytometric analysis showing reduced Stat3 tyrosine phosphorylation in peripheral blood cells from individuals with NS in both the quiescent state and after IL-6 stimulation. Panel b, statistical analysis of six NS patients with confirmed PTPN11 mutations and six sex- and age-matched normal controls (p < 0.04, statistics performed using unpaired Student's t test). Panel c, Western blot analyses examining basal and IL-6- (50 ng/ml) stimulated STAT3 phosphorylation and total STAT3 protein level in nucleated peripheral blood cells from control and individuals with NS. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Error bars represent S.D. Asterisks indicate the difference is statistically significant.
Activated Tyr(P)-Stat3 Level Is Significantly Lower in NS Patients
To further investigate whether reduced STAT3 function is associated with the pathogenesis of NS in humans, we collected peripheral blood samples from five NS patients with sequence confirmed PTPN11 gain-of-function mutations and six age- and sex-matched individuals lacking any congenital disease diagnosis and assayed baseline and IL-6-stimulated Tyr(P)-STAT3 (Tyr705) levels using flow cytometric and Western blot analysis. In the quiescent state, Tyr(P)-STAT3 is significantly higher in cells obtained from control individuals compared with that from NS patients, which persisted after 30 min of IL-6 treatment (Fig. 2B). These findings demonstrate a tight association between significantly lower activated STAT3 levels and the NS phenotype in patients bearing PTPN11 mutations. Taken together with previous work showing Stat3 activation is enhanced in Shp2-deficient cells (33–35), these findings strongly suggest that Shp2 serves as an upstream phosphatase to negatively regulate Stat3 activation.
Mutant Shp2 Inhibits EGF-stimulated Stat3 Activation and DNA Binding
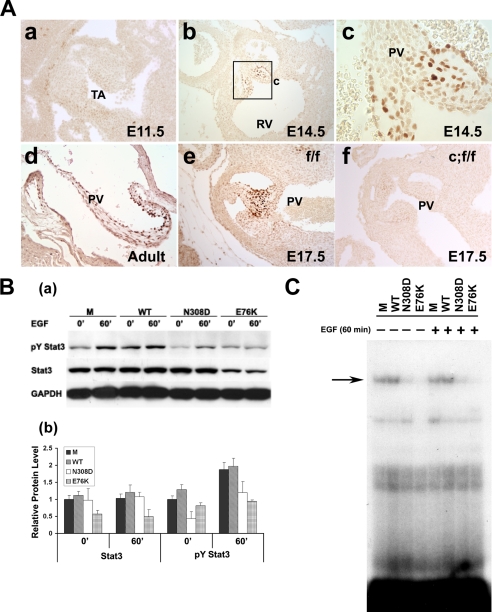
Given the apparent role of Stat3 in the hematopoietic phenotype of NS patients, we next investigated the potential role of Stat3 in cardiac valve development because NS patients commonly manifest congenital heart defects such as pulmonic valve stenosis. Using immunohistological staining to examine the cellular distribution of activated Stat3 in valvular tissues during normal development, we found that Tyr(P)-Stat3 expression is highly restricted to the developing valvular tissues in normal mouse embryos (Fig. 3A), suggesting that dysregulated Stat3 expression or function may contribute to the abnormal valve development. Signaling from the EGFR has been shown to play a critical role in valvulogenesis and Shp2 has been shown to play a positive role in EGFR-stimulated regulation of cellular proliferation and apoptosis during valvular remodeling (37–39). Given that EGFR signaling is a key regulator for the Stat3 signaling pathway (40, 41), our observation of activated Stat3 expression in the developing cardiac valves provides a potential clue for linking EGFR signaling with Shp2 and Stat3 dysregulation in the pathogenesis of pulmonary stenosis in NS patients. To test whether gain-of-function Shp2 mutants negatively regulate Stat3 activation in response to EGF, we retrovirally transduced MIEG3 (control vector), WT Shp2, Shp2N308D (the most common PTPN11 mutation found in NS), or Shp2E76K (a somatic mutation found in JMML) into NIH3T3 cells. Transduced EGFP-expressing cells were collected using FACS. After overnight serum deprivation, transduced NIH3T3 cells were treated with 50 ng/ml EGF and characterized by Western blot and EMSA to determine the level of Tyr(P)-Stat3 (Tyr705) and DNA binding activity, respectively (Fig. 3, B and C). EGF stimulation induced elevated levels of Tyr(P)-Stat3 in both MIEG3- and WT Shp2-expressing NIH3T3 cells. However, cells transduced with either Shp2N308D or Shp2E76K demonstrated reduced activation of Stat3 (Fig. 3B). More interestingly, the Shp2E76K mutant, which had a much greater phosphatase activity when compared with the Shp2N308D mutant (9), had a greater impact in Stat3 inactivation. The protein level of Stat3 was markedly reduced in Shp2E76K cells when compared with that of Shp2N308D-transduced cells. This finding suggests that the degree of Stat3 inactivation depends on the activity of the phosphatase present in various Shp2 gain-of-function mutants. This Shp2-phosphatase activity-dependent inactivation of Stat3 strongly suggested a role for Stat3 in Shp2-mediated signaling pathway. Given that Tyr(P)-Stat3 is known to be a positive regulator for Stat3 cellular accumulation (42), this dramatic reduction of the total Stat3 protein level in the Shp2E76K-transduced cells is most likely due to a stronger inhibition of the Stat3 phosphorylation level. EMSA further revealed that expression of either Shp2N308D or Shp2E76K remarkably reduced Stat3 DNA binding activity irrespective of EGF stimulation (Fig. 3C). These findings further suggest that Stat3 is potentially relevant to aberrant valvulogenesis in individuals with NS.
FIGURE 3.
Stat3 activation in valvulogenesis, and Shp2 gain-of-function mutants inhibit the activation of Stat3 in response to EGF. A, immunohistochemistry staining of activated Stat3 (pY Stat3) in developing and mature PV of wild type mice. Nuclear brown signals are positive staining. Activated Stat3 is apparent in E14.5 embryonic heart and is restricted to the developing leaflets (A, panels a to c), and remains into adult stage (panel d). Activated Stat3 is present in the PV of control embryo hearts at E17.5 (panel e), absent in PV from Stat3f/f/nestin-cre embryo heart at the same stage (panel f). B, panel a, after serum deprivation overnight, Western blot analysis was used to examine basal and EGF-stimulated (50 ng/ml) Stat3 tyrosine phosphorylation in NIH3T3 cells expressing Shp2 mutants (N308D and E76K) or controls (MIEG3 or WT Shp2). Panel b, density quantification of the protein band in B, panel a. C, EMSA to examine Stat3 DNA binding activity using the m67-SIE probe and nuclear extracts from NIH3T3 cells transduced with MIEG3 (M), WT Shp2 (WT), Shp2N308D (N308D), or Shp2E76K (E76K). Error bars represent S.D.
Conditional Ablation of Stat3 Gives Rise to Pulmonary Stenosis
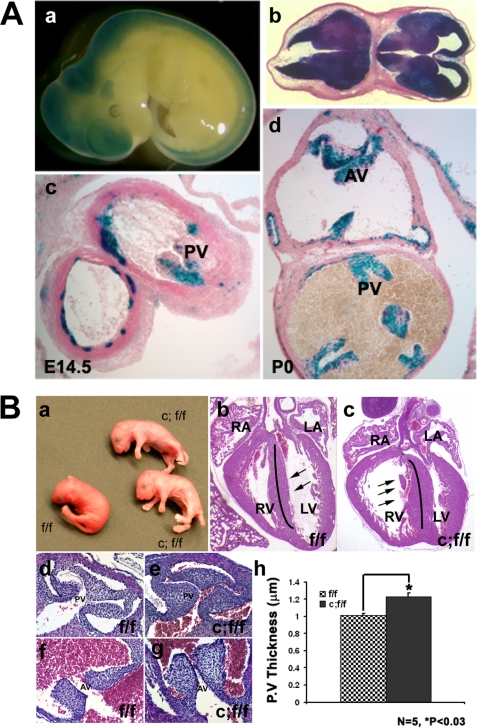
In an independent study to address the role of Stat3 in brain development and function, we generated Stat3f/f/nestin-cre mice (26). Surprisingly, shortly following birth, the neonatal pups appeared pale, quickly become cyanotic (Fig. 4B, panel a), and died within a few hours due to respiratory failure (26). Because this phenotype suggested abnormal development within the cardiovascular system, we analyzed nestin-directed cre recombinase activity using nestin-cre/Rosa26R mice and found that cre was not only expressed in the central nervous system, but also in the developing pulmonary, aortic, and atrioventricular valves (Fig. 4A). Specifically, Fig. 4A, c and d, shows that cre activity in the heart is associated with the developing leaflets of pulmonary and aortic valves. Immunostaining confirmed that Stat3 was ablated in developing pulmonary valves in Stat3f/f/nestin-cre mice compared with control littermates (Fig. 3A, e and f).
FIGURE 4.
Conditional deletion of Stat3 with nestin-cre induces pulmonary stenosis due to the thickening of leaflets. A, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside(X-gal) staining of nestin-cre/Rosa26R mouse embryos and newborns indicates the cre activity in the developing mouse embryos. A, panels a and b, central nervous system is positive for cre activity in the mouse embryos at E10.5. Panels c and d, cre activity was found in the pulmonary valves (PV) and the aortic valves (AV) at E14.5 (A, panel c) and P0 (B, panel d). B, ablation of Stat3 with nestin-cre leads to pulmonary stenosis caused by the thickening and enlargement of leaflets. B, panel a, newborn of Stat3f/f (f/f) and Stat3f/f/Nestin-cre (c;f/f) mice. B, panel b, hematoxylin and eosin staining of the transverse section of heart from control (f/f) and STAT3f/f/Nestin-cre (c;f/f) newborn mice (P0). Note that Stat3 ablation leads to right ventricle dilation in Stat3f/f/Nestin-cre mice. B, panels d–g, hematoxylin and eosin staining of sections of the pulmonary valve and aorta valve regions in control (B, panels d and f) and Stat3f/f/Nestin-cre mutant (B, panels e and g) newborn mice. LV, left ventricle; RV, right ventricle. B, panel h, quantification of pulmonary valve thickness in Stat3f/f/Nestin-cre and control hearts (P0) (n = 6, p < 0.03, Student's paired t test; 2 tails).
Gross examination and histological analysis indicated that the lung of Stat3f/f/nestin-cre mice inflated normally without obvious defects in development or formation. However, cardiac morphological and histological analysis demonstrated that the right ventricles of all Stat3f/f/ nestin-cre mutant hearts were significantly dilated and the ventricular septum was hypertrophied. In many cases, the right ventricular dilation was severe enough to compress the left ventricle (Fig. 4B, b and c). These findings suggested obstruction of blood flow from the right ventricle with resultant ventricular hypertension and dilation. Analysis by serial sectioning to follow the morphology of the pulmonary artery indicated that the pulmonary valve leaflets (PV) in the Stat3f/f/nestin-cre mice were significantly enlarged and thickened (Fig. 4B, d–h) when compared with the controls. Quantitative measurement of the size of the PV leaflets confirmed this observation (Fig. 4B, h). Although the valve thickening may not be the sole cause of death for the Stat3f/f/nestin-cre mice, this unique abnormal valvular defect closely resembles the valvular defect in NS patients and the NS-D61G mouse model (8).
Lack of Stat3 Does Not Affect ERK Activation in Response to GM-CSF
It has been shown that Shp2 mutations found in NS and JMML patients induce hyperactivation of Ras and lead to prolonged ERK activation in response to GM-CSF, EGF, and other cytokines in several cell types (8, 9, 12, 27, 31). One possible interpretation for the NS-like phenotype in Stat3 mutant mice is that Stat3 ablation may enhance Ras-ERK activation. However, Western blot and immunostaining demonstrated that ablation of Stat3 did not affect the level of ERK activation in either bone marrow-derived macrophage cells (Fig. 5A) or pulmonary valve tissues (data not shown), which implies that the loss of Stat3 function or expression may not directly impact the ERK-mediated pathway. Interestingly, a previous study had demonstrated that deletion of Stat3 in bone marrow cells can induce an enhanced and prolonged ERK activation in response to G-CSF (43). As BM cells are a mixed cell population, further analysis will be necessary to determine which cell type deficient in Stat3 has enhanced the ERK activation (43). Furthermore, mouse embryonic fibroblast cells derived from Shp2D61G mutant mice demonstrated a normal ERK activation in response to multiple growth factors, whereas an enhanced ERK activation was observed in other cells types and tissues in Shp2D61G mutant mice (8), suggesting a cell type or cytokine-dependent event involved in the cross-talk between Stat3- and ERK-mediated pathways.
FIGURE 5.
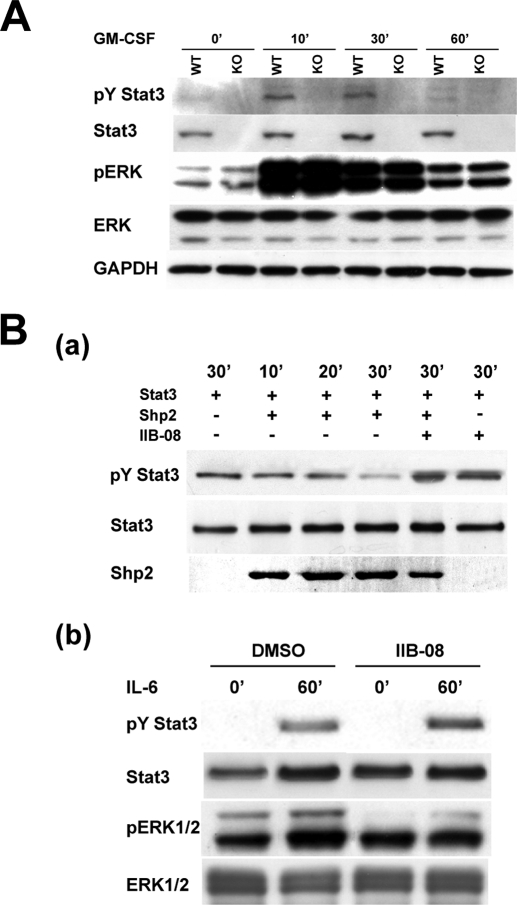
Shp2 dephosphorylates Tyr(P)-Stat3 and Shp2-Stat3-mediated signaling critically contributes to the pathogenesis of NS. A, Western blot analysis of Stat3-deficient bone marrow-derived macrophage progenitors treated with GM-CSF to assess Stat3 and ERK activation. B, panel a, purified constitutively active Shp2 protein (catalytic domain) was capable of dephosphorylating Tyr(P)-Stat3. Stat3 protein was immunoprecipitated from IL-6-treated Raw 264.7 cell extract, and then incubated with purified recombinant Shp2 protein (active form, 2 μg) for the indicated time with or without preincubation of the small molecule Shp2 phosphatase inhibitor IIB-08 (100 μm). Panel b, Shp2 inhibitor IIB-08 inhibits ERK activation, whereas enhancing the Tyr(P)-Stat3 level, in IL-6 induced Raw 264.7 cells. Raw 264.7 cells were pretreated with the Shp2 inhibitor IIB-08 (10 μm), or dimethyl sulfoxide (DMSO) for 60 min, and then incubated with IL-6 (20 ng/ml) for 60 min before being subjected to lysis and Western blot analysis.
Shp2 Dephosphorylates Tyr(P)-Stat3
To understand how Shp2 regulates Stat3 phosphorylation, we performed an in vitro phosphatase assay to investigate whether Shp2 is able to directly dephosphorylate Tyr(P)-Stat3. Fig. 5B, a, shows that purified Shp2 protein is capable of dephosphorylating Tyr(P)-Stat3 and a specific Shp2 phosphatase inhibitor, IIB-08 (see “Materials and Methods”), blocks the Shp2-mediated Tyr(P)-Stat3 dephosphorylation. Moreover, IIB-08 enhances IL-6-induced Stat3 phosphorylation in Raw 264.7 cells, whereas attenuating the ERK activity from the known positive role in ERK signaling of Shp2 (Fig. 5B, b). Collectively, these results strongly suggest that Tyr(P)-Stat3 is a substrate for Shp2 phosphatase.
Stat3 Overexpression Reduces the Sensitivity to GM-CSF Stimulation in Shp2E76K-transduced Cells
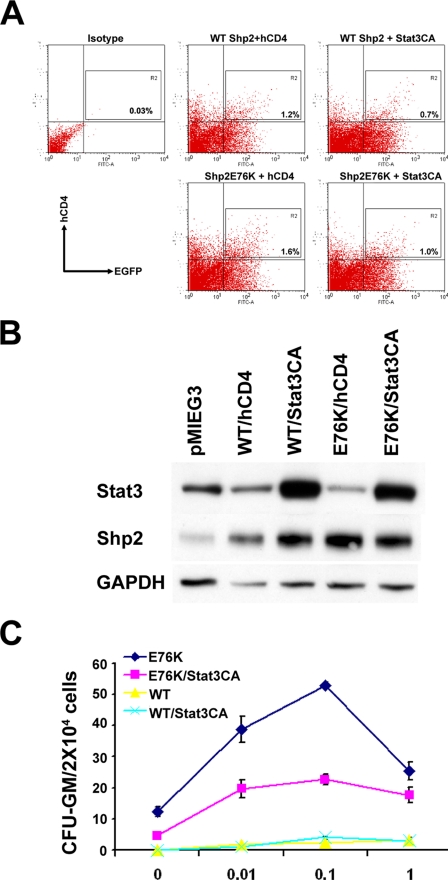
JMML patients and murine hematopoietic progenitors ectopically expressing gain-of-function Shp2 mutants demonstrate increased sensitivity to GM-CSF (8, 27, 30–32). To further determine whether inhibition of Stat3 crucially contributes to this phenotype induced by gain-of-function Shp2 mutations, we performed co-transduction experiments using constitutively active Stat3 (Stat3CA) (44) and Shp2E76K in murine BM cells. Bone marrow low density mononuclear cells were isolated and co-transduced with WT Shp2-pMIEG3-EGFP or Shp2E76K-pMIEG3-EGFP in combination with pMIEG3-hCD4 or Stat3CA-pMIEG3-hCD4. Following transduction, cells were sorted for EGFP and human CD4 antigen expression using FACS to enrich for transduced double positive cells (Fig. 6A). Western blot analysis was used to confirm the Shp2 and Stat3 overexpression in virus-transduced and FACS-enriched EGFP and CD4 expressing cells (Fig. 6B). Using methylcellulose colony assays with increasing concentrations of GM-CSF (0–1 ng/ml), we found that, in sharp contrast to Shp2E76K-expressing cells, Stat3CA/Shp2E76K co-expressing cells had a dramatic drop in their sensitivity to GM-CSF (Fig. 6C). This result strongly indicates that dysregulation of Stat3 expression and/or function plays a pivotal role in mutant Shp2-induced pathogenesis in both NS and JMML.
FIGURE 6.
Stat3 overexpression reduces the sensitivity to GM-CSF stimulation in Shp2E76K-transduced cells. A, bone marrow low density mononuclear cells were isolated and co-transduced with WT Shp2 or Shp2E76K pMIEG3-EGFP in combination with pMIEG3-hCD4 or pMIEG3-Stat3CA-hCD4. Following transduction, cells were sorted for EGFP or human CD4 antigen expression using FACS to enrich for transduced double positive cells. B, Western blotting showing Shp2 and Stat3 overexpression in virus-transduced and FACS-enriched EGFP and CD4 expressing cells. C, methylcellulose-based colony forming assay demonstrating that Stat3CA expression corrects the gain-of-function Shp2-induced hematopoietic progenitor hypersensitivity to GM-CSF. Shown are representative data from two independent experiments that demonstrate similar results. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Error bars represent S.D.
DISCUSSION
Numerous studies have demonstrated dysregulation of the RAS-MAPK signaling cascade as a common pathogenetic feature of several overlapping, yet distinct, congenital syndromes (1, 45, 46). However, the variable phenotypic aspects of each syndrome, even within NS patients, imply that dysregulation of ancillary signaling pathways, in addition to the RAS-MAPK cascade, may also contribute to the observed anomalies, thus culminating in the unique aspects of each disorder.
Pulmonic stenosis is commonly observed among individuals with mutations in the PTPN11 allele (18). Recently, studies using several mutant mice lacking HB-EGF expression have revealed that HB-EGF, in conjunction with ErbB receptors, has a critical role in normal cardiac valve formation at both cushion formation and tissue remodeling stages (37). In addition, Shp2 has been shown to play a positive role in EGFR-stimulated regulation of cellular proliferation and apoptosis during valvular remodeling (38). Our current study revealed that tyrosine-phosphorylated Stat3 is specifically expressed in developing valve tissues, and conditional ablation of Stat3 leads to pulmonary stenosis. Furthermore, Shp2 gain-of-function mutations, N308D and E76K, can both down-regulate Stat3 activation by EGF in 3T3 fibroblast cells, further suggesting that Stat3 plays a critical role in Shp2 and EGFR-mediated signaling. Consistent with our finding, a recent study also demonstrated another functional connection between Shp2 and Stat3 in neural progenitor differentiation. The gp130/Stat3 signaling pathway was shown to be critical for multipotent precursors to adapt the glial differentiation fate over neuronal fate (47, 48). Gauthier and colleagues (49) demonstrated that genetic knockdown of Shp2 in cultured cortical precursors or in the embryonic cortex led to the inhibition of basal neurogenesis, whereas enhancing precocious astrocyte formation. Conversely, expression of Shp2D61G promoted neuronal differentiation, whereas reducing astrogenesis, again suggesting a tight correlation between Stat3 and Shp2 in different development processes.
Previously, several lines of evidence suggested that Shp2 is a negative regulator for Stat protein activation. For instance, Shp2-deficient embryonic stem cells demonstrated an elevated Tyr(P)-Stat3 level in response to leukemia inhibitory factor (35). Another work showed that the disruption of the Shp2 binding tyrosyl residues on gp130 (Tyr759) resulted in prolonged Stat3 activation (33). More interestingly, Wang and colleagues (50) have reported that Shp2 is able to interact with and dephosphorylate Stat5A. However, this remains controversial as Stat5 activation has been shown to be elevated in Shp2E76K-transduced bone marrow macrophage cells in response to IL-3 (31). In our current study, we demonstrate that two gain-of-function Shp2 mutants (Shp2N308D and Shp2E76K) were able to inactivate Stat3. Among them, Shp2N308D is one of the most common NS germline mutations, whereas Shp2E76K is only found in JMML patients as a somatic mutation. Previously, Gelb and colleagues (9) demonstrated that Shp2E76K possessed a significantly higher phosphatase activity when compared with Shp2N308D. Interestingly, we found that Shp2E76K is more potent in inactivating Stat3 when compared with Shp2N308D. This Shp2-phosphatase activity-dependent inactivation of Stat3 suggests that Shp2 is an upstream phosphatase for Stat3. Consistent with this, biochemical analyses demonstrated that Shp2 is able to dephosphorylate Stat3, and this activity can be inhibited by a Shp2-specific small molecular inhibitor.
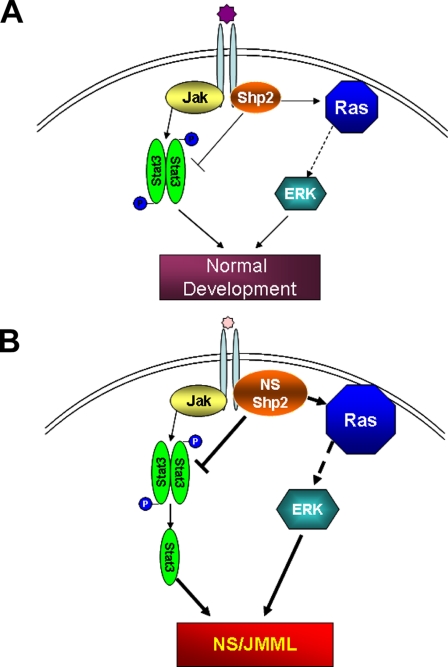
Our study did not exclude the importance of the RAS-MAPK-mediated pathway in contributing to NS and JMML. In fact, we believe that the negative Shp2-Stat3 and positive Shp2-RAS pathways are functionally synergistic. For example, IL-3 is known to support mast cell differentiation in normal hematopoietic progenitors. We have observed that Shp2D61Y and Shp2E76K hematopoietic progenitors give rise to significantly skewed monocytic differentiation in response to IL-3 (supplemental Fig. S1).5 Interestingly, the Stat3-deficient hematopoietic progenitors demonstrate only modest monocytic differentiation when cultured under the same conditions. This observation of these similar, yet distinctive, phenotypes suggests that the inactivation of Stat3 synergizes with the dysregulation of the RAS-MAPK cascade in giving rise to the clinical features of NS and/or JMML (Fig. 7).
FIGURE 7.
Schematic diagram of dual-signaling pathways regulated by Shp2. In normal physiological conditions, Stat3 activity is under the positive and negative regulation of Janus kinases and Shp2, respectively (A). Shp2 gain-of-function mutations lead to the hyperactivation of the RAS/ERK signaling pathway and the excessive inactivation of Stat3, which synergistically promotes the pathogenesis of Noonan syndrome and/or JMML (B).
One of the major goals in our study is to demonstrate whether down-regulation of Stat3 activity functionally contributes to NS and/or JMML pathogenesis. To determine this, we performed rescue experiments by transducing constitutively active Stat3 to Shp2E76K-transduced bone marrow cells and assaying for GM-CSF hypersensitivity. Our results clearly show that Stat3CA is able to significantly reduce GM-CSF sensitivity in Shp2E76K-transduced bone marrow cells, which provides strong evidence that interrupting the balance between Shp2 and Stat3 activation may be one of the major factors contributing to the pathogenesis of mutant Shp2-mediated NS and/or JMML pathogenesis. Taken together, our findings have important implications not only for understanding the molecular basis of Shp2-mediated NS and JMML pathogenesis, but also for developing eventual therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Drs. Reuben Kapur and Tony Firulli for critical review of the manuscript and members of the Shou and Fu laboratories for valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL81092 (to W. S.), HL70259 (to W. S.), HL85098 (to W. S. and M. P.), HL82981 (to R. J. C.), and CA69202 (to Z. Y. Z.). This work was also supported in part by the Indiana University Department of Pediatrics, Protocol Development Team.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Z. Yang and R. J. Chan, submitted for publication.
- NS
- Noonan syndrome
- JMML
- juvenile myelomonocytic leukemia
- MAPK
- mitogen-activated protein kinase
- IL
- interleukin
- FACS
- fluorescence-activated cell sorting
- GM-CSF
- granulocyte-macrophage colony stimulating factor
- EMSA
- electrophoretic mobility shift assay
- EGFP
- enhanced green fluorescent protein
- PV
- pulmonary valve
- Stat3CA
- constitutively active Stat3
- ERK
- extracellular signal-regulated kinase
- STAT
- signal transducers and activators of transcription
- PTP
- protein-tyrosine phosphatase
- WT
- wild type.
REFERENCES
- 1.Tartaglia M., Gelb B. D. (2005) Annu. Rev. Genomics Hum. Genet. 6, 45–68 [DOI] [PubMed] [Google Scholar]
- 2.Bader-Meunier B., Tchernia G., Miélot F., Fontaine J. L., Thomas C., Lyonnet S., Lavergne J. M., Dommergues J. P. (1997) J. Pediatr. 130, 885–889 [DOI] [PubMed] [Google Scholar]
- 3.Choong K., Freedman M. H., Chitayat D., Kelly E. N., Taylor G., Zipursky A. (1999) J. Pediatr. Hematology/Oncology 21, 523–527 [PubMed] [Google Scholar]
- 4.Tartaglia M., Mehler E. L., Goldberg R., Zampino G., Brunner H. G., Kremer H., van der Burgt I., Crosby A. H., Ion A., Jeffery S., Kalidas K., Patton M. A., Kucherlapati R. S., Gelb B. D. (2001) Nat. Genet. 29, 465–468 [DOI] [PubMed] [Google Scholar]
- 5.Tartaglia M., Kalidas K., Shaw A., Song X., Musat D. L., van der Burgt I., Brunner H. G., Bertola D. R., Crosby A., Ion A., Kucherlapati R. S., Jeffery S., Patton M. A., Gelb B. D. (2002) Am. J. Hum. Genet. 70, 1555–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Z. Q., Yu D. H., Park M., Marshall M., Feng G. S. (2000) Mol. Cell Biol. 20, 1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng G. S. (1999) Exp. Cell Res. 253, 47–54 [DOI] [PubMed] [Google Scholar]
- 8.Araki T., Mohi M. G., Ismat F. A., Bronson R. T., Williams I. R., Kutok J. L., Yang W., Pao L. I., Gilliland D. G., Epstein J. A., Neel B. G. (2004) Nat. Med. 10, 849–857 [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M., Niemeyer C. M., Fragale A., Song X., Buechner J., Jung A., Hählen K., Hasle H., Licht J. D., Gelb B. D. (2003) Nat. Genet. 34, 148–150 [DOI] [PubMed] [Google Scholar]
- 10.Loh M. L., Vattikuti S., Schubbert S., Reynolds M. G., Carlson E., Lieuw K. H., Cheng J. W., Lee C. M., Stokoe D., Bonifas J. M., Curtiss N. P., Gotlib J., Meshinchi S., Le Beau M. M., Emanuel P. D., Shannon K. M. (2004) Blood 103, 2325–2331 [DOI] [PubMed] [Google Scholar]
- 11.Kratz C. P., Niemeyer C. M., Castleberry R. P., Cetin M., Bergsträsser E., Emanuel P. D., Hasle H., Kardos G., Klein C., Kojima S., Stary J., Trebo M., Zecca M., Gelb B. D., Tartaglia M., Loh M. L. (2005) Blood 106, 2183–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fragale A., Tartaglia M., Wu J., Gelb B. D. (2004) Hum. Mutat. 23, 267–277 [DOI] [PubMed] [Google Scholar]
- 13.Keilhack H., David F. S., McGregor M., Cantley L. C., Neel B. G. (2005) J. Biol. Chem. 280, 30984–30993 [DOI] [PubMed] [Google Scholar]
- 14.Roberts A. E., Araki T., Swanson K. D., Montgomery K. T., Schiripo T. A., Joshi V. A., Li L., Yassin Y., Tamburino A. M., Neel B. G., Kucherlapati R. S. (2007) Nat. Genet. 39, 70–74 [DOI] [PubMed] [Google Scholar]
- 15.Pandit B., Sarkozy A., Pennacchio L. A., Carta C., Oishi K., Martinelli S., Pogna E. A., Schackwitz W., Ustaszewska A., Landstrom A., Bos J. M., Ommen S. R., Esposito G., Lepri F., Faul C., Mundel P., López Siguero J. P., Tenconi R., Selicorni A., Rossi C., Mazzanti L., Torrente I., Marino B., Digilio M. C., Zampino G., Ackerman M. J., Dallapiccola B., Tartaglia M., Gelb B. D. (2007) Nat. Genet. 39, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 16.Razzaque M. A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M., Fujiwara Y., Matsushima M., Mizuno K., Tokuyama M., Hirota H., Muneuchi J., Higashinakagawa T., Matsuoka R. (2007) Nat. Genet. 39, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 17.Schubbert S., Zenker M., Rowe S. L., Böll S., Klein C., Bollag G., van der Burgt I., Musante L., Kalscheuer V., Wehner L. E., Nguyen H., West B., Zhang K. Y., Sistermans E., Rauch A., Niemeyer C. M., Shannon K., Kratz C. P. (2006) Nat. Genet. 38, 331–336 [DOI] [PubMed] [Google Scholar]
- 18.Sarkozy A., Carta C., Moretti S., Zampino G., Digilio M. C., Pantaleoni F., Scioletti A. P., Esposito G., Cordeddu V., Lepri F., Petrangeli V., Dentici M. L., Mancini G. M., Selicorni A., Rossi C., Mazzanti L., Marino B., Ferrero G. B., Silengo M. C., Memo L., Stanzial F., Faravelli F., Stuppia L., Puxeddu E., Gelb B. D., Dallapiccola B., Tartaglia M. C. (2009) Hum. Mutat. 30, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tartaglia M., Gelb B. D. (2009) in Monographs in Human Genetics (Zenker M. ed) Vol. 17, pp. 20–39, Karger Press, Basel, Switzerland [Google Scholar]
- 20.Bromberg J., Darnell J. E., Jr. (2000) Oncogene 19, 2468–2473 [DOI] [PubMed] [Google Scholar]
- 21.Levy D. E., Lee C. K. (2002) J. Clin. Invest. 109, 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamero A. M., Potla R., Wegrzyn J., Szelag M., Edling A. E., Shimoda K., Link D. C., Dulak J., Baker D. P., Tanabe Y., Grayson J. M., Larner A. C. (2006) J. Biol. Chem. 281, 16238–16244 [DOI] [PubMed] [Google Scholar]
- 23.Kritikou E. A., Sharkey A., Abell K., Came P. J., Anderson E., Clarkson R. W., Watson C. J. (2003) Development 130, 3459–3468 [DOI] [PubMed] [Google Scholar]
- 24.Welte T., Zhang S. S., Wang T., Zhang Z., Hesslein D. G., Yin Z., Kano A., Iwamoto Y., Li E., Craft J. E., Bothwell A. L., Fikrig E., Koni P. A., Flavell R. A., Fu X. Y. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1879–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laouar Y., Welte T., Fu X. Y., Flavell R. A. (2003) Immunity 19, 903–912 [DOI] [PubMed] [Google Scholar]
- 26.Gao Q., Wolfgang M. J., Neschen S., Morino K., Horvath T. L., Shulman G. I., Fu X. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4661–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan R. J., Leedy M. B., Munugalavadla V., Voorhorst C. S., Li Y., Yu M., Kapur R. (2005) Blood 105, 3737–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang H. C., Zhang S., Thieu V. T., Slee R. B., Bruns H. A., Laribee R. N., Klemsz M. J., Kaplan M. H. (2005) Immunity 22, 693–703 [DOI] [PubMed] [Google Scholar]
- 29.Asao H., Fu X. Y. (2000) J. Biol. Chem. 275, 867–874 [DOI] [PubMed] [Google Scholar]
- 30.Emanuel P. D., Bates L. J., Castleberry R. P., Gualtieri R. J., Zuckerman K. S. (1991) Blood 77, 925–929 [PubMed] [Google Scholar]
- 31.Mohi M. G., Williams I. R., Dearolf C. R., Chan G., Kutok J. L., Cohen S., Morgan K., Boulton C., Shigematsu H., Keilhack H., Akashi K., Gilliland D. G., Neel B. G. (2005) Cancer Cell 7, 179–191 [DOI] [PubMed] [Google Scholar]
- 32.Schubbert S., Lieuw K., Rowe S. L., Lee C. M., Li X., Loh M. L., Clapp D. W., Shannon K. M. (2005) Blood 106, 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtani T., Ishihara K., Atsumi T., Nishida K., Kaneko Y., Miyata T., Itoh S., Narimatsu M., Maeda H., Fukada T., Itoh M., Okano H., Hibi M., Hirano T. (2000) Immunity 12, 95–105 [DOI] [PubMed] [Google Scholar]
- 34.Zhang E. E., Chapeau E., Hagihara K., Feng G. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16064–16069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan R. J., Johnson S. A., Li Y., Yoder M. C., Feng G. S. (2003) Blood 102, 2074–2080 [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt I., Palleschi A., Petrucci T. C., Sorcini M., Schoch C., Foa R., Emanuel P. D., Gelb B. D. (2006) Am. J. Hum. Genet. 78, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwamoto R., Mekada E. (2006) Cell Struct. Funct. 31, 1–14 [DOI] [PubMed] [Google Scholar]
- 38.Chen B., Bronson R. T., Klaman L. D., Hampton T. G., Wang J. F., Green P. J., Magnuson T., Douglas P. S., Morgan J. P., Neel B. G. (2000) Nat. Genet. 24, 296–299 [DOI] [PubMed] [Google Scholar]
- 39.Qu C. K., Yu W. M., Azzarelli B., Feng G. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8528–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo H. W., Hsu S. C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J. Y., Hung M. C. (2005) Cancer Cell 7, 575–589 [DOI] [PubMed] [Google Scholar]
- 41.Zhong Z., Wen Z., Darnell J. E., Jr. (1994) Science 264, 95–98 [DOI] [PubMed] [Google Scholar]
- 42.Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. (2007) Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamezaki K., Shimoda K., Numata A., Haro T., Kakumitsu H., Yoshie M., Yamamoto M., Takeda K., Matsuda T., Akira S., Ogawa K., Harada M. (2005) Stem Cells 23, 252–263 [DOI] [PubMed] [Google Scholar]
- 44.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 45.Chan R. J., Feng G. S. (2007) Blood 109, 862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kratz C. P., Niemeyer C. M., Zenker M. (2007) J. Mol. Med. 85, 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D. A., Rozovsky I., Stahl N., Yancopoulos G. D., Greenberg M. E. (1997) Science 278, 477–483 [DOI] [PubMed] [Google Scholar]
- 48.Rajan P., McKay R. D. (1998) J. Neurosci. 18, 3620–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gauthier A. S., Furstoss O., Araki T., Chan R., Neel B. G., Kaplan D. R., Miller F. D. (2007) Neuron 54, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y., Wen R., Yang S., Schuman J., Zhang E. E., Yi T., Feng G. S., Wang D. (2003) J. Biol. Chem. 278, 16520–16527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.