Abstract
The binding of the adaptor protein APPL1 to adiponectin receptors is necessary for adiponectin-induced AMP-activated protein kinase (AMPK) activation in muscle, yet the underlying molecular mechanism remains unknown. Here we show that in muscle cells adiponectin and metformin induce AMPK activation by promoting APPL1-dependent LKB1 cytosolic translocation. APPL1 mediates adiponectin signaling by directly interacting with adiponectin receptors and enhances LKB1 cytosolic localization by anchoring this kinase in the cytosol. Adiponectin also activates another AMPK upstream kinase Ca2+/calmodulin-dependent protein kinase kinase by activating phospholipase C and subsequently inducing Ca2+ release from the endoplasmic reticulum, which plays a minor role in AMPK activation. Our results show that in muscle cells adiponectin is able to activate AMPK via two distinct mechanisms as follows: a major pathway (the APPL1/LKB1-dependent pathway) that promotes the cytosolic localization of LKB1 and a minor pathway (the phospholipase C/Ca2+/Ca2+/calmodulin-dependent protein kinase kinase-dependent pathway) that stimulates Ca2+ release from intracellular stores.
Adiponectin, an adipokine abundantly expressed in adipose tissue, exhibits anti-diabetic, anti-inflammatory, and anti-atherogenic properties and hence is a potential therapeutic target for various metabolic diseases (1–3). The beneficial effects of adiponectin are mediated through the direct interaction of adiponectin with its cell surface receptors, AdipoR1 and AdipoR2 (4, 5). Adiponectin increases fatty acid oxidation and glucose uptake in muscle cells by activating AMP-activated protein kinase (AMPK)3 (4, 6), which depends on the interaction of AdipoR1 with the adaptor protein APPL1 (Adaptor protein containing Pleckstrin homology domain, Phosphotyrosine binding domain, and Leucine zipper motif) (5). However, the underlying mechanisms by which APPL1 mediates adiponectin signaling to AMPK activation and other downstream targets remain unclear.
AMPK is a serine/threonine protein kinase that acts as a master sensor of cellular energy balance in mammalian cells by regulating glucose and lipid metabolism (7, 8). AMPK is composed of a catalytic α subunit and two noncatalytic regulatory subunits, β and γ. The NH2-terminal catalytic domain of the AMPKα subunit is highly conserved and contains the activating phosphorylation site (Thr172) (9). Two AMPK variants, α1 and α2, exist in mammalian cells that show different localization patterns. AMPKα1 subunit is localized in non-nuclear fractions, whereas the AMPKα2 subunit is found in both nucleus and non-nuclear fractions (10). Biochemical regulation of AMPK activation occurs through various mechanisms. An increase in AMP level stimulates the binding of AMP to the γ subunit, which induces a conformational change in the AMPK heterotrimer and results in AMPK activation (11). Studies have shown that the increase in AMPK activity is not solely via AMP-dependent conformational change, rather via phosphorylation by upstream kinases, LKB1 and CaMKK. Dephosphorylation by protein phosphatases is also important in regulating the activity of AMPK (12).
LKB1 has been considered as a constitutively active serine/threonine protein kinase that is ubiquitously expressed in all tissues (13, 14). Under conditions of high cellular energy stress, LKB1 acts as the primary AMPK kinase through an AMP-dependent mechanism (15–17). Under normal physiological conditions, LKB1 is predominantly localized in the nucleus. LKB1 is translocated to the cytosol, either by forming a heterotrimeric complex with Ste20-related adaptor protein (STRADα/β) and mouse protein 25 (MO25α/β) or by associating with an LKB1-interacting protein (LIP1), to exert its biological function (18–22). Although LKB1 has been shown to mediate contraction- and adiponectin-induced activation of AMPK in muscle cells, the underlying molecular mechanisms remain elusive (15, 23).
CaMKK is another upstream kinase of AMPK, which shows considerable sequence and structural homology with LKB1 (24–26). The two isoforms of CaMKK, CaMKKα and CaMKKβ, encoded by two distinct genes, share ∼70% homology at the amino acid sequence level and exhibit a wide expression in rodent tissues, including skeletal muscle (27–34). Unlike LKB1, AMPK phosphorylation mediated by CaMKKs is independent of AMP and is dependent only on Ca2+/calmodulin (35). Hence, it is possible that an LKB1-independent activation of AMPK by CaMKK exists in muscle cells. However, whether and how adiponectin stimulates this pathway in muscle cells are not known.
In this study, we demonstrate that in muscle cells adiponectin induces an APPL1-dependent LKB1 translocation from the nucleus to the cytosol, leading to increased AMPK activation. Adiponectin also activates CaMKK by stimulating intracellular Ca2+ release via the PLC-dependent mechanism, which plays a minor role in activation of AMPK. Taken together, our results demonstrate that enhanced cytosolic localization of LKB1 and Ca2+-induced activation of CaMKK are the mechanisms underlying adiponectin-stimulated AMPK activation in muscle cells.
EXPERIMENTAL PROCEDURES
Plasmids, Adiponectin, Chemicals, and Antibodies
The cDNAs encoding full-length human APPL1 was described previously (5). The cDNA encoding amino acids 1–427 of LKB1 was cloned by PCR from a mouse cDNA library and subcloned into the mammalian expression vector pBEX1 (36), in-frame at its COOH terminus with a sequence encoding the hemagglutinin (HA) tag, or the pcDNA 3.1 myc/His A plasmid (Invitrogen), in-frame at its COOH terminus with a sequence encoding the Myc tag. Plasmids encoding YFP-fused wild type and the SL-26 mutant of LKB1 were kindly provided by Dr. Dario R. Alessi (37). The plasmids encoding GST-APPL1 (CT) (amino acids 455–693) and the anti-APPL1 antibody were described previously (5, 38). The globular form of adiponectin was produced in Escherichia coli as described previously (5). Monoclonal antibodies to HA tag (Covance, Emeryville, CA), β-tubulin 2.1 (Sigma), Myc tag (produced in-house with the Myc 1–9E10.2 cell clone from ATCC), and LKB1 (Upstate) were used in the experiments. STO-609 was obtained from Sigma; U73122 was from Santa Cruz Biotechnology (Santa Cruz, CA). All other antibodies were obtained from Cell Signaling Technologies (Danvers, MA).
Cell Lines, Cell Culture, and Cellular Fractionation
Conditions for culturing C2C12 cells, the APPL1-suppressed C2C12 cells, scramble control C2C12 cells, and HeLa cells are described in our previous studies (5, 38). The experimental procedure for cellular fractionation is essentially the same as described previously (39).
In Vitro Binding Studies and Co-immunoprecipitation
The experimental procedures for in vitro binding and co-immunoprecipitation in C2C12 cells are essentially the same as described previously (5). For detection of the interaction between APPL1 and LKB1 in mouse tissue, in vitro incubation of skeletal muscle with adiponectin was performed as described by Kuoppamaa et al. (40). In brief, skeletal muscle from 3-month-old mice was excised, cut into small pieces, and oxygenated for 30 min in Krebs-Henseleit buffer (KHB), containing 5 mm HEPES, 5 mm glucose, 15 mm mannitol, and 0.1% bovine serum albumin. The muscle samples were then incubated without or with globular adiponectin (2.5 μg/ml) in KHB buffer at 35 °C for 10, 20, and 30 min. The muscle samples were immediately frozen in liquid nitrogen at the end of the treatment period. For protein extraction, the frozen samples were homogenized in ice-cold homogenization buffer (50 mm HEPES, pH 7.6, 150 mm NaCl, 20 mm sodium pyrophosphate, 20 mm β-glycerophosphate, 10 mm NaF, 2 mm sodium orthophosphate, 2 mm EDTA, 1% Nonidet P-40, 10% glycerol, 2 mm phenylmethylsulfonyl fluoride, 1 mm MgCl2, 1 mm CaCl2, 10 μg/ml leupeptin, and 10 μg/ml aprotinin), kept on ice for 30 min, and centrifuged at 14,000 rpm for 20 min. The protein concentration in the supernatant was determined by the Bradford assay. APPL1 was immunoprecipitated from 1 mg of the crude tissue protein with an antibody specific for APPL1. Immunoprecipitated APPL1 and co-immunoprecipitated LKB1 were detected by Western blotting with specific antibodies for these proteins.
Western Blot and Statistical Analyses
The expression and phosphorylation levels of proteins were detected by Western blot of cell lysates or immunoprecipitation with specific antibodies. Quantification of the relative increase in protein phosphorylation (expressed as percentage of basal phosphorylation, arbitrarily set as 1.0) was performed using the Scion Image Alpha 4.0.3.2 program (Scion Corp.) and normalized for the amount of protein expression in each experiment. Statistical evaluation of the data were done using one-way analysis of variance.
Immunofluorescence Studies and Statistical Analyses
The experimental procedures for immunofluorescence studies on the cellular translocation of LKB1 are essentially the same as described in our previous study (5). The percentage of cytosolic LKB1 is calculated as P = C/(N + C), where N represents cells in which LKB1 is localized predominantly in the nucleus and C represents cells with predominantly cytosolic LKB1. The fold change of cytosolically localized LKB1 with treatment is calculated as F = Ptreated/Pcontrol. Statistical evaluations of the data were done using one-way analysis of variance. *, p < 0.05; **, p < 0.01.
Generation of LKB1 Knockdown Cells
LKB1 short hairpin RNA constructs cloned in pSM2 vector (catalog number RMM1766-96740219, Open Biosystems) were stably expressed in C2C12 cells and selected using puromycin, as described before (5). The cells stably expressing pSM2 vector generated by the same method were used as the control.
Ca2+ Signal Measurements
Ca2+ imaging experiments were performed as described previously (41). In brief, C2C12 myoblasts were seeded on Delta T dishes (Bioptechs Inc.) and incubated with a fluorescent Ca2+-sensitive dye (3 μm, Fluo-4 AM, Molecular Probes, catalog number F14202) at room temperature for 30 min. After two careful washes with phosphate-buffered saline and 20 min in fresh Dulbecco's modified Eagle's medium, the cells were imaged. Images were acquired with a confocal laser-scanning microscope (Bio-Rad, MRC-600) attached to an inverted microscope (Nikon TE200) using ND filter 1 at the rate of 1 frame/s. We used a 20× water 0.45 NA objective (Plan Fluor, Nikon) for 10-min recordings. Loading and imaging were carried out in recording media at 37 °C (12.78 g/liter Dulbecco's modified Eagle's medium, 5% fetal bovine serum, 25 μm HEPES, pH 7.2, no phenol red). Adiponectin (1 μg/ml) was used to initiate Ca2+ release. For experiments using RNA interference cells, cells were bathed in warm Rat Ringer solution after loading (135 mm NaCl, 5.4 mm KCl, 5 mm HEPES, pH 7.2). Ringer solution was prepared with 1.8 mm CaCl2 except for a zero Ca2+ Ringer solution, which also contained 1 mm EGTA. For experiments with inhibitor U73122, C2C12 cells were preincubated with 3 μm U73122 for 1 h before the cells were incubated with Fluo-4 AM. Images were analyzed with ImageJ (rsb.info.nih.gov) software. Images were not filtered for analysis or display. Ca2+ increases are reported as ΔF/F = (F − Frest)/Frest, where Frest is the resting level of fluorescence, and F is fluorescence intensity at any time.
RESULTS
Adiponectin-stimulated Cytosolic Translocation of LKB1 Is Important for AMPK Activation in C2C12 Cells
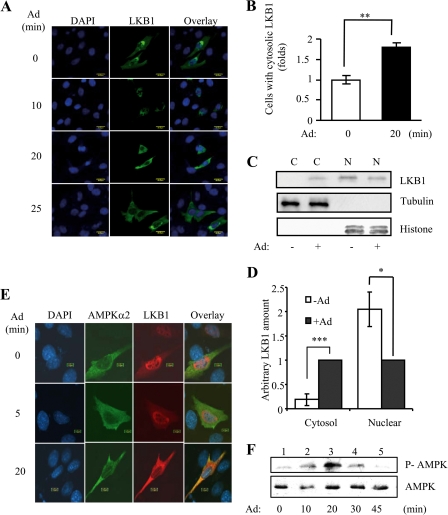
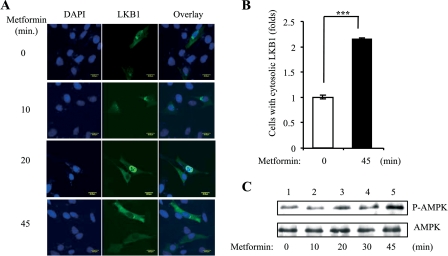
To determine whether cytosolic localization of LKB1 plays a role in adiponectin-stimulated AMPK activation, we first examined the cellular localization of LKB1 in C2C12 myoblasts. Under basal conditions, both overexpressed (Fig. 1A) and endogenous (Fig. 1C) LKB1 are localized mainly in the nucleus. In response to adiponectin stimulation, overexpressed LKB1 translocated from the nucleus to the cytosol in a time-dependent manner (Fig. 1, A and B). Similarly, adiponectin treatment led to a translocation of endogenous LKB1 from the nucleus to the cytosol (Fig. 1, C and D). Overexpressed AMPKα2 is localized in the cytosol, and adiponectin treatment has no effect on AMPKα2 localization (Fig. 1E). Treating C2C12 myotubes with adiponectin led to a time-dependent phosphorylation of AMPKα2, and the maximal phosphorylation was observed at 20 min (Fig. 1F). The correlation between LKB1 cytosolic translocation and AMPKα2 phosphorylation suggests a mechanism by which AMPKα2 is activated by adiponectin in C2C12 cells. In C2C12 cells, metformin also induced a time-dependent LKB1 cytosolic translocation that correlated with AMPK activation (Fig. 2). These findings suggest that cytosolic translocation of LKB1 could be a common mechanism for AMPK activation in response to these insulin sensitizers.
FIGURE 1.
Adiponectin-stimulated cytosolic translocation of LKB1 is important for AMPK activation in C2C12 cells. A, time course of adiponectin-induced LKB1 translocation. Confocal microscopy images depict the localization of the Myc-tagged LKB1 (green), stained with Myc antibody, in C2C12 myoblasts treated with or without adiponectin (Ad, 1 μg/ml) as indicated. The cell nuclei were stained with DAPI (blue). Scale bar, 20 μm. B, quantification of cells with cytosolic LKB1 shown in A. The error bars represent mean ± S.E. from three independent experiments. **, p < 0.01. C, cell fractionation of endogenous LKB1. After serum starvation and treatment with adiponectin (Ad, 1 μg/ml) for 20 min, C2C12 myoblasts were subjected to fractionation to obtain the nuclear (N) and cytoplasmic (C) fractions. The LKB1 protein was detected with an antibody specific to LKB1 (1st panel). The protein levels of tubulin (a marker of cytosolic protein) or histone (a marker of nuclear protein) in the fractions were detected and used as markers to indicate successful fractionation. D, quantification of endogenous LKB1 in cytosol and nucleus in response to adiponectin stimulation shown in C. The error bars represent mean ± S.E. from three independent experiments. ***, p < 0.001; *, p < 0.1. E, LKB1 co-localized with AMPKα2 in response to adiponectin stimulation. Confocal images depicting the localization of HA-tagged LKB1 and Myc-tagged AMPKα2 in C2C12 myoblasts treated with or without adiponectin (Ad, 1 μg/ml) for 0–20 min are shown. The localization and expression of LKB1 (red, 3rd column) or AMPKα2 (green, 2nd column) were determined with HA and Myc antibodies, respectively. The cell nuclei were stained with DAPI (blue, 1st column). Overlays of LKB1, AMPKα2, and nuclei are shown in the 4th column. Scale bar, 10 μm. F, time course for adiponectin-stimulated AMPK activation. After serum starvation, C2C12 myoblasts were treated with adiponectin (Ad, 1 μg/ml) as indicated. The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel) and AMPK protein (2nd panel) levels was detected by Western blot analysis with specific antibodies.
FIGURE 2.
Metformin-stimulated cytosolic translocation of LKB1 and AMPK activation in C2C12 cells. A, time course of metformin-induced LKB1 translocation. Confocal microscopy images depict the localization of the Myc-tagged LKB1 (green, 2nd column), stained with Myc antibody, in C2C12 myoblasts treated with or without metformin (500 μm). The cell nuclei were stained with DAPI (blue, 1st column). Scale bar, 20 μm. B, quantification of cells with cytosolic LKB1 shown in A. The error bars represent mean ± S.E. from three independent experiments. ***, p < 0.001. C, time course for metformin-stimulated AMPK activation. After serum starvation, C2C12 myoblasts were treated with metformin (500 μm) as indicated. The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel) and AMPK protein (2nd panel) levels were detected by Western blot analysis with specific antibodies.
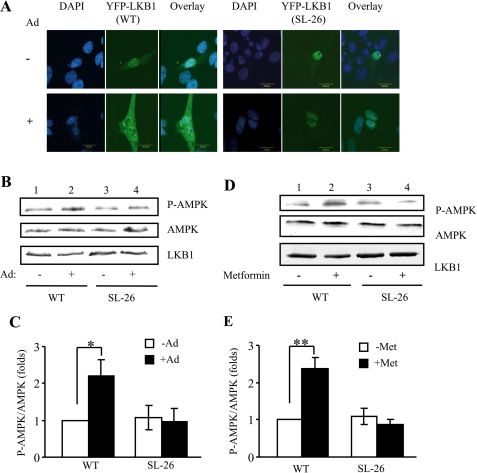
To test whether LKB1 cytosolic translocation is essential for adiponectin-stimulated AMPK activation, we examined adiponectin-stimulated localization of LKB1SL-26, a constitutively nucleus-localized but kinase-active LKB1 mutant originally identified in Peutz-Jeghers syndrome patients (13). In C2C12 myoblasts, overexpressed LKB1SL-26 is exclusively localized in the nucleus, and adiponectin treatment did not induce translocation of LKB1SL-26 to cytosol (Fig. 3A). In addition, overexpression of LKB1SL-26 inhibited adiponectin- and metformin-stimulated AMPK phosphorylation in C2C12 myoblasts (Fig. 3, B–E). Taken together, these results demonstrate that enhanced cytosolic localization of LKB1 provides a mechanism by which adiponectin activates AMPK in muscle cells.
FIGURE 3.
Cytosolic translocation of LKB1 is essential for AMPK phosphorylation. A, effect of adiponectin on LKB1SL-26 translocation. Confocal microscopy images depict the localization of the YFP-tagged LKB1 wild type (WT) (green, left panel) and YFP-tagged LKB1SL-26 (green, right panel) in C2C12 myoblasts treated with or without adiponectin (Ad, 1 μg/ml) for 20 min. The cell nuclei were stained with DAPI (blue). Scale bar, 20 μm. Serum-starved C2C12 myoblasts overexpressing LKB1-WT or LKB1SL-26 were treated with or without adiponectin (1 μg/ml for 20 min) (B and C) or metformin (500 μm for 45 min) (D and E). The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel), AMPK protein (2nd panel), and YFP-LKB1 (3rd panel) levels was detected by Western blot analysis with specific antibodies. The error bars represent mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01.
Adiponectin-induced LKB1 Subcellular Translocation Is Dependent on APPL1
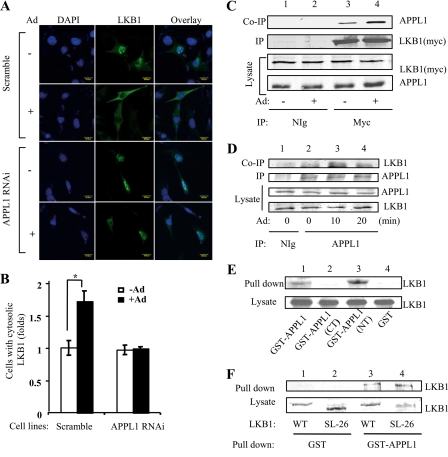
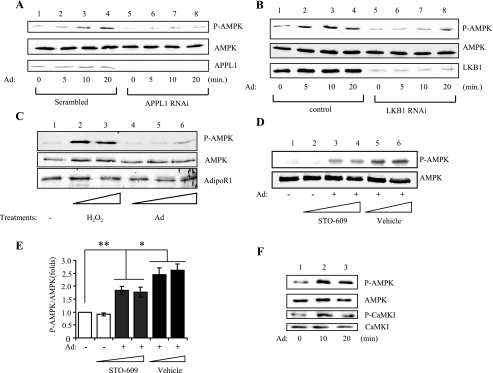
The adaptor protein APPL1 is critical for adiponectin-stimulated AMPK activation in muscle cells (5). To determine whether APPL1 plays a role in adiponectin-stimulated LKB1 subcellular translocation, we examined LKB1 cellular localization in control C2C12 and APPL1-suppressed C2C12 myoblasts (5). Under basal conditions, overexpressed LKB1 was predominantly nuclearly localized in the scramble control C2C12 and APPL1-suppressed C2C12 myoblasts (Fig. 4A). Interestingly, in APPL1-suppressed C2C12 cells, adiponectin-stimulated LKB1 cytoplasmic translocation is greatly reduced compared with that in the scramble control cells (Fig. 4, A and B).
FIGURE 4.
Adiponectin-induced LKB1 subcellular translocation is dependent on APPL1. A, APPL1 is required for adiponectin-induced LKB1 translocation. Confocal microscopy images depict the localization of the Myc-tagged LKB1 in C2C12 APPL1 short hairpin RNA stable cells or scramble control cells treated with or without adiponectin (Ad, 1 μg/ml) for 20 min. The localization of LKB1 (green) was determined with an antibody to the Myc tag. The cell nuclei were stained with DAPI (blue). Scale bar, 20 μm. B, quantification of cells with cytosolic LKB1 shown in A. The cells with cytosolic LKB1 were counted and shown as a graphic representation. The error bars represent mean ± S.E. from three independent experiments. *, p < 0.05. C, LKB1 interacts with APPL1 in cells. C2C12 myoblasts transiently expressing Myc-tagged LKB1 were serum-starved and treated with or without adiponectin (Ad, 1 μg/ml) for 10 min. Immunoprecipitated (IP) LKB1 with an anti-Myc monoclonal antibody (2nd panel), co-immunoprecipitated (Co-IP) endogenous APPL1 (1st panel), and the input of LKB1 and APPL1 proteins (3rd to 4th panels) were detected with specific antibodies as indicated. D, adiponectin stimulates the interaction of LKB1 with APPL1 in skeletal muscle tissue. Endogenous APPL1 was immunoprecipitated from the mouse skeletal muscle tissue treated without or with adiponectin (Ad, 2.5 μg/ml) as indicated. Co-immunoprecipitated LKB1 (1st panel), immunoprecipitated APPL1 (2nd panel), and the input of APPL1 and LKB1 proteins (3rd and 4th panel) were detected with specific antibodies as indicated. E, LKB1 interacts with the BAR domain of APPL1 in vitro. Cell lysates of C2C12 myotubes were incubated with GST-APPL1 (amino acids 1–709), GST-APPL1 (CT) (amino acids 455–693, PTB domain), GST-APPL1 (NT) (amino acids 5–270, BAR domain), and GST beads. Bound LKB1 (1st panel) and LKB1 in the lysate (2nd panel) were detected by Western blot analysis using an antibody specific to LKB1. F, APPL1 interacts with nuclear LKB1 in vitro. C2C12 myoblasts overexpressing YFP-tagged wild type (WT) or SL-26 mutant of LKB1 were pulled down with GST or GST-APPL1 fusion proteins. The bound LKB1 and LKB1 expression control (lysate) were detected with anti-green fluorescent protein antibody.
To determine the mechanism by which APPL1 regulates LKB1 subcellular translocation, we tested if APPL1 and LKB1 interact in cells. Co-immunoprecipitation experiments showed that endogenous APPL1 interacts with Myc-tagged LKB1 in C2C12 myoblasts, and the interaction was stimulated by adiponectin (Fig. 4C). Similar results were obtained in mouse skeletal muscle cells where adiponectin treatment enhanced the association of APPL1 and LKB1 (Fig. 4D). Affinity binding experiments demonstrated that LKB1 could interact with APPL1 in vitro, and the NH2-terminal Bin1/amphiphysin/Rvs167 (BAR) domain of APPL1 is involved in this interaction (Fig. 4E). In agreement with this result, our yeast two-hybrid study showed that the NH2-terminal BAR domain of APPL1 interacted with LKB1 (data not shown). However, we detected no interaction between APPL1 and AMPK, although under a similar condition APPL1 formed a complex with Akt (data not shown), consistent with the finding of others (43). These results suggest that the interaction with LKB1 may provide a mechanism by which APPL1 mediates adiponectin signaling to AMPK activation. To test whether nuclear and cytosolic LKB1 have different binding affinity to APPL1, we examined the binding of APPL1 to wild type and the SL-26 form of LKB1, a constitutively nuclear localized LKB1 mutant that has no response to adiponectin stimulation (Fig. 3A). We found that APPL1 bound with similar affinity to both the wild type and SL-26 mutant of LKB1 in vitro (Fig. 4F), suggesting that the stimulatory effect of adiponectin on the interaction between APPL1 and LKB1 could be mediated by promoting LKB1 cytosolic translocation. However, we cannot rule out the possibility that adiponectin stimulation causes APPL1 conformational change or post-translational modification, which allows APPL1 to bind LKB1 with higher affinity in cells. Nevertheless, our results show that adiponectin stimulation alters the nuclear/cytosolic ratio of LKB1 and allows APPL1 anchoring LKB1 in the cytosol to efficiently activate AMPK. As LKB1 exists as a complex with STRAD and MO25 in cells (21, 44), we tested whether adiponectin has any effect on LKB1-STRAD-MO25 complex formation. Our data indicated that adiponectin had no effect on the association of LKB1 with STRAD and MO25, and APPL1 did not show a direct interaction with either STRADα or MO25β in the presence or absence of adiponectin (data not shown).
Existence of an APPL1/LKB1-independent Mechanism for AMPK Activation by Adiponectin
We previously found that completely suppressing APPL1 expression levels in C2C12 cells significantly reduced adiponectin-stimulated AMPK phosphorylation (5). However, with longer stimulation, adiponectin was able to stimulate AMPK phosphorylation in the APPL1-suppressed cells, although to a lesser extent (Fig. 5A), indicating the presence of an APPL1-independent mechanism for AMPK activation. To test this, we generated LKB1-suppressed C2C12 cells. In LKB1 knockdown C2C12 myotubes, adiponectin stimulated AMPK phosphorylation to a lesser extent as compared with control cells (Fig. 5B). A similar observation was made in HeLa cells, which lack endogenous LKB1 (Fig. 5C). These results suggest the existence of an APPL1/LKB1-independent pathway that is able to mediate the stimulatory effect of adiponectin on AMPK activation.
FIGURE 5.
Adiponectin-stimulated AMPK activation is partially mediated through CaMKK with an APPL1/LKB1-independent mechanism. A, AMPK phosphorylation in APPL1-suppressed C2C12 cells. APPL1-suppressed or scramble control C2C12 myoblasts were treated with adiponectin (Ad, 1 μg/ml) as indicated. The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel) and the protein levels of AMPK (2nd panel) and APPL1 (3rd panel) were detected by Western blot analysis with specific antibodies. RNAi, RNA interference. B, AMPK phosphorylation in LKB1-suppressed C2C12 myoblasts. LKB1-suppressed or the control C2C12 myoblasts were treated with adiponectin (Ad, 1 μg/ml) as indicated. The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel), the protein levels of AMPK (2nd panel), and LKB1 (3rd panel) were detected by Western blot analysis with specific antibodies. C, AMPK phosphorylation in HeLa cells. HeLa cells were serum-starved and treated with or without H2O2 (500 and 1000 μm as positive control, lanes 2 and 3, respectively) or adiponectin (Ad, 0.5, 1, and 2 μg/ml, lanes 4–6, respectively) for 15 min. The Thr172 phosphorylation of AMPK (P-AMPK, 1st panel), AMPK protein (2nd panel), and AdipoR1 protein (3rd panel, as a loading control) levels were detected by Western blot analysis with specific antibodies. D, CaMKK inhibitor partially blocks adiponectin-stimulated AMPK activation. C2C12 myotubes were pretreated with STO-609 (0.2, 0.5, or 1.0 μg/ml for 6 h in lanes 2–4, respectively) or vehicle control (lanes 5 and 6), followed by adiponectin (Ad) stimulation (lanes 3–6, 1 μg/ml for 20 min). The phosphorylation (1st panel) and protein (2nd panel) levels of AMPK in cell lysates were detected by Western blot with specific antibodies as indicated. E, quantification of AMPK phosphorylation shown in D. The error bars represent mean ± S.E. from four independent experiments. *, p < 0.05; **, p < 0.01. F, adiponectin stimulates CaMKK activity in skeletal muscle tissue. Mouse skeletal muscle tissue was treated without or with adiponectin (Ad, 2.5 μg/ml) as indicated. The homogenates from the tissue were analyzed for P-AMPK at Thr172 (1st panel), AMPK protein (2nd panel), P-CaMKI at Thr177 (3rd panel, a marker of CaMKK activity), and CaMKI protein (4th panel) were detected with specific antibodies as indicated.
Both CaMKKα and CaMKKβ act as upstream kinases for AMPK (24–26). Hence we investigated whether these kinases play a role in mediating adiponectin-stimulated AMPK activation in muscle cells. Treating C2C12 myotubes with the CaMKK inhibitor, STO-609, partially blocked the stimulatory effect of adiponectin on AMPK phosphorylation (Fig. 5D, 1st panel, lanes 5 and 6 versus lanes 3 and 4) (Fig. 5E), suggesting the involvement of CaMKK in adiponectin-stimulated AMPK activation. The activation of CaMKK in response to adiponectin stimulation was further confirmed in mouse skeletal muscle tissues, in which adiponectin stimulated the phosphorylation of CaMKI, a substrate of CaMKK (Fig. 5F).
Adiponectin Induces Ca2+ Release in a PLC-dependent and an APPL1-independent Manner
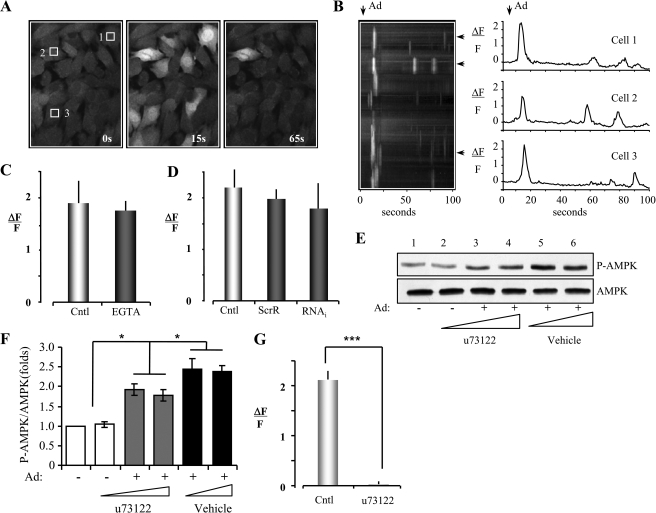
Increasing intracellular Ca2+ concentrations, which activates CaMKK (25, 29, 30), correlates with increased AMPK activity in LKB1-deficient cells (24–26). Consequently, we tested whether adiponectin stimulates Ca2+ release in C2C12 myoblasts. Cells were loaded with the Ca2+ indicator dye Fluo-4 AM and imaged with fluorescence microscopy (Fig. 6A). The fluorescent intensity of Fluo-4 AM (ΔF/F) in ∼50% of the cells rapidly increased over 2-fold within 15 s of treating the cells with adiponectin (Fig. 6, A and B), whereas there is no fluorescent signal in the cells treated with vehicle control (phosphate-buffered saline) (data not shown). The adiponectin-induced increase in Ca2+ signal was not affected by treating C2C12 myoblasts with the Ca2+ chelator EGTA (Fig. 6C), suggesting that adiponectin-stimulated Ca2+ release is from an intracellular Ca2+ store. Interestingly, no significant difference in adiponectin-induced Ca2+ release was observed between the APPL1-suppressed C2C12 myoblasts and the scramble control cells (Fig. 6D). To investigate the potential molecular mechanism by which adiponectin stimulates intracellular Ca2+ release, we tested the effect of the PLC inhibitor, U73122, on adiponectin-stimulated AMPK activation. As shown in Fig. 6E, U73122 partially blocked adiponectin-induced AMPK phosphorylation. U73122 treatment led to a 30% reduction of adiponectin-induced AMPK phosphorylation compared with adiponectin treatment control (Fig. 6, E and F), which is similar to the inhibitory effect by the CaMKK inhibitor (STO-609) (Fig. 6F versus Fig. 5E). These results suggest that adiponectin activates PLC to produce inositol triphosphate (IP3) in the cytosol, which in turn activates IP3 receptors and leads to Ca2+ release from the ER (46). Consistent with this model, we found that adiponectin-stimulated Ca2+ release was completely blocked in the presence of U73122 (Fig. 6G), strongly indicating that the ER was the sole source of Ca2+ release. Together, these results indicate that adiponectin can also activate AMPK via the APPL1/LKB1-independent but PLC/Ca2+/CaMKK-dependent pathway by releasing intracellular Ca2+ from the ER via IP3 receptor activation.
FIGURE 6.
Adiponectin induces intracellular Ca2+ release from ER in a PLC-dependent and APPL1-independent manner. A, images of cultured C2C12 myoblasts loaded with the Ca2+ indicator Fluo-4 AM before (0 s) and after (15 and 65 s) addition of adiponectin (1 μg/ml) to the recording chamber. B, spatial-temporal stack of images (1 frame per s) for the entire field presented in A plus single trace Ca2+ responses of three individual cells indicated by the white squares in A. C, histogram plot of adiponectin-induced peak Ca2+ responses in zero (EGTA) versus normal extracellular Ca2+ (Cntl). The error bars represent mean ± S.D. from three independent experiments. D, histogram plot of adiponectin-induced peak Ca2+ responses for APPL1 short hairpin RNA cells (RNA interference (RNAi)) versus scrambled RNA interference (ScrR) or wild type C2C12 myoblasts (Cntl). The error bars represent mean ± S.D. from three independent experiments. E, PLC inhibitor partially blocks adiponectin-stimulated AMPK activation. C2C12 myotubes were pretreated with u73122 (1, 1, and 3 μm for 6 h in lanes 2–4, respectively) or vehicle control (lanes 5 and 6), followed by adiponectin (Ad) stimulation (lanes 3–6, 1 μg/ml for 20 min). The phosphorylation (1st panel) and protein (2nd panel) levels of AMPK in cell lysates were detected by Western blot with specific antibodies as indicated. F, quantification of AMPK phosphorylation shown in E. The error bars represent mean ± S.E. from four independent experiments. *, p < 0.05. G, histogram plot of adiponectin-induced peak Ca2+ responses for no inhibitor treatment (Cntl) versus pretreatment of the C2C12 myoblasts with u73122 (3 μm for 1 h before adding adiponectin). The error bars represent mean ± S.D. from three independent experiments. ***, p < 0.001.
DISCUSSION
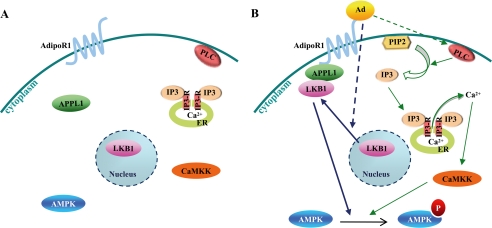
We have recently found that the adaptor protein APPL1 interacts directly with adiponectin receptors and positively mediates adiponectin signaling to activate AMPK in muscle cells (5). In this study, we investigated the molecular mechanism for adiponectin-mediated AMPK activation in C2C12 cells. Adiponectin induces AMPK activation through two different signaling pathways as follows: the APPL1/LKB1-dependent major pathway by promoting the cytosolic localization of LKB1, and the PLC/Ca2+/CaMKK-dependent minor pathway by releasing Ca2+ from intracellular stores (Fig. 7).
FIGURE 7.
A model of adiponectin-stimulated AMPK activation in muscle cells. A, under basal conditions, LKB1 is localized in the nucleus and APPL1, CaMKK, and inactive AMPK in the cytosol. B, adiponectin stimulation induces LKB1 translocation from the nucleus into cytosol, associates with APPL1, and phosphorylates AMPK. Adiponectin also activates PLC, which subsequently produces IP3 and stimulates IP3 receptor on the ER membrane. The release of Ca2+ from the ER leads to the stimulation of CaMKK, which activates AMPK in an APPLl-independent manner.
Adiponectin exists either as multimers of full-length adiponectin or as globular adiponectin (2, 47, 48). In this study, we used the globular adiponectin because this form has a higher affinity for muscle cells than full-length adiponectin (6). Studies have shown that acute treatment of mice with globular adiponectin increases fatty acid oxidation in muscle (47), and overexpression of globular adiponectin inhibits the progression of atherosclerosis in vivo (50).
One of the major findings in this study is that the APPL1/LKB1-dependent pathway plays a major role in mediating adiponectin-stimulated AMPK activation. Our results show that LKB1 cytosolic translocation is essential for adiponectin-induced AMPK activation, because overexpression of a constitutively nucleus-localized but enzymatically active mutant of LKB1, LKB1SL-26, blocked the effects of adiponectin on AMPK activation (Fig. 3, A–C). This notion is supported by the finding that metformin-induced AMPK activation in C2C12 cells also involves cytosolic translocation of LKB1 (Fig. 2 and Fig. 3, D and E). Cytosolic translocation of LKB1 appears to be a common mechanism to activate AMPK, because AMPK activation by metformin and peroxynitrite in endothelial cells also involves LKB1 cytosolic translocation (51–53). Our study demonstrates for the first time that adiponectin stimulates the cytosolic translocation of LKB1 in C2C12 cells.
Suppression of APPL1 expression inhibited LKB1 cytosolic translocation (Fig. 4, A and B) and significantly attenuated AMPK activation in response to adiponectin stimulation (Fig. 5A), suggesting that APPL1 is essential for adiponectin-induced and LKB1-catalyzed AMPK activation (5). APPL1 interacts with the majority of its associating proteins through its PTB domain, whereas the BAR domain is implicated only in the binding of APPL2 and Rab5 (54–56). In this study, we identified LKB1 as a new binding partner of APPL1 and showed that the BAR domain of APPL1 is involved in this interaction (Fig. 4E). It is possible that, in addition to its role in mediating adiponectin signaling by interacting with AdipoR1 and AdipoR2, APPL1 functions as an anchoring protein to tether LKB1 in the cytosol, which leads to subsequent phosphorylation of AMPK (Fig. 7). To date it remains elusive how the function of constitutively active LKB1 is regulated in cells (57). Although multiple phosphorylation sites have been identified in LKB1, most of the studies showed that phosphorylation at these sites have no effect on LKB1 activity and/or function (51, 57, 58). Xie et al. (51) recently identified Ser428 as a PKCζ-mediated phosphorylation site in human LKB1 and showed that phosphorylation at this site regulates metformin-induced LKB1 activity and its cytosolic localization in endothelial cells. However, these findings were disputed by a very recent study showing that phosphorylation at the residue equivalent to Ser428 of human LKB1 had no effect on the activity of mouse LKB1 to phosphorylate its downstream targets (58). LKB1 function in cells is modulated by its cellular localization. Several LKB1-associated proteins such as STRAD and MO25 have been reported to regulate LKB1 subcellular localization (21, 22, 44, 59). Binding to STRAD anchors LKB1 in the cytosol where the LKB1-STRAD complex is stabilized by MO25 (21). However, we found that APPL1 did not interact with MO25 or STRAD in C2C12 cells, suggesting the presence of an alternative mechanism by which APPL1 mediates adiponectin- or metformin-induced LKB1 cytosolic translocation.
Accumulating evidence suggests that LKB1 is a major, but not the only, upstream kinase of AMPK in skeletal muscle cells. One of the other AMPK upstream kinases is CaMKK (24–26). Interestingly, adiponectin has been reported to activate AMPK through CaMKKβ in human umbilical vein endothelial cells (60). In this study, we demonstrated that adiponectin is able to activate CaMKK and AMPK in C2C12 myoblasts by inducing intracellular Ca2+ release. This result is in agreement with the findings of others who showed that AMPK could be activated by oxytocin-induced or the thyroid hormone triiodothyronine-induced Ca2+ release in C2C12 myoblasts (42, 45). Although the precise biochemical mechanism by which adiponectin induces Ca2+ release remains to be established, this activation process appears to be APPL1-independent but involves PLC-mediated IP3 production (Fig. 7). Increased IP3 production by PLC in response to adiponectin stimulation could trigger IP3 receptor activation, which in turn induces Ca2+ release from the ER (46). Adiponectin receptors contain seven transmembrane domains but have been suggested to be structurally and functionally different from typical G protein-coupled receptors (4). The findings that adiponectin activates PLC and promotes IP3 production indicate that this conclusion may need to be revisited. It is possible that adiponectin-induced and IP3-mediated Ca2+ release is because of the interaction of adiponectin with another receptor subtype. Alternatively, adiponectin receptors may couple with a G protein and activate PLC in a manner similar to the classic G protein-coupled receptors. Future studies will be needed to test these possibilities.
We demonstrated in this study that adiponectin-stimulated activation of AMPK in C2C12 cells is mainly through the APPL1/LKB1-dependent pathway (Fig. 3C and Fig. 4B), whereas the PLC/Ca2+/CaMKK-dependent pathway seems to play a minor role in the activation process (Fig. 5E and Fig. 6F). AMPK is well documented as an energy sensor, and it is not surprising that the activity of this kinase is subject to a dynamic and specific regulation by multiple signaling pathways in cells. It is possible that the APPL1/LKB1-dependent pathway and the PLC/Ca2+/CaMKK-dependent pathway are activated by adiponectin simultaneously in muscle cells. Alternatively, the activation of these distinct pathways may depend on the specific condition required for normal cell metabolism and function. For example, it has been shown that LKB1 is a primary AMPK kinase in skeletal muscle under conditions of high cellular energy stress (4). On the other hand, calcium signaling may play a major role in AMPK activation under more gentle contraction conditions (49). Thus, the involvement of both signaling pathways in adiponectin-induced AMPK activation could provide a mechanism to specifically and dynamically regulate AMPK activity and function in response to environment and nutrition requirements (26).
In summary, we have uncovered two different mechanisms by which adiponectin activates AMPK in C2C12 cells, which involves enhanced LKB1 cytosolic translocation and PLC/Ca2+-mediated CaMKK activation (Fig. 7). We have also demonstrated the presence of APPL1-dependent and -independent pathways mediating the stimulatory effect of adiponectin. Our results thus provide new insight into the mechanisms by which growth factors or pharmacological drugs activate AMPK, a promising therapeutic target for the treatment of diabetes, inflammation, and cardiovascular diseases.
Acknowledgments
We thank Derong Hu and Crystal Ramirez for excellent technical assistance, and Dr. Victoria Frohlich (Associate Director, Digital Optical Imaging Facility, University of Texas Health Science Center, San Antonio) for assistance in confocal microscopy studies. We also thank Dr. Dario Alessi for providing YFP-fused LKB1 constructs, and Drs. Nicolas Musi and Ning Zhang for helping with the isolation of skeletal muscle tissues from mouse.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK69930 (to L. Q. D.), RO1 DK76902 (to F. L.), T32 AG021890 (to J. C. E.), and PPG AG19316 (to J. D. L.). This work was also supported by a career development award from the American Diabetes Association (to L. Q. D.).
- AMPK
- AMP-activated protein kinase
- PLC
- phospholipase C
- CaMKK
- Ca2+/calmodulin-dependent protein kinase kinase
- ER
- endoplasmic reticulum
- IP3
- inositol triphosphate
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- YFP
- yellow fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Scherer P. E., Williams S., Fogliano M., Baldini G., Lodish H. F. (1995) J. Biol. Chem. 270, 26746–26749 [DOI] [PubMed] [Google Scholar]
- 2.Kadowaki T., Yamauchi T. (2005) Endocr. Rev. 26, 439–451 [DOI] [PubMed] [Google Scholar]
- 3.Mao X., Hong J. Y., Dong L. Q. (2006) Mini Rev. Med. Chem. 6, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M., Murakami K., Ohteki T., Uchida S., Takekawa S., Waki H., Tsuno N. H., Shibata Y., Terauchi Y., Froguel P., Tobe K., Koyasu S., Taira K., Kitamura T., Shimizu T., Nagai R., Kadowaki T. (2003) Nature 423, 762–769 [DOI] [PubMed] [Google Scholar]
- 5.Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ-Roberts C. Y., Hong J. Y., Kim R. Y., Liu F., Dong L. Q. (2006) Nat. Cell Biol. 8, 516–523 [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., Eto K., Akanuma Y., Froguel P., Foufelle F., Ferre P., Carling D., Kimura S., Nagai R., Kahn B. B., Kadowaki T. (2002) Nat. Med. 8, 1288–1295 [DOI] [PubMed] [Google Scholar]
- 7.Hue L., Rider M. H. (2007) Essays Biochem. 43, 121–137 [DOI] [PubMed] [Google Scholar]
- 8.Towler M. C., Hardie D. G. (2007) Circ. Res. 100, 328–341 [DOI] [PubMed] [Google Scholar]
- 9.Hawley S. A., Davison M., Woods A., Davies S. P., Beri R. K., Carling D., Hardie D. G. (1996) J. Biol. Chem. 271, 27879–27887 [DOI] [PubMed] [Google Scholar]
- 10.Salt I., Celler J. W., Hawley S. A., Prescott A., Woods A., Carling D., Hardie D. G. (1998) Biochem. J. 334, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. (2004) J. Clin. Invest. 113, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witczak C. A., Sharoff C. G., Goodyear L. J. (2008) Cell. Mol. Life Sci. 65, 3737–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Höglund P., Järvinen H., Kristo P., Pelin K., Ridanpää M., Salovaara R., Toro T., Bodmer W., Olschwang S., Olsen A. S., Stratton M. R., de la Chapelle A., Aaltonen L. A. (1998) Nature 391, 184–187 [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto K., Göransson O., Hardie D. G., Alessi D. R. (2004) Am. J. Physiol. Endocrinol. Metab. 287, E310–E317 [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto K., McCarthy A., Smith D., Green K. A., Grahame, Hardie D., Ashworth A., Alessi D. R. (2005) EMBO J. 24, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley S. A., Selbert M. A., Goldstein E. G., Edelman A. M., Carling D., Hardie D. G. (1995) J. Biol. Chem. 270, 27186–27191 [DOI] [PubMed] [Google Scholar]
- 17.Sanders M. J., Grondin P. O., Hegarty B. D., Snowden M. A., Carling D. (2007) Biochem. J. 403, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiainen M., Ylikorkala A., Mäkelä T. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith D. P., Spicer J., Smith A., Swift S., Ashworth A. (1999) Hum. Mol. Genet. 8, 1479–1485 [DOI] [PubMed] [Google Scholar]
- 20.Tiainen M., Vaahtomeri K., Ylikorkala A., Mäkelä T. P. (2002) Hum. Mol. Genet. 11, 1497–1504 [DOI] [PubMed] [Google Scholar]
- 21.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. (2003) EMBO J. 22, 5102–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith D. P., Rayter S. I., Niederlander C., Spicer J., Jones C. M., Ashworth A. (2001) Hum. Mol. Genet. 10, 2869–2877 [DOI] [PubMed] [Google Scholar]
- 23.Imai K., Inukai K., Ikegami Y., Awata T., Katayama S. (2006) Biochem. Biophys. Res. Commun. 351, 595–601 [DOI] [PubMed] [Google Scholar]
- 24.Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 25.Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., Witters L. A. (2005) J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 26.Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., Carling D. (2005) Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 27.Sakagami H., Saito S., Kitani T., Okuno S., Fujisawa H., Kondo H. (1998) Brain Res. Mol. Brain Res. 54, 311–315 [DOI] [PubMed] [Google Scholar]
- 28.Tokumitsu H., Enslen H., Soderling T. R. (1995) J. Biol. Chem. 270, 19320–19324 [DOI] [PubMed] [Google Scholar]
- 29.Edelman A. M., Mitchelhill K. I., Selbert M. A., Anderson K. A., Hook S. S., Stapleton D., Goldstein E. G., Means A. R., Kemp B. E. (1996) J. Biol. Chem. 271, 10806–10810 [DOI] [PubMed] [Google Scholar]
- 30.Anderson K. A., Means R. L., Huang Q. H., Kemp B. E., Goldstein E. G., Selbert M. A., Edelman A. M., Fremeau R. T., Means A. R. (1998) J. Biol. Chem. 273, 31880–31889 [DOI] [PubMed] [Google Scholar]
- 31.Vinet J., Carra S., Blom J. M., Harvey M., Brunello N., Barden N., Tascedda F. (2003) Brain Res. Mol. Brain Res. 111, 216–221 [DOI] [PubMed] [Google Scholar]
- 32.Witczak C. A., Fujii N., Hirshman M. F., Goodyear L. J. (2007) Diabetes 56, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 33.Jensen T. E., Rose A. J., Jørgensen S. B., Brandt N., Schjerling P., Wojtaszewski J. F., Richter E. A. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1308–E1317 [DOI] [PubMed] [Google Scholar]
- 34.McGee S. L., Mustard K. J., Hardie D. G., Baar K. (2008) J. Physiol. 586, 1731–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. (2006) J. Biol. Chem. 281, 32207–32216 [DOI] [PubMed] [Google Scholar]
- 36.Bram R. J., Hung D. T., Martin P. K., Schreiber S. L., Crabtree G. R. (1993) Mol. Cell. Biol. 13, 4760–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sapkota G. P., Boudeau J., Deak M., Kieloch A., Morrice N., Alessi D. R. (2002) Biochem. J. 362, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong L. Q., Zhang R. B., Langlais P., He H., Clark M., Zhu L., Liu F. (1999) J. Biol. Chem. 274, 8117–8122 [DOI] [PubMed] [Google Scholar]
- 39.Lim M. A., Kikani C. K., Wick M. J., Dong L. Q. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14006–14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuoppamaa H., Skrobuk P., Sihvo M., Hiukka A., Chibalin A. V., Zierath J. R., Koistinen H. A. (2008) Diabetes Metab. Res. Rev. 24, 554–562 [DOI] [PubMed] [Google Scholar]
- 41.Camacho P., Lechleiter J. D. (1993) Science 260, 226–229 [DOI] [PubMed] [Google Scholar]
- 42.Lee E. S., Uhm K. O., Lee Y. M., Kwon J., Park S. H., Soo K. H. (2008) Regul. Pept. 151, 71–74 [DOI] [PubMed] [Google Scholar]
- 43.Mitsuuchi Y., Johnson S. W., Sonoda G., Tanno S., Golemis E. A., Testa J. R. (1999) Oncogene 18, 4891–4898 [DOI] [PubMed] [Google Scholar]
- 44.Baas A. F., Boudeau J., Sapkota G. P., Smit L., Medema R., Morrice N. A., Alessi D. R., Clevers H. C. (2003) EMBO J. 22, 3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamauchi M., Kambe F., Cao X., Lu X., Kozaki Y., Oiso Y., Seo H. (2008) Mol. Endocrinol. 22, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikoshiba K., Hattori M. (2000) Sci. STKE. 51, PE1. [DOI] [PubMed] [Google Scholar]
- 47.Fruebis J., Tsao T. S., Javorschi S, Ebbets-Reed D., Erickson M. R., Yen F. T., Bihain B. E., Lodish H. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waki H., Yamauchi T., Kamon J., Kita S., Ito Y., Hada Y., Uchida S., Tsuchida A., Takekawa S., Kadowaki T. (2005) Endocrinology 146, 790–796 [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen S. B., Rose A. J. (2008) Front. Biosci. 13, 5589–5604 [DOI] [PubMed] [Google Scholar]
- 50.Yamauchi T., Kamon J., Waki H., Imai Y., Shimozawa N., Hioki K., Uchida S., Ito Y., Takakuwa K., Matsui J., Takata M., Eto K., Terauchi Y., Komeda K., Tsunoda M., Murakami K., Ohnishi Y., Naitoh T., Yamamura K., Ueyama Y., Froguel P., Kimura S., Nagai R., Kadowaki T. (2003) J. Biol. Chem. 278, 2461–2468 [DOI] [PubMed] [Google Scholar]
- 51.Xie Z., Dong Y., Scholz R., Neumann D., Zou M. H. (2008) Circulation 117, 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z., Dong Y., Zhang M., Cui M. Z., Cohen R. A., Riek U., Neumann D., Schlattner U., Zou M. H. (2006) J. Biol. Chem. 281, 6366–6375 [DOI] [PubMed] [Google Scholar]
- 53.Song P., Xie Z., Wu Y., Xu J., Dong Y., Zou M. H. (2008) J. Biol. Chem. 283, 12446–12455 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R. G., Zerial M. (2004) Cell 116, 445–456 [DOI] [PubMed] [Google Scholar]
- 55.Deepa S. S., Dong L. Q. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E22–E36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nechamen C. A., Thomas R. M., Dias J. A. (2007) Mol. Cell. Endocrinol. 260–262, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alessi D. R., Sakamoto K., Bayascas J. R. (2006) Annu. Rev. Biochem. 75, 137–163 [DOI] [PubMed] [Google Scholar]
- 58.Fogarty S., Hardie D. G. (2009) J. Biol. Chem. 284, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boudeau J., Scott J. W., Resta N., Deak M., Kieloch A., Komander D., Hardie D. G., Prescott A. R., van Aalten D. M., Alessi D. R. (2004) J. Cell Sci. 117, 6365–6375 [DOI] [PubMed] [Google Scholar]
- 60.Hattori Y., Nakano Y., Hattori S., Tomizawa A., Inukai K., Kasai K. (2008) FEBS Lett. 582, 1719–1724 [DOI] [PubMed] [Google Scholar]