SUMMARY
Hepatitis C virus (HCV) chronically infects 3% of the world’s population, and complications from HCV are the leading indication for liver transplantation. Given the need for better anti-HCV therapies, one strategy is to identify and target cellular cofactors of the virus lifecycle. Using a genome-wide siRNA library, we identified 96 human genes that support HCV replication, with a significant number of them being involved in vesicle organization and biogenesis. Phosphatidylinositol 4-kinase PI4KA and multiple subunits of the COPI vesicle coat complex were among the genes identified. Consistent with this, pharmacologic inhibitors of COPI and PI4KA blocked HCV replication. Targeting hepcidin, a peptide critical for iron homeostasis, also affected HCV replication, which may explain the known dysregulation of iron homeostasis in HCV infection. The host cofactors for HCV replication identified in this study should serve as a useful resource in delineating new targets for anti-HCV therapies.
INTRODUCTION
Hepatitis C virus (HCV) is an RNA virus with the remarkable ability to establish a chronic infection in 70%–80% of exposed humans. As many as 3% of the world’s population are chronically infected with HCV. Of that number, as many as 30% will develop cirrhosis within 20 years of initial infection, which subsequently may lead to liver failure and/or hepatocellular carcinoma. As a result, HCV-related liver disease is now the leading indication for liver transplantation worldwide. The best current therapy for chronic HCV infection is the combination of peginterferon and ribavirin. Unfortunately, these agents are associated with a high rate of side effects and produce sustained virologic response in only about half of treated individuals. Although small molecule inhibitors of the HCV NS3-4A serine protease and NS5B RNA-dependent RNA polymerase are in clinical development, monotherapy with direct antiviral agents leads to the rapid emergence of viral resistance mutations (Reesink et al., 2006) because of the error-prone nature of the viral RNA polymerase.
Another approach to control HCV is to identify the host cofactors that support the viral lifecycle. Like all viruses, hepatitis C is dependent on host proteins for viral entry, uncoating, replication, virion assembly, and egress. Targeting these cofactors may impose a higher barrier to viral resistance and also offers the possibility of blocking the HCV lifecycle at multiple complementary steps. For instance, a small-molecule inhibitor of cyclophilin B, which has been shown to interact with and positively regulate the viral RNA polymerase (Watashi et al., 2005), potently inhibits HCV replication in vivo (Flisiak et al., 2008). In addition to cyclophilin B, to date, a number of other cellular proteins have been identified as HCV cofactors (Ng et al., 2007; Randall et al., 2007; Supekova et al., 2008). In this study, we describe the results of a whole-genome siRNA library screen designed to identify host proteins that support HCV replication.
RESULTS
Whole-Genome siRNA Screen for Host Cofactors of HCV Replication
We screened for host proteins involved in HCV replication using the Huh7/Rep-Feo subgenomic genotype 1b HCV replicon (Tanabe et al., 2004) that we had previously adapted to high-throughput screening of small molecule libraries (Kim et al., 2007). This replicon is an RNA molecule that encodes the HCV nonstructural proteins (NS3 through NS5B) that are sufficient to mediate viral replication as well as a firefly luciferase-neomycin phosphotransferase fusion protein (Figure S1). Firefly luciferase activity is a linear function of replicon RNA copy number and therefore permits sensitive and rapid quantitation of HCV replication (Tanabe et al., 2004). We optimized siRNA transfection conditions using a luciferase siRNA (Figure S2).
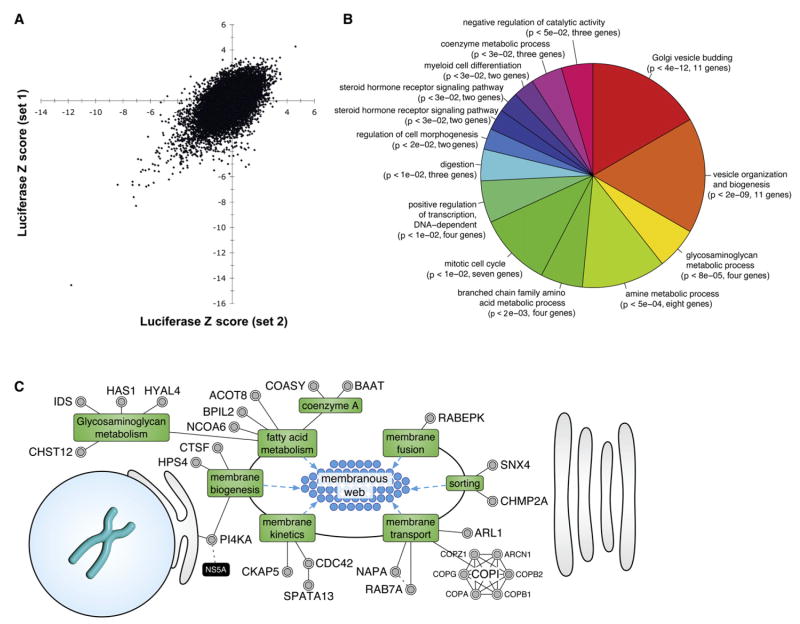
The Huh7/Rep-Feo replicon cell line was then used to screen a whole-genome siRNA library containing 21,094 siRNA pools targeting the entire human NCBI RefSeq transcript database. Each pool is made up of four individual siRNA duplexes, each directed against a different sequence within the target transcript. Firefly luciferase activity was measured in duplicate plates, as was cellular ATP content, as an indicator of cell viability. Figure 1A depicts the Z scores of the luciferase activities from the duplicate screening plates across all of the screening pools. A number of pools that strongly suppressed HCV replication in duplicate wells can be identified in the lower left quadrant, while very few strongly discordant values are seen. A complete summary of the whole-genome screen of all 21,094 pools is given in Table S1.
Figure 1. Bioinformatics of the RNAi Screen.
(A) Replicate Z scores from the primary screen. Each point represents the Z score of an individual siRNA pool in one experimental pool plotted against the Z score of the same pool in the experimental replicate well. A Z score is the number of standard deviations of the experimental luciferase activity above the median plate value. Negative Z scores indicate inhibition of HCV replication.
(B) Gene ontology biological process terms significantly (p < 0.05) overrepresented by the genes that scored in the screen. The terms are ordered clockwise by ascending p values.
(C) Functional mapping of HCV dependency factors to lipid- and membrane-related processes that support the formation of a membranous web structure needed for viral replication. Dotted gray lines indicate protein-protein interactions; dotted blue lines indicate processes that impinge on membranous web structures; green boxes indicate cellular processes; and black boxes refer to HCV proteins. Supporting references are listed in Table S4.
Using a false discovery rate-based approach for hit selection (Storey, 2002), we identified 236 pools (1.1% of the library) that inhibited HCV replication with a q value threshold of 0.10 without significant toxicity. These pools entered the second round of screening, in which the four component siRNAs in each pool were individually rescreened. Of these 236 pools, 24 targeting ribosomal subunits were not evaluated further, as it is known that the HCV polyprotein is translated by the host translational machinery (Otto and Puglisi, 2004).
Of the remaining 212 pools, 186 pools (87.7%) had at least one siRNA decrease HCV replication by: 2 SD compared to a nontargeting control siRNA. Ninety-six pools met a threshold of at least two siRNAs out of four decreasing HCV replication by: 2 SD (Table S2). We identified only 13 siRNA pools that increased viral replication with a threshold of q < 0.10 (Table S1); the strongest effect was only a 2.7-fold increase, and these hits were not further evaluated.
The 96 HCV cofactors were functionally analyzed using Gene Ontology (GO) biological process terms. Seventy-nine genes had at least one GO term associated, and overall, 254 terms were mapped to these genes. Using a hypergeometric distribution, 51 terms were significantly (p < 0.05) overrepresented and mapped to 37 genes. The significant terms were clustered, and the term with the lowest p value in each cluster was used to represent that cluster (Table S3). The most significantly enriched processes were Golgi vesicle binding and vesicle organization and biogenesis (Figure 1B). Both of these terms included the six members of the COPI complex.
To obtain a more comprehensive view of the processes represented by the 96 genes, a database framework was created to integrate information from multiple data sources (see Supplemental Data) and the scientific literature to identify the subcellular localization and associated functions of each gene. The results of this procedure are illustrated in Figures 1C and S3, with supporting references listed in Table S4. Twenty-seven genes (28%) were associated with the processes of membrane biogenesis, sorting, and trafficking. In addition, six genes were identified as affecting the cellular RNA machinery, including SERBP1, HNRPAB, and NOP5/NOP58.
There was little overlap between the results of our screen and three other limited RNAi screens for HCV replication, which screened ~4000 (Ng et al., 2007), 510 (Supekova et al., 2008), and 62 genes (Randall et al., 2007). We therefore retested 11 siRNA pools (TBXA2R, SLC12A4, SLC12A5, VRK1, CSK1, JAK1, DDX3X, STAT3, ELAVL1, DICER1, and HSPBP1), which represented the top hits from each of the prior screens in Huh7/Rep-Feo cells, and which did not score in our primary screen (Figure S4). These pools were retested in 96-well format. Strong (>75%) inhibition of Huh7/Rep-Feo replication was observed only with TBXA2R silencing. The TBXA2R siRNA pool did inhibit Huh7/Rep-Feo replication by 43% in the primary screen (Table S1), though this degree of inhibition did not fall within the predefined q < 0.10 threshold for secondary screening. Moderate inhibition (50%–75%) was seen with CSK, DDX3X, HSPBP1, and SLC12A5 silencing, while < 50% inhibition was observed with DICER1, ELAVL1, JAK1, SLC12A4, STAT3, and VRK1 silencing.
Despite the apparent inhibition of HCV replication by siRNAs targeting SLC12A5, we were unable to identify any expression of SLC12A5 in Huh7/Rep-Feo cells (data not shown), which is consistent with prior studies showing that SLC12A5 is a neuron-specific transcript and is not expressed in the liver (Payne et al., 1996; Uvarov et al., 2007). This indicates that the SLC12A5 siRNA pool used by Ng et al. and also used in this experiment exerts its effect on HCV replication through an offtarget effect.
In addition to the hits discussed below, we retested seven of our top-ranking hits (CHMP2A, CKAP5, NAPA, RAB7A, RABEPK, SERBP1, and TRIM62) in the OR6 replicon cell line (Figure S5). The OR6 replicon contains a full-length genotype 1b HCV genome and a Renilla luciferase reporter gene (Figure S1), in contrast to the subgenomic Rep-Feo replicon. These genes were chosen by inspection from the list of hits based on their known or proposed roles in membrane trafficking (CHMP2A, NAPA, RAB7A, RABEPK), in RNA trafficking or translation (CKAP5, SERBP1), and in the lifecycle of other viruses (TRIM proteins in HIV infection). All except for TRIM62 were reconfirmed using the criterion of at least two siRNAs causing at least 2-fold inhibition of HCV replication, though three of the four TRIM62 siRNA duplexes tested did inhibit OR6 replication by at least40%.
The PI 4-Kinase PI4KA Is Essential for HCV Replication
A high-ranking hit in our siRNA screen was PI4KA, also known as PIK4CA or PI4KIIIα. PI4KA encodes a 230 kDa protein and is widely expressed (Balla et al., 1997); however, the function of this enzyme in mammalian cells has not been well defined. There are four known phosphatidylinositol (PI) 4-kinases in mammalian cells that catalyze the production of PI 4-phosphate from PI (Balla and Balla, 2006). It has been proposed that the four mammalian PI 4-kinases have distinct functions that are mediated largely by their localization to different intracellular compartments (Balla and Balla, 2006). PI4KA is believed to be predominantly localized to the ER (Wong et al., 1997), though a fraction may also be found at the Golgi (Nakagawa et al., 1996).
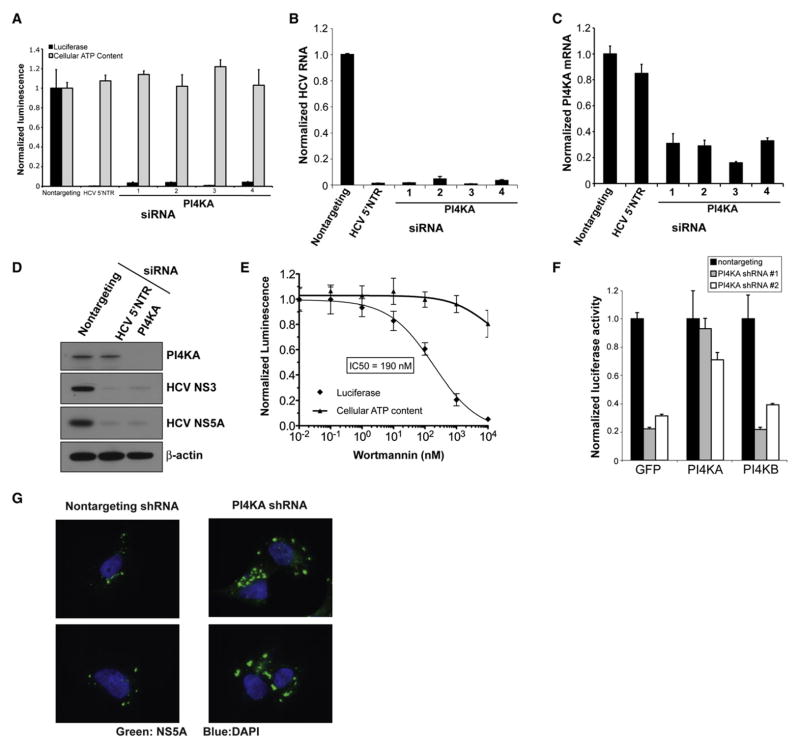
PI4KA silencing by four individual siRNA duplexes strongly inhibited HCV replication in the OR6 full-length replicon cell line (Figure 2A) without detectable cytotoxicity, as determined by cellular ATP content. PI4KA depletion also strongly inhibited viral replication in the genotype 2a JFH1 infectious HCV model (Wakita et al., 2005) by over 2 log10 (Figure 2B). All four siRNA duplexes depleted PI4KA mRNA transcript levels (Figure 2C); moreover, siRNA duplex #3 was the most potent for both PI4KA transcript silencing as well as for inhibition of HCV replication. Immunoblotting for HCV nonstructural proteins in the OR6 cell line confirmed that PI4KA depletion was associated with the loss of HCV nonstructural proteins (Figure 2D, lane 3), comparable to the effect of directly targeting the replicon RNA with an HCV-specific siRNA (Figure 2D, lane 2).
Figure 2. PI4KA Is Essential for Hepatitis C Virus Replication.
(A) PI4KA silencing with four individual siRNA duplexes blocks replication of the full-length HCV replicon OR6 (black bars) without significant cytotoxicity as measured by cellular ATP content (gray bars). Values were obtained from quadruplicate wells in two independent experiments and are mean ± SD.
(B) siRNAs against PI4KA block replication of the infectious JFH1 HCV strain. Cells were infected with JFH1 48 hr posttransfection, and HCV RNA was quantified by quantitative PCR (qPCR) at 24 hr postinfection. Values were obtained from triplicate qPCR replicates from duplicate wells and are mean ± SD.
(C) The magnitude of HCV inhibition parallels the degree of PI4KA silencing. PI4KA transcripts were quantified by qPCR 48 hr posttransfection. Values are mean± SD.
(D) PI4KA silencing depletes HCV nonstructural proteins in OR6 replicon cells as determined by immunoblotting with the indicated antibodies. Cell lysates were prepared from OR6 cells transfected with the indicated siRNAs for 72 hr.
(E) Wortmannin inhibits HCV replication with an IC50 consistent with a type III PI 4-kinase. OR6 replicon cells were treated with the indicated concentrations of wortmannin for 24 hr. Replicate wells were assayed for luciferase activity (diamonds) or cellular ATP content (triangles). Values were obtained from quadruplicate wells in two independent experiments and are mean ± SD.
(F) PI4KA shRNAs block HCV replication and can be rescued by an RNAi-resistant PI4KA construct but not by PI4KB. OR6 replicon cells were transduced with MMLV retroviral vectors encoding GFP, PI4KA, or PI4KB cDNAs lacking the 3′UTR. The cells were then transduced 24 hr later with lentiviral vectors encoding a nontargeting shRNA (black bars) or two different shRNAs targeting the PI4KA 3′UTR (gray and white bars). Cells were assayed 96 hr after lentiviral transduction. Values were obtained from quadruplicate wells in two independent experiments and are mean ± SD.
(G) PI4KA silencing induces the formation of abnormally large NS5A-positive membrane structures. UHCVcon57.3 cells were transduced with the indicated shRNA constructs 4 days prior to induction of HCV polyprotein expression by withdrawal of tetracycline from the growth medium. Twenty-four hours after induction, cells were fixed and stained with a monoclonal antibody to HCV NS5A (green) and counterstained with DAPI to highlight cell nuclei (blue).
Two of the four PI 4-kinases, PI4KA and PI4KB, are inhibited by wortmannin, although at IC50s much higher (~50–300 nM) (Balla and Balla, 2006) than for PI 3-kinases (~1–5 nM); no selective PI 4-kinase inhibitors have been published. The two other PI 4-kinases are wortmannin insensitive. We found that HCV replication in OR6 replicon cells was inhibited by wortmannin with an IC50 of 190 nM, which is consistent with the reported IC50 of PI4KA (Figure 2E) and further demonstrates that PI4KA’s enzymatic activity is required for its role in HCV replication. Wortmannin treatment at 10 nM, a concentration that effectively inhibits PI 3-kinases, had no significant effect on HCV replication. There was no significant cytotoxicity with wortmannin treatment for 24 hr at concentrations up to 1 μM. Moreover, PI4KA silencing reduced the observed IC50 of wortmannin by over 3-fold (Figure S6), supporting our hypothesis that wortmannin inhibits HCV signaling by inhibiting PI4KA activity.
Finally, we generated two independent short hairpin RNA (shRNA) lentiviral constructs targeting the 3′UTR of PI4KA. As expected, both PI4KA shRNAs blocked HCV replication in OR6 replicon cells, while a nontargeting shRNA had no effect on HCV replication (Figure 2F). We then generated a shRNA-resistant, full-length PI4KA expression construct lacking the 3′UTR targeted by our shRNA constructs as well as a construct expressing the other type III PI 4-kinase, PI4KB. Expression of shRNA-resistant, full-length PI4KA rendered OR6 replicon cells resistant to both PI4KA shRNAs. However, the effect of silencing endogenous PI4KA on HCV replication could not be rescued by exogenous expression of PI4KB, demonstrating that the dependence of HCV on PI4KA is highly specific.
We then sought to ascertain the function of PI4KA in HCV replication. Given the reported localization of PI4KA to the ER, we hypothesized that it might have a role in the generation of HCV nonstructural protein-associated membranes. However, this hypothesis could not be readily tested in the HCV replicon or JFH1 infectious viral systems because PI4KA silencing potently inhibits HCV replication and thus HCV protein expression. Therefore, we studied the effect of PI4KA silencing on the localization of HCV nonstructural proteins in the UHCVcon57.3 cell line (Egger et al., 2002), which is stably transformed with a construct encoding the full-length HCV polyprotein under the control of a tet-off promoter. Expression of the HCV polyprotein is therefore uncoupled from viral replication in this cell line. Induction of HCV polyprotein expression in cells transduced with a nontargeting shRNA results in small punctate membranous structures that stain positive for HCV NS5A by immunofluorescence (Figure 2G). However, PI4KA silencing in UHCVcon57.3 cells followed by induction of HCV polyprotein expression led to the formation of abnormally large NS5A-positive structures, indicating that PI4KA is required for the correct formation of the small membranous structures that have been shown by others to be the sites of viral RNA replication (Gosert et al., 2003).
The COPI Coat Complex Is Required for Replication Early in the Viral Lifecycle
The COPI coat is a multisubunit complex composed of the multimeric coatomer complex and the ARF GTPase. COPI-coated vesicles mediate retrograde retrieval of ER-resident proteins from the Golgi and also play a role in intra-Golgi vesicle trafficking (Béthune et al., 2006). Six coatomer subunits scored in the primary siRNA screen (Tables S1–S2). Another high-ranking hit in the primary screen, CDC42, binds to coatomer via γ-COP (Wu et al., 2000).
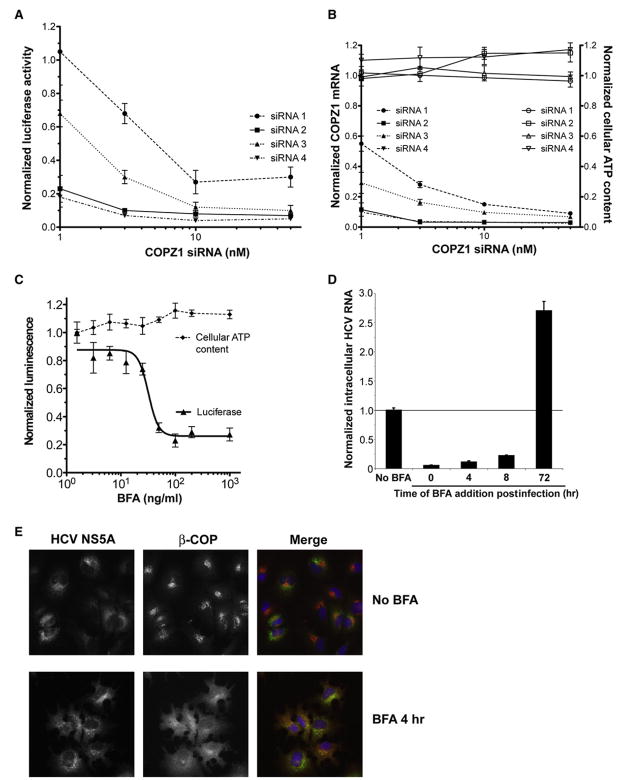
We characterized the effect of COPZ1 silencing in further detail. Four individual siRNA duplexes against COPZ1 all inhibited HCV replication in the full-length OR6 replicon system in a concentration-dependent manner (Figure 3A). There was no cytotoxicity observed under the conditions used in this experiment (Figure 3B), and all four siRNA duplexes depleted COPZ1 mRNA in a concentration-dependent manner. Of note, the potency of the siRNA duplexes for COPZ1 silencing paralleled their potency for inhibition of HCV replication.
Figure 3. The COPI Complex Supports HCV Replication Early in the Viral Lifecycle.
(A) Individual siRNA duplexes against COPZ1 block replication of the full-length HCV replicon OR6 in a concentration-dependent manner. OR6 cells were transfected with the indicated siRNA duplexes for 64 hr and then assayed for luciferase activity. Values were obtained from quadruplicate wells in two independent experiments and are mean ± SD.
(B) Individual siRNA duplexes against COPZ1 deplete COPZ1 transcripts in a dose-dependent manner without detectable cytotoxicity. OR6 cells transfected with the indicated siRNA duplexes as in (A) were assayed for cellular ATP content (open symbols) or for COPZ1 transcript levels by quantitative real-time PCR (closed symbols). Values were obtained from triplicate qPCR measurements from duplicate wells in two independent experiments and are mean ± SD.
(C) Brefeldin A (BFA), a pharmacologic inhibitor of COPI function, blocks replication of the OR6 full-length replicon. OR6 cells were treated with the indicated concentrations of BFA for 24 hr and then assayed for luciferase activity (triangles) or cellular ATP content (diamonds). Values were obtained in quadruplicate and are mean ± SD.
(D) BFA blocks replication of the infectious JFH1 isolate early in viral infection and blocks HCV secretion late in infection. BFA was added at 100 ng/mL to Huh7.5.1 cells at the indicated times relative to the time of JFH1 infection. Intracellular HCV RNA was quantified by qPCR at 20 hr of BFA treatment and normalized to HCV RNA from mock treated cells. Values were obtained from triplicate qPCR replicates from duplicate wells and are mean ± SD.
(E) Preformed NS5A-positive structures are resistant to BFA treatment. OR6 cells were treated with 100 μg/mL BFA (lower panels) or 1% ethanol (upper panels) for 4 hr and then processed for immunofluorescence for HCV NS5A (left panels) and HCV NS5A (middle panels). Nuclei were counterstained with DAPI (blue). BFA causes dissociation of β-COP from the Golgi apparatus within minutes of treatment.
To further confirm that COPI activity is required for HCV replication, we employed the pharmacologic COPI inhibitor brefeldin A (BFA), which blocks COPI coat assembly by inhibiting ARF-specific guanine nucleotide exchange factors. BFA inhibited replication of the OR6 full-length replicon in a dose-dependent fashion, again without measurable cytotoxicity over a 24 hr period (Figure 3C).
We then examined the ability of BFA to inhibit replication of the cell-culture-infectious HCV strain JFH1. Consistent with previous reports (Gastaminza et al., 2008), BFA treatment of Huh7.5.1 cells that had already been infected for 3 days resulted in an increase in intracellular viral RNA (Figure 3D). However, in contrast to the effect of BFA on established HCV infection, BFA treatment of Huh7.5.1 cells at the time of JFH1 infection potently inhibited viral replication. BFA effectively inhibited HCV replication when added at up to 8 hr postinfection, indicating that its effect on blocking HCV replication occurs after viral entry; maximal internalization and fusion of HCV pseudoparticles occur by 3 hr of virus-cell contact (Meertens et al., 2006). Furthermore, the observed increase in intracellular HCV RNA when BFA is added in established HCV infection indicates that the ability of BFA to block HCV replication early in viral infection is not due to nonspecific cellular toxicity or global inhibition of RNA synthesis. Immunofluorescence staining of HCV NS5A and β-COP demonstrated that preformed NS5A-positive structures in OR6 HCV replicon cells were resistant to short-term BFA treatment under conditions that induce the rapid dissociation of COPI from the Golgi complex (Figure 3E), suggesting that COPI is not actively required for the dynamic maintenance of established HCV replication complexes as it is for the maintenance of the Golgi complex.
Hepcidin Is a Cofactor for HCV Replication
Hepcidin, encoded by the HAMP (hepcidin antimicrobial peptide) gene, is a peptide hormone that is a key regulator of systemic iron homeostasis by inhibiting the iron transporter ferroportin (Nemeth et al., 2004b). Chronic HCV infection has long been recognized to be associated with elevated serum iron indices as well as with increased hepatic iron stores. This screen identified an unexpected effect of hepcidin silencing on HCV replication.
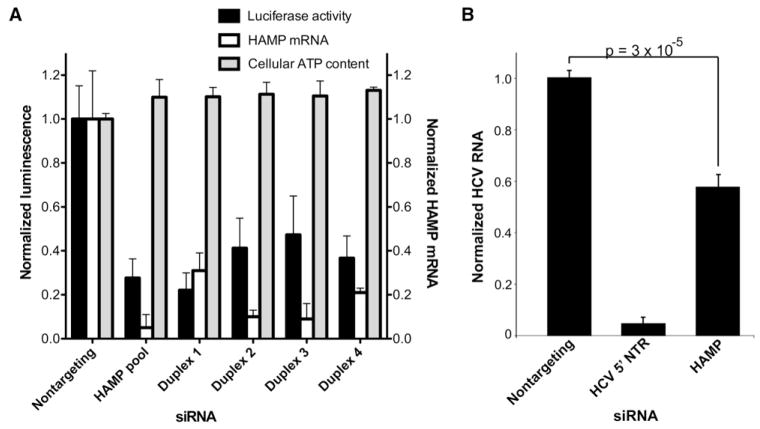
HAMP silencing in the OR6 full-length replicon model by any of four individual siRNA duplexes or with the HAMP siRNA pool inhibited HCV replication (Figure 4A) without measurable cytotoxicity over 72 hr. We also found that HAMP silencing significantly decreased JFH1 replication, though to a lesser magnitude than in the two replicon systems (Figure 4B). Preliminary studies using a dicistronic reporter construct to measure HCV IRES-dependent translation and cap-dependent translation (Itsui et al., 2006) found inhibition of both IRES- and cap-dependent translation in HAMP-silenced Huh7 cells (data not shown). Though this finding remains to be confirmed by additional methodologies, it suggests that HAMP perturbation may affect HCV replication by altering cellular translation at a more global level in the hepatocyte.
Figure 4. The Iron Regulatory Hormone Hepcidin Supports HCV Replication.
(A) HAMP silencing blocks replication of the full-length HCV replicon OR6. OR6 cells were transfected with the indicated siRNAs for 72 hr and then assayed for luciferase activity (black bars), HAMP transcript levels (white bars), and cellular ATP content (gray bars). Values were obtained from quadruplicate wells in three independent experiments and are mean ± SD.
(B) HAMP silencing blocks replication of the infectious JFH1 HCV strain in Huh7 cells. Two-tailed p value was calculated by indepdendent two-sample t test. Values were obtained from triplicate qPCR replicates from duplicate wells in two independent experiments and are mean ± SD.
DISCUSSION
Functional genomic approaches allow for the unbiased identification of HCV host cofactors independent of any preconceived models of the HCV lifecycle or any assumptions about gene function. This technology has been successfully applied to the identification of novel host dependency factors for HIV (Brass et al., 2008). We have conducted a whole-genome RNAi screen for host proteins that support HCV replication and were able to functionally confirm the significance of several of these cofactors in the fully infectious JFH1 HCV strain.
HCV, like other positive-strand RNA viruses, replicates on altered host membranes, which in the case of HCV have been called “membranous webs” (Egger et al., 2002; Gosert et al., 2003). The mechanisms by which these membranous webs form are largely unknown. Here, we identified 27 host genes that play a role in lipid metabolism, membrane biogenesis, kinetics, and trafficking; all are likely to be key processes in the formation and maintenance of the membranous web structure.
PI4KA is an attractive “druggable” target for anti-HCV therapeutics. Both genotype 1 and 2 HCV models exhibit a very strong dependency on PI4KA function; furthermore, PI4KA silencing is well tolerated in cultured cells. Further evidence that PI4KA has a direct function in HCV replication comes from a study (Ahn et al., 2004) that identified an interaction between HCV NS5A and PI4KA in a yeast two-hybrid screen. The development of selective PI4KA inhibitors will facilitate the study of this protein’s cellular function as well as its role in HCV replication, and it may also yield therapies for chronic HCV infection.
The dependence of HCV replication on the COPI coatomer complex is shared by poliovirus and Drosophila C virus (Cherry et al., 2006; Maynell et al., 1992). All three of these RNA viruses replicate on host membrane-derived compartments, and we speculate that the assembly of these viral replication compartments is somehow dependent on COPI. Further support for this hypothesis comes from the finding that γ-COP and its interacting partner, CDC42, are significantly enriched on detergent-insoluble membranes from HCV replicon-expressing cells (Mannová et al., 2006). These data would suggest that COPI is directly involved in membranous web formation and that the block of HCV replication by COPI inhibition is not an indirect effect of disrupting Golgi-ER vesicle trafficking.
Finally, we identified hepcidin as a cofactor for HCV replication. Chronic hepatitis C infection has long been recognized to be associated with increased serum iron and transferrin saturation as well as hepatic iron accumulation; moreover, hepatic hepcidin mRNA expression is increased in patients with chronic HCV infection (Aoki et al., 2005). Hepcidin transcription is stimulated by iron overload as well as by inflammation through IL-6 (Nemeth et al., 2004a), which is elevated in patients with chronic HCV. The identification of hepcidin as a HCV replication cofactor points to a molecular basis for the well-known clinical association between chronic HCV infection and dysregulation of iron homeostasis. Moreover, the potent upregulation of hepcidin transcription by IL-6 potentially creates a positive feedback loop between chronic inflammation and HCV replication.
An important limitation of our study is that the replicon model permits identification of host cofactors of replication but not of other stages of the viral lifecycle. Future studies using fully infectious, cell-culture-adapted HCV strains will be able to study other aspects of the HCV lifecycle, such as viral entry, uncoating, virion assembly, and secretion.
The host cofactors identified in this screen had little overlap with those identified in three previous limited siRNA screens for HCV host cofactors (Ng et al., 2007; Randall et al., 2007; Supekova et al., 2008). Similarly, relatively little overlap has been observed among the hits in three genome-wide RNAi screens for HIV host cofactors (Brass et al., 2008; König et al., 2008; Zhou et al., 2008). There are many factors that must be considered when comparing our studies with prior limited siRNA screens. First, the siRNA sequences used in our screen were the same as for the Ng et al. screen, but were different from those used in the Supekova et al. and Randall et al. screens, likely resulting in different knockdown efficiencies and off-target effects for each gene screened.
Second, the HCV models were different in each screen. Randall et al. used the J6/JFH1 genotype 2a infectious virus, while the other screens used different subgenomic genotype 1b replicons. Silencing of the host protein DDX3X, which binds to the HCV core protein, suppresses subgenomic HCV replication much less efficiently than full-length HCV replication (Ariumi et al., 2007). Furthermore, genotype-specific differences in host cofactor dependency could also potentially account for differences in screens using different HCV genotypes.
Third, the methods of siRNA transfection and siRNA concentration and the duration of silencing varied among the screens, thus biasing the screens toward protein with shorter half-lives (in the Supekova et al. screen) or longer half-lives (Ng et al.). Randall et al. tested 62 genes, which is a sufficiently small number that siRNA could be introduced by electroporation, and the duration of silencing could be optimized for each gene tested; both methods are impractical for whole-genome screens.
Fourth and finally, the threshold for identifying significant hits for secondary validation varied among the different screens. For example, TBXA2R would have been selected as a hit in our primary screen had we used the Ng et al. threshold of 30% inhibition. However, using this lower threshold in our screen would have resulted in over 1800 pools for secondary screening, which would have been an impractical number of hits for deconvolution.
The strength of whole-genome siRNA screens is their ability to identify roles for genes and biological pathways in biological processes free of a priori hypotheses regarding their function. Our screen has identified many candidate host cofactors for HCV replication, some of which may offer new therapeutic avenues for the treatment of chronic infection. Further studies of these cofactors may yield important mechanistic insights into the formation and maintenance of the membrane-associated HCV replication complex. Furthermore, we have identified a possible molecular link between iron homeostasis, chronic inflammation, and chronic HCV infection. However, because of the very significant false-negative (variability in protein depletion owing to variation in siRNA efficacy and protein half-life) and false-positive (off-target effects) rates inherent to high-throughput siRNA screens, no single screen can be expected to yield a truly comprehensive map of all of the host genes that are involved in a particular process. Such maps will require the integration of multiple RNAi screens with complementary technologies.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Anti-NS5A (7-D4) and NS3 (9-G2) monoclonal antibodies were obtained from ViroGen (Cambridge, MA). Anti-β-COP and anti-PI4KA polyclonal antibodies were obtained from Abcam (Cambridge, MA) and Cell Signaling Technology (Danvers, MA), respectively. Anti-β-actin monoclonal antibody (AC-74), BFA, and wortmannin were obtained from Sigma-Aldrich (St. Louis, MO).
siRNA Transfection
siRNA was introduced into Huh7/Rep-Feo cells by reverse transfection using HiPerFect Transfection Reagent (QIAGEN; Valencia, CA) according to the manufacturer’s instructions. Nontargeting siRNA and siRNA targeting firefly luciferase were obtained from QIAGEN and Ambion (Austin, TX), respectively. siRNA targeting the HCV 5′NTR was synthesized according to the si313 sequence (Chevalier et al., 2007). For pilot and validation experiments, control siRNAs were spotted onto 96- or 384-well plates. HiPerFect was diluted in Opti-MEM I (Invitrogen; Carlsbad, CA) and then added to the prespotted siRNAs to allow lipid-siRNA complexes to form. Trypsinized Huh7/Rep-Feo cells were added to the preformed lipid-siRNA complexes at 5000 cells/well (96-well format) or 2000 cells/well (384-well format) and then incubated for 72 hr. Firefly luciferase was assayed using Bright-Glo reagent (Promega; Madison, WI) and a 2103 EnVision Ultra Sensitive Luminescence Plate Reader (PerkinElmer; Waltham, MA).
High-Throughput Screening
Screening was performed at the Institute of Chemistry and Cell Biology screening facility at Harvard Medical School. The library used for this screen was the siARRAY Human Genome siRNA Library (Dharmacon; Lafayette, CO). Each well in the library contains a pool of four siRNAs designed to target four different sequences within a single human transcript. All of the unique human genes in the NCBI RefSeq database are included in this set of 21,094 genes. Each pool is predicted to silence target gene expression at the mRNA level by at least 75%.
A Bravo Liquid Handling Platform (Velocity11; Menlo Park, CA) was used to transfer siRNAs from 384-well library stock plates to quadruplicate screening plates. Diluted HiPerFect was then added to the siRNA with a WellMate Microplate liquid dispenser (Matrix Technologies; Hudson, NH). Upon formation of siRNA-lipid complexes, 2000 trypsinized Huh7/Rep-Feo cells were added to each well. Plates were incubated at 5% CO2, 37°C for 72 hr, whereupon two screening plates were assayed for luciferase activity using Bright-Glo reagent, and two were assayed for cell viability by cellular ATP content using CellTiter-Glo reagent (Promega) according to the manufacturer’s instructions.
Luminescence values were converted to Z scores and then p values (see Supplemental Data for further details). For hit selection, we used Storey’s false discovery rate-based approach to multiple-hypothesis testing in order to control the false-positive error rate without inflating the false-negative error rate (Storey, 2002). A q value of 0.10, corresponding to a false-discovery rate of 0.10, was used as the threshold for selecting hit siRNA pools from the primary screen for siRNA deconvolution. The threshold of 0.10 means that 10% of the total number of hit pools selected are expected to be false-positive results. Pools that generated significant cell toxicity (>2-fold decrease in CellTiter-Glo signal) were rejected, as were pools that targeted ribosomal subunits or putative transcripts that had been removed by the most recent RefSeq revision.
The four individual siRNA duplexes comprising each hit siRNA pool were then individually picked and spotted into separate wells for retesting in the Huh/Rep-Feo replicon system. Transfection and luminescence assays were performed exactly as described for the primary screen.
Gene Ontology, Subcellular Localization, and Gene Annotation
Please refer to the Supplemental Data for details on the bioinformatics methods used.
Secondary Validation in Full-Length HCV Replicon Cells
Selected genes that were associated with HCV replication in Huh7/Rep-Feo subgenomic replicon cells were tested in OR6 full-length replicon cells expressing the entire genotype 1b HCV polypeptide downstream of a Renilla luciferase reporter gene (Ikeda et al., 2005). siRNA duplexes were spotted into 96-well plates to a final concentration of 50 nM. Diluted HiPerFect Transfection Reagent was added to each well, followed by plating of OR6 cells. Renilla luciferase activity was normalized to cellular ATP content as determined by CellTiter-Glo.
JFH1 Infection and Quantitative Real-Time PCR
Infectious genotype 2a JFH1 HCV was prepared as previously described (Wakita et al., 2005). Huh7.5.1 and Huh7 cells were reverse transfected in 96-well plates with siRNA duplexes under the same conditions as OR6 cells. siRNA-transfected cells were then infected with JFH1 virus at an moi of ~0.2. Total cellular and viral RNA was isolated postinfection using RNeasy Mini columns (QIAGEN) with on-column DNase digestion, reverse transcribed by random priming with the High Capacity cDNA Reverse Transcription Kit (Applied Bio-systems; Foster City, CA), and then quantitated by real-time PCR using the DyNAmo HS SYBR Green qPCR kit (Finnzyme; Espoo, Finland). Efficiency-corrected relative quantitation (Pfaffl, 2001) was used with GAPDH as an internal control. All primer sequences used are given in Table S5.
Construction of shRNA Lentiviral and MMLV Retroviral Expression Constructs
shRNAs designed to target the 3′UTR of the PI4KA transcript (Table S5) were cloned into the lentiviral shRNA vector pLKO.1 (Moffat et al., 2006) and confirmed by sequencing. These constructs or a nontargeting shRNA construct (pLKO.1-scramble; Addgene; Cambridge, MA) were cotransfected with the packaging vectors psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259) into 293T cells to generate VSV-G pseudotyped lentiviral particles.
Full-length (PI4K230) and truncated (PI4K92) N-terminal myc-tagged PI4KA constructs in pcDNA3.1/zeo(+) were subcloned into pFB (Stratagene; La Jolla, CA), a MMLV retroviral vector. VSV-G pseudotyped retroviral particles were generated by transfection of pFB expression constructs into the 293GPG packaging cell line (Ory et al., 1996). Lentiviral and retroviral supernatants were harvested 48 and 72 hr posttransfection, 0.45 μm filtered, and stored at −80°C. Target cells were infected with lentiviral or retroviral particles for 4 hr in the presence of 8 μg/mL polybrene (Sigma).
Immunofluorescence Staining
UHCVcon57.3 cells were plated on gelatin-coated glass coverslips and transduced with a lentiviral vector encoding a nontargeting shRNA or an shRNA against PI4KA. After induction of HCV polyprotein expression by tetracycline withdrawal, the cells were fixed with 3% paraformaldehyde, permeabilized with 0.3% Triton X-100 in PBS, and then blocked with 10% FBS/1% BSA in PBS with 0.1% Triton X-100. Coverslips were incubated with a NS5A monoclonal antibody overnight at 4°C, washed, and then incubated with Alexa 594 donkey anti-mouse secondary antibody for 1 hr at RT. Coverslips were mounted with Fluoromount-G (SouthernBiotech; Birmingham, AL) and viewed on an epifluorescence microscope.
Supplementary Material
Supplementary Table 1 | Host genes for which ≥ individual siRNA duplexes inhibit HCV replication by ≥ 2 SD
Supplementary Table 2 | Host cofactors that support HCV replication at a threshold of ≥ 2 siRNA duplexes inhibiting replication by ≥ 3 SD
Supplementary Table 3 | Gene ontology biological process analysis of HCV host dependency factors
Supplementary Table 4 | Analysis of transcription factor binding sites over-represented in 1Kb promoters of HCV cofactors
Supplementary Table 5 | Supporting references for placing genes in Figures 1C and Supplementary Figure 3.
Supplementary Table 6 | RNAi and primer sequences
Acknowledgments
Huh7.5.1, OR6, and UHCVCon57.3 cell lines were gifts from Doctor Francis Chisari (Scripps Institute; La Jolla, CA), Doctors N. Kato and M. Ikeda (Okayama University Graduate School of Medicine; Okayama, Japan), and Doctor Darius Moradpour (University of Lausanne; Lausanne, Switzerland), respectively. The JFH1 strain was a gift of Doctor T. Wakita (Tokyo Metropolitan Institute for Neuroscience; Tokyo, Japan). PI4KA constructs were obtained from Doctor Paul van Bergen en Henegouwen (Universiteit Utrecht; Netherlands). We would like to acknowledge the technical assistance provided by Doctor Caroline Shamu, Stewart Rudnicki, Sean Johnston, and David Wrobel at the ICCB-Longwood screening facility at the Harvard Medical School. This publication was made possible by grants AI080122 (to A.W.T.), AI062773 and DK043351 (to R.J.X.), and AI069939 and DK078772 (to R.T.C.) from the National Institutes of Health as well as by a Tosteson Postdoctoral Fellowship from the Massachusetts Biomedical Research Corporation (to A.W.T.) and grants from the American Gastroenterological Association and the American Liver Foundation (to L.F.P.).
Footnotes
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, six figures, and five tables and can be found online at http://www.cell.com/cellhostandmicrobe/supplemental/S1931-3128(09)00061-4.
References
- Ahn J, Chung KS, Kim DU, Won M, Kim L, Kim KS, Nam M, Choi SJ, Kim HC, Yoon M, et al. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J Biochem Mol Biol. 2004;37:741–748. doi: 10.5483/bmbrep.2004.37.6.741. [DOI] [PubMed] [Google Scholar]
- Aoki CA, Rossaro L, Ramsamooj R, Brandhagen D, Burritt MF, Bowlus CL. Liver hepcidin mRNA correlates with iron stores, but not inflammation, in patients with chronic hepatitis C. J Clin Gastroenterol. 2005;39:71–74. [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Abe KI, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD box RNA helicase is required for hepatitis C virus (HCV) RNA replication. J Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla T, Downing GJ, Jaffe H, Kim S, Zólyomi A, Catt KJ. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J Biol Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- Béthune J, Wieland F, Moelleken J. COPI-mediated transport. J Membr Biol. 2006;211:65–79. doi: 10.1007/s00232-006-0859-7. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Saulnier A, Benureau Y, Fléchet D, Delgrange D, Colbère-Garapin F, Wychowski C, Martin A. Inhibition of hepatitis C virus infection in cell culture by small interfering RNAs. Mol Ther. 2007;15:1452–1462. doi: 10.1038/sj.mt.6300186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisiak R, Horban A, Gallay P, Bobardt M, Selvarajah S, Wiercinska-Drapalo A, Siwak E, Cielniak I, Higersberger J, Kierkus J, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun. 2005;329:1350–1359. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- Itsui Y, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Koyama T, Takeda Y, Nakagawa M, Kakinuma S, Sekine Y, et al. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J Viral Hepat. 2006;13:690–700. doi: 10.1111/j.1365-2893.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- Kim SS, Peng LF, Lin W, Choe WH, Sakamoto N, Schreiber SL, Chung RT. A cell-based, high-throughput screen for small molecule regulators of hepatitis C virus replication. Gastroenterology. 2007;132:311–320. doi: 10.1053/j.gastro.2006.10.032. [DOI] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannová P, Fang R, Wang H, Deng B, McIntosh MW, Hanash SM, Beretta L. Modification of host lipid raft proteome upon hepatitis C virus replication. Mol Cell Proteomics. 2006;5:2319–2325. doi: 10.1074/mcp.M600121-MCP200. [DOI] [PubMed] [Google Scholar]
- Maynell LA, Kirkegaard K, Klymkowsky MW. Inhibition of poliovirus RNA synthesis by brefeldin. A J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol. 2006;80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J Biol Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004a;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004b;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Payne JA, Stevenson TJ, Donaldson LF. Molecular characterization of a putative K-Cl cotransporter in rat brain. A neuronal-specific isoform J Biol Chem. 1996;271:16245–16252. doi: 10.1074/jbc.271.27.16245. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reesink HW, Zeuzem S, Weegink CJ, Forestier N, van Vliet A, van de Wetering de Rooij J, McNair L, Purdy S, Kauffman R, Alam J, et al. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology. 2006;131:997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Royal Stat Soc B. 2002;64:479–498. [Google Scholar]
- Supekova L, Supek F, Lee J, Chen S, Gray N, Pezacki JP, Schlapbach A, Schultz PG. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J Biol Chem. 2008;283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Sakamoto N, Enomoto N, Kurosaki M, Ueda E, Maekawa S, Yamshiro T, Nakagawa M, Chen CH, Kanazawa N, et al. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon- alpha. J Infect Dis. 2004;189:1129–1139. doi: 10.1086/382595. [DOI] [PubMed] [Google Scholar]
- Uvarov P, Ludwig A, Markkanen M, Pruunsild P, Kaila K, Delpire E, Timmusk T, Rivera C, Airaksinen MS. A novel N-terminal isoform of the neuron-specific K-Cl cotransporter KCC2. J Biol Chem. 2007;282:30570–30576. doi: 10.1074/jbc.M705095200. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Wong K, Meyers R, Cantley LC. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J Biol Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- Wu WJ, Erickson JW, Lin R, Cerione RA. The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature. 2000;405:800–804. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 | Host genes for which ≥ individual siRNA duplexes inhibit HCV replication by ≥ 2 SD
Supplementary Table 2 | Host cofactors that support HCV replication at a threshold of ≥ 2 siRNA duplexes inhibiting replication by ≥ 3 SD
Supplementary Table 3 | Gene ontology biological process analysis of HCV host dependency factors
Supplementary Table 4 | Analysis of transcription factor binding sites over-represented in 1Kb promoters of HCV cofactors
Supplementary Table 5 | Supporting references for placing genes in Figures 1C and Supplementary Figure 3.
Supplementary Table 6 | RNAi and primer sequences