Abstract
Niemann-Pick C1-like 1 (NPC1L1) plays a critical role in the enterohepatic absorption of free cholesterol. Cellular cholesterol depletion induces the transport of NPC1L1 from the endocytic recycling compartment to the plasma membrane (PM), and cholesterol replenishment causes the internalization of NPC1L1 together with cholesterol via clathrin-mediated endocytosis. Although NPC1L1 has been characterized, the other proteins involved in cholesterol absorption and the endocytic recycling of NPC1L1 are largely unknown. Most of the vesicular trafficking events are dependent on the cytoskeleton and motor proteins. Here, we investigated the roles of the microfilament and microfilament-associated triple complex composed of myosin Vb, Rab11a, and Rab11-FIP2 in the transport of NPC1L1 from the endocytic recycling compartment to the PM. Interfering with the dynamics of the microfilament by pharmacological treatment delayed the transport of NPC1L1 to the cell surface. Meanwhile, inactivation of any component of the myosin Vb·Rab11a·Rab11-FIP2 triple complex inhibited the export of NPC1L1. Expression of the dominant-negative mutants of myosin Vb, Rab11a, or Rab11-FIP2 decreased the cellular cholesterol uptake by blocking the transport of NPC1L1 to the PM. These results suggest that the efficient transport of NPC1L1 to the PM is dependent on the microfilament-associated myosin Vb·Rab11a·Rab11-FIP2 triple complex.
Cholesterol homeostasis in human bodies is maintained through regulated cholesterol synthesis, absorption, and excretion. Intestinal cholesterol absorption is one of the major pathways to maintain cholesterol balance. NPC1L1 (Niemann-Pick C1-like protein 1), a polytopic transmembrane protein highly expressed in the intestine and liver, is required for dietary cholesterol uptake and biliary cholesterol reabsorption (1–4). Genetic or pharmaceutical inactivation of NPC1L1 significantly inhibits cholesterol absorption and confers the resistance to diet-induced hypercholesterolemia (1, 2, 4). Ezetimibe, an NPC1L1-specific inhibitor, is currently used to prevent and treat cardiovascular diseases (5).
Human NPC1L1 contains 1,332 residues with 13 transmembrane domains (6). The third to seventh transmembrane helices constitute a conserved sterol-sensing domain (4, 7). NPC1L1 recycles between the endocytic recycling compartment (ERC)3 and the plasma membrane (PM) in response to the changes of cholesterol level (8). ERC is a part of early endosomes that is involved in the recycling of many transmembrane proteins. It is also reported that ERC is a pool for free cholesterol storage (9). When cellular cholesterol concentration is low, NPC1L1 moves from the ERC to the PM (8, 10). Under cholesterol-replenishing conditions, NPC1L1 and cholesterol are internalized together and transported to the ERC (8). Disruption of microfilament, depletion of the clathrin·AP2 complex, or ezetimibe treatment can impede the endocytosis of NPC1L1, thereby decreasing cholesterol internalization (8, 10, 11).
The microfilament (MF) system, part of the cytoskeleton network, is required for multiple cellular functions such as cell shape maintenance, cell motility, mitosis, protein secretion, and endocytosis (12, 13). The major players in the microfilament system are actin fibers and motor proteins (14). Actin fibers form a network that serves as the tracks for vesicular transport (15, 16). Meanwhile, the dynamic assembly and disassembly of actin fibers and the motor proteins provides the driving force for a multitude of membrane dynamics including endocytosis, exocytosis, and vesicular trafficking between compartments (15, 16).
Myosins are a large family of motor proteins that are responsible for actin-based mobility (14). Class V myosins (17, 18), comprising myosin Va, Vb, and Vc, are involved in a wide range of vesicular trafficking events in different mammalian tissues. Myosin Va is expressed mainly in neuronal tissues (19, 20), whereas myosins Vb and Vc are universally expressed with enrichment in epithelial cells (21, 22). Class V myosins are recruited to their targeting vesicles by small GTPase proteins (Rab) (23). Rab11a and Rab11 family-interacting protein 2 (Rab11-FIP2) facilitate the binding of myosin Vb to the cargo proteins of endocytic recycling vesicles (24–28).
Myosin Vb binds Rab11a and Rab11-FIP2 through the C-terminal tail (CT) domain. The triple complex of myosin Vb, Rab11a, and Rab11-FIP2 is critical for endocytic vesicular transport and the recycling of many proteins including transferrin receptor (29), AMPA receptors (30), CFTR (28), GLUT4 (31, 32), aquaporin-2 (26), and β2-adrenergic receptors (33). The myosin Vb-CT domain (24) competes for binding to Rab11a and Rab11-FIP2 and functions as a dominant-negative form. Expression of the CT domain substantially impairs the transport of vesicles. Deficient endocytic trafficking is also observed in cells expressing the GDP-locked form of Rab11a (S25N) (34) or a truncated Rab11-FIP2, which competes for the rab11a binding (35).
Here we investigated the roles of actin fibers and motor proteins in the cholesterol-regulated endocytic recycling of NPC1L1. Using pharmaceutical inactivation, dominant-negative forms, and an siRNA technique, we demonstrated that actin fibers and myosin Vb·Rab11a·Rab11-FIP2 triple complex are involved in the export of NPC1L1 to the PM and that this intact MF-associated triple complex is required for efficient cholesterol uptake. Characterization of the molecules involved in the recycling of NPC1L1 may shed new light upon the mechanism of cholesterol absorption.
EXPERIMENTAL PROCEDURES
Materials
The reagents used include horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgG (from Jackson ImmunoResearch Laboratories); filipin, cytochalasin D, latrunculin B, jasplakinolide, nocodazole, and colchicine (from Sigma-Aldrich); sulfosuccinimidyl 6-(biotinamido)hexanoate and NeutrAvidin-agarose (from Pierce); methyl-β-cyclodextrin (CDX) (from Cyclodextrin Technologies Development, Inc.); and other reagents from previously described sources (8).
Plasmids
The plasmids expressing the EGFP-tagged myosin Vb full-length, myosin Va, and myosin Vb C-terminal dominant-negative tails were generously provided by Dr. Alaa El-Husseini (University of British Columbia, Canada). Myosin Vb-CTΔRBD was generated as described previously (25). Human myosin Vc-CT (residues 912–1742), Rab11a, and Rab11-FIP2 were PCR-amplified from the cDNA of HEK293 cells and inserted in-frame into pEGFP-C1 or other vectors with the indicated tags described in the text. The truncated Rab11-FIP2 encoding residues 446–511 was subcloned from the plasmids containing the full-length Rab11-FIP2 and inserted into pEGFP-C1.
Cell Culture
CRL-1601 (McArdle RH7777 rat hepatoma cell), and CRL-1601/NPC1L1-EGFP, which stably expresses NPC1L1-EGFP, were grown in a monolayer at 37 °C in 5% CO2. The cells were maintained in medium A (Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% fetal bovine serum. For CRL-1601/NPC1L1-EGFP, an additional 200 μg/ml G418 was supplemented. The cholesterol-depleting medium was medium A supplemented with 5% lipoprotein-deficient serum, 10 μm compactin, 50 μm mevalonate, and 1.5% CDX. Cholesterol-replenishing medium contained medium A supplemented with 5% lipoprotein-deficient serum, 10 μm compactin, 50 μm mevalonate, and different concentrations of cholesterol-CDX. The cholesterol·CDX inclusion complexes were prepared as described previously (36).
Transfection, Co-immunoprecipitation, and Immunoblotting
Transfection of cells with FuGENE HD (Roche Applied Science) was performed according to the manufacturer's manual. Conditions of the drug incubations are described in the legend for Fig. 6. At the end of incubation, triplicate samples for each treatment were harvested and lysed in lysis buffer (PBS containing 5 mm EGTA, 5 mm EDTA, 0.5% digitonin, and protease inhibitors). Whole cell lysate was incubated with anti-EGFP beads and rotated at 4 °C for 2 h. The agarose beads were then washed five times with lysis buffer. Proteins bound to the beads were eluted by elution buffer (0.2 m glycine, pH 2.9), and the eluent was mixed with SDS-PAGE loading buffer and incubated at 37 °C for 30 min before being analyzed on SDS-PAGE. Immunoblotting was carried out as described previously (8).
FIGURE 6.
The binding of NPC1L1 to Rab11a and myosin Vb is regulated by cholesterol depletion. A, plasmids encoding NPC1L1-EGFP and Rab11a-T7 were co-transfected into CRL-1601 cells. 48 h later, the cells were depleted of cholesterol by treatment with 1% CDX for various time durations. Then the cells were harvested, and immunoprecipitation (IP) was performed by pulling down EGFP-tagged NPC1L1 with anti-EGFP antibody-coupled agarose. Immunoblot (IB) analysis was carried out using the indicated antibodies. B, plasmids encoding NPC1L1-T7 and EGFP-myosin Vb were co-transfected into CRL-1601 cells. 48 h later, the cells were depleted of cholesterol by treatment 1% CDX for various durations of time. Then the cells were harvested, and immunoprecipitation was performed by pulling down EGFP-tagged myosin Vb with anti-EGFP antibody-coupled agarose. Immunoblot analysis was carried out using the indicated antibodies. Results shown are representative of three independent experiments.
Cell Surface Biotinylation Assay
The cells were washed twice with ice-cold PBS and treated with 1 mg/ml sulfosuccinimidyl 6-(biotinamido)hexanoate in PBS for 40 min at 4 °C. The cells were washed with ice-cold buffer A (20 mm Tris-HCl, pH 8.0, and 150 mm NaCl) and incubated in buffer A for 15 min at room temperature. Then, cells were collected by scraping and lysed in buffer B (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1% Nonidet P-40). Each sample of cell lysates was incubated with 100 μl of 50% (v/v) NeutrAvidin-agarose and rotated overnight at 4 °C. The agarose beads were then washed three times with buffer B. Biotinylated proteins binding to the beads were eluted by incubation with SDS-PAGE sample buffer at 37 °C for 30 min and analyzed by SDS-PAGE followed by Western blot.
RNA Interference
Duplexes of siRNA were synthesized by GenePharma (Shanghai, China). The sequences of the siRNAs targeting rat myosin Vb, Rab11a, and Rab11-FIP2 were 5′-GGTCCAGGACTTAGAAGCT-3′, 5′-GCAAGAGTACC3 ATTGGAGT-3′, and 5′-GGTGGCAATCAATCTCAAT-3′, respectively. The siRNA against VSV-G was described previously and used as a negative control. Transfection of siRNA was carried out as described previously (37).
Filipin Staining and Fluorescence Quantification
Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with PBS. A fresh 5 mg/ml filipin stock solution prepared in ethanol was diluted to a concentration of 50 μg/ml with PBS containing 10% fetal bovine serum. Cells were stained with this diluted filipin solution in the dark for 30 min at room temperature followed by three washes with PBS. Filipin signals of stained cells were analyzed with a Leica TCS SP5 confocal microscope equipped with a two-photon laser using an excitation wavelength of 720 nm. In each experiment, images of the same channel were acquired at identical laser output, gain, and offset. Fluorescence quantification was carried out as described previously (8). In brief, two circles, one outlining the whole cell and the other beneath the plasma membrane, were drawn manually. After subtraction of the background, the fluorescence intensities in each circle were measured by Image-Pro Plus, version 5.02, and taken as the total and intracellular fluorescence intensity. The intensity of each cell was arbitrarily defined as 1, against which the intracellular signals were normalized. For every time point, 50 cells were randomly selected and calculated.
RESULTS
MF Is Required for the Cholesterol-regulated Endocytic Recycling of NPC1L1
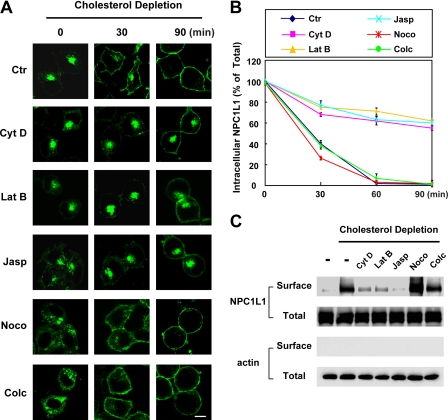
Our previous studies have shown that disruption of MF by cytochalasin D inhibits the endocytic recycling of NPC1L1 between the PM and the ERC, whereas disruption of the microtubules by nocodazole shows no effect (8). Because the transport of intracellular vesicles is a dynamic process that requires the polymerization and depolymerization of MF, we further investigated the roles of MF in the vesicular transport of NPC1L1. Cytochalasin D (38) (to depolymerize MF), latrunculin B (39) (to inhibit actin polymerization), and jasplakinolide (40) (to stabilize the MF) affected MF with different mechanisms; they were all tested in our experiments to exclude nonspecific effects. Both nocodazole (41) and colchicine (42) depolymerized the microtubules and were used as controls here. We first tested how the translocation of NPC1L1 to the PM upon cholesterol depletion was affected by different drugs. CRL1601/NPC1L1-EGFP cells, which stably express NPC1L1-EGFP fusion proteins, were pretreated with the indicated compounds to disturb the cytoskeleton. Then the cells were incubated in cholesterol-depleting medium along with the indicated drugs, and the localization of EGFP-NPC1L1 was analyzed at different time points. In control cells, NPC1L1-EGFP gradually moved to the plasma membrane (Fig. 1A, Ctr (top row)). However, in the cells treated with MF inhibitors, the translocation of NPC1L1-EGFP to PM was reduced (Fig. 1A, rows 2–4). Quantitative analysis confirmed that NPC1L1 nearly completely relocalized from intracellular vesicles to the PM in control cells after 90 min of cholesterol depletion, whereas more than 50% of NPC1L1 remained in the ERC of cytochalasin D-, latrunculin B-, and jasplakinolide-treated cells (Fig. 1B). In cells treated with nocodazole or colchicine, the perinuclear localization of NPC1L1-EGFP was dispersed. This effect was probably due to the disruption of ERC structure by these drugs, as ERC has been reported to associate with the microtubules and disruption of the microtubules interrupts the organization of ERC (9, 24, 43, 44). However, NPC1L1 still efficiently relocated to the PM after cholesterol-depletion (Fig. 1, A, rows 5 and 6, and B). The effects of different compounds on the transport of NPC1L1 were also validated by surface biotinylation assay (Fig. 1C).
FIGURE 1.
The transport of NPC1L1 to the PM requires microfilament integrity and dynamics. A, CRL-1601/NPC1L1-EGFP cells were pretreated with the indicated drugs for 30 min to disturb the cytoskeleton. These compounds were present throughout all time durations. The cells were then incubated in cholesterol-depleting medium to reduce cellular cholesterol for various time durations. At the indicated time points, the cells were fixed and examined by confocal microscopy. Images show the cells representative of the whole cell population at each indicated time point. Ctr, control; Cyt D, cytochalasin D; Lat B, latrunculin B; Jasp, jasplakinolide; Noco, nocodazle; Colc, colchicine. Bar, 10 μm. B, quantification of the intracellular localization of NPC1L1-EGFP shown in A. Error bars represent standard deviations. C, CRL-1601/NPC1L1-EGFP cells were treated as shown in A. After depletion of cholesterol for 30 min, the cells were collected and subjected to a surface biotinylation assay. Immunoblot analysis was carried out using the indicated antibodies. Results shown are representative of three independent experiments.
We next tested the MF dependence of cholesterol-induced NPC1L1-EGFP endocytosis. Nocodazole and colchicine had no obvious effect on the internalization of NPC1L1-EGFP, although the intracellular distribution of NPC1L1-EGFP was dispersed (supplemental Fig. S1B, compare rows 5 and 6 with row 1). However, cytochalasin D, latrunculin B, and jasplakinolide reduced the endocytosis of NPC1L1-EGFP (supplemental Fig. S1B, compare rows 2–4 with row 1). These results indicate that the cholesterol-regulated recycling of NPC1L1 is dependent on the integrity as well as the dynamics of the MF network.
Expression of the Myosin Vb-CT Domain Inhibits Translocation of NPC1L1 to the PM
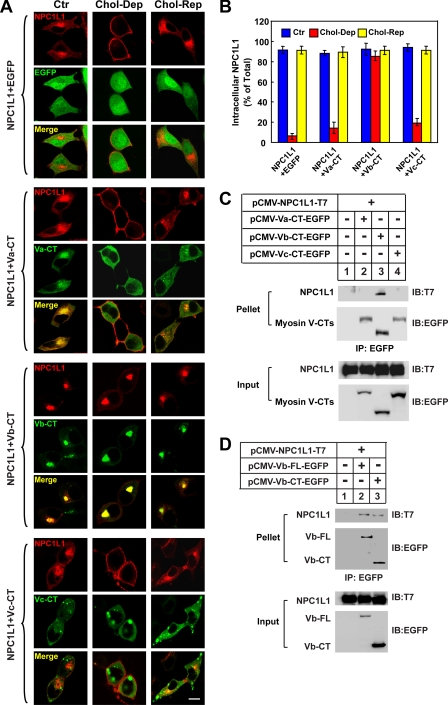
Myosins are the mechanoenzymatic proteins that move along the MF powered by ATP hydrolysis (45). The myosin V class, comprising myosin Va, Vb, and Vc, was recently shown to be required for the endocytic recycling of a series of receptors (17, 18, 21–27, 30). To test whether the class V myosins are involved in the vesicular transport of NPC1L1, we co-expressed the EGFP-tagged CT domains of myosins Va, Vb, and Vc with RFP-tagged NPC1L1. The myosin V-CT domains lack the motor domain and function as dominant-negative mutants. Western blotting showed that similar levels of myosin V-CTs (supplemental Fig. S2, upper row) and NPC1L1-RFP (supplemental Fig. S2, middle row) were expressed. We then examined the localization and recycling of NPC1L1 in the presence of different myosin V-CTs. In control cells, NPC1L1 localized in the ERC at steady state (Fig. 2A, first set of rows, Ctr). When cells were depleted of cholesterol for 60 min, NPC1L1 moved to the PM (Fig. 2A, first set, Chol-Dep). Then, replenishment of cholesterol promoted the endocytosis of NPC1L1 to intracellular vesicles (Fig. 2A, first set, Chol-Rep). Expression of the myosin Vb-CT led to a condensed ERC where it co-localized with NPC1L1 (Fig. 2A, third set, Ctr). Furthermore, myosin Vb-CT inhibited the transport of NPC1L1 to the PM when cells were depleted of cholesterol (Fig. 2A, third set, Chol-Dep). On the contrary, expression of either myosin Va-CT or myosin Vc-CT had no dramatic effects on the localization and recycling of NPC1L1 (Fig. 2A, second and fourth sets), although myosin Va-CT co-localized with NPC1L1 at steady state (Fig. 2A, second set, Ctr). Fluorescence quantification showed that only approximately less than 20% of the NPC1L1 localized in the intracellular vesicles in the control or myosin Va-CT- or Vc-CT-expressing cells after cholesterol depletion (Fig. 2B, Chol-Dep). However, myosin Vb-CT caused more than 80% of the NPC1L1 to be retained within the cells when cholesterol was depleted (Fig. 2B, Chol-Dep). Co-immunoprecipitation experiments showed that NPC1L1 interacted with both myosin Vb-CT (Fig. 2, C and D) and full-length myosin Vb (Fig. 2D, Vb-FL), whereas no interaction was detected between NPC1L1 and myosin Va-CT or Vc-CT (Fig. 2C). Taken together, these results suggested that myosin Vb is specifically involved in the transport of NPC1L1 to the PM induced by cholesterol depletion.
FIGURE 2.
The C-terminal tail of Myosin Vb inhibits the transport of NPC1L1 to PM. A, the EGFP-tagged CTs of myosin Va, Vb, and Vc were co-expressed with NPC1L1-RFP in CRL-1601 cells. 48 h after transfection, the cells were either depleted of cholesterol for 60 min (Chol-Dep) or replenished with cholesterol for 60 min (Chol-Rep) following cholesterol depletion. Then the cells were fixed and examined by confocal microscopy. Images show the cells representative of the whole cell population at each indicated time point. Ctr, control; Bar, 10 μm. B, quantification of the intracellular localization of NPC1L1-RFP shown in A. Error bars represent standard deviations. C, EGFP-tagged CTs of myosin Va, Vb, and Vc were co-expressed individually with NPC1L1-T7 in CRL-1601 cells. 48 h after transfection, the cells were collected, and immunoprecipitation (IP) was performed by pulling down EGFP-tagged myosin V-CT variants with anti-EGFP-agarose. IB, immunoblot. D, EGFP-tagged myosin Vb-CT and full-length (FL) myosin Vb were co-expressed individually with NPC1L1-T7 in CRL-1601 cells. 48 h after transfection, the cells were collected, and EGFP-tagged myosin Vb variants were immunoprecipitated by using anti-EGFP-agarose. Immunoblot analysis was carried out using the indicated antibodies. Results shown are representative of four independent experiments.
Involvement of Rab11a and Rab11-FIP2 in Translocation of NPC1L1 toward the PM
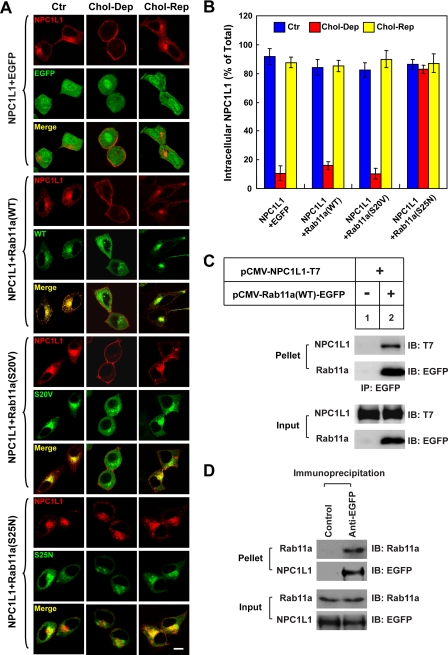
Myosin Vb associates with Rab11a and Rab11-FIP2 through the CT domain (24, 26), and Rab11a/Rab11-FIP2 facilitates the loading of myosin Vb to cargo vesicles. Deletion of the Rab11a binding site in myosin Vb-CT abolishes its dominant-negative effect on the export of GluR1 to the cell surface (25, 39). NPC1L1 co-localized with Rab11a in the ERC and deletion of the Rab11a binding site in myosin Vb-CT (named myosin Vb-CTΔRBD) neutralized its dominant-negative effect on the translocation of NPC1L1 to the PM (supplemental Fig. S3A, third set of rows), suggesting that Rab11a and Rab11-FIP2 may be involved in the transport of NPC1L1 to the PM. To test this hypothesis, we co-expressed the EGFP-tagged wild type (WT), the GTP-locked form (S20V), and the GDP-locked form (S25N) of rab11a, respectively, with NPC1L1-RFP (Fig. 3A). Rab11a(S25N) cannot exchange GDP with GTP and works as a dominant-negative mutant form. At steady state, both Rab11a(WT) and Rab11a(S20V) co-localized with NPC1L1 in the ERC (Fig. 3A, second and third sets of rows, Ctr), whereas Rab11a(S25N) and NPC1L1 only partially co-localized (Fig. 3A, fourth set, Ctr). When cholesterol levels were changed, NPC1L1 in Rab11a(WT)- or Rab11a(S20V)-co-expressing cells recycled between the PM and the ERC. In contrast, the transport of NPC1L1 to PM was inhibited in Rab11a (S25N) expressing cells (Fig. 3A, fourth set, Chol-Dep). Quantification of NPC1L1 localization illustrated that after cholesterol depletion, less than 20% of the NPC1L1 resided within the cells RFP or Rab11a(WT)- or Rab11a(S20V)-expressing cells (Fig. 3B, Chol-Dep). However, more than 85% of the NPC1L1 was restrained in intracellular vesicles in the cells expressing Rab11a(S25N), the dominant-negative mutant of Rab11a (Fig. 3B, Chol-Dep). Meanwhile, a co-immunoprecipitation experiment confirmed the interaction between transfected NPC1L1 and the transfected Rab11a (Fig. 3C) as well as the interaction between the transfected NPC1L1 and the endogenous Rab11a (Fig. 3D). Therefore, we concluded that Rab11a binds NPC1L1 and facilitates the efficient transport of NPC1L1 to the PM in response to cholesterol depletion.
FIGURE 3.
GDP-locked form of Rab11a (S25N) inhibits the transport of NPC1L1 to the PM. A, EGFP-tagged Rab11a(WT), Rab11a(S20V), and Rab11a(S25N) were co-expressed individually with NPC1L1-RFP in CRL-1601 cells. 48 h after transfection, the cells were either depleted of cholesterol for 60 min (Chol-Dep) or replenished with cholesterol for 60 min (Chol-Rep) following cholesterol depletion. Then the cells were fixed and examined by confocal microscopy. Images show cells that are representative of the whole cell population at each indicated time point. Ctr, control. Bar, 10 μm. B, quantification of the intracellular localization of NPC1L1-RFP shown in A. Error bars represent standard deviations. C, the EGFP-tagged Rab11a was co-expressed individually with NPC1L1-T7 in CRL-1601 cells. 48 h after transfection, cells were collected and immunoprecipitation (IP) performed by incubating cell lysates with anti-EGFP-agarose. Immunoblot (IB) analysis was carried out using the indicated antibodies. D, CRL1601/NPC1L1-EGFP cells were harvested and subjected to IP with control or anti-EGFP agarose. IB analysis was carried out using the indicated antibodies. Results shown are representative of three independent experiments.
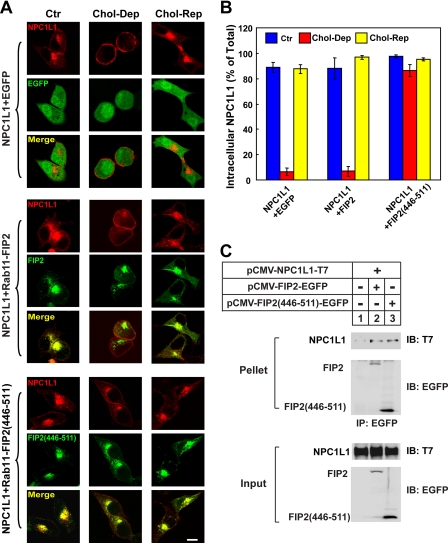
Similar experiments were performed using the dominant-negative form of Rab11-FIP2 containing residues 446–511 (named Rab11-FIP2-(446–511)), which compete for Rab11 binding (46). Fluorescence imaging showed that NPC1L1 co-localized with Rab11-FIP2 in the ERC (Fig. 4A, second set of rows, Ctr), and the cholesterol depletion-induced transport of NPC1L1 to the PM was inhibited in Rab11-FIP2-(446–511)-expressing cells (Fig. 4A, third set, Chol-Dep). Quantification also confirmed that the intracellularly localized NPC1L1 in Rab11-FIP2-(446–511)-expressing cells (∼82%) was significantly higher than that of the control (∼4%) or Rab11-FIP2-expressing cells (∼5%) (Fig. 4B). Subsequent co-immunoprecipitation experiments verified the association between NPC1L1 and Rab11-FIP2 (Fig. 4C).
FIGURE 4.
Rab11-FIP2-(446–511) blocks the transport of NPC1L1 to PM. A, NPC1L1-RFP were co-expressed with EGFP-tagged Rab11-FIP2 and Rab11-FIP2-(446–511) individually in CRL-1601 cells. 48 h after transfection, the cells were either depleted of cholesterol for 60 min (Chol-Dep) or replenished with cholesterol for 60 min (Chol-Rep) following cholesterol depletion. Then the cells were fixed and examined by confocal microscopy. Images show cells that are representative of the whole cell population at each indicated time point. Ctr, control. Bar, 10 μm. B, quantification of the intracellular localization of NPC1L1-RFP shown in A. Error bars represent standard deviations. C, EGFP-tagged Rab11-FIP2 and Rab11-FIP2-(446–511) were co-expressed individually with NPC1L1-T7 in CRL-1601 cells. 48 h after transfection, EGFP-tagged Rab11-FIP2 variants were immunoprecipitated (IP) with anti-EGFP antibody-coupled agarose. Immunoblot (IB) analysis was carried out using the indicated antibodies. Results shown are representative of three independent experiments.
Myosin Vb has been reported to associate with Rab8a-positive vesicles (31, 47). Therefore, Rab8a was tested similarly by co-transfection and imaging analysis. However, none of the WT or GTP- or GDP-locked forms of Rab8a had an obvious effect on the recycling of NPC1L1 protein (supplemental Fig. S4). Therefore, it is unlikely that Rab8a is involved in the cholesterol-regulated recycling of NPC1L1.
The Endocytic Recycling Triple Complex Is Required for Translocation of NPC1L1 to the PM
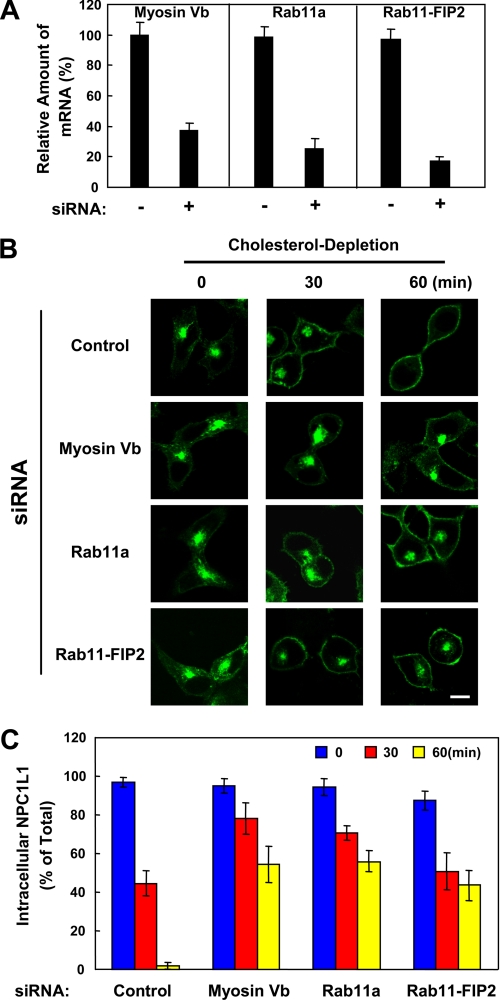
Next, we examined the requirement of endogenous myosin Vb, Rab11a, and Rab11-FIP2 in the transport of NPC1L1 to the PM using siRNA-mediated gene silencing. Real-time PCR analysis showed that the mRNA levels of myosin Vb, Rab11a, and Rab11-FIP2 were reduced by ∼60, ∼75, and ∼80%, respectively (Fig. 5A). Then we analyzed the transport of NPC1L1-EGFP to the PM induced by cholesterol depletion following RNA interference. As shown in Fig. 5, B and C, knockdown of myosin Vb, Rab11a, or Rab11-FIP2 caused retention of ∼50% of the NPC1L1 in intracellular vesicles compared with control cells after cholesterol depletion for 60 min. To rule out the possible off-target effect of siRNAs, two additional siRNAs against each component of the triple complex were tested, and consistent results were obtained (data not shown). Thus, the transport of NPC1L1 to the PM relies on the myosin Vb·Rab11a·Rab11-FIP2 endocytic recycling triple complex, and depletion of any one of the components inhibits the transport of NPC1L1 to PM.
FIGURE 5.
siRNA-mediated silencing of endogenous myosin Vb, Rab11a, or Rab11-FIP2 attenuates the transport of NPC1L1 to PM. A, real-time PCR showing the knockdown efficiency of myosin Vb, Rab11a, or Rab11-FIP2. B, CRL1601/NPC1L1-EGFP cells were transfected with the indicated siRNAs. 72 h after transfection, the cells were depleted of cholesterol for different durations of time. At each indicated time point, the cells were fixed and examined by confocal microscopy. Images show the cells representative of the whole cell population at each indicated time point. Bar, 10 μm. C, quantification of the intracellular localization of NPC1L1-EGFP shown in B. Error bars represent standard deviations. Results shown are representative of three independent experiments.
Cholesterol-regulated Binding of NPC1L1 to Rab11a and Myosin Vb
Because the myosin Vb·Rab11a· Rab11-FIP2 complex facilitated the cholesterol depletion-induced transport of NPC1L1 to the PM, we then tested whether the interaction between NPC1L1 and the triple complex was regulated by cholesterol. Consistent with the earlier results (Fig. 3C), NPC1L1 associated with Rab11a at steady state (Fig. 6A, lane 3). After 10 min of CDX treatment, the interaction between NPC1L1 and Rab11a gradually decreased (Fig. 6A, lanes 4–8), suggesting that Rab11a gradually dissociated from NPC1L1 during cholesterol depletion. On the other hand, the binding of NPC1L1 to myosin Vb did not change during the first 20 min after CDX treatment (Fig. 6B, lanes 3–5) but was drastically reduced following the 20-min time point (Fig. 6B, lanes 6–8). Our data indicate that Rab11a and myosin Vb dissociated sequentially from NPC1L1.
Disturbing the Endocytic Recycling Triple Complex Inhibits NPC1L1-mediated Cholesterol Uptake
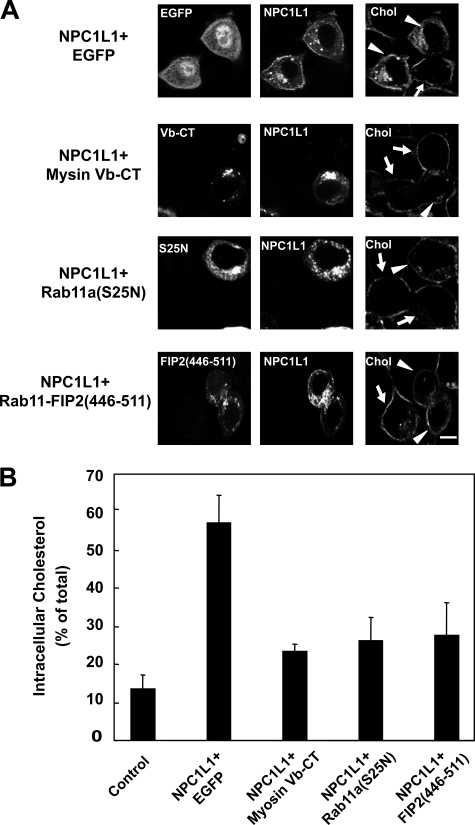
Our previous studies showed that NPC1L1 facilitates cholesterol uptake via its vesicular endocytosis from the PM (8). Therefore, localization of NPC1L1 on the PM is a prerequisite for cholesterol uptake. To further elucidate the role of the endocytic recycling triple complex in NPC1L1-mediated cholesterol uptake, NPC1L1-RFP was co-expressed with EGFP-tagged myosin Vb-CT, Rab11a(S25N), or Rab11-FIP2-(446–511) in rat hepatocyte CRL1601 cells. After transfection, the cells were depleted of cholesterol followed by cholesterol replenishment. Then the cells were fixed, stained with filipin, and imaged by two-photon confocal microscopy to analyze cholesterol uptake. As shown in Fig. 7A, cells expressing NPC1L1-RFP and EGFP (indicated by arrowheads) took up much more cholesterol than the untransfected control cells (indicated by arrows). However, in myosin Vb-CT-, Rab11a(S25N)-, and Rab11-FIP2(446–511)-expressing cells, the uptake of cholesterol was similar to the untransfected cells. Quantification showed that after cholesterol replenishment, the cholesterol internalization in EGFP-expressing cells (∼60%) was 4 times that in the control cells (∼15%), whereas expression of myosin Vb-CT, Rab11a(S25N), or Rab11-FIP2-(446–511) reduced cholesterol internalization mediated by NPC1L1 (Fig. 7B). These data indicate that inhibiting the myosin Vb·Rab11a·Rab11-FIP2 triple complex blocks the transport of NPC1L1 to the PM and decreases NPC1L1-mediated cholesterol uptake.
FIGURE 7.
Myosin Vb-CT, Rab11a(S25N), or Rab11-FIP2-(446–511) impairs the NPC1L1-mediated cholesterol uptake. A, NPC1L1-RFP was co-expressed with EGFP-tagged myosin Vb-CT, Rab11a(S25N), and Rab11-FIP2-(446–511) individually in CRL-1601 cells. 48 h after transfection, the cells were depleted of cholesterol followed by cholesterol replenishment for 60 min. Then the cells were fixed, stained with filipin, and examined by two-photon confocal microscopy. Images show the cells representative of the whole cell population at each indicated time point. Bar, 10 μm. Arrows indicate the cells with no expression of the transfected genes. Arrowheads indicate the cells expressing the transfected genes. B, quantification of the intracellular cholesterol of the cells shown in A. Error bars represent standard deviations. Results shown are representative of two independent experiments.
DISCUSSION
Here we investigated the role of the MF and the myosin Vb·Rab11a·Rab11-FIP2 triple complex in cholesterol-regulated endocytic recycling of NPC1L1. First, the integrity and dynamics of the MF are critical for the transport of NPC1L1. Second, the endocytic recycling triple complex, consisting of the MF-interacting motor myosin Vb, the small GTPase Rab11a, and the adaptor Rab11-FIP2, mediates the transport of NPC1L1 to the PM. Inactivation of the triple complex inhibits the export of NPC1L1 and NPC1L1-mediated cholesterol uptake.
Interestingly, our data support that not only the integrity but also the dynamics of actin fibers are important for the transport of NPC1L1, suggesting that the MF may function more than as the tracks for cargo motility. The MF network is highly dynamic and may participate in multiple steps of vesicle trafficking including scission, transport, and fusion. The polymerization and depolymerization of the MF may provide the mechanical force for each step. Meanwhile, many proteins are involved in the remodeling of the MF, such as the ARP2/3 (48) complex that initiates MF nucleation and facilitates vesicle transport. Based on our previous result that ARP2/3 is associated with NPC1L1, it is very likely that MF remodeling factors such as ARP2/3 also play a role in NPC1L1 recycling and cholesterol uptake (8).
Myosin Vb has been found to be responsible for the endocytic shuttling and sorting of proteins including transferrin receptor, CFTR, AMPA receptors, BSEP, GLUT4, aquaporin-2, and β2-adrenergic receptor (18, 24–33). Our finding that the myosin Vb·Rab11a·Rab11-FIP2 complex is also required for NPC1L1 recycling further supports the hypothesis that myosin Vb and its effectors function mainly in the traffic of transmembrane proteins between the PM and recycling endosomes. In addition, myosin Vb and Rab11a have been shown to be critical for the bile canaliculus formation in polarized hepatocytes (49). In fact, human NPC1L1 is also highly expressed in the liver in addition to the intestine. We and others have found that NPC1L1 localizes on the apical membrane of cultured hepatocytes, which resemble the canalicular membrane (data not shown) (8, 50). It is very likely that in human liver, myosin Vb, together with Rab11a and Rab11-FIP2, targets NPC1L1 to the canalicular membrane to reabsorb the free cholesterol from bile.
Although the MF system is required for the recycling of NPC1L1 between the PM and ERC, the role of the myosin Vb·Rab11a·Rab11-FIP2 triple complex in the internalization of NPC1L1 has not been determined here. Upon disrupting the myosin Vb·Rab11a·Rab11-FIP2 complex, NPC1L1 did not relocalize to the PM after cholesterol depletion in our experimental system, and therefore we could not study the internalization of NPC1L1 under cholesterol repletion conditions. Because it has been shown that the triple complex is localized in endocytic recycling endosomes, whereas NPC1L1 presents on the PM after cholesterol depletion, it is unlikely that the myosin Vb·Rab11a·Rab11-FIP2 triple complex is involved directly in the endocytosis of NPC1L1.
Loss-of-function mutations of myosin Vb can cause microvillus inclusion disease (MVID) (51), which is characterized by lack of microvilli on the surface of intestinal enterocytes and accumulation of microvilli in intracellular vacuolar structures. The vesicles that are otherwise targeted to the apical or basolateral membranes become aberrantly localized in the patient's enterocytes, suggesting the importance of myosin Vb in the correct targeting of endocytic vesicles in the intestine. Although the distribution of NPC1L1 in the intestinal cells of MVID patients has not been studied, it is likely that the localization of NPC1L1 and cholesterol absorption are both disrupted in MVID enterocytes, as myosin Vb is required for the targeting of NPC1L1. More work is required to address the physiological role of the myosin Vb·Rab11a·Rab11-FIP2 triple complex in cholesterol absorption in animals.
Myosin Vb and Rab11a showed different trends when they dissociated from NPC1L1 during cholesterol depletion (Fig. 6), implying that Rab11a and myosin Vb may associate with NPC1L1 in different ways during the translocation of NPC1L1 to the PM. We also found that the Rab11a binding domain in myosin Vb-CT is required for the dominant-negative effect (supplemental Fig. S3). Taking all of our findings together, the following model is conceivable. Rab11a and Rab11-FIP2 binds to NPC1L1 at the steady state and bridges the interaction between NPC1L1 and myosin Vb. When the cellular cholesterol level drops, Rab11a/Rab11-FIP2 dissociates from the NPC1L1·myosin Vb complex, which allows the export of NPC1L1·myosin Vb toward the PM in a MF-dependent manner. After arriving at the PM, myosin Vb is separated and released. The interaction between NPC1L1 and myosin Vb or Rab11a/Rab11-FIP2 may be direct or may be mediated by other factors. Additional studies are needed to address this question.
In summary, our data provide evidence showing that the myosin Vb·Rab11a·Rab11-FIP2 triple complex is required for the transport of NPC1L1 to the PM and plays an important role in cholesterol uptake. This finding enriches our knowledge of the molecular pathway of cholesterol absorption and provides a basis for developing novel cholesterol absorption inhibitors.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of myosin Va and Vb plasmids from Dr. Alaa El-Husseini (University of British Columbia, Canada). We thank Yu-Xiu Qu, Su-Zhe Pan, Qing Li, and Chun-Geng Yi for technical assistance and Dr. Wei Qi for critical reading of the manuscript.
This work was supported by Grants 2009CB919000 and 2007AA09Z402 from the Ministry of Science and Technology of China, 30670432 and 90713025 from the National Natural Science Foundation of China, and 08JC1421300 and 08431900500 from the Shanghai Science and Technology Committee.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ERC
- endocytic recycling compartment
- MF
- microfilament
- NPC1L1
- Niemann-Pick C1-like 1
- PM
- plasma membrane
- WT
- wild type
- CT
- C-terminal tail
- MVID
- microvillus inclusion disease
- CDX
- methyl-β-cyclodextrin
- siRNA
- small interfering RNA
- EGFP
- enhanced green fluorescent protein
- PBS
- phosphate-buffered saline
- RFP
- red fluorescent protein.
REFERENCES
- 1.Davies J. P., Scott C., Oishi K., Liapis A., Ioannou Y. A. (2005) J. Biol. Chem. 280, 12710–12720 [DOI] [PubMed] [Google Scholar]
- 2.Davis H. R., Jr., Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., Detmers P. A., Graziano M. P., Altmann S. W. (2004) J. Biol. Chem. 279, 33586–33592 [DOI] [PubMed] [Google Scholar]
- 3.Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. (2007) J. Clin. Invest. 117, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M. P. (2004) Science 303, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 5.Davis H. R., Veltri E. P. (2007) J. Atheroscler. Thromb. 14, 99–108 [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Chu B. B., Ge L., Li B. L., Yan Y., Song B. L. (2009) J. Lipid Res, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J. P., Levy B., Ioannou Y. A. (2000) Genomics 65, 137–145 [DOI] [PubMed] [Google Scholar]
- 8.Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. (2008) Cell Metab. 7, 508–519 [DOI] [PubMed] [Google Scholar]
- 9.Maxfield F. R., McGraw T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 10.Yu L., Bharadwaj S., Brown J. M., Ma Y., Du W., Davis M. A., Michaely P., Liu P., Willingham M. C., Rudel L. L. (2006) J. Biol. Chem. 281, 6616–6624 [DOI] [PubMed] [Google Scholar]
- 11.Chang T. Y., Chang C. (2008) Cell Metab. 7, 469–471 [DOI] [PubMed] [Google Scholar]
- 12.Prentki M., Chaponnier C., Jeanrenaud B., Gabbiani G. (1979) J. Cell Biol. 81, 592–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segawa A., Yamashina S. (1989) Cell Struct. Funct. 14, 531–544 [DOI] [PubMed] [Google Scholar]
- 14.Dantzig J. A., Liu T. Y., Goldman Y. E. (2006) Ann. N.Y. Acad. Sci. 1080, 1–18 [DOI] [PubMed] [Google Scholar]
- 15.Apodaca G. (2001) Traffic 2, 149–159 [DOI] [PubMed] [Google Scholar]
- 16.Schafer D. A. (2002) Curr. Opin. Cell Biol. 14, 76–81 [DOI] [PubMed] [Google Scholar]
- 17.Bridgman P. C. (2004) J. Neurobiol. 58, 164–174 [DOI] [PubMed] [Google Scholar]
- 18.Langford G. M. (2002) Traffic 3, 859–865 [DOI] [PubMed] [Google Scholar]
- 19.Tilelli C. Q., Martins A. R., Larson R. E., Garcia-Cairasco N. (2003) Neuroscience 121, 573–586 [DOI] [PubMed] [Google Scholar]
- 20.Walikonis R. S., Jensen O. N., Mann M., Provance D. W., Jr., Mercer J. A., Kennedy M. B. (2000) J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez O. C., Cheney R. E. (2002) J. Cell Sci. 115, 991–1004 [DOI] [PubMed] [Google Scholar]
- 22.Zhao L. P., Koslovsky J. S., Reinhard J., Bähler M., Witt A. E., Provance D. W., Jr., Mercer J. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10826–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passeron T., Bahadoran P., Bertolotto C., Chiaverini C., Buscà R., Valony G., Bille K., Ortonne J. P., Ballotti R. (2004) FASEB J. 18, 989–991 [DOI] [PubMed] [Google Scholar]
- 24.Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Jr., Mercer J. A., Bähler M., Goldenring J. R. (2001) Mol. Biol. Cell 12, 1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisé M. F., Wong T. P., Trinh A., Hines R. M., Liu L., Kang R., Hines D. J., Lu J., Goldenring J. R., Wang Y. T., El-Husseini A. (2006) J. Biol. Chem. 281, 3669–3678 [DOI] [PubMed] [Google Scholar]
- 26.Nedvetsky P. I., Stefan E., Frische S., Santamaria K., Wiesner B., Valenti G., Hammer J. A., 3rd, Nielsen S., Goldenring J. R., Rosenthal W., Klussmann E. (2007) Traffic 8, 110–123 [DOI] [PubMed] [Google Scholar]
- 27.Volpicelli L. A., Lah J. J., Fang G., Goldenring J. R., Levey A. I. (2002) J. Neurosci. 22, 9776–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swiatecka-Urban A., Talebian L., Kanno E., Moreau-Marquis S., Coutermarsh B., Hansen K., Karlson K. H., Barnaby R., Cheney R. E., Langford G. M., Fukuda M., Stanton B. A. (2007) J. Biol. Chem. 282, 23725–23736 [DOI] [PubMed] [Google Scholar]
- 29.Provance D. W., Jr., Addison E. J., Wood P. R., Chen D. Z., Silan C. M., Mercer J. A. (2008) BMC Cell Biol. 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., Edwards J. G., Riley N., Provance D. W., Jr., Karcher R., Li X. D., Davison I. G., Ikebe M., Mercer J. A., Kauer J. A., Ehlers M. D. (2008) Cell 135, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikura S., Klip A. (2008) Am. J. Physiol. Cell Physiol. 295, C1016–C1025 [DOI] [PubMed] [Google Scholar]
- 32.Kessler A., Tomas E., Immler D., Meyer H. E., Zorzano A., Eckel J. (2000) Diabetologia 43, 1518–1527 [DOI] [PubMed] [Google Scholar]
- 33.Millman E. E., Zhang H., Zhang H., Godines V., Bean A. J., Knoll B. J., Moore R. H. (2008) Traffic 9, 1958–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996) J. Cell Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jagoe W. N., Lindsay A. J., Read R. J., McCoy A. J., McCaffrey M. W., Khan A. R. (2006) Structure 14, 1273–1283 [DOI] [PubMed] [Google Scholar]
- 36.Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., DeBose-Boyd R. A., Tang J. J., Li B. L., Song B. L. (2007) Cell Metab. 6, 115–128 [DOI] [PubMed] [Google Scholar]
- 37.Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 38.Dubinsky W. P., Mayorga-Wark O., Schultz S. G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9421–9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibbon B. C., Kovar D. R., Staiger C. J. (1999) Plant Cell 11, 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie L., Forer A. (2008) Cell Motil. Cytoskeleton 65, 876–889 [DOI] [PubMed] [Google Scholar]
- 41.Lee J. C., Field D. J., Lee L. L. (1980) Biochemistry 19, 6209–6215 [DOI] [PubMed] [Google Scholar]
- 42.Caperta A. D., Delgado M., Ressurreiçca̋o F., Meister A., Jones R. N., Viegas W., Houben A. (2006) Protoplasma 227, 147–153 [DOI] [PubMed] [Google Scholar]
- 43.Yamashiro D. J., Tycko B., Fluss S. R., Maxfield F. R. (1984) Cell 37, 789–800 [DOI] [PubMed] [Google Scholar]
- 44.Vossenkämper A., Nedvetsky P. I., Wiesner B., Furkert J., Rosenthal W., Klussmann E. (2007) Am. J. Physiol. Cell Physiol. 293, C1129–C1138 [DOI] [PubMed] [Google Scholar]
- 45.Taft M. H., Hartmann F. K., Rump A., Keller H., Chizhov I., Manstein D. J., Tsiavaliaris G. (2008) J. Biol. Chem. 283, 26902–26910 [DOI] [PubMed] [Google Scholar]
- 46.Lindsay A. J., McCaffrey M. W. (2002) J. Biol. Chem. 277, 27193–27199 [DOI] [PubMed] [Google Scholar]
- 47.Roland J. T., Kenworthy A. K., Peranen J., Caplan S., Goldenring J. R. (2007) Mol. Biol. Cell 18, 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollard T. D. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 49.Wakabayashi Y., Dutt P., Lippincott-Schwartz J., Arias I. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15087–15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu L. (2008) Curr. Opin. Lipidol. 19, 263–269 [DOI] [PubMed] [Google Scholar]
- 51.Müller T., Hess M. W., Schiefermeier N., Pfaller K., Ebner H. L., Heinz-Erian P., Ponstingl H., Partsch J., Röllinghoff B., Köhler H., Berger T., Lenhartz H., Schlenck B., Houwen R. J., Taylor C. J., Zoller H., Lechner S., Goulet O., Utermann G., Ruemmele F. M., Huber L. A., Janecke A. R. (2008) Nat. Genet. 40, 1163–1165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.