Abstract
Human polymerase kappa (hPol κ) is one of four eukaryotic Y-class DNA polymerases and may be an important element in the cellular response to polycyclic aromatic hydrocarbons such as benzo[a]pyrene, which can lead to reactive oxygenated metabolite-mediated oxidative stress. Here, we present a detailed analysis of the activity and specificity of hPol κ bypass opposite the major oxidative adduct 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxoG). Unlike its archaeal homolog Dpo4, hPol κ bypasses this lesion in an error-prone fashion by inserting mainly dATP. Analysis of transient-state kinetics shows diminished “bursts” for dATP:8-oxoG and dCTP:8-oxoG incorporation, indicative of non-productive complex formation, but dATP:8-oxoG insertion events that do occur are 2-fold more efficient than dCTP:G insertion events. Crystal structures of ternary hPol κ complexes with adducted template-primer DNA reveal non-productive (dGTP and dATP) alignments of incoming nucleotide and 8-oxoG. Structural limitations placed upon the hPol κ by interactions between the N-clasp and finger domains combined with stabilization of the syn-oriented template 8-oxoG through the side chain of Met-135 both appear to contribute to error-prone bypass. Mutating Leu-508 in the little finger domain of hPol κ to lysine modulates the insertion opposite 8-oxoG toward more accurate bypass, similar to previous findings with Dpo4. Our structural and activity data provide insight into important mechanistic aspects of error-prone bypass of 8-oxoG by hPol κ compared with accurate and efficient bypass of the lesion by Dpo4 and polymerase η.
DNA damage incurred by a multitude of endogenous and exogenous factors constitutes an inevitable challenge for the replication machinery, and various mechanisms exist to either remove the resulting lesions or bypass them in a more or less mutation-prone fashion (1). Error-prone polymerases are central to trans-lesion synthesis across sites of damaged DNA (2, 3). Four so-called Y-class DNA polymerases have been identified in humans, Pol η,4 Pol ι, Pol κ, and Rev1, which exhibit different activities and abilities to replicate past a flurry of individual lesions (4, 5). Homologs have also been identified and characterized in other organisms, notably DinB (Pol IV) in Escherichia coli (6–8), Dbh in Sulfolobus acidocaldarius (9, 10), and Dpo4 in Sulfolobus solfataricus (11, 12). A decade of investigations directed at the structural and functional properties of bypass polymerases have significantly improved our understanding of this class of enzymes (5, 13). A unique feature of Y-class polymerases, compared with the common right-handed arrangement of palm, thumb, and finger subdomains of high fidelity (i.e. A-class) DNA polymerases (14), is a “little finger” or “PAD” (palm-associated domain) subdomain that plays a crucial role in lesion bypass (12, 15–21). In addition to the little finger subdomain at the C-terminal end of the catalytic core, both Rev1 and Pol κ exhibit an N-terminal extension that is absent in other translesion polymerases. The N-terminal extension in the structure of the ternary (human) hPol κ·DNA·dTTP complex folds into a U-shaped tether-helix-turn-helix “clasp” that is located between the thumb and little finger domains, allowing the polymerase to completely encircle the DNA (18). Although the precise role of the clasp for lesion bypass by hPol κ remains to be established, it is clear that this entity is functionally important, because mutant enzymes with partially or completely removed clasps exhibit diminished catalytic activity compared with the full-length catalytic core (hPol κ N1–526) or a core lacking the N-terminal 19 residues (hPol κ N19–526; the construct used for crystal structure determination of the ternary complex (18)).
7,8-Dihydro-8-oxo-2′-deoxyguanosine (8-oxoG), found in both lower organisms and eukaryotes, is a major lesion that is a consequence of oxidative stress. The lesion is of relevance not only because of its association with cancer (22, 23), but also in connection with aging (24), hepatitis (25), and infertility (26). It is far from clear which DNA polymerases bypass 8-oxoG most often in a cellular context, but given the ubiquitous nature of the lesion it seems likely that more than one enzyme could encounter the lesion. Replicative polymerases commonly insert dATP opposite template 8-oxoG, with the lesion adopting the preferred syn conformation (e.g. 27, 28). It was recently found that the translesion polymerase Dpo4 from S. solfataricus synthesizes efficiently past 8-oxoG, inserting ≥95% dCTP > dATP opposite the lesion (29, 30). The efficient and low error bypass of the 8-oxoG lesion by Dpo4 is associated to a large extent with an arginine residue in the little finger domain (17). In the crystal structure of the ternary Dpo4·DNA·dCTP complex, the side chain of Arg-332 forms a hydrogen bond to the 8-oxygen of 8-oxoG, thus shifting the nucleoside conformational equilibrium toward the anti state and enabling a Watson-Crick binding mode with the incoming dCTP (30). The efficient and accurate replication of templates bearing 8-oxoG by yeast Pol η (31, 32) may indicate similarities between the bypass reactions catalyzed by the archaeal and eukaryotic enzymes. In contrast, bypass synthesis opposite 8-oxoG by human Pol κ is error-prone, resulting in efficient incorporation of A (33–35). The inaccurate bypass of 8-oxoG is thought to contribute to the deleterious effects associated with the lesion. These observations indicate different behaviors of the eukaryotic trans-lesion Pol κ and its archaeal “homolog” Dpo4 vis-à-vis the major oxidative stress lesion 8-oxoG. A mechanistic understanding of human DNA polymerases that bypass 8-oxoG in an error-prone fashion, such as hPol κ, is therefore of great interest.
To elucidate commonalities and differences between the trans-8-oxoG syntheses of S. solfataricus Dpo4, yeast Pol η, and hPol κ, we carried out a comprehensive analysis of the bypass activity for the latter with template·DNA containing the 8-oxoG lesion, including pre-steady-state and steady-state kinetics of primer extension opposite and beyond 8-oxoG and LC-MS/MS assays of full-length extension products. We determined crystal structures of ternary hPol κ-(8-oxoG)DNA-dGTP and hPol κ-(8-oxoG)DNA-dATP complexes, apparently the first for any complex with adducted DNA for the κ enzyme reported to date. Our work demonstrates clear distinctions between genetically related translesion polymerases and provides insights into the origins of the error-prone reactions opposite 8-oxoG catalyzed by Y-family DNA polymerases.
EXPERIMENTAL PROCEDURES
Materials
All unlabeled dNTPs were obtained from Amersham Biosciences (Piscataway, NJ), and [γ-32P]ATP was purchased from PerkinElmer Life Sciences. All oligonucleotides used in this work were synthesized by Midland Certified Reagent Co. (Midland, TX) and purified using high-performance liquid chromatography by the manufacturer, with analysis by matrix-assisted laser desorption time-of-flight MS. The primer/template sequences used are listed in supplemental Table S1.
hPol κ Catalytic Core Protein Expression and Purification
The human polymerase κ construct comprising residues 19–526 (hPol κ19–526) was generated by ATG Laboratories (Eden Prairie, MN) and cloned into the pBG101 vector (Center for Structural Biology, Vanderbilt University, to generate a His-GST fusion protein with a PreScission protease cleavage site. In our work, both the hPol κ construct and the purification protocol are different from that described by Lone et al. (18), but the fragment cloned (residues 19–526) matches theirs. In the published work the hPol κ19–526 fragment was cloned into pBJ842 to generate yeast-expressible GST fusion proteins, and the GST-tagged protein was expressed in yeast strain BJ5464, whereas we relied on an His-GST fusion protein expressible in E. coli. The His-GST-hPol κ19–526 fusion protein was expressed in E. coli BL21 Gold cells (Stratagene). Cells were grown at 37 °C and 250 rpm for 3 h, followed by induction for 3 h (37 °C and 250 rpm) by addition of isopropyl β-d-1-thiogalactopyranoside (1 mm), and finally harvested by centrifugation. Buffer containing 50 mm Tris-HCl (pH 7.4), 0.5 m NaCl, 10% glycerol (v/v), 5 mm β-mercaptoethanol, lysozyme (1 mg/ml), and a protease inhibitor mixture (Roche Applied Science) was added to the harvested pellet. The suspension was sonicated, and supernatant was recovered from an ultracentrifugation step (35,000 × g, 1 h, 4 °C). The protein was purified by two affinity steps, using nickel-Sepharose (Amersham Biosciences), followed by glutathione-Sepharose 4B (Amersham Biosciences) beads. Briefly, the protein was bound to a nickel-chelating column in buffer containing 50 mm Tris-HCl (pH 7.4), 0.5 m NaCl, 10% glycerol (v/v), and 5 mm β-mercaptoethanol. The column was washed with 40–60 mm imidazole, and the protein eluted as a single peak in 400 mm imidazole. Following dialysis, the protein was added to the GST column in buffer containing 50 mm Tris-HCl (pH 7.4), 0.15 m NaCl, 10% glycerol (v/v), and 5 mm β-mercaptoethanol. After washing, the protein was then cleaved from the His-GST tag by treatment with PreScission protease (Amersham Biosciences) on the GST column, according to the methods suggested by the manufacturer. The mutant enzymes were prepared using a QuikChange site-directed mutagenesis kit (Stratagene); all other methods employed were the same as those used for wild-type enzyme. Both wild-type and mutant enzymes were purified to >95% purity (supplemental Fig. S1).
Full-length Extension Assay
A 32P-labeled primer was annealed to template oligonucleotide by heating a 1:1 solution of oligonucleotide to 95 °C for 5 min and then slow cooling to room temperature. The primer was then incubated with hPol κ and extended in the presence of a mixture of all four dNTPs. Each reaction was initiated by adding dNTP·Mg2+ (1 mm of each dNTP and 5 mm MgCl2) solution to a preincubated hPol κ·DNA complex (100 nm hPol κ and 200 nm DNA). The reaction was carried out at 37 °C in 50 mm Tris-HCl buffer (pH 7.4) containing 5 mm dithiothreitol (DTT), 100 μg ml−1 bovine serum albumin (BSA), and 10% (v/v) glycerol. At the indicated time points, 4-μl aliquots were quenched with 36 μl of a 95% formamide/20 mm EDTA/0.1% bromphenol blue (w/v)/0.1% xylene cyanol (w/v) solution and were separated by electrophoresis on a 20% polyacrylamide (w/v)/7 m urea gel.
Steady-state Kinetics
hPol κ-catalyzed single-nucleotide incorporation was measured over a range of dNTP concentrations. All reactions were carried out at 37 °C in 50 mm Tris-HCl buffer (pH 7.4) containing 5.0 mm DTT, 50 μg ml−1 BSA, and 10% glycerol (v/v). hPol κ (1 nm) was preincubated with radiolabeled DNA (100 nm), and the reaction was initiated by adding dNTP·MgCl2. Aliquots were quenched with 500 mm EDTA (pH 9.0) after varying incubation times. Substrate and product DNA were separated by electrophoresis on a 20% polyacrylamide (w/v)/7 m urea gel. The products were then visualized using a PhosphorImager and quantitated using Quantity OneTM software (Bio-Rad). The initial portion of the velocity curve was fit to a linear equation in the program Prism (GraphPad, San Diego, CA). The resulting velocity was plotted as a function of dNTP concentration and then fit to a hyperbola, correcting for enzyme concentration, to obtain estimates of kcat and Km,dNTP.
Transient-state Kinetics
All pre-steady-state experiments were performed using a KinTek RQF-3 model chemical quench-flow apparatus (KinTek Corp., Austin, TX) with 50 mm Tris-HCl (pH 7.4) buffer in the drive syringes. All experiments were carried out at 37 °C in a buffer containing 50 mm Tris-HCl buffer (pH 7.4) containing 5 mm DTT, 100 μg ml−1 BSA, and 10% (v/v) glycerol. Polymerase catalysis was stopped by the addition of 500 mm EDTA (pH 9.0). Substrate and product DNA was separated by electrophoresis on a 20% polyacrylamide (w/v)/7 m urea gel. The products were then visualized using a PhosphorImager and quantitated using Quantity OneTM software. Results obtained under single-turnover conditions were fit to Equation 1,
where A is the product formed in first binding event, kobs is the rate constant defining polymerization under the conditions used for the experiment being analyzed, and t is time. Results obtained under enzyme limiting conditions were fit to Equation 2,
where A is the product formed in first binding event, kobs is the rate constant defining polymerization under the conditions used for the experiment being analyzed, kss is steady-state velocity of nucleotide incorporation, and t is time.
LC-MS/MS Analysis of Oligonucleotide Products from hPol κ19–526 Reactions
hPol κ19–526 (5 μm) was preincubated with primer/template DNA (10 μm), and the reaction was initiated by addition of dNTP (1 mm each) and MgCl2 (10 mm) in a final volume of 100 μl. hPol κ19–526 catalysis was allowed to proceed at 37 °C for 4 h in 50 mm Tris-HCl buffer (pH 7.8 at 25 °C) containing 50 mm NaCl, 1 mm DTT, 50 μg ml−1 BSA, and 10% glycerol (v/v). The reaction was terminated by extraction of the remaining dNTPs using a size-exclusion chromatography column (Bio-Spin 6 chromatography column, Bio-Rad). Concentrated stocks of Tris-HCl, DTT, and EDTA were added to restore the concentrations to 50 mm, 5 mm, and 1 mm, respectively. Next, E. coli uracil DNA glycosylase (20 units, Sigma-Aldrich) was added, and the solution was incubated at 37 °C for 6 h to hydrolyze the uracil residue on the extended primer. The reaction mixture was then heated at 95 °C for 1 h in the presence of 0.25 m piperidine, followed by removal of the solvent by centrifugation under vacuum. The dried sample was resuspended in 100 μl of H2O for MS analysis.
LC-MS/MS analysis (30, 36) was performed on an Acquity UPLC system (Waters, Milford, MA) connected to a Finnigan LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, MA), operating in the electrospray (ESI) negative ion mode. An Acquity UPLC BEH octadecylsilane (C18) column (1.7 μm, 1.0 mm × 100 mm) was used with the following LC conditions: Buffer A contained 10 mm NH4CH3CO2 plus 2% CH3CN (v/v) and buffer B contained 10 mm NH4CH3CO2 plus 95% CH3CN (v/v). The following gradient program was used with a flow rate of 150 μl min−1: 0–2.5 min, linear gradient from 100% A to 95%A/5% B (v/v); 2.5–6.0 min, linear gradient to 75% A/25% B (v/v); 6–6.5 min, linear gradient to 100% B; 6.5–8.0 min, hold at 100% B; 8.0–9.0 min, linear gradient to 100% A; 9.0–12.0 min, hold at 100% A. The temperature of the column was maintained at 50 °C. Samples were injected with an autosampler system. ESI conditions were as follows: source voltage, 4 kV; source current, 100 μA; auxiliary gas flow rate setting, 20; sweep gas flow rate setting, 5; sheath gas flow setting, 34; capillary voltage, −49 V; capillary temperature, 350 °C; and tube lens voltage, −90 V. MS/MS conditions were as follows: normalized collision energy, 35%; activation Q, 0.250; and activation time, 30 ms. The calculations of the CID fragmentations of oligonucleotide sequences were done using a program linked to the Mass Spectrometry Group of Medicinal Chemistry at the University of Utah.
Crystallization Conditions
Primer and template sequences used in crystallization were the same as those used for the crystallization of S. solfataricus Dpo4 (30): 13-mer primer, 5′-GGG GGA AGG ATT C-3′; and 18-mer template, 5′-TCA C(8-oxoG)G AAT CCT TCC CCC-3′.
For the hPol κ19–526-dGTP complex, the hPol κ19–526 protein stock solution, containing 350 mm NaCl, 50 mm HEPES (pH 7.3), 10% (v/v) glycerol, 5 mm β-mercaptoethanol, and 1 mm tris(2-carboxyethyl)phosphine, was mixed with the annealed DNA duplex (ratio of protein:DNA was 1:1.2) and supplemented with 10 mm CaCl2 and 10 mm dGTP. Crystals appeared under precipitate in sitting drops, containing a mixture of equal volumes of protein and reservoir solutions. The reservoir solution contained 18% poly(ethylene) glycol 5000 monomethyl ether (w/v), 0.2 m ammonium acetate, 0.1 m NaCl, and 20 mm MES buffer (pH 6.5). The crystals were large, but mostly of an irregular morphology and branching off into adjacent specimen.
The hPol κ19–526·dATP complex was crystallized as described above for the hPol κ19–526·dGTP complex, except that the protein stock solution contained 1× phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.5 mm KH2PO4, pH 7.4), 10% glycerol (v/v), 1 mm EDTA, 1 mm DTT, 5 mm β-mercaptoethanol, 0.1% Nonidet P-40 (v/v), 5 mm CaCl2, and 5 mm dATP. Only a few crystals grew from a drop equilibrated against a reservoir containing 22% polyethylene glycol 5000 monomethyl ether, 0.2 m ammonium acetate, 0.1 m NaCl, and 20 mm MES (pH 6.5).
X-ray Diffraction Data Collection, Structure Determination, and Refinement
X-ray diffraction data frames of cryo-cooled crystals of hPol κ19–526-dGTP and hPol κ19–526-dATP complexes were collected on the 21-ID-G and 21-ID-F beamlines, respectively, of the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). Some 30 crystals of the hPol κ19–526·dGTP complex were tested for diffraction, and the best one diffracted up to 3.16-Å resolution. Crystals of the hPol κ19–526·ATP complex diffracted to a lower resolution of 3.7 Å. Human Pol κ19–526·dGTP diffraction frames were processed and scaled with HKL2000 (37) and hPol κ19–526·ATP with XDS (38). The selected data statistics are summarized in Table 1.
TABLE 1.
Crystal data and refinement statistics for hPol κ·DNA·dNTP complexes
| Structure | hPolκ19–526·dGTP | hPolκ19–526·dATP | ||
|---|---|---|---|---|
| X-ray source | APS, LS-CAT | APS, LS-CAT | ||
| Beamline | 21-ID-G | 21-ID-F | ||
| Detector | MAR-300 | MAR-225 | ||
| Wavelength (Å) | 1.00 | 1.00 | ||
| Temperature (K) | 110 | 110 | ||
| No. of crystals | 1 | 1 | ||
| Space group | C2221 | C2221 | ||
| Unit cell (a, b, c; Å) | 165.48,217.63,117.96 | 167.83,220.85,119.21 | ||
| Resolution range (Å) | 50.0–3.16 | 50.0–3.7 | ||
| Highest resolution shella | (3.36-3.16) | (3.93-3.70) | ||
| No. of measurements | 238,204 (17,090) | 124,035 (18,804) | ||
| No. of unique reflections | 35,030 (4,619) | 23,041 (3,419) | ||
| Redundancy | 6.8 (3.7) | 5.4 (5.5) | ||
| Completeness (%) | 95.0 (76.1) | 96.9 (86.4) | ||
| Rmergeb (%) | 14.8 (54.0) | 14.9 (62.3) | ||
| Signal to noise (〉I/σI〉) | 11.0 (2.73) | 10.4 (3.37) | ||
| Solvent content (%) | 74.6 | 75.7 | ||
| Model composition (asymmetric unit) | ||||
| Monomer | A | B | A | B |
| Amino acid residues range | 33–224/281–520 | 33–223/282–519 | 33–224/282–520 | 33–223/282–520 |
| Template nucleotides range | 2–18 | 2–16 | 2–18 | 2–16 |
| Primer nucleotides range | 1–13 | 3–13 | 1–13 | 3–13 |
| No. Ca2+ ions | 2 | 2 | 2 | 2 |
| No. dGTP | 1 | 1 | ||
| No. dATP | 1 | 1 | ||
| No. water molecules | 30 | 13 | ||
| Rfc (%) | 24.1 | 23.6 | ||
| Rfreed (%) | 27.8 | 27.3 | ||
| Temperature factors | ||||
| from Wilson plot (Å2) | 63.5 | 62.5 | ||
| mean isotropic (Å2) | 96.7 | 95.5 | ||
| r.m.s.d. in temperature factors | ||||
| Bonded main chain atoms (Å2) | 1.2 | 1.2 | ||
| Bonded side chain atoms (Å2) | 1.7 | 1.4 | ||
| r.m.s. standard deviation from ideal values | ||||
| Bond lengths (Å) | 0.008 | 0.009 | ||
| Bond angles (°) | 1.4 | 1.5 | ||
| Dihedral angles (°) | 21.7 | 22.0 | ||
| Improper angles (°) | 1.8 | 1.0 | ||
a Values in parentheses correspond to the highest resolution shells.
b Rmerge = Σhkl Σj = 1,N|〈Ihkl[〉 − Ihkl,j|/Σhkl Σj = 1,N|Ihkl,j|, where the outer sum (hkl) is taken over the unique reflections.
c Rf = Σhkl ‖Fohkl| − k|Fchkl‖/Σhkl |Fohkl|, where |Fohkl| and |Fchkl| are the observed and calculated structure factor amplitudes, respectively.
d Rfree idem, for the set of reflections (5% of the total) omitted from the refinement process.
The structure of the hPol κ19–526·dGTP complex was determined by molecular replacement using the program PHASER (39) and a monomer of hPol κ19–526 (pdb accession code 2OH2 (18)) as the search model. The program TURBO-FRODO5 was used for visualization and model building into σA electron density maps, composed with the modified σA coefficients (40). The dGTP molecule in the syn conformation and two Ca2+ ions were positioned into unambiguous, positive difference Fourier electron density at the active site of both monomers per asymmetric unit. The 8-oxoG residue initially modeled in the anti conformation does not properly fit the σA electron density map (Fig. S2A, left panel). Therefore, the syn conformation of 8-oxoG was chosen based on the shape of the density obtained from an improved averaged electron density map calculated with DM (41), using the 2-fold NCS averaging (Fig. S2A, middle panel). Several rounds of refinement were carried out with CNS (42), including energy minimization, simulated annealing with torsion angles, and individual isotropic temperature factors refinement. Both, 8-oxoG and dGTP were kept in the syn conformation until the end of the refinement (Fig. S2A, right panel).
For the structure determination of the hPol κ19–526·ATP complex by molecular replacement, the final coordinates of the hPol κ19–526·dGTP complex without dNTP and solvent molecules served as the initial model, owing to the similarities between the unit cell constants of the two complexes (Table 1). The orientation of the model found in the initial solution was optimized by several rounds of rigid-body refinement. As in the case of the hPol κ19–526·dGTP complex, an average electron density map was calculated with DM to distinguish between the syn and anti orientations of the dATP and 8-oxoG residues at the active site (supplemental Fig. S2, B and C, respectively, middle panels). The average density map indicated unambiguously that the 8-oxoG base should be in the syn orientation (supplemental Fig. S2B). Interpretation of the electron density was not straightforward in the case of dATP; due to the low resolution of the data, it is difficult to distinguish between the syn and anti conformations. However, taking into account the relative orientations of hydrogen-bond donors and acceptors of the 8-oxoG and dATP residues (supplemental Fig. S2, B and C), we reasoned that dATP is in anti conformation, thus pairing with 8-oxoG in the Hoogsteen mode.
The quality and stereochemistry of the models were inspected using PROCHECK (43), and the standard procedures were inspected in CNS (42). The crystallographic figures were prepared with PyMOL.6 Final coordinates and structure factors for the crystallographic models of the two hPol κ complexes have been deposited in the Protein Data Bank (http://www.rcsb.org). The PDB ID codes are 2w7o for hPol κ·dGTP (r2w7osf for structure factors) and 2w7p for hPol κ·ATP (r2w7psf for the structure factors).
RESULTS
Extension of Oligonucleotide Primers by hPol κ in the Presence of all Four dNTPs
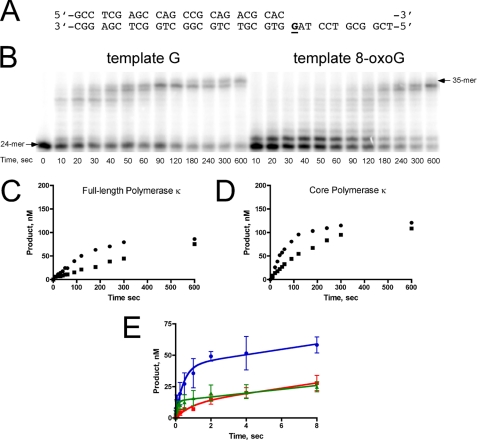
hPol κ catalysis opposite unmodified DNA- and 8-oxoG-modified template DNA and extension past the modification were allowed to proceed in the presence of all four dNTPs (Fig. 1). Full-length hPol κ can extend the 24/36-mer primer/template DNA (supplemental Table S1), but it is actually slower than the truncated core domain. The hPol κ core domain (residues N19–526; hPol κ19–526) can extend an unmodified 24/36-mer primer/template DNA substrate in ∼3 min under the conditions tested. When 8-oxoG is in the template strand, hPol κ catalysis is slowed for both the full-length and truncated forms of the enzyme, but extension does occur. A larger fraction of the substrate persists as a 25-mer for both forms of the enzyme, which indicates less efficient extension beyond the 8-oxoG-modified base. Product formation also appears to be more distributive for 8-oxoG-modified template, which is evident in the less pronounced sigmoidicity of the product curves (Fig. 1, C and D).
FIGURE 1.
Steady-state and pre-steady-state kinetics of hPol κ-catalyzed insertion opposite and extension past 8-oxoG. A, sequence of 24/36-mer primer/template DNA used in full-length extension analysis, where G indicates template G or 8-oxoG. B, DNA synthesis by full-length hPol κ (100 nm) on either unmodified or 8-oxoG modified template DNA (200 nm) was allowed to proceed in the presence of a 4 mm dNTP mix. C, formation of 35-mer extension product is plotted as a function of time for full-length hPol κ-catalyzed extension of 24/36-mer DNA containing G (circles) or 8-oxoG (squares). D, formation of 35-mer extension product is plotted as a function of time for core hPol κ19–526-catalyzed extension of 24/36-mer DNA containing G (circles) or 8-oxoG (squares). E, pre-steady-state analysis of core hPol κ19–526-catalyzed nucleotide incorporation opposite 8-oxoG-modified DNA indicates that a non-productive form dominates the first binding event. hPol κ-catalyzed (50 nm) incorporation of dNTP (1 mm) opposite 24/36-mer template DNA (100 nm): dCTP:G (blue circles), dCTP:8-oxoG (red squares), dATP:8-oxoG (green triangles). The results for extension were fit to Equation 2, yielding the following kinetic parameters: dCTP:G, A = 42 ± 3 nm, kobs = 2.3 ± 0.4 s−1, kss = 2.1 ± 0.7 s−1; dCTP:8-oxoG, A = 11 ± 3 nm, kobs = 1.1 ± 0.4 s−1, kss = 2.1 ± 0.4 s−1; dATP:8-oxoG, A = 14 ± 1 nm, kobs = 9.5 ± 2.8 s−1, kss = 1.5 ± 0.3 s−1. The experiments were performed in triplicate. The error bars represent the standard deviation from the mean.
Kinetic Analysis of hPol κ Catalysis Opposite 8-oxoG
The relative catalytic efficiency of insertion opposite 8-oxoG by hPol κ has been measured previously using steady-state analysis (33). We measured these values with this particular oligonucleotide to compare enzyme and substrate differences, and the values were similar to those reported previously (supplemental Table S2). hPol κ is nearly as efficient at incorporation of dATP opposite 8-oxoG as it is incorporating dCTP opposite unmodified G, once again negating the concept that the enzyme functions only as an extender. Incorporation of dCTP opposite 8-oxoG is inhibited ∼10-fold relative to dATP:8-oxoG pairing, with a higher Km,dNTP being the primary difference between the two incorporation events.
Pre-steady-state analysis of hPol κ-catalyzed insertion of dATP and dCTP opposite 8-oxoG was performed. Under enzyme-limiting conditions, dCTP incorporation opposite G exhibits an essentially full burst in product amplitude (84%), in contrast to what was reported previously by others (44). The data presented by our group here and elsewhere (45) clearly show that hPol κ is fully active and forms productive complexes with unmodified DNA. By way of comparison, both dATP and dCTP incorporation opposite 8-oxoG result in diminished burst amplitudes relative to unmodified dCTP:G (Fig. 1D). The product amplitudes are ∼20% of the burst observed with unmodified DNA, indicating that ∼80% of the hPol κ·8-oxoG complexes are in a non-productive form during the first catalytic event. The rates observed indicate that dATP is incorporated opposite 8-oxoG ∼9-fold faster than dCTP, consistent with the steady-state results.
Next, transient-state kinetic experiments were performed under conditions where enzyme was in excess of DNA substrate. The rate constant defining the maximal forward rate of catalysis, kpol, and the apparent nucleotide binding affinity, Kd,dNTP, were then measured by varying the concentration of dNTP in the reaction mixture (Table 2). As with enzyme-limiting conditions, the excess-enzyme experiments show that hPol κ converts >70% of substrate to product in the pre-steady state. During the first binding event, hPol κ is ∼140-fold more efficient at incorporation of dATP opposite 8-oxoG relative to incorporation of dCTP opposite the lesion. Notably, the product amplitude for 8-oxoG-containing substrates remained near 30% of the total substrate present in solution even when enzyme was in 4-fold excess of substrate, again indicative of non-productive ternary complexes. Next-base extension of both A:8-oxoG and C:8-oxoG pairings was also tested. Incorporation of the next correct nucleotide proceeded at a moderately slower rate for the both 8-oxoG pairs (supplemental Fig. S3) relative to what was measured with unmodified DNA. Although the rates for extension of C:8-oxoG and A:8-oxoG pairings were similar, the amplitude of product turned over in the first binding event was less for the C:8-oxoG pairing.
TABLE 2.
Pre-steady-state kinetic results
| Primer/template | dNTP | kPol | Kd,dNTP | ΔEfficiency relative to dCTP:G |
|---|---|---|---|---|
| s−1 | μm | |||
| -G- | dCTP | 10.3 ± 0.8 | 36 ± 14 | |
| 8-oxoG- | dCTP | 0.40 ± 0.04 | 90 ± 35 | 65-fold less |
| -8oxoG- | dATP | 8.2 ± 0.2 | 13 ± 3 | 2-fold better |
LC-MS/MS Analysis of Full-length Extension Products
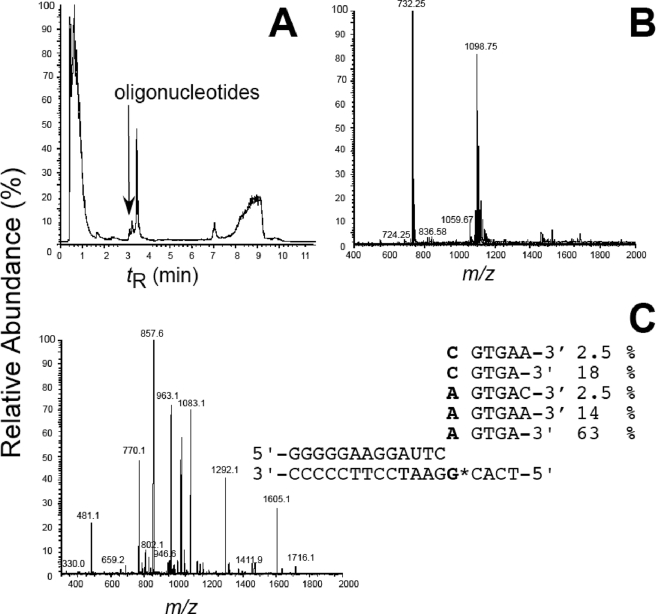
Unambiguous identification of full-length extension products resulting from hPol κ19–526 catalysis was carried out using an LC-MS/MS approach described previously (36). Five ions were observed in the MS spectrum that corresponded to extension products (Fig. 2). The CID fragmentation pattern of the major ion in the spectrum is consistent with the sequence 5′-pTCAGTGA-3′, which corresponds to the insertion of A opposite 8-oxoG and accurate full-length extension of the primer (Fig. 2). The identities of the other products were confirmed by CID MS/MS (supplemental Figs. S4–S7). Of the products observed, ∼80% correspond to insertion of dATP opposite 8-oxoG, and the remaining ∼20% were dCTP insertion opposite 8-oxoG, consistent with the kinetic analyses.
FIGURE 2.
LC-MS analysis of hPol κ19–526-catalyzed full-length extension products confirm that mutagenic insertion of dATP is favored. A, total ion current trace of products derived from hPol κ-catalyzed extension of 13/18-mer DNA containing 8-oxoG in the template. B, ESI mass spectrum of the oligonucleotide peaks that elute at tR 3.2 min. C, CID mass spectrum of ion m/z 1099 (where G* denotes 8-oxoG) and a summary of the products observed in the reaction mixture.
X-ray Crystallography of Ternary hPol κ19–526·DNA·dNTP Complexes
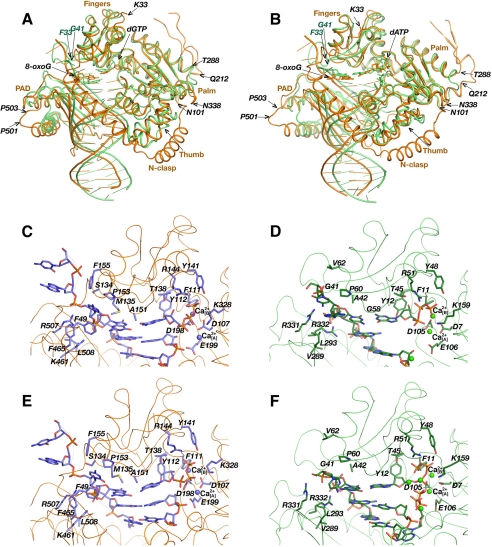
The hPol κ19–526 protein was crystallized in complex with an 18-mer template/13-mer primer DNA duplex identical to that previously used for structure determination of complexes between the Dpo4 polymerase from S. solfataricus and 8-oxoG-containing DNA (primer sequence, 5′-GGG GGA AGG ATT C-3′; and template sequence, 3′-CCC CCT TCC TAA GG*C ACT-5′) (supplemental Table S1 and Fig. 2) (30). The 8-oxoG adduct (G*) is located at position 5 in the above template strand to generate a pair with the incoming dNTP at the active site of the polymerase. Ca2+ was used in place of Mg2+ for the crystallization of ternary protein·DNA·dNTP complexes, and trials were conducted with each of the four dNTPs. However, crystals could only be grown with either dGTP or dATP. Both the hPol κ19–526·DNA·dGTP and hPol κ19–526·DNA·ATP complexes (hereafter referred to as hPol κ19–526·dGTP and hPol κ19–526·ATP, respectively) crystallized in space group C2221, with two independent molecules (named A and B) per asymmetric unit (Table 1). In both structures, complex A is better ordered than complex B (supplemental Fig. S2), and all analyses below were therefore carried out using the A complexes. In the hPol κ19–526·dGTP complex A encompasses residues 33–224 and 281–520 and complex B encompasses residues 33–223 and 282–519. The hPol κ19–526·ATP complexes include residues 33–224 and 282–520 in complex A and residues 33–223 and 282–520 in complex B (Fig. 3, A and B, and Table 1). In both structures, only the 5′-terminal template nucleoside is missing (tT1) from the DNA duplex in complex A. On the other hand, five residues are missing from the DNA in complex B in both the hPol κ19–526·dGTP and the hPol κ19–526·ATP structures (tT1, tC17, tC18, pG1, and pG2).
FIGURE 3.
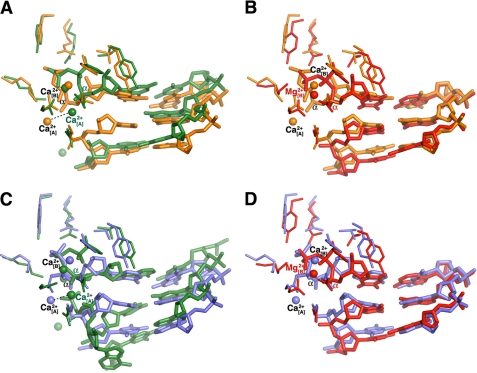
Similarities and differences between the hPol κ19–526·DNA-dNTP and Dpo4·DNA·dNTP complexes. A, superimposition of the hPol κ19–526·dGTP complex (orange) and the analogous Dpo4·dGTP complex (green; pdb accession code 2c22) (30). B, superimposition of the hPol κ19–526·ATP complex (orange) and the analogous Dpo4·ATP complex (green; pdb accession code 2c2d) (30). The active sites in the hPol κ19–526·dGTP (C) and Dpo4·dGTP (D) complexes. The active sites in the hPol k19–526·ATP (E) and Dpo4·ATP (F) complexes. The DNA and the protein active site residues are displayed as thin sticks colored by atom, and Ca2+ ions are represented as spheres.
Similarities and Differences in the Three-dimensional Architectures of hPol κ19–526 and S. solfataricus Dpo4
Our use of a DNA primer-template construct identical to that previously employed in the structural analysis of native (12) and 8-oxoG-adducted Dpo4·DNA complexes (30) for determining crystal structures of hPol κ19–526 complexes facilitates a comparison between the architectures of the two proteins. The three-dimensional structure of hPol κ19–526 is very similar to that of Dpo4 (Fig. 3, A and B). The proteins superimpose with a root mean square deviation (r.m.s.d.) of 1.7 Å between the positions of Cα atoms. The orientations of the palm, thumb, and finger domains match quite well, with some differences arising as a result of insertions or deletions in the region of three loops. Thus, hPol κ19–526 contains a loop with one additional residue (Asn-338), and, in addition, features a long extension (residues Asn-212 to Thr-288; hPol κ numbering scheme) in the thumb. On the other hand, Dpo4 has a longer loop in the fingers domain (Fig. 3, A and B) (residues Phe-33 to Gly-41; Dpo4 numbering scheme, pdb accession code 2c2d (30)). The little finger domains also exhibit similar architectures with the exception of a 3-residue longer loop in hPol κ (Pro-501 to Pro-503). The role played by the long extension from Asn-212 to Thr-288 in hPol κ is not clear, because this region is disordered and partly absent from the structure. The major difference between the structural architectures of the two proteins is constituted by the hPol κ N-terminal extension (residues Lys-33 to Asn-101), referred to as N-clasp (18) (Fig. 3, A and B). The presence of this N-clasp allows hPol κ to fully encircle the DNA, whereby the little finger domain adopts an orientation that is slightly more detached from the DNA in comparison with the Dpo4·DNA complex. The N-clasp, which interacts with the unpaired portion of the template strand, may substitute for the function of the Phe-33 to Gly-41 loop in Dpo4 that is absent in hPol κ and helps guide the 5′-unpaired template strand toward the active site in the former.
The Active Site of hPol κ19–526 Only Accommodates One Template Residue at a Time
The structure of the hPol κ19–526·dGTP complex reveals that only one template nucleotide (8-oxoG) is located inside the active site and that it pairs with dGTP at the insertion position (Fig. 3, A and C). This arrangement corresponds to the so-called “Type I” complex of Dpo4 (12). It differs from the active-site configuration observed in the Dpo4 complex with the same 8-oxoG-modified DNA duplex and dGTP (Fig. 3, A and D; pdb accession code 2c22 (30)). In that so-called “Type II” complex, the template 8-oxoG was skipped, and instead, the 5′-adjacent cytosine that was simultaneously accommodated inside the active site, paired with dGTP.
The hPol κ19–526·ATP structure also shows a configuration with only the template 8-oxoG positioned inside the active site and opposite dATP (Fig. 3, B and E). However, compared with the corresponding Dpo4·ATP complex where two dATPs were observed in the active site (Fig. 3, B and F; pdb accession code 2c2d (30)), the hPol κ19–526·ATP complex features only one dATP molecule.
The inability by hPol κ of accepting a second template nucleotide inside the active site is a result of the rather constrained space there. In hPol κ19–526, the unpaired template strand is guided into the active site by interactions with residues from the finger and little finger domains and the N-clasp. The template residue tC4 is sandwiched between Pro153 from the fingers domain and Phe49 from the N-clasp, while Met-135 guards the entry into the active site (Fig. 3, C and E). The positively charged residues Arg-507 and Lys-461 interact with the phosphate groups of the tC4 and 8-oxoG residues, respectively. Residues Phe-155, Ser-134, Pro-153, Met-135, and Ala-151 from the fingers domain in hPol κ that line the path taken by the template strand (Fig. 3, C and E) have longer side chains than the corresponding residues in Dpo4 (Val-62, Gly-41, Pro-60, Ala-42, and Gly-58 (Fig. 3, D and F). The resulting steric constraints provide unfavorable conditions for the simultaneous accommodation of two template residues into the active site of hPol κ.
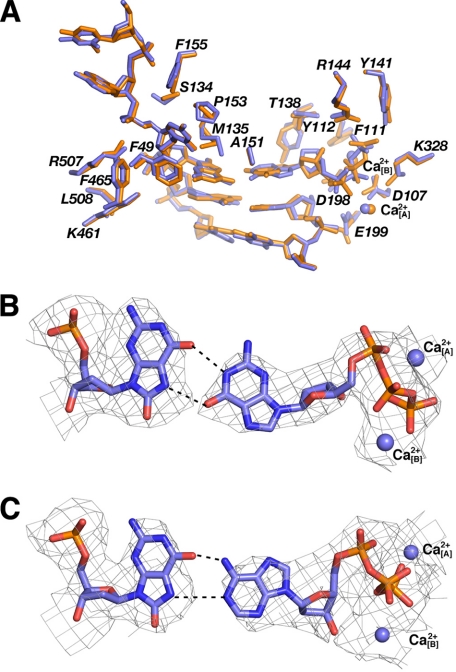
Base Pairing Modes at the Active Site of hPol κ
The superimposition of the hPol κ19–526·dGTP and hPol κ19–526·ATP complexes shows that the active site organization of the two complexes is very similar with the exception of the conformation of the dNTP nucleotides (Fig. 4A). The active-site geometry of hPol κ19–526 forces the 8-oxoG to pair with the incoming dGTP in an unusual way. Both 8-oxoG and dGTP are in the syn conformation (Fig. 4B and supplemental Fig. S2; see “Experimental Procedures” regarding the determination of the nucleoside conformations), and they pair via two hydrogen bonds formed between O6(8oxoG)…N1H(dGTP) and N7H(8-oxoG)…O6(dGTP). It appears that hPol κ19–526 does not possess a residue at the edge of the active site that can influence the syn/anti equilibrium of the 8-oxoG nucleoside, as was seen in the corresponding Dpo4·dGTP structure (17, 30). There, 8-oxoG is left unpaired and Arg-332 helps stabilize 8-oxoG in anti conformation via a hydrogen bond to its O8 oxygen (Fig. 3, C and D). The hPol κ19–526 residue corresponding to Arg-332 of Dpo4 is Leu-508. Above the primer strand, dGTP phosphates interact with residues Tyr-112, Arg-144, and Lys-328, which are conserved in Dpo4 as well as in all other Y-family polymerases (18). However, the dGTP phosphates adopt a “boat-like” orientation in hPol κ19–526 compared with the chair conformation seen in the Dpo4·dGTP complex (Figs. 3C, 3D, and 5A).
FIGURE 4.
Base pairing modes at the hPol κ19–526 active site. A, superimposition of the hPol κ19–526·dGTP (orange) and hPol κ19–526·ATP (blue) active sites. B, pairing between 8-oxoG and dGTP in the hPol κ19–526·dGTP complex. C, pairing between 8-oxoG and dATP in the hPol κ19–526·ATP complex. In B and C the 3Fo − 2Fc. Fourier sum electron density is represented by thin wires, contoured at the 1σ level, and pairs of Ca2+ ions are represented by blue spheres.
FIGURE 5.
Comparison of the DNA and dNTP configurations at the active sites. Pairs of protein complexes were superimposed, and DNA, dNTP, and active site residues surrounding the dNTP are shown in stick representation. Metal ions, Ca2+ in the case of hPol κ19–526 (current work) and Dpo4 (30), and Mg2+ for hPol κ19–526 (pdb accession code 2oh2) (18) are represented by spheres. A, superimposition of hPol κ19–526·dGTP (yellow) and S. solfataricus Dpo4·dGTP (green; pdb accession code 2c22) (30). B, superimposition of hPol κ19–526·dGTP (yellow) and hPol κ19–526 in complex with unmodified DNA and dTTP (red; pdb accession code 2oh2) (18). C, superimposition of hPol κ19–526·ATP (blue) and Dpo4·ATP (green; pdb accession code 2c2d) (30). D, superimposition of hPol κ19–526·ATP (blue) and hPol κ19–526 in complex with unmodified DNA and dTTP (red; pdb accession code 2oh2) (18).
Two calcium ions were observed at the active site of the hPol κ19–526·dGTP complex. One Ca2+ ion occupies the position corresponding to the “metal B” in Dpo4. The other Ca2+ ion is ∼5.1 Å away from the “metal A” position seen with the Dpo4·dGTP complex (Figs. 3C, 3D, and 5A). Also, the α-phosphate of the dGTP in hPol κ19–526 is located at 3.8 Å from its position in the Dpo4·dGTP complex. The same feature was observed in the superimposition of the hPol κ19–526·dGTP complex and hPol κ19–526 in complex with unmodified DNA and dTTP (18), where the distance between the α-phosphate positions of dNTPs is 4.1 Å (Fig. 5B). Therefore, in hPol κ19–526·dGTP, the dGTP α-phosphate and the Ca2+ corresponding to metal A lie farther away (∼8.2 Å (mol. A), 7.2 Å (mol. B), 8.6 Å (mol. A), and 7.0 Å (mol. B), respectively) from the 3′-OH of the pC14 primer than in other complexes (Fig. 5, A and B). The metal A distance to the dGTP α-phosphate is ∼4.3 Å. The relative spacings of Ca2+, dGTP phosphate, and primer 3′-OH in the structure are characteristic of a non-productive complex and are consistent with our kinetic data studies that demonstrated no incorporation of dGTP into the growing primer chain.
In the hPol κ19–526·ATP structure, dATP pairs with 8-oxoG in a Hoogsteen fashion (Figs. 3E and 5C and supplemental Fig. S2B; see “Experimental Procedures” regarding the determination of the nucleoside conformations). The 8-oxoG nucleoside unambiguously adopts a syn conformation, thus exposing its Hoogsteen face toward the incoming dATP. However, the decision between the syn and anti orientations of dATP was more challenging, because the electron density at the current resolution limit does not permit one to clearly distinguish between the two. Deoxyadenosine, in the standard anti orientation, can form two hydrogen bonds with syn 8-oxoG (Fig. 4C and supplemental Fig. S2B). On the other hand, deoxyadenosine in the syn orientation opposite syn 8-oxoG leads to only a single hydrogen bond (supplemental Fig. S2C). Considering that the in vitro primer extension data revealed the preferential incorporation of dATP over dCTP (Figs. 1 and 2) and dGTP (Fig. 2; no incorporation) and taking into account our earlier observation that adenosine is in the anti orientation when it pairs with syn 8-oxoG at the active site of Dpo4 (Fig. 3F), we settled on an anti orientation of dATP at the active site of hPol κ19–526 (Fig. 3E). However, regardless of a syn or an anti orientation of the base, the positions of the dATP α-phosphate and the metal A Ca2+ are relatively far from the 3′-OH of the primer (∼6.9 Å and 7.6 Å, respectively), a situation reminiscent of the position of dGTP in the hPol κ19–526·dGTP structure (Figs. 3C, 3E, and 4A). The metal A lies within ∼4.0–4.4 Å distance from the dATP α-phosphate. The orientation of dATP phosphates in a boat-like orientation in the hPol κ19–526·ATP complex bears resemblance to the conformation of dATP observed in structure of the Dpo4·ATP complex (Figs. 3E, 3F, and 5C). The distances between the α-phosphate positions of the dATPs and the metal A positions of Ca2+ ions in the two structures are 2.0 Å and 4.3 Å, respectively. The larger relative spacing between reactive groups at the active site of the hPol κ19–526·ATP complex holds also in comparisons with structures of other polymerase complexes with either native base pairs or 8-oxoG:dATP pairs at the active site (Fig. 5, C and D).
Kinetic Analysis of Mutant hPol κ Catalysis Opposite 8-oxoG
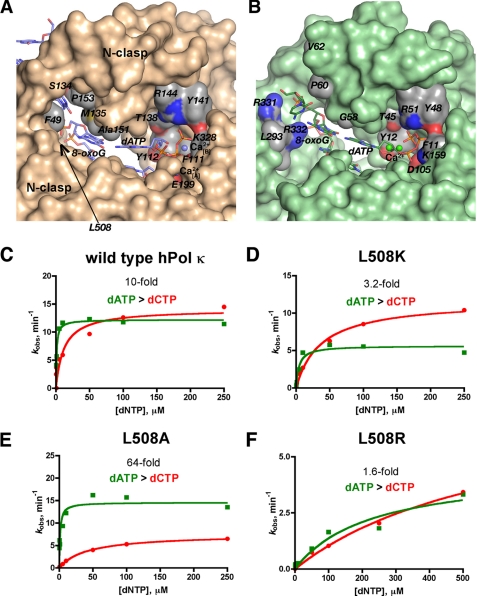
The availability of crystal structures with hPol κ bound to 8-oxoG-modified DNA, as well as previous studies with other Y-family members (i.e. Dpo4 and Pol η) allowed us to further investigate the mechanistic features that facilitate error-prone bypass of 8-oxoG by hPol κ and accurate nucleotide insertion opposite 8-oxoG by Dpo4 and yeast Pol η. Superimposition of the little finger domains of Dpo4, hPol κ, and yeast Pol η reveals that both Dpo4 and yeast Pol η possess a positive center (Arg-332 for Dpo4 and Lys-498 for Pol η) near the template 8-oxoG, whereas hPol κ has a hydrophobic residue, Leu-508. Previous work from our group has shown that an electrostatic interaction between Arg-332 and 8-oxoG is important in facilitating accurate and efficient bypass of 8-oxoG by Dpo4 (17). To test whether or not we could modulate the accuracy of hPol κ bypass of 8-oxoG, we generated three hPol κ19–526 mutants: L508K, L508R, and L508A.
Steady-state kinetic analysis of the three mutants showed that the catalytic efficiency opposite unmodified DNA was unperturbed for L508K and L508A, but the L508R mutant showed a decrease in catalytic efficiency of ∼29-fold for dCTP insertion opposite G (supplemental Table S2). Nucleotide incorporation opposite 8-oxoG was measured for the three mutants (Fig. 6, C–F). As expected the L508K mutant shows a trend toward more accurate bypass of 8-oxoG, primarily due to a decrease in the catalytic efficiency for dATP insertion opposite 8-oxoG (Fig. 6D). The L508A mutant was the most error-prone enzyme tested, inserting dATP opposite 8-oxoG 64-fold more efficiently than dCTP opposite 8-oxoG and 2.2-fold better than dCTP:G insertion events (Fig. 6E). The L508R mutant inserts dCTP and dATP equally well opposite 8-oxoG, but the overall catalytic efficiency was severely perturbed because of a high Km,dNTP value.
FIGURE 6.
Molecular surface representations of the hPol κ19–526 and Dpo4 active sites. The two active sites are depicted in the same orientation following a superimposition of the two enzyme structures. A, the hPol κ19–526·ATP surface is colored in orange with the exception of portions around selected active site residues that are colored by atom (carbon in gray, sulfur in yellow, oxygen in red, and nitrogen in blue). The Leu-508 residue is obscured behind other residues, but its position is indicated with an arrow. B, the Dpo4·ATP (pdb accession code 2c2d) (30) surface is shown in green with the exception of portions around selected active site residues that are colored by atom (see panel A for color code). C–F, plots of kobs versus [dNTP] are shown to compare steady-state incorporation catalytic efficiencies of dCTP (red circles) and dATP (green squares) for wild-type hPol κ and the mutant enzymes. Each panel shows the -fold kinetic preference of the enzyme for dATP over dCTP during insertion opposite 8-oxoG.
Pre-steady-state analyses of the mutant enzymes were performed, and they also showed some interesting trends. For example, the L508K mutant insertion of dATP and dCTP opposite 8-oxoG proceeds at essentially the same rate (kp), which is in contrast to the faster rate of dATP incorporation opposite 8-oxoG by the wild-type hPol κ enzyme (Fig. 1D). L508A, on the other hand, incorporated dATP ∼100-fold faster than dCTP opposite 8-oxoG (supplemental Fig. S8). Although the L508R mutant did exhibit a burst in product formation during insertion of dCTP opposite G, the burst phase was lost when 8-oxoG was present in the template.
DISCUSSION
Many studies have focused on the ability of Y-family DNA polymerases to bypass DNA adducts, but far fewer reports have elucidated the mechanistic determinants that make one enzyme better suited than another to bypass specific forms of damage. We have undertaken a careful examination of the structural features and activities that define hPol κ bypass of arguably the most common form of oxidative damage to the genome to better understand the molecular features that influence mutation events that may occur during periods of increased oxidative stress or diminished repair of oxidative lesions. Consistent with previous reports (33, 35), hPol κ displays ∼10-fold kinetic preference for dATP over dCTP during insertion opposite 8-oxoG. It is noteworthy that the hPol κ19–526 core polymerase is consistently more efficient than the full-length enzyme, in line with previous observations by others based on steady-state kinetics for extension of matched and mismatched DNA primer termini (18). Judging from single-base incorporations, the core protein is about five times more efficient at inserting either dATP or dCTP opposite 8-oxoG than full-length hPol κ, in line with previous results showing higher activity for the core enzyme from the Prakash and Aggarwal groups (18), as well as a report on the bypass properties of the DinB homologue from Arabidopsis thaliana documenting increased activity of the core Pol κ from the plant kingdom (46). Transient-state kinetic analysis of the insertion events showed that hPol κ can insert dATP opposite 8-oxoG with equal efficiency to dCTP insertion opposite G. However, most of the binding events that occur between hPol κ- and 8-oxoG-modified template DNA are non-productive, as evidenced by a reduced burst amplitude for both dCTP and dATP insertion opposite 8-oxoG (Fig. 1).
We determined the crystal structures of ternary complexes of hPol κ19–526 with either dGTP or dATP paired with the template 8-oxoG. In both active sites the nucleoside triphosphates as well as the metal A ion are not optimally positioned for primer extension. In the hPol κ19–526·ATP complexes, the 3′-oxygen of the terminal primer residue and the phosphorus atom of the α-phosphate are spaced slightly tighter (∼6.9 Å in both molecules) relative to the corresponding distances in the hPol κ19–526·dGTP complexes (mol. A: 8.2 Å; mol. B: 7.2 Å). However in both complexes these distances are rather long compared with the ideal values of 3.4–4.0 Å observed in complexes where the hydroxyl group of the primer is poised for nucleophilic attack (47). Similarly the metal A ion distance to the 3′-terminal OH of the primer ranges between 7.0 Å and 8.6 Å in hPol κ19–526·ATP and ·dGTP structures, far longer than the ideal distance of 2.2 Å necessary for a Mg2+ ion to assist in the reaction (47). Even considering the fact that the complexes were crystallized in the presence of Ca2+ and not Mg2+ and differences to ideal active site alignments are thus expected, the distances between the principal participants in the reaction (3′-OH, NTP α-phosphate and metal A ion) are large and consistent with non-productive configurations. In the case of the complex with dGTP, the structural data are in-line with the inability of the polymerase to insert G opposite 8-oxoG. The model of the dATP complex is of lower resolution, but a rearrangement is also needed to get the 3′-terminal OH of the primer close enough to the α-phosphate of dATP (the nucleotide adopts the standard anti orientation opposite 8-oxoG in syn (Fig. 4C)) for the nucleophilic attack to occur. Although the crystallographic models represent an inactive state with regard to the first catalytic step, the structural data confirm the preferred accommodation of the 8-oxoG adduct in the syn conformation by hPol κ. Despite repeated efforts, we were unable to grow crystals of the ternary complex with dCTP at the active site, the structure of which would presumably have revealed a standard Watson-Crick pairing mode between the incoming nucleotide triphosphate and 8-oxoG.
The observation that hPol κ is error-prone during insertion opposite 8-oxoG differs significantly from the results of similar studies of 8-oxoG bypass synthesis by yeast Pol η (31, 32) and the S. solfataricus Dpo4 polymerase (30). Dpo4 exhibits 19-fold higher efficiency for dCTP > dATP incorporation opposite 8-oxoG and a 4-fold higher efficiency of extension following an 8-oxoG:C pair compared with 8-oxoG:A (29, 30). In fact, the activation energy for dCTP incorporation opposite 8-oxoG is actually lower than opposite G (4.2 versus 8.4 kcal mol−1) (30). The lowered activation barrier to dCTP insertion opposite 8-oxoG was attributed in part to an important interaction between the O8 atom and Arg-332 in the little finger domain of Dpo4 (17). It is noteworthy that Arg-332 in Dpo4 is capable of tracking the O atom of 8-oxoG and thus influence the syn/anti equilibrium of the adducted nucleoside in the replicative and post-replicative positions. These hydrogen bonding interactions by Arg-332 underscore a key difference between the two polymerases, namely the ability of Dpo4 to accommodate two template bases at its active site and the limitation to a single template residue at the active site of hPol κ.
We generated three hPol κ mutants to better understand the molecular features that influence nucleotide incorporation opposite 8-oxoG. The first mutant, L508K, exhibited a kinetic profile of nucleotide insertion opposite 8-oxoG that tends toward more accurate bypass of 8-oxoG (Fig. 6 and supplemental Table S2). The rate of dATP insertion opposite 8-oxoG (kp) was decreased by the L508K substitution (supplemental Fig. S8). The L508K mutant also exhibits a decreased steady-state catalytic efficiency for dATP insertion opposite 8-oxoG, which suggests that a lysine at position 508 of hPol κ serves to repel the syn orientation through interactions between the exocyclic amino group of 8-oxoG. Based on the kinetic parameters, the repulsion between Lys-508 and the exocyclic amino group of guanine inhibits insertion of dATP more than it stabilizes the anti conformation of 8-oxoG during insertion of dCTP.
In contrast to hPol κ L508K, the L508A mutant showed a trend toward even more error-prone insertion opposite 8-oxoG (Fig. 6 and supplemental Table S2). Strikingly, hPol κ L508A inserts dATP 2.2-fold more efficiently than it inserts dCTP opposite G, a fact that is further supported by a faster rate of dATP insertion in transient-state experiments. It seems that at least some element of hPol κ bypass of 8-oxoG is determined by simple steric hindrance, because removal of the leucine side chain allows better accommodation of the syn 8-oxoG:dATP Hoogsteen pair. It has been postulated that steric bulk at C8 can weaken dG:dC base pairs by destabilizing the anti conformation of dG (49). However, hPol κ appears to be able to further enhance the intrinsic preference by 8-oxoG for the syn conformation.
The third hPol κ mutant, L508R, exhibits reduced catalytic efficiency for unmodified DNA and 8-oxoG containing templates (supplemental Table S2). However, both dCTP and dATP are inserted with approximately equal efficiency opposite 8-oxoG, which is similar to the results obtained for L508K. The loss of catalytic efficiency is difficult to explain in the absence of further data. The much higher Km,dNTP, observed for both dCTP and dATP insertion opposite 8-oxoG, as well as for dCTP insertion opposite G, might indicate a decreased ability of hPol κ L508R to accommodate the incoming dNTP in the polymerase active site, but at this point such an assertion is speculative.
The hPol κ L508K and L508A mutants exhibit kinetic trends that are very similar to those reported for mutant forms of the model DNA polymerase from bacteriophage T7 (48). In the study with T7exo− it was shown that mutating Lys-536 in the finger domain to alanine increased the ratio of dATP insertion relative to dCTP insertion opposite 8-oxoG from 33% A to 95% A, presumably by removing an electrostatic barrier to the syn orientation of 8-oxoG, which allows Hoogsteen pairing to occur between the lesion and dATP. The current evidence clearly indicates that a single well positioned positive center can greatly influence the accuracy of 8-oxoG bypass for different classes of DNA polymerases.
The influence of the N-clasp during hPol κ-catalyzed bypass of 8-oxoG is unknown, but it is possible that the presence of this additional domain is one factor that results in the formation of non-productive complexes during bypass of 8-oxoG. A similar conclusion was derived from modeling studies of Pol κ bypass of benzo[a]pyrene-derived N6 dA adducts (49). The same work proposed that the N-clasp stabilized bulky minor-groove adducts by promoting anti-oriented N2 dG benzo[a]pyrene adducts, which is consistent with kinetic studies from our group showing that Pol κ bypasses of N-2 dG adducts of increasing size accurately and efficiently (45). It has been shown previously that hPol κ68–526, which lacks the N-clasp, has a greatly reduced catalytic efficiency (18). Therefore, a slight perturbation in the N-clasp position caused by the presence of 8-oxoG could conceivably result in catalytically inactive complexes. Alternatively, it may be that the active site of hPol κ is more sterically constrained than either Dpo4 or yeast Pol η. As noted above, there is a region of the finger domain in which hPol κ has bulkier side chains (Phe-155, Ser-134, Pro-153, Met-135, and Ala-151) than the corresponding residues in Dpo4 (Val-62, Gly-41, Pro-60, Ala-42, and Gly-58). The steric differences place more constraints upon hPol κ catalysis than Dpo4. Further supporting the influence of sterics during 8-oxoG bypass, it is interesting to note that the burst amplitude observed for hPol κ L508A-catalyzed insertion of dCTP opposite 8-oxoG is very similar to the burst observed in the control experiment (supplemental Fig. S8), which indicates that the slight increase in space near the templating 8-oxoG allows a greater fraction of productive complexes to form in the first binding event.
Although the kinetic and structural data presented here are informative in many ways, it remains difficult to pinpoint exactly what drives the error-prone nature of hPol κ-catalyzed bypass of 8-oxoG. The kinetic data with the Leu-508 mutants are consistent with a role for electrostatic interactions between the little finger and the template DNA in determining nucleotide selectivity, but electrostatics are not the sole means of modulating syn/anti equilibria of 8-oxoG. The reduced burst amplitude observed in pre-steady-state experiments indicates that 8-oxoG is not easily accommodated in the active site of hPol κ (Fig. 1D), despite the fact that this polymerase shares strong structural homology with Dpo4. It is possible that the active site of hPol κ is more rigid than that of Dpo4. The failure of hPol κ to accommodate a Type II complex (i.e. −1 frameshift deletion), instead pairing syn 8-oxoG with syn dGTP, is also supportive of the idea that the structural dynamics of hPol κ are more limited than for Dpo4 and explains why dGTP incorporation is less favorable than dATP insertion opposite 8-oxoG.
The α-helices comprising the N-clasp sit atop the major groove near the polymerase active site and make contact with both the finger and little finger domains of hPol κ (Fig. 6A). The N-clasp might constrain conformational fluctuations between the finger and little finger domains, which would limit the ability of 8-oxoG to shift between syn and anti modes. Notably, the little finger domain is translocated ∼50 Å away from the DNA binding cleft when the N-clasp is absent (50) and removing the N-clasp severely reduces the catalytic efficiency of hPol κ (18). These data are consistent with the view that the N-clasp stabilizes the orientation of the little finger domain relative to the DNA. As 8-oxoG approaches the polymerase active site it is presumably in the more thermodynamically stable syn orientation, because it is in single-stranded DNA. The aforementioned constraints imparted by structural features that are unique to hPol κ could in essence serve to “lock down” syn-oriented 8-oxoG as it enters the cleft between the little finger and finger domains, thereby promoting error-prone insertion of dATP opposite the lesion. Indeed, Met-135 is positioned on top of syn 8-oxoG and follows the long dimension of the adducted base (supplemental Fig. S9). The average distance between the Cg-Sd-Ce elbow from this Met and the 8-oxoG base plane in the dATP and dGTP complexes varies somewhat but is as short as 4 Å. A similar stacking interaction cannot be established with 8-oxoG in the anti orientation, and it is conceivable that Met-135 thereby influences the syn/anti equilibrium of 8-oxoG. A structural alignment reveals that Ala-42 corresponds to hPol κ Met-135 in Dpo4 (supplemental Fig. S9). However, unlike the longer side chain of Met, the methyl group of Ala is unlikely to affect the orientation of 8-oxoG at the active site of Dpo4 in a significant manner. It can reasonably be proposed that structural constraints imposed by the N-clasp, which are not explicitly observed by either kinetic or structural results but which can be inferred from a combination of the two experimental approaches, act in coordination with specific electrostatic and steric features of the little finger domain and the active site to facilitate error-prone insertion of dATP opposite 8-oxoG by the Y-family DNA polymerase hPol κ.
Supplementary Material
Acknowledgments
We are grateful to S. Pattanayek for help with computer system administration and to Drs. P. S. Pallan, R. Pattanayek, and Z. Wawrzak for assistance with x-ray diffraction data collection at the Advanced Photon Source (APS), Argonne, IL. A. I. wishes to thank Dr. R. Fotedar for continued support. We thank K. Trisler for assistance in preparation of the manuscript. Vanderbilt University is a member institution of the Life Sciences Collaborative Access Team at Sector 21 of the APS.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 ES05355 (to M. E.), P30 ES000267 (to F. P. G. and M. E.), R01 ES010375 (to F. P. G.), and F32 CA119776 (to R. L. E.). Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02–06CH11357.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S2 and Figs. S1–S9.
The atomic coordinates and structure factors (codes 2w7o and 2w7p) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
C. Cambillau and A. Roussel (1997) Turbo Frodo, Version Open GL.1, Université Aix-Marseille II, Marseille, France.
W. L. DeLano (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA.
- Pol
- (DNA) polymerase
- hPol
- human polymerase
- BSA
- bovine serum albumin
- CID
- collision-induced dissociation
- Dpo4
- S. solfataricus DNA polymerase IV
- DTT
- dithiothreitol
- ESI
- electrospray ionization
- LC
- liquid chromatography
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- 8-oxoG
- 7,8-dihydro-8-oxo-2′-deoxyguanosine
- r.m.s.d.
- root mean square deviation
- MES
- 2-(N-morpholino)ethanesulfonic acid.
REFERENCES
- 1.Friedberg E. C., Walker G. C., Siede W., Wood R. D., Shultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, 2nd Ed., ASM Press, Washington, D. C. [Google Scholar]
- 2.Friedberg E. C., Fischhaber P. L., Kisker C. (2001) Cell 107, 9–12 [DOI] [PubMed] [Google Scholar]
- 3.Lehmann A. R. (2006) Mol. Cell 24, 493–495 [DOI] [PubMed] [Google Scholar]
- 4.Guengerich F. P. (2006) Chem. Rev. 106, 420–452 [DOI] [PubMed] [Google Scholar]
- 5.Prakash S., Johnson R. E., Prakash L. (2005) Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 6.Kim S. R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., Sofuni T., Nohmi T., Ohmori H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi S., Valentine M. R., Pham P., O'Donnell M., Goodman M. F. (2002) J. Biol. Chem. 277, 34198–34207 [DOI] [PubMed] [Google Scholar]
- 8.Zhou B. L., Pata J. D., Steitz T. A. (2001) Mol. Cell 8, 427–437 [DOI] [PubMed] [Google Scholar]
- 9.Potapova O., Grindley N. D., Joyce C. M. (2002) J. Biol. Chem. 277, 28157–28166 [DOI] [PubMed] [Google Scholar]
- 10.Silvian L. F., Toth E. A., Pham P., Goodman M. F., Ellenberger T. (2001) Nat. Struct. Biol. 8, 984–989 [DOI] [PubMed] [Google Scholar]
- 11.Boudsocq F., Iwai S., Hanaoka F., Woodgate R. (2001) Nucleic Acids Res. 29, 4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H., Boudsocq F., Woodgate R., Yang W. (2001) Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 13.Yang W., Woodgate R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steitz T. A. (1999) J. Biol. Chem. 274, 17395–17398 [DOI] [PubMed] [Google Scholar]
- 15.Alt A., Lammens K., Chiocchini C., Lammens A., Pieck J. C., Kuch D., Hopfner K. P., Carell T. (2007) Science 318, 967–970 [DOI] [PubMed] [Google Scholar]
- 16.Boudsocq F., Kokoska R. J., Plosky B. S., Vaisman A., Ling H., Kunkel T. A., Yang W., Woodgate R. (2004) J. Biol. Chem. 279, 32932–32940 [DOI] [PubMed] [Google Scholar]
- 17.Eoff R. L., Irimia A., Angel K. C., Egli M., Guengerich F. P. (2007) J. Biol. Chem. 282, 19831–19843 [DOI] [PubMed] [Google Scholar]
- 18.Lone S., Townson S. A., Uljon S. N., Johnson R. E., Brahma A., Nair D. T., Prakash S., Prakash L., Aggarwal A. K. (2007) Mol. Cell 25, 601–614 [DOI] [PubMed] [Google Scholar]
- 19.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2005) Science 309, 2219–2222 [DOI] [PubMed] [Google Scholar]
- 20.Nair D. T., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K. (2004) Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 21.Trincao J., Johnson R. E., Escalante C. R., Prakash S., Prakash L., Aggarwal A. K. (2001) Mol. Cell 8, 417–426 [DOI] [PubMed] [Google Scholar]
- 22.Degan P., Shigenaga M. K., Park E. M., Alperin P. E., Ames B. N. (1991) Carcinogenesis 12, 865–871 [DOI] [PubMed] [Google Scholar]
- 23.Malins D. C., Haimanot R. (1991) Cancer Res. 51, 5430–5432 [PubMed] [Google Scholar]
- 24.Fraga C. G., Shigenaga M. K., Park J. W., Degan P., Ames B. N. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4533–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimoda R., Nagashima M., Sakamoto M., Yamaguchi N., Hirohashi S., Yokota J., Kasai H. (1994) Cancer Res. 54, 3171–3172 [PubMed] [Google Scholar]
- 26.Fraga C. G., Motchnik P. A., Shigenaga M. K., Helbock H. J., Jacob R. A., Ames B. N. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11003–11006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogg M., Wallace S. S., Doublié S. (2005) Curr. Opin. Struct. Biol. 15, 86–93 [DOI] [PubMed] [Google Scholar]
- 28.Hsu G. W., Ober M., Carell T., Beese L. S. (2004) Nature 431, 217–221 [DOI] [PubMed] [Google Scholar]
- 29.Rechkoblit O., Malinina L., Cheng Y., Kuryavyi V., Broyde S., Geacintov N. E., Patel D. J. (2006) PLoS Biol. 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang H., Irimia A., Choi J. Y., Angel K. C., Loukachevitch L. V., Egli M., Guengerich F. P. (2006) J. Biol. Chem. 281, 2358–2372 [DOI] [PubMed] [Google Scholar]
- 31.Carlson K. D., Washington M. T. (2005) Mol. Cell. Biol. 25, 2169–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haracska L., Yu S. L., Johnson R. E., Prakash L., Prakash S. (2000) Nat. Genet. 25, 458–461 [DOI] [PubMed] [Google Scholar]
- 33.Haracska L., Prakash L., Prakash S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16000–16005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohashi E., Ogi T., Kusumoto R., Iwai S., Masutani C., Hanaoka F., Ohmori H. (2000) Genes Dev. 14, 1589–1594 [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J. S., Geacintov N. E., Wang Z. (2000) Nucleic Acids Res. 28, 4138–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang H., Goodenough A. K., Choi J. Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., Guengerich F. P. (2005) J. Biol. Chem. 280, 29750–29764 [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. (1988) J. Appl. Crystallogr. 21, 916–924 [Google Scholar]
- 39.McCoy A. J. (2007) Acta Crystallogr. D. Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vellieux F. M., Read R. J. (1997) Methods Enzymol. 277, 18–53 [DOI] [PubMed] [Google Scholar]
- 41.Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 [Google Scholar]
- 42.Brünger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 44.Carlson K. D., Johnson R. E., Prakash L., Prakash S., Washington M. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi J. Y., Angel K. C., Guengerich F. P. (2006) J. Biol. Chem. 281, 21062–21072 [DOI] [PubMed] [Google Scholar]
- 46.García-Ortiz M. V., Roldán-Arjona T., Ariza R. R. (2007) FEBS J. 274, 3340–3350 [DOI] [PubMed] [Google Scholar]
- 47.Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. (2006) Structure 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brieba L. G., Eichman B. F., Kokoska R. J., Doublié S., Kunkel T. A., Ellenberger T. (2004) EMBO J. 23, 3452–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia L., Geacintov N. E., Broyde S. (2008) Nucleic Acids Res. 36, 6571–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uljon S. N., Johnson R. E., Edwards T. A., Prakash S., Prakash L., Aggarwal A. K. (2004) Structure 12, 1395–1404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.