Abstract
ATP-driven proteolysis plays a major role in regulating the bacterial cell cycle, development and stress responses. In the nitrogen-fixing symbiosis with host plants, Sinorhizobium meliloti undergoes a profound cellular differentiation including endoreduplication of the genome. The regulatory mechanisms governing the alterations of the S. meliloti cell cycle in planta are largely unknown. Here, we report the characterization of two cpdR homologs, cpdR1 and cpdR2, of S. meliloti that encode single-domain response regulators. In Caulobacter crescentus, CpdR controls the polar localization of the ClpXP protease, thereby mediating the regulated proteolysis of key protein(s), such as CtrA, involved in cell-cycle progression. The S. meliloti cpdR1-null mutant can invade the host cytoplasm, however, the intracellular bacteria are unable to differentiate into bacteroids. We show that S. meliloti CpdR1 has a polar localization pattern and a role in ClpX positioning similar to C. crescentus CpdR, suggesting a conserved function of CpdR proteins among f¿-proteobacteria. However, in S. meliloti, free-living cells of the cpdR1-null mutant show a striking morphology of irregular coccoids and aberrant DNA replication. Thus, we demonstrate that CpdR1 mediates the coordination of cell-cycle events, which are critical for both the free-living cell division and the differentiation required for the chronic intracellular infection.
Introduction
Rhizobia are α-proteobacteria with the remarkable ability to form a nitrogen-fixing symbiosis with compatible legume hosts (reviewed in Broughton et al., 2000; Gibson et al., 2008; Jones et al., 2007). The symbiosis is based on chronic infection coordinated by the exchange of signal molecules between the symbiotic partners (reviewed in Jones et al., 2007; Kobayashi and Broughton, 2008; Perret et al., 2000). The process of infection of a plant host by rhizobia is comprised of multiple developmental stages, in which the bacteria modulate their cell proliferation in concert with the development of host cells (reviewed in Foucher and Kondorosi, 2000; Oke and Long, 1999). In the free-living stage, rhizobia cells divide in an asymmetric manner, producing two daughters of different sizes (Hallez et al., 2004; Lam et al., 2003). Upon recognition of a compatible host plant, rhizobia enter the root through host-derived tubes called infection threads (reviewed in Gage and Margolin, 2000; Gage, 2004). The extension of infection threads is synchronized with the colonization of rhizobia by the restriction of bacterial cell division and the collective movement of bacteria (Fournier et al., 2008; Gage, 2002). At the tip of the infection thread, the bacteria are released into the nodule cell by a process analogous to phagocytosis (reviewed in Brewin, 1998; Jones et al., 2007): the bacteria are engulfed with host-derived membranes (called peri-bacteroid membranes) and sealed off into membrane-bound vesicles (called symbiosomes) within the host cytoplasm, where the oxygen tension is reduced by leghemoglobin. The microoxic condition induces expression of genes encoding the nitrogenase and genes essential for microoxic respiration in rhizobia (reviewed in Kaminski et al., 1998). At the same time, the bacteria that have been newly released from the infection thread generally divide several times and then cease to divide upon initiation of nitrogen fixation (reviewed in Oke and Long, 1999). At this stage, the bacteria chronically infecting the host cytoplasm are referred to as bacteroids and act like plant organelles, where metabolites from both partners are interchanged (reviewed in Prell and Poole, 2006).
Sinorhizobium meliloti induces nodules of an indeterminate type on plants of Medicago, Melilotus and Trigonella genera (reviewed in Jones et al., 2007). In such indeterminate nodules, the bacterial symbionts undergo an even more striking cellular differentiation (Mergaert et al., 2006). In the process of the bacteroid development, DNA replication is repeated several times without cell division, resulting in the formation of elongated cells containing up to 24 copies of the genome. Fully differentiated bacteroids possess permeabilized cell envelopes and lose the ability to resume growth (Kobayashi et al., 2001; Mergaert et al., 2006). Although genes required for the symbiotic chronic infection have been actively studied in S. meliloti (reviewed in Gibson et al., 2008; Jones et al., 2007), the mechanism(s) governing the differentiation processes is largely unknown.
In several bacterial species, regulated proteolysis by ClpXP has been shown to control vital cellular processes (reviewed in Dougan et al., 2002; Jenal and Hengge-Aronis, 2003) including cell division (Jenal and Fuchs, 1998), cell differentiation (Liu et al., 1999) and nitrogen fixation (Rodriguez et al., 2006). ClpXP is an AAA+ protease, which is composed of the ClpX ATPase and the ClpP peptidase (reviewed in Baker and Sauer, 2006; Sauer et al., 2004). Substrates are initially recognized by ClpX and are subsequently unfolded and transferred to ClpP for degradation. In Caulobacter crescentus, a non-symbiotic member of α-proteobacteria, ClpXP proteolytically modulates the cellular level of CtrA (Chien et al., 2007; Gorbatyuk and Marczynski, 2005; Jenal and Fuchs, 1998; McGrath et al., 2006), an essential response regulator which regulates DNA replication and cell division (collectively, the progression of the cell cycle) (Laub et al., 2000; 2002; Quon et al., 1996; 1998). At least partially due to this role, ClpXP is essential for viability in C. crescentus (Jenal and Fuchs, 1998). C. crescentus shares several regulatory components (such as CtrA) involved in cell-cycle progression and differentiation with S. meliloti and its pathogenic relative Brucella species (Barnett et al., 2001; Bellefontaine et al., 2002; Hallez et al., 2004). It is therefore plausible that the ClpXP-mediated proteolysis might play a critical role in S. meliloti and its chronic infection.
Interestingly, during C. crescentus cell cycle, ClpXP dynamically localizes to the cell pole and the cell-division plane, providing temporal and spatial specificity to the proteolysis of substrates (McGrath et al., 2006). The proper localization of ClpXP is directed by another response regulator CpdR, which physically interacts with ClpXP, and is dependent on the phosphorylation state of CpdR (Iniesta et al., 2006). When CpdR is unphosphorylated, it localizes to the cell pole, thereby mediating ClpXP localization to the cell pole. When CpdR is phosphorylated, it and ClpXP are not localized to the cell pole and consequently CtrA is not degraded. The phosphorylation (thus, inactivation) of CpdR is mediated by the CckA-ChpT phosphorelay, in which the histidine kinase CckA phosphorylates the histidine phosphotransferase ChpT, which in turn phosphorylates CpdR (Biondi et al., 2006). The same phosphorelay also phosphorylates CtrA, which is in this case activated by the phosphorylation. Thus, CpdR mediates the fine-tuning of the cell-cycle progression in C. crescentus by modulating the ClpXP-mediated proteolysis of CtrA (Biondi et al., 2006; Iniesta et al., 2006; 2008).
In this report, we investigated the role of CpdR in S. meliloti and present the first indication that proteolytic regulation and cell-cycle progression is critical for the chronic intracellular infection. The S. meliloti chromosome encodes two cpdR homologs (SMc04044 and SMc00720), designated cpdR1 and cpdR2. SMc04044 and SMc00720 do not appear to be in operons containing other ORFs (a tRNA-Val gene is located 285 bp downstream of SMc04044 and the insertion sequence ISRm22 is located downstream of SMc00720 in the opposite direction). Putative single domain-response regulators encoded by cpdR1 and cpdR2 share 61% and 46% amino-acid sequence identity with the CpdR protein of C. crescentus in the two pairwise comparisons, respectively, and share 42% amino-acid sequence identity with each other. We found that both cpdR homologs could be disrupted, while the clpX homolog was essential in S. meliloti. Only the cpdR1 mutant was symbiotically defective. Our examination of these mutants revealed that CpdR1 function is required for proper morphogenesis in free-living cells and for differentiation into bacteroids. We also found that CpdR1 functions to couple DNA replication to cell division in S. meliloti. In contrast to the C. crescentus cpdR mutant, which is poised at the G1 phase of the cell cycle, the S. meliloti cpdR1 mutant accumulates more than two copies of genomic DNA, demonstrating the plasticity of this regulatory network among α-proteobacteria. Thus, in S. meliloti, cell division in the free-living stage and the bacteroid differentiation are controlled, in part, by CpdR1.
Results
Expression profiles of cpdR and clpX homologs in free-living and in planta cells of S. meliloti
To examine the role of the CpdR homologs in S. meliloti, we first determined their transcriptional expression during free-living growth and symbiosis. We also compared the expression profiles of the cpdR homologs and clpX by measuring their transcriptional expression in parallel. Chromosomal loci of cpdR genes (cpdR1 and cpdR2) and clpX were transcriptionally fused with uidA by inserting pJH104, an integration vector carrying promoter-less uidA (for cpdR1 and cpdR2) (Ferguson et al., 2005) or a uidA-Nmr cassette (for clpX) (Metcalf and Wanner, 1993), downstream of each ORF (ATG of uidA were located 23, 23 and 59 bp downstream of the stop codons of cpdR1, cpdR2 and clpX, respectively), yielding strains Rm1021cpdR1uidA, Rm1021cpdR2uidA and Rm1021clpXuidA.
To measure expression in the free-living state, exponentially growing cells were inoculated into fresh M9 minimal media and β-glucuronidase activities of S. meliloti strains were monitored 1, 16, 24, 40, 48, 72 and 90 hours post subculture (Fig. 1, panel A). Both cpdR1 and cpdR2 fusions were expressed weakly throughout the growth phases. On the other hand, the expression of the clpX fusion was enhanced when cells entered stationary phase.
Fig. 1. Expression of S. meliloti cpdR and clpX homologs in free-living cells and bacteroids.
Panels A represents the levels of β-glucuronidase activity (given in Miller’s units) for the transcriptional level of cpdR1, cpdR2 and clpX transcriptionally fused to uidA, respectively. The wild-type S. meliloti Rm1021 showed no β-glucuronidase activity (data not shown). Assays were performed at 1, 16, 24, 40, 48, 72 and 90 hours after subculture. The values reported represent the means of three independent experiments with standard errors (error bars). A growth curve of a representative strain (monitored by 1cmOD600) is also shown (red curves). Panels B to D represent histochemical localization of β-glucuronidase activity in nodule hand-sections. Nodules were harvested from alfalfa plants infected with S. meliloti strains used in Panel A. β-glucuronidase activity was visualized as blue precipitates of the chromogenic substrate 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc). A total of 30–40 nodules from five plants were examined for each fusion. The meristematic zone of nodule is marked with white asterisks.
In order to study gene expression during symbiosis, nodules elicited on alfalfa by the strains carrying uidA fusions were sectioned and stained for β-glucuronidase activity (Fig. 1, panel B–C). S. meliloti induces formation of indeterminate-type nodules with persistent meristems (which are marked with asterisks in Fig. 1, panel B–C). Expression of clpX and the cpdR fusions occurs throughout the nodule. This is consistent with the possibility that the CpdR proteins as well as ClpX are present throughout symbiotic development and could potentially play a role in multiple stages of symbiosis.
CpdR1 localizes to cell poles
Since our assay with uidA-transcriptional fusions indicated that two cpdR homologs are transcribed (albeit at a low level) in S. meliloti, we asked if both of the CpdR proteins share the same function with C. crescentus CpdR; localization to the cell pole and recruitment of ClpXP. To this end, the localization of CpdR1 and CpdR2 was examined. We fused yfpmut2A206K (encoding a monomeric derivative of YFP, and referred to herein as yfp) to cpdR1 and cpdR2 under the control of their native promoters on the low-copy vector pTH1227 (Cheng et al., 2007). Resulting plasmids, p-cpdR1-yfp and p-cpdR2-yfp, carrying cpdR-yfp fusion genes were introduced into the wild-type S. meliloti strain Rm1021.
In the log-phase cells, a single CpdR1-YFP focus was visible above the background fluorescence in ~6% of cells (n=1399) (Fig. 2A–C; Table 1). In C. crescentus, it has been reported that the CpdR focus only appears at particular stages of the cell cycle (the swarmer-to-stalked cell transition and the predivisional stage) (Iniesta et al., 2006). Since the S. meliloti cultures were not synchronized, it is reasonable to speculate that the cultures consisted of a heterogeneous cell population, where ~6% of cells were at the cell-cycle stage(s) specific for polar localization of CpdR1. The formation of CpdR1-YFP foci was also observed in ~5% (n = 1273) of cells in stationary phase, although the YFP foci signal was faint compared to the foci intensity of cells in log phase (Fig. 2D–F; Table 1). Of the foci that formed, ~100% (n = 245) of the CpdR1-YFP foci were localized at the cell poles (Supplemental Table S1) and we did not detect any cells with more than one focus (this is similar to C. crescentus) (Table 1).
Fig. 2. Subcellular localization of CpdR1 and CpdR2 in free-living S. meliloti cells.
Panels A to L represent subcellular localization of CpdR1-YFP (A to F) and CpdR2-YFP (G to L) fusion proteins in free-living S. meliloti by epi-fluorescence microscopy. S. meliloti strains carrying fusion genes were grown in M9 medium supplemented with succinate. Cells were examined in the logarithmic phase (A to C and G to I) or the stationary phase (D to F and J to L). Localization of YFP-fusion proteins was visualized in living cells as represented in green (B, E, H and K). The cell membrane was stained by FM4-64 as represented in red (A, D, G and J). Epi-fluorescence views of different filters were merged to analyze relative localization (C, F, I and L). No focus was observed in cells of the wild-type S. meliloti strain Rm1021 (data not shown). Scale bar represents 2 μm.
Table 1.
Formation of CpdR-YFP foci in S. meliloti Rm1021
| % of cells with n foci |
|||||
|---|---|---|---|---|---|
| Growth phase | No. of cells | 0 | 1 | 2≤ | |
| CpdR1-YFP | Logarithmic | 1399 | 94 | 6 | ND |
| Stationary | 1273 | 95 | 5 | ND | |
| CpdR2-YFP | Logarithmic | 534 | 100 | ND | ND |
| Stationary | 561 | 99 | <1 | ND | |
We also investigated the localization of CpdR2-YFP in S. meliloti. We found that the formation of CpdR2-YFP foci was observed in only a small subpopulation of stationary-phase cells (~0.4% of cells, n = 561; Table 2). In both log- and stationary-phase cultures, some of cells have brighter CpdR2-YFP signals throughout the cell than other cells (Fig. 2G–L). It should be noted that each YFP fusion was transcribed from the native promoters of cpdR1 and cpdR2 respectively, supporting our results with uidA fusions showing that cpdR1 and cpdR2 were expressed throughout the growth phases. The active localization of CpdR1 to cell poles suggested that CpdR1 shares a similar function with C. crescentus CpdR, while the significance of CpdR2 localization remains unclear.
Table 2.
Formation of highly branched cells in S. meliloti Rm1021 strains
| % of cells with morphology |
||||
|---|---|---|---|---|
| Strains | No. of cells | rod | elongated | highly branched |
| Rm1021(pTH1227) | 413 | 99 | 1< | ND |
| Rm1021minCDE−(pTH1227) | 453 | 99 | 1< | ND |
| Rm1021(p-cpdR1D53A) | 551 | 82 | 12 | 6 |
| Rm1021minCDE−(p-cpdR1D53A) | 405 | 90 | 10 | ND |
“rod”, 1–3 μm rod-shape with two poles; “elongated”, cells longer than 4 μm with two to three poles; “highly branched”, cells with more than three poles.
S. meliloti cpdR1 and cpdR2 are not essential, while clpX provides an essential function
To further examine the function of cpdR1 and cpdR2 in S. meliloti, both were disrupted by inserting a spectinomycin/streptomycin resistance gene (for cpdR1) (Fellay et al., 1987) or a uidA-Cmr cassette (for cpdR2) (Metcalf and Wanner, 1993), generating strains Rm1021ΩcpdR1 and Rm1021cpdR2::uidA. The cpdR1-null mutant grew much slower than the parental strain, while the growth of the cpdR2-null mutant was virtually indistinguishable from that of the parental strain (data not shown). This further supports the idea that the two cpdR homologs have non-identical roles in S. meliloti. In addition, we found that clpX is essential for viability of S. meliloti, as is the case for C. crescentus (Jenal and Fuchs, 1998). We attempted to generate a clpX-null allele in S. meliloti. The plasmid, pJQ-clpX−, a suicide vector pJQ200-SK (Quandt and Hynes, 1993) derivative in which the clpX ORF was disrupted by insertion of a neomycin resistance (Nmr) marker (Fellay et al., 1987), was integrated into the chromosomal clpX locus by single-crossover. Counter-selection for the double-crossover in the resulting strain was performed with derivatives that contained either a plasmid carrying the functional copy of clpX (p-clpX+) or the empty vector pTH1227. The disruption of chromosomal clpX occurred only in the presence of p-clpX+ (data not shown), indicating that clpX encodes an essential function in S. meliloti. Similar procedures were employed to show that ctrA and ccrM, genes encoding proteins critical for cell-cycle progression, are also essential in S. meliloti (Barnett et al., 2001; Wright et al., 1997). We conclude that CpdR1 has an important role during the free-living growth of S. meliloti and that clpX is essential under the growth conditions examined in this study. We speculate that the role of CpdR in free-living S. meliloti likely involves the regulation of ClpXP protease as reported for C. crescentus CpdR (Iniesta et al., 2006).
CpdR1 is critical for the bacteroid differentiation
To test whether the cpdR homologs are important for symbiosis, alfalfa seedlings were inoculated with the cpdR1- or cpdR2-null mutants. Four weeks post inoculation, alfalfa plants infected with the wild type grew to a shoot length of 11.4 ± 0.3 cm with pink-colored and elongated nodules on the roots (Fig. 3A). In contrast, plants inoculated with the cpdR1-null mutant appeared to be starved of nitrogen, having short shoots (2.9 ± 0.1 cm) with yellowed leaves (Fig. 3A). Nodules induced by the cpdR1-null mutant were white-colored, small and non-elongated globular shape. The size and appearance of the shoots of the plants were indistinguishable from those of the mock-inoculated plants, which had no nodules on their roots. This could indicate that the nodules induced by the cpdR1-null mutant lacked nitrogen-fixing activity (Fig. 3A). No significant difference was observed in nodule number or in plant growth between plants infected with the wild type and the cpdR2-null mutant (Fig. 3A).
Fig. 3. CpdR1 is critical for symbiosis between S. meliloti and alfalfa.
(A) The cpdR1-null mutant elicits ineffective nodules on alfalfa roots. The symbiotic defect can be complemented by plasmid-born cpdR1+, the cpdR1-yfpmut2 fusion gene and C. crescentus cpdR but not by cpdR1D53A or vector controls (pTH1227 and pLXM-GFP). The disruption of cpdR2 has neither a positive nor a negative effect on growth of plants relative to that of the wild-type S. meliloti strain Rm1021. Growth of alfalfa plants were examined four weeks after inoculation. The plant heights are the lengths of shoots (cm). Error bars indicate standard errors. At least 20 plants were examined for each strain. (B to E) In nodules elicited by the wild-type S. meliloti strain Rm1021 viewed by transmission electron microscopy (B and D), cells from the nitrogen fixing zone are packed with bacteroids and contained a large central vacuole (v). Nodules elicited by the cpdR1-null mutant (C and E) contain plant cells filled with bacteria of irregular-coccoid morphology and have large amyloplasts (a). Scale bars represent 2 μm.
A more striking phenotype of the cpdR1-null mutant was observed by electron microscopy. Fig. 3B and D shows a plant cell from the nitrogen-fixing zone of a nodule containing the wild-type S. meliloti strain Rm1021. These bacteroids generally have elongated or Y-shaped morphology and are five to ten times longer than the free-living cells. Fig. 3C and E show the corresponding region of a nodule containing the cpdR1-null mutant. The host cells contain numerous intracellular bacteria, indicating that the cpdR1-null mutant is not especially impaired in the early stages of nodule invasion through the root hair or in invading the cytoplasm of the host cell. However, the morphology of these intracellular bacteria is diverse. Although some cells are abnormally enlarged, most of the bacteria appear to be highly irregular spherically-shaped (coccoid) cells much smaller than the bacteroids of the wild-type S. meliloti Rm1021 and have refractory cytoplasm indicative of aborting bacteroids (Campbell et al., 2002). The host cells also contained large amyloplasts. Amyloplasts are starch deposits that are normally present in nodule cells early in development and are absorbed upon S. meliloti infection of alfalfa. Thus, in S. meliloti, CpdR1 is critical for establishing the chronic intracellular infection, particularly, at the stage of bacteroid differentiation.
The symbiotic deficiency of the cpdR1-null mutant was fully complemented by p-cpdR1+, a plasmid carrying the wild-type cpdR1+, and also by p-cpdR1-yfp, but not by the empty vector pTH1227. In C. crescentus, it has been shown that the activity of CpdR is regulated by phosphorylation (Iniesta et al., 2006). Only the unphosphorylated CpdR localizes to the cell pole and is capable of directing the polar localization of ClpXP. To understandthe role of CpdR phosphorylation status in symbiosis, we generated an allele of cpdR1, cpdR1D53A, that codes for an alanine at position 53 instead of the conserved aspartic acid. In C. crescentus, CpdRD51A, the analog of S. meliloti CpdR1D53A, was shown to prevent phosphorylation of the protein, resulting in a constitutively active form that directs ClpXP to the cell pole throughout the cell cycle (Iniesta et al., 2006). Introduction of the cpdR1D53A allele did not restore the symbiotic deficiency of the cpdR1-null mutant (Fig. 3A), indicating that the fine-tuning of CpdR1 phosphorylation status is important for symbiosis. Heterologous expression of C. crescentus cpdR in the S. meliloti cpdR1-null background complements the symbiotic deficiency as well as the growth defect of the free-living cells, indicating that the CpdR function is conserved between S. meliloti and C. crescentus. We conclude that S. meliloti CpdR1 phosphorylation is critical for its function and that C. crescentus cpdR can complement the S. meliloti cpdR1 defect indicating a conserved role of CpdR among α-proteobacteria.
CpdR1 is required for proper morphogenesis of S. meliloti
The abnormal morphology of the intracellular cells of the cpdR1-null mutant in alfalfa led us to examine the cell morphology in the free-living stage. As free-living cells, the wild-type S. meliloti are rod shaped (Fig. 4A). Strikingly, free-living cells of the cpdR1-null mutant are highly irregular swollen or coccoid shape with the size generally three to four times larger than the size of wild-type S. meliloti cells (Fig. 4B). Some of cells were further enlarged and branched with single or multiple asymmetric septa (data not shown). The morphology, and the size of the free-living cells of the cpdR1-null mutant, was similar to those of the in planta intracellular cells. This suggests that the cpdR1-null mutant could not initiate the differentiation process into bacteroid form. Introduction of the wild-type cpdR1+ to the cpdR1-null mutant restored normal morphology (Fig. 4C), while introduction of the C. crescentus cpdR only partially rescued the morphological defect (Fig. 4D). The S. meliloti cpdR1-null mutant expressing C. crescentus CpdR appeared to be rod shaped with, occasionally, single or multiple small branches (Fig. 4D). Introduction of the cpdR1D53A allele into the cpdR1-null mutant failed to rescue the morphology of the mutant and exacerbated slow growth phenotype (Fig. 4F; data not shown). Moreover, expression of cpdR1D53A in the wild-type S. meliloti Rm1021 transformed ~6% (n = 551) of the cells into highly branched cells with more than three poles (Fig. 4G; Table 2). Such cells were not detected in the wild-type S. meliloti carrying the empty vector (n = 413) or in the wild-type S. meliloti alone (Table 2). Thus, CpdR1 function is critical for the proper morphogenesis during both free-living growth and during bacteroid differentiation. The morphology of the cpdR2-null mutant was indistinguishable from the wild-type S. meliloti Rm1021 (Fig. 4E), indicating that cpdR2 is not important for cell morphogenesis in the free-living form.
Fig. 4. Cell morphology of S. meliloti strains.
DIC view of the wild-type S. meliloti stain Rm1021 (A), the cpdR1-null mutant (B), the cpdR1-null mutant carrying p-cpdR1+ (C) or p-CccpdR+ (D), the cpdR2-null mutant (E), the cpdR1-null mutant (F) or the wild-type strain (G) carrying p-cpdR1D53A, the minCDE-null mutant (H), the minCDEcpdR1 double mutant (I) and the minCDE-null mutant carrying p-cpdR1D53A. Cells were grown in LBMC medium to logarithmic phase and processed for DIC microscopy. Scale bars represent 2 μm.
The morphological phenotype caused by CpdR1D53A is Min dependent
During the examination of morphology of free-living cells, we noticed that the morphology of the cpdR1-null mutant is similar to the morphology of the minE-disrupted mutant of S. meliloti (Cheng et al., 2007). The MinE protein forms, together with MinC and MinD, the Min cell-division inhibitor system. The Min system is well studied in E. coli, where it directs cell division to the mid-cell by negatively regulating the formation of the Z ring (reviewed in Margolin, 2005). MinC is the inhibitor of Z-ring formation, which is recruited to the membrane by MinD and induced to oscillate by MinE. Although the minE single mutant shows a morphological defect, the minCDE triple mutant has wild-type morphology in S. meliloti, indicating that the phenotype of the minE mutant is likely due to the nonspecific action of MinC and MinD (Cheng et al., 2007; Fig. 4H; Table 2). Overexpression of MinD or MinCDE in S. meliloti generates highly branched cells (Cheng et al., 2007), which is also similar to the morphology we observe with S. meliloti Rm1021 expressing CpdR1D53A (Fig. 4G).
The similarity in morphology between the cpdR1-null mutant and the minE-null mutant, as well as between the CpdR1D53A-expressing strain and Min-overexpressing strains, led us to hypothesize that cpdR affects proper activity of the Min system. To test this hypothesis, minCDE were disrupted in the cpdR-null and cpdRD53A backgrounds. Disruption of minCDE did not restore the morphology of the cpdR1-null mutant (Fig. 4I), indicating that the morphological defect of the cpdR1-null mutant is independent of the Min system. However, when minCDE was disrupted in S. meliloti Rm1021(p-cpdR1D53A), highly branched cell (with more than three poles) was not detected any longer, while elongated cells (with two or three poles) were still observed (Fig. 4J; Table 2). Thus, the morphogenesis of highly branched cells by CpdR1D53A appears to depend, in part, on the Min system.
CpdR1 affects the formation of ClpX-YFP foci
C. crescentus cpdR can partially complement the S. meliloti cpdR1-null mutant, suggesting that the molecular function is conserved for both gene products. In C. crescentus, the role of CpdR in directing ClpX to the cell pole has been demonstrated (Iniesta et al., 2006). To examine the role of CpdR1 in the ClpX localization, we fused yfp to the genomic copy of clpX in S. meliloti. The strain, Rm1021clpX-yfp, which carries the clpX-yfp fusion gene as the only copy of clpX, is viable. The growth rate and the cell morphology of the clpX-yfp strain was indistinguishable from the wild-type S. meliloti strain Rm1021, indicating that the ClpX-YFP fusion protein is functional. In logarithmic-phase growth, ~9% (n = 1005) of cells had one focus of ClpX-YFP visible above the background fluorescence (Fig. 5A–C; Table 3). In stationary-phase cells, ClpX-YFP foci formed in ~88% (n = 594) of cells (Fig. 5D–F; Table 3). Most of the cells containing ClpX-YFP foci had one focus per cell (Table 3). This observation is interesting, since CpdR1-YFP foci were observed in ~5% (n = 1273) of cells in stationary phase (Table 1). Thus, in stationary-phase cells, most of the population of cells with ClpX-YFP foci do not correspond to the population of cells with CpdR1-YFP foci. In both growth phases, ClpX-YFP foci were preferentially formed at the cell poles. We found that ~78% (n = 134) and ~97% (n = 150) of ClpX-YFP foci were located at the poles in logarithmic- and stationary-phase cells, respectively (Table S2). Interestingly, ClpX-YFP foci were observed in less than ~1% of mature bacteroids (n = 579). Taken together, these observations indicate that ClpX-YFP foci formation is growth-phase regulated.
Fig. 5. Subcellular localization of ClpX in cells of S. meliloti strains.
Panels A to R represent subcellular localization of the ClpX-YFP fusion protein in free-living S. meliloti strains (A to F and J to R) or bacteroids (G to I). Localization of ClpX-YFP in free-living cells of the wild-type S. meliloti strain Rm1021 (A to F), the cpdR1-null mutant (J to O) and the wild-type S. meliloti strain Rm1021 carrying p-cpdR1D53A (P to R) was examined by epi-fluorescence microscopy essentially in the same way as in Figure 2. Panel G to H represent subcellular localization of ClpX-YFP fusion protein in bacteroids of the wild-type S. meliloti Rm1021 by epi-fluorescence/DIC microscopy. Nodules were harvested from alfalfa plants infected with Rm1021clpX-yfp and immediately crushed. Long, blanched bacteroid cells were visually distinguished from non-differentiated S. meliloti cells or plant-derived materials (G). In contrast to the free-living cells, the membrane of bacteroids could not be stained by FM4-64 (data not shown). Localization of ClpX-YFP was visualized in bacteroids as represented in green (H). Epi-fluorescence view and DIC view were merged to analyze relative localization (I). Scale bars represent 2 μm.
Table 3.
Growth phase-dependent formation of ClpX-YFP foci in S. meliloti strains
| Percentage of cells with n foci |
|||||
|---|---|---|---|---|---|
| S. meliloti strain | Growth phase | No. of cells | 0 | 1 | 2≤ |
| Rm1021 | Logarithmic | 1005 | 90 | 9 | 1 |
| Stationary | 594 | 12 | 84 | 4 | |
| Bacteroid | 579 | 99 | 1 | ND | |
| Rm1021cpdR1ΩSpr | Logarithmic | 545 | 94 | 5 | <1 |
| Stationary | 1004 | 88 | 11 | <1 | |
| Rm1021cpdR2::uidA-Cmr | Logarithmic | 938 | 91 | 8 | <1 |
| Stationary | 869 | 21 | 76 | 3 | |
| Rm1021(p-cpdR1D53A) | Logarithmic | 798 | 91 | 8 | 1 |
| Rm1021(p-cpdR2D52A) | Logarithmic | 677 | 88 | 11 | 1 |
To assess the role of two CpdR homologs in directing ClpX to the cell pole, the localization of ClpX-YFP was examined in the cpdR1- and cpdR2-null mutants. Compared to the wild-type, a smaller population of the cpdR1-null cells showed ClpX-YFP foci (Fig. 5J–O; Table 3). We found that the percentage of stationary-phase cells with ClpX-YFP foci was reduced ~eight-fold in the cpdR1-null mutant (Table 3). In C. crescentus, it has been reported that ClpX localization to the cell-division plane is independent of CpdR function (Iniesta et al., 2006). It has to be noted that, since we cannot distinguish the cell-division plane of S. meliloti (it is difficult to determine by DIC microscopy whether two proximal cells are currently dividing or just placed side by side), we can only score the location of foci as “polar” when they are located at the fully formed pole of a cell (Supplementary Table S2). The decreased formation of ClpX foci in the cpdR1-null mutant further supports the hypothesis that CpdR1 plays a conserved role in the polar localization of ClpX in S. meliloti. We could not score the location of ClpX foci in the cpdR1-null mutant, because the “poles” were not obvious in the irregular coccoid cells. In the cpdR1-null mutant, we also noticed that the foci intensity of ClpX-YFP was reduced relative to that of the foci formed in the wild-type background. In the wild-type background, ClpX-YFP foci were readily captured by an exposure for 240 milliseconds, while an exposure of 6000 milliseconds was necessary to capture distinct foci in the cpdR1-null mutant. This observation suggests that the loss of CpdR1 function pleiotropically affects the ClpX expression and/or the accumulation of ClpX-YFP at the cell poles.
We also examined the effect of CpdR1D53A on ClpX localization in S. meliloti. In the wild-type S. meliloti Rm1021 expressing CpdR1D53A, cell morphologies are abnormal (see Fig. 4G), but the formation and the localization of ClpX-YFP foci nonetheless showed common features with background Rm1021: a minority of cells formed a ClpX focus, and of those that did, the foci were at cell poles (Fig. 5P–R; Table S2). These observations differ from results in C. crescentus (Iniesta et al., 2006), where the expression of CpdRD51A resulted in ~100% polar localization of ClpX foci. This difference is probably because we expressed plasmid-born cpdR1D53A under the control of a tac promoter in the wild-type S. meliloti Rm1021, where the wild-type cpdR gene is also present. In the C. crescentus strain, however, cpdRD51A replaces the wild-type cpdR locus and was expressed from the native cpdR promoter (Iniesta et al., 2006). We did not examine localization of ClpX-YFP in the cpdR1-null mutant expressing CpdR1D53A, since the morphology of the cells was severely defective (data not shown). In the cpdR2-null mutant or the wild-type S. meliloti expressing CpdR2D53A, cells formed ClpX-YFP foci of intensity indistinguishable from the foci in wild-type cells, although the population of cells with ClpX-YFP foci was slightly affected compared to the wild-type S. meliloti background (Fig. S1; Table 3). We conclude that CpdR1 is important for proper subcellular localization of ClpX-YFP in S. meliloti.
CpdR1 functions to coordinate initiation of DNA replication with cell-cycle progression
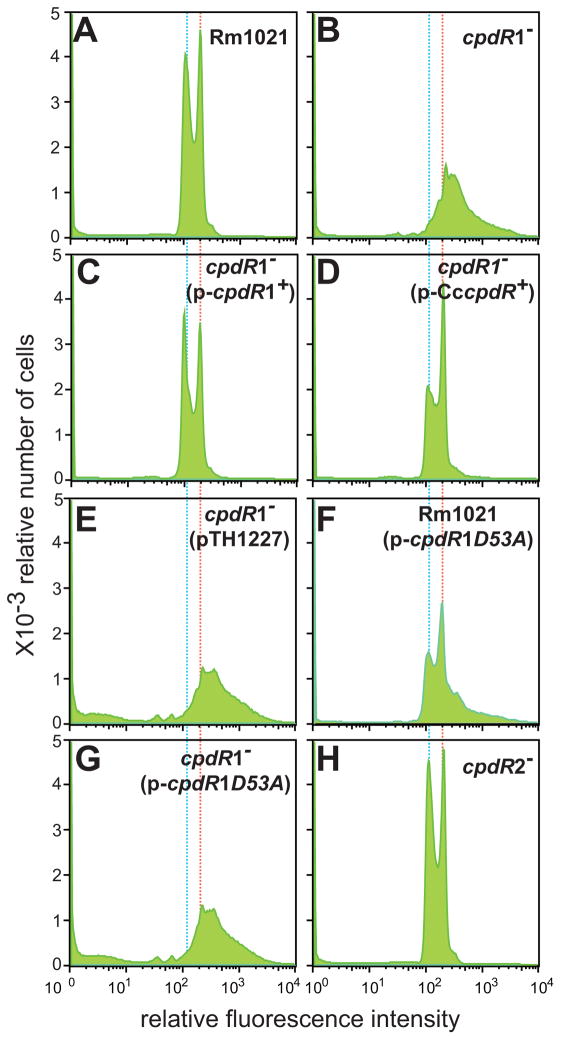
In S. meliloti, it has been shown that the larger cell volume correlates with the polyploid state of the cell (Mergaert et al., 2006). The enlarged cell volume of the cpdR1-null mutant led us to examine the DNA content of S. meliloti cells. Flow cytometry is a well-established tool for studying the cell cycle in synchronized cultures of C. crescentus (Winzeler and Shapiro, 1995). It has also been used in S. meliloti to examine the state of chromosomal replication in cell populations (Mergaert et al., 2006; Wright et al., 1997). As reported previously (Mergaert et al., 2006), in the wild-type S. meliloti strain Rm1021, DNA content distribution of an exponentially growing cell population is composed of two peaks (Fig. 6A). The first (left) peak indicates the number of cells with 1C DNA content (marked with blue-dotted line). The second peak is positioned around twice the relative fluorescence intensity of the first peak and represents the cells with 2C DNA content (marked with red-dotted line). In the cpdR1-null mutant, one broad peak representing cells with more than one genome (mainly 2~5C) equivalents were observed (Fig. 6B), indicating that most cells had multiple copies of the genome. Thus, in the cpdR1-null mutant, aberrant DNA replication uncoupled from cell division seems to have occurred. Introduction of the wild-type cpdR1+ restored proper progression of cell cycle in the cpdR1-null mutant (Fig. 6C). The profile of DNA content distribution of the cpdR1-null mutant expressing C. crescentus CpdR was similar to the profile in the wild-type, although the population with 2C DNA content was larger than the population with 1C DNA content (Fig. 6D).
Fig 6. Flow cytometry analyses of S. meliloti strains.
The DNA contents of S. meliloti strains were measured in a population of 50,000 cells with a Becton Dickinson FACScan machine at 530 nm. Relative cell number (y axis) is plotted against the Cytox Green fluorescence signal (arbitrary units). 1C and 2C of the genome equivalents were marked with blue-and red-dotted lines, respectively.
This observation, together with the partially-rescued morphology of the S. meliloti cpdR1-null mutant by C. crescentus cpdR, suggests that C. crescentus CpdR is partially functional in S. meliloti. C. crescentus CpdR likely has a different affinity for S. meliloti ChpT and ClpX, causing a slight perturbation in progression of the S. meliloti cell cycle. In the wild-type S. meliloti expressing CpdR1D53A, the population with 2C DNA content was also larger than the population with 1C DNA content (Fig. 6F). In this strain, moreover, populations with 2~5C DNA content were also detected, likely corresponding to the cells with elongated or highly branched morphology. The DNA content profile of the cpdR1-null mutant expressing CpdR1D53A was indistinguishable from the cpdR1-null mutant with the empty vector or the mutant alone (Fig. 6E and G). Thus, expression of CpdR1D53A also causes aberrant cell-cycle progression and the effect of CpdR1D53A expression requires the presence of wild-type cpdR1+. The cpdR2-null mutant showed DNA content profile indistinguishable from the wild-type S. meliloti Rm1021 (Fig. 6H). These data suggest that the function of CpdR1 is critical for proper coupling of DNA replication with cell division in S. meliloti.
Discussion
In this study, we demonstrate that CpdR plays a critical role in S. meliloti. In the free-living stage, cells of the cpdR1-null mutant showed an irregular coccoid morphology instead of the rod shape of the parental strain. The cells are generally three to four times larger in volume than wild-type cells. In S. meliloti, it has been shown that the larger cell volume correlates with the polyploid state of a cell (Mergaert et al., 2006). Strikingly, the cpdR1-null mutant tends to accumulate higher copies (generally 2~5C) of genomic DNA. Thus, in S. meliloti, the loss of CpdR1 function results in uncoupling between DNA replication and cell division, causing a higher genomic content and enlarged spherical morphology of the mutant cells.
Moreover, we demonstrate that CpdR1 is required for symbiosis between S. meliloti and the host plant alfalfa. Upon inoculation in alfalfa, the cpdR1-null mutant is capable of inducing the formation of nodules on the root and capable of invading the cytoplasm of nodule cells. However, it appears that nitrogen fixation does not occur in the resulting nodules. In these nodules, the intracellular bacteria remain morphologically similar to the free-living cells of the cpdR1-null mutant and do not differentiate into wild-type bacteroids, which are normally larger and elongated. In wild-type S. meliloti, elongation of the bacteria during bacteroid maturation is a result of repeated DNA replication without cell division (endoreduplication) (Mergaert et al., 2006). The inability of intracellular cells to differentiate into bacteroids indicates that, in the cpdR1-null mutant, normal endoreduplication required for bacteroid differentiation is not initiated. Thus, in the cpdR1-null mutant, the loss of proper cell-cycle control is likely a common cause of both the aberrant morphology in the free-living stage and a defect in bacteroid differentiation. In other words, the same regulatory mechanism involving CpdR1, which couples initiation of DNA replication to cell division in free-living stage, also affects the endoreduplication during bacteroid development.
What is the molecular function of CpdR1 critical for coordinating cell-cycle progression? In C. crescentus, CpdR directs ClpXP complex to the cell pole, thereby mediating regulated proteolysis of ClpXP substrates (Iniesta et al., 2006). The cross-species complementation of S. meliloti CpdR1 with C. crescentus CpdR strongly suggests a conserved role for CpdR homologs between α-proteobacterial species. In agreement with this idea, we observed that loss of CpdR1 function attenuates the polar localization of ClpX-YFP.
It is therefore likely that the phenotype of the cpdR1-null mutant is caused by a lack of proper proteolysis by ClpXP. In C. crescentus, CtrA is an important substrate of ClpXP (Chien et al., 2007; Gorbatyuk and Marczynski, 2005; Jenal and Fuchs, 1998). CtrA controls, directly or indirectly, critical cell-cycle regulated genes including those involved in polar morphogenesis, DNA replication initiation, DNA methylation, cell division, and cell wall metabolism (Laub et al., 2002). CtrA is conserved in S. meliloti and it has been shown that C. crescentus ctrA can substitute for S. meliloti ctrA for viability (Barnett et al., 2001). CtrA protein of S. meliloti has a conserved ClpX-recognition signal at the C terminus (Flynn et al., 2003; Chien et al., 2007), suggesting that CtrA is also a substrate for ClpXP in S. meliloti. Modified abundance of CtrA affects cell-cycle progression (Quon et al., 1998) and at least partially accounts for aberrant cell-cycle progression in the absence of cpdR1 in S. meliloti.
Regarding cell-cycle progression, the phenotype of the cpdR1-null mutant fundamentally differs in S. meliloti from the phenotype reported in the C. crescentus cpdR-null mutant. In C. crescentus, the cpdR-null mutant also showed an aberrant morphology; an abnormal location of the cell-division plane and straight-cell morphology rather than the crescentoid morphology of the wild-type (Iniesta et al., 2006). It has also been reported that disruption of C. crescentus cpdR results in decreased motility, a longer generation time and, in a subpopulation, elongation of the cell (Skerker and Laub, 2005). In S. meliloti, on the other hand, we found that a disruption of cpdR1 results in significant enlargement of the cell size and spherical cell shape in most of the cells. More importantly, the loss of CpdR function does not cause polyploidy in C. crescentus (Duerig et al., 2009).
The different phenotypes resulted from loss of CpdR function on DNA replication likely reflects the plasticity of the regulation network involving CtrA in α-proteobacterial species (Hallez et al., 2004). In C. crescentus, phosphorylated CtrA (CtrA~P) binds to five distinct sites in the origin of replication to repress replication initiation by blocking the access of proteins for DNA replication initiation (Quon et al., 1998; Siam and Marczynski, 2000). CpdR directs ClpXP to the pole, where ClpXP degrades the CtrA~P, allowing for de-repression of DNA replication initiation. In the absence of CpdR, the ClpXP protease is not positioned at the pole and, consequently, CtrA~P is not degraded, keeping the replication origin physically blocked and DNA replication initiation repressed. Yet, CpdR is not essential for viability most likely because CtrA~P could still be inactivated by dephosphorylation.
In S. meliloti, on the other hand, no conserved CtrA-binding site has been detected in the replication origin (Hallez et al., 2004). In Brucella abortus, an intracellular pathogen closely related to S. meliloti, the replication origin also lacks a functional CtrA-binding site and was shown not to be bound by B. abortus CtrA in vitro (Bellefontaine et al., 2002). From these observations, it has been proposed that, in S. meliloti and B. abortus, DNA replication initiation is not regulated by the binding of CtrA to the replication origin (Bellefontaine et al., 2002; Hallez et al., 2004). It is therefore likely that, in the absence of CpdR function, the replication origin is available for DNA replication initiation even though CtrA is not proteolytically regulated. In agreement with this model, we found that, in S. meliloti, the loss of CpdR1 function does not result in repression of DNA replication initiation and can even provoke DNA replication in an unregulated manner.
We find it likely that genes regulated by CtrA are misregulated by the stabilization of CtrA and at least partially responsible for the aberrant cell-cycle progression in the cpdR1-null mutant. In S. meliloti and C. crescentus, it has been reported that overexpression of ccrM, a CtrA-target gene encoding a DNA methyltransferase, results in the loss of control of DNA replication and aberrant cell morphology, phenotypes similar to the cpdR1-null mutant (Wright et al., 1996; 1997; Zweiger et al., 1994). Although the accumulation of CcrM protein was not observed in the C. crescentus cpdR mutant (Iniesta et al., 2006), it is possible that ccrM is overexpressed in the S. meliloti cpdR1 mutant, contributing to its phenotype. In the S. meliloti symbiotic plasmid pSymA, putative CtrA-binding sites have been detected in promoter regions of repA2 (Hallez et al., 2004), which encodes a putative replication protein A. Unregulated expression of repA2 could also be the cause of the aberrant DNA replication in the cpdR1-null mutant.
In S. meliloti and B. abortus, two and four CtrA-binding motifs were detected in an upstream region of the minCDE operon, respectively, while minCDE is absent in C. crescentus genome (Bellefontaine et al., 2002; Hallez et al., 2004; Cheng et al., 2007). When CpdR1D53A, the constitutively active variant of CpdR1, is expressed in S. meliloti, minCDE is probably de-repressed, causing the over-branched cell morphology of the strain. The over-branched cell phenotype was only seen in cells expressing CpdR1D53A in the presence of wild-type cpdR1+. It is unclear how the effect of CpdR1D53A expression requires the wild-type cpdR1+ in S. meliloti.
In addition to the perturbation in cell-cycle progression, abundance of other ClpXP substrates as a consequence of reduced ClpXP localization may contribute to the phenotype of the cpdR1-null mutant. It has been shown that ClpXP plays a major role in protein quality control by degrading SsrA-tagged proteins (Gottesman et al., 1998; Herman et al., 1998; Keiler et al., 1996). The SsrA tag is a peptide that is added co-translationally to the C termini of nascent polypeptides in stalled translation complexes, and targets these proteins for degradation by ClpXP and other proteases (Keiler et al., 1996). It is possible that SsrA-tagged proteins are not properly degraded by ClpXP in the absence of CpdR1, contributing to the pleiotropic phenotype of the cpdR1-null mutant. In agreement with this idea, the Bradyrhizobium japonicum sra locus, which encodes a ssrA homolog, is essential for symbiosis with the host plant (Ebeling et al., 1991).
We observed that the majority (~88%) of cells contain ClpX-YFP foci in stationary phase cells, while ClpX-YFP foci were observed in around ~10% of cells in logarithmic phase. Such increased formation of ClpX foci in stationary phase has not been reported in other organisms. The localization of ClpX has been examined in C. crescentus and the Gram-positive bacteria Bacillus subtilis (Kain et al., 2008; Kirstein et al., 2008; McGrath et al., 2006; Simmons et al., 2008). In the case of C. crescentus, however, the localization was not examined in stationary phase. In B. subtilis, a cpdR homolog has not been identified, and ClpX-GFP forms foci preferentially at the poles in approximately half of cells (Simmons et al., 2008). The percentage of cells with ClpX foci remains constant from logarithmic phase to stationary phase (L.A. Simmons, unpublished). Thus, our observation suggests that, in S. meliloti, ClpXP-dependent proteolysis plays an important role during stationary phase, in addition to its role in the cell-cycle progression. In E. coli, it has been reported that cultures of the strains lacking functional ClpP or ClpX displayed a more rapid loss of viability during extended stationary phase than the wild type (Weichart et al., 2003). It is interesting to note that the cpdR1-null mutant displays decreased foci formation of ClpX also in stationary phase cells, suggesting that CpdR1 function is required for the ClpX localization in both growth phases.
Thus, further study is required to determine the molecular function of CpdR1 and ClpXP in S. meliloti. Moreover, the genome of S. meliloti encodes the second homolog of cpdR, cpdR2, and three clpP homologs. Although we could not detect a phenotype of the cpdR2-null mutant, CpdR, ClpX and ClpP proteins may form a network, which regulates various physiological processes in the complex life cycle of S. meliloti. As we discussed above, since the regulatory network involving CtrA in S. meliloti and Brucella species share a similar structure (Hallez et al., 2004), the CpdR protein may also have a critical role in coordinating cell-cycle progression in Brucella species. It would be interesting to examine whether CpdR is also critical for the chronic infection of Brucella species.
Experimental procedures
Microbiological techniques
Strains and plasmids used in this study are listed in Supplemental Table S3. E. coli recombinants were grown at 37°C on Luria-Bertani media/medium (Sambrook et al., 1989). S. meliloti strain Rm1021 and its derivatives were raised at 30°C in/on M9 succinate medium with biotin or LBMC (LB supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2). Gentamycin (Gm), neomycin (Nm), streptomycin (Sp), streptomycin (Sm), and tetracycline (Tet) were added at concentrations of 50, 200, 50, 500 and 15 μg ml−1, respectively. Plasmids were mobilized into S. meliloti strain Rm1021 and derivative strains by tri-parental mating using pRK600 as the helper plasmid (Ditta et al., 1980). Pfu turbo (Stratagene) was used in all PCR reaction followed by cloning. The amplified products were cloned into pCR-Blunt II-TOPO (Invitrogen) and verified by sequencing the inserts.
Construction of insertional disruptions of clpX and cpdR homologs
To construct the insertional disruption of cpdR1 (SMc04044), cpdR2 (SMc00720) and clpX, we amplified each full-length ORF with flanking regions by PCR using Rm1021 genomic DNA and the following primer pairs: for cpdR1 (5′-CTCGAGAAACGTGGCGCTGCGGAACAGTTCATCGACGAAAT-3′/5′-CTGCAGTTCCGTCGAGCGCGCTAAGGCGTTGAAATAACGAA-3′); for cpdR2 (5′-CCCGGGACCGAATCCAAGGGCGTCAGGATCACCAAGGATAT-3′/5′-TCTAGAGCGCTACGGCCTTGCGGATGTCGGGCAGTGAAAAT-3′); for clpX (5′-TTGCGGCAAGAGCCAGCATGAAGTCCGC-3′/5′-ATGCTGTTGCAGACCTTGCGACCTGCCC-3′). Underlined nucleotides correspond to mismatches introduced to create selected restriction sites. To disrupt cpdR1, cpdR2 and clpX, a Smr/Spr cassette (from pHP45Ω; Fellay et al., 1987), an uidA-Cmr cassette (from pWM4; Metcalf and Wanner, 1993) and a Kmr/Nmr cassette (from pHP45Ω-Km; Fellay et al., 1987) were inserted into the internal NruI, SalI and BglII sites, respectively. DNA fragments containing disrupted cpdR and clpX homologs were sub-cloned into pJQ200-SK (Quandt and Hynes, 1993), and the resulting plasmids were mobilized into Rm1021 or its derivatives by tri-parental mating. Transconjugants were first selected on LBMC plates containing Sm and Gm, then on LBMC plates containing appropriate antibiotics to select the antibiotic-resistance markers in the disrupted genes and 5% (w/v) sucrose to select for double crossovers. Replacement of wild-type clpX and cpdR genes with its disrupted loci was confirmed by PCR.
Construction of plasmids for expression of CpdR proteins, ClpX and CpdR variants
The cpdR1, cpdR2 and clpX genes were PCR amplified by using Rm1021 genomic DNA and the following primer pairs: for cpdR1 (5′-CTCGAGAAGAAGTAGCCACGGCCAGATATGACTGCGAAAAT-3′/5′-CTGCAGATATGTTCGCTGCTCAGGCGGCCAGCATCTTGTT-3′); for cpdR2 (5′-CTCGAGAAAAAATACGGGAGGCCATGATGGCGAAAATCCTGATCA-3′/5′-CTGCAGCTATGCATGAATTTTCCCCGGACAGCCCTGCCGCTT-3′); for clpX; (5′-GGATCCGGAAGGAAGTGGAAATGAGCAAGGTCAGCGGTA-3′/5′-GCATGCTGTGGCCTCAAGCCGAAACGTTGGTCTTCTCCT-3′). The cloned fragments were sub-cloned in pTH1227, the low-copy-number vector for S. meliloti carrying lacIq and the tac promoter (Cheng et al., 2007). The resulting plasmids, carrying cpdR homologs or clpX under the control of tac promoter, were mobilized into Rm1021 by tri-parental mating. The cloned cpdR1 and cpdR2 were converted into cpdR1D53A and cpdR2D52A by site-directed mutagenesis (Stratagene) with following primer pairs: for cpdR1D53A (5′-CCCTTTTCGCTTCTCCTGACCGCCATCGTCATGCCGGAGATGGAC-3′/5′-GTCCATCTCCGGCATGACGATGGCGGTCAGGAGAAGCGAAAAGGG-3′); for cpdR2D52A (5′-CCGTTCGACCTCTTGCTATCCGCCATCCGGATGCCGGTCATGGAC-3′/5′-GTCCATGACCGGCATCCGGATGGCGGATAGCAAGAGGTCGAACGG-3′). Underlined nucleotides correspond to mismatches introduced to create the site-directed mutation. The mutated genes were verified by sequencing and cloned in pTH1227 as the same way for the wild-type genes.
Construction of chromosomal uidA-transcriptional fusions
To construct cpdR-uidA transcriptional fusions, ORFs of cpdR1 and cpdR2 were amplified by PCR using the following primer pairs: cpdR1uidA (5′-ACTAGTTCCCCCATCGGAACCCTTTTTACCATGTCCGGTTCTT-3′/5′-CTCGAGTCAGGCGGCCAGCATCTTGTTGACCTCGTTGACGAGGT-3′); cpdR2uidA (5′-ACTAGTAGTGGTGACGGGCGACGCAACCTAACTTGGCTCGT-3′/5′-CTCGAGTCAGGCGGCCAGTGCGAGCGCTACGGCCTTGCGGA-3′). The cloned fragments were sub-cloned into the suicide uidA reporter vector pJH104. Resulting plasmids were mobilized into Rm1021 by tri-parental mating. Transconjugants were selected on LBMC plates containing Sm and Nm. It must be noted that derivatives of pJH104 were inserted into directly (0 bp) downstream of native gene loci by single crossover recombination without disrupting the genes (ATG of the uidA gene is located 23 bp downstream of the stop codon of each cpdR homolog). Although the insertion of pJH104 derivatives results in an additional copy of cpdR1 and cpdR2, the second copy of cpdR genes lack an upstream promoter. Insertions of pJH104 were confirmed by PCR. To construct a clpX-uidA transcriptional fusion, the clpX ORF and the downstream region of clpX were amplified separately by PCR using following primer pairs: clpXuidA (5′-TCTAGAGGAAGGAAGTGGAAATGAGCAAGGTCAGCGGTA-3′/5′-GGATCCTGTGGCCTCAAGCCGAAACGTTGGTCTTCTCCT-3′); the downstream region of clpX (5′-CCCGGGAGGAGAAGACCAACGTTTCGGCTTGAGGCCACA-3′/5′-CTCGAGTCTCGGCACGCGACCGTCCTTCGACCAGAACCT-3′). Two fragments were sub-cloned together in pJQ200-SK and combined with uidA-Nmr cassette from pWM6 (Metcalf and Wanner, 1993). The resulting pJQ200-SK derivative carries clpX followed by uidA-Nmr cassette and the downstream sequence of native clpX locus (ATG of the uidA gene is located 59 bp downstream of the stop codon of clpX). The resulting plasmid was integrated into Rm1021 by double crossover as described above. Thus, in resulting strains, uidA is inserted downstream of the stop codons of chromosomal cpdR1, cpdR2 and clpX, being transcribed as the single transcriptional units with cpdR homologs and clpX.
β-glucuronidase assay in free-living Rm1021 strains
The relative expression level of each uidA transcriptional fusion was determined by assaying β-glucuronidase activity as previously described (Jefferson et al., 1986) with the following modifications. The assay buffer was supplemented with 10 mM EDTA (pH 8.0) and 0.1% sarcosyl, and the enzyme assays were performed with 5 mM p-nitrophenyl β-D-glucuronide substrate (Sigma). The β-glucuronidase activity was normalized to the cell density (1cmOD600) and represented in Miller’s unit (Miller, 1972).
Nodulation assay and β-glucuronidase assay in nodules
Seedlings of alfalfa (Medicago sativa cv. Iroquois: Agway Inc., Plymouth IN, USA) were inoculated with Rm1021 derivatives on Petri dishes containing Jensen agar as described previously (Leigh et al., 1985; Pellock et al., 2000). Four-week-old plants were examined for the symbiotic phenotypes. For β-glucuronidase assay, four-weeks-old nodules were excised, hand-sectioned longitudinally in half, and incubated in X-gluc staining buffer [1 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide, 0.1 M Na-phosphate buffer (pH 7), 10 mM EDTA, 0.5 mM K-Ferricyanide, 0.5 mM K-Ferrocyanide, 0.1% Triton X-100] at 37°C for overnight. Plants inoculated with Rm1021 derivatives containing pJH104 insertions showed a wild-type phenotype for nodulation.
Construction for tagging CpdR1, CpdR2 and ClpX with YFP
For tagging CpdR1 and CpdR2 with YFP, we fused plasmid-born cpdR1 and cpdR2 with yfpmut2A206K, encoding the A206K missense mutation generating monomeric YFP, of pKL183 (Lemon and Grossman, 2000). cpdR homologs (including the promoter region and the entire ORF except the stop codon) were amplified by PCR using the following primer pairs: cpdR1-yfpmut2 (5′-AAGCTTGATCGAGGCGCGACTGGTCGAGAAAGAACTGGGGA-3′/5′-CTCGAGGGCGGCCAGCATCTTGTTGACCTCGTTGACGAGGT-3′); cpdR2-yfpmut2 (5′-AAGCTTAGTGGTGACGGGCGACGCAACCTAACTTGGCTCGT-3′/5′-CTCGAGGGCGGCCAGTGCGAGCGCTACGGCCTTGCGGATGT-3′). The cloned fragments were sub-cloned into the HindIII-XhoI sites of pTH1227, removing lacIq and the tac promoter from pTH1227, and combined with the XhoI-PstI fragment containing yfpmut2A206K. The resulting pTH1227 derivatives, carrying the cpdR1- or cpdR2-yfpmut2A206K fusion gene under the control of native promoters of cpdR1 or cpdR2, were mobilized into the wild-type Rm1021 by tri-parental mating. For tagging ClpX with YFP, we fused chromosomal clpX with yfpmut2A206K. The 3′ region of clpX coding sequence was amplified by PCR using the following primer pair: clpX-yfpmut2 (5′-GGGCCCTCGACAAGATTTCCCGTAAGTCCGACAACCCGT-3′/5′-GTCGACAGCCGAAACGTTGGTCTTCTCCTCGGAACGCT-3 ′). The cloned fragments was sub-cloned in pJQ200-SK and combined with the XhoI-PstI fragment containing yfpmut2A206K. The resulting pJQ200-SK derivative, carrying the 3′ region of clpX fused to yfpmut2A206K, was integrated into the native clpX locus by single-cross over, disrupting the wild-type clpX locus. The resulting S. meliloti strain carries the clpX-yfpmut2 fusion gene as only one copy of functional clpX. We noticed that S. meliloti carrying sacB gene is symbiotically defective and therefore isolated a spontaneous sacB− strain for in planta assay.
Live cell microscopy
Aliquots of cells grown in M9 medium supplemented with succinate were stained with the vital membrane dye FM4-64 (Molecular Probes). The following Chroma filter sets were used: 41029 for YFP and 41002C for FM4-64. Exposure time for CpdR1-YFP, CpdR2-YFP and ClpX-YFP fusion protein was 1000, 1000 and 250 milliseconds, respectively. In the case of ClpX-YFP in the cpdR1-null background, 6000 milliseconds of exposure was required to capture the distinctive foci. Images were acquired, colorized, and merged using OpenLab software (Improvision). To assess the effect of the growth phases in free-living cells, aliquots of M9 cultures of stationary phase (1cmOD600 of >1.5) or log-phase (1cmOD600 of 0.4–0.6) were observed under the microscope. For bacteroids, whole alfalfa plants on Petri dishes containing Jensen agar were harvested four weeks after inoculation. Nodules were immediately harvested, crushed and bacteroids were observed by microscopy. Positions of foci were scored after colorization and merging of microscopic images. All data presented here are cumulative from at least two independent experiments, each of which gave nearly identical results.
Flow cytometry
For flow cytometry analyses, S. meliloti strains were grown to the mid-logarithmic phase in LBMC. The cells were fixed in 90% ethanol for 16 hours at 4°C, incubated in the sodium citrate buffer (50mM sodium citrate with 3.3 μg ml−1 RNase H) for 3 hours at 50°C and stained by Cytox Green (1:6000 diluted in the sodium citrate buffer) (Molecular Probes). For each flow cytometry experiment, the DNA content was measured in a population of 50,000 cells with a Becton Dickinson FACScan machine at 530 nm. The data were collected and analyzed by using the FlowJo software (Tree Star).
Supplementary Material
Acknowledgments
We are indebted to Mary Lou Pardue (Massachusetts Institute of Technology) and Alan D. Grossman (Massachusetts Institute of Technology) for the use of their microscopes. We are grateful to Jiujun Cheng and Turlough M. Finan (McMaster University) for providing us pTH1218 and pTH1227. We wish to thank Judith Carlin and Marianne White for their help. The authors thank Mary Ellen Wiltrout for insightful discussions and kind suggestions; Celeste Peterson, Glenn Paradis and Michiko E. Taga for help with flow cytometry; Melanie Barker-Berkmen for help with the microscopy.
This work was supported by National Institutes of Health grant GM31010 (to G.C.W.), National Cancer Institute (NCI) Grant CA21615-27 (to G.C.W), MIT Center for Environmental Health Sciences NIEHS P30 ES002109, JSPS Postdoctoral Fellowships for Research Abroad (to H.K.) and a postdoctoral fellowship from the NCI (to L.A.S.) and a Jane Coffin Childs Memorial Fund Fellowship (to P.C.) and NIH Grant 5K99GM084157-01 (to P.C.). G.C.W. is an American Cancer Society Research Professor.
References
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol. 2001;183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol. 2002;43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Brewin NJ. Tissue and cell invasion by Rhizobium: The structure and development of infection threads and symbiosis. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae. Dordrecht, Boston, London: Kluwer Acad Press; 1998. pp. 417–429. [Google Scholar]
- Broughton WJ, Jabbouri S, Perret X. Keys to symbiotic harmony. J Bacteriol. 2000;182:5641–5652. doi: 10.1128/jb.182.20.5641-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GRO, Reuhs BL, Walker GC. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci U S A. 2002;99:3938–3943. doi: 10.1073/pnas.062425699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Sibley CD, Zaheer R, Finan TM. A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology. 2007;153:375–387. doi: 10.1099/mic.0.2006/001362-0. [DOI] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeling S, Kundig C, Hennecke H. Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J Bacteriol. 1991;173:6373–6382. doi: 10.1128/jb.173.20.6373-6382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson GP, Datta A, Carlson RW, Walker GC. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol Microbiol. 2005;56:68–80. doi: 10.1111/j.1365-2958.2005.04536.x. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Foucher F, Kondorosi E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol Biol. 2000;43:773–786. doi: 10.1023/a:1006405029600. [DOI] [PubMed] [Google Scholar]
- Gage DJ. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol. 2002;184:7042–7046. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Margolin W. Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol. 2000;3:613–617. doi: 10.1016/s1369-5274(00)00149-1. [DOI] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci U S A. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. Embo J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Hengge-Aronis R. Regulation by proteolysis in bacterial cells. Curr Opin Microbiol. 2003;6:163–172. doi: 10.1016/s1369-5274(03)00029-8. [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski PA, Batut J, Boistard P. A survey of symbiotic nitrogen fixation by rhizobia. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae. Dordrecht, Boston, London: Kluwer Acad Press; 1998. pp. 431–460. [Google Scholar]
- Kain J, He GG, Losick R. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol. 2008;190:6749–57. doi: 10.1128/JB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Strahl H, Moliere N, Hamoen LW, Turgay K. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol. 2008;70:682–94. doi: 10.1111/j.1365-2958.2008.06438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Broughton WJ. Fine-tuning of symbiotic genes in rhizobia: Flavonoid signal transduction cascade. In: Dilworth MJ, James EK, Sprent JI, Newton WE, editors. Nitrogen Fixation: Origins, Applications, and Research Progress Volume 7: Nitrogen-fixing Legume Symbiosis. Springer; Netherlands: 2008. pp. 117–152. [Google Scholar]
- Kobayashi H, Sunako M, Hayashi M, Murooka Y. DNA synthesis and fragmentation in bacteroids during Astragalus sinicus root nodule development. Biosci Biotechnol Biochem. 2001;65:510–515. doi: 10.1271/bbb.65.510. [DOI] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Cosby WM, Zuber P. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset AE, Barloy-Hubler F, Galibert F, Kondorosi A, Kondorosi E. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci U S A. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Wanner BL. Construction of new beta-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- Miller JF. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1972. [Google Scholar]
- Oke V, Long SR. Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol. 1999;2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Cheng HP, Walker GC. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J Bacteriol. 2000;182:4310–4318. doi: 10.1128/jb.182.15.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H, Mendoza A, Cruz MA, Holguin G, Glick BR, Bashan Y. Pleiotropic physiological effects in the plant growth-promoting bacterium Azospirillum brasilense following chromosomal labeling in the clpX gene. FEMS Microbiol Ecol. 2006;57:217–225. doi: 10.1111/j.1574-6941.2006.00111.x. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R, Marczynski GT. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. Embo J. 2000;19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Grossman AD, Walker GC. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J Bacteriol. 2008;190:6758–6768. doi: 10.1128/JB.00590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J Bacteriol. 2003;185:115–125. doi: 10.1128/JB.185.1.115-125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.