Abstract
Mutations that enhance the response to double-stranded RNA (dsRNA) have revealed components of the RNA interference (RNAi) pathway or related small RNA pathways. To explore these small RNA pathways, we screened for Caenorhabditis elegans mutants displaying an enhanced response to exogenous dsRNAs. Here we describe the isolation of mutations in two adjacent, divergently transcribed open reading frames (eri-6 and eri-7) that fail to complement. eri-6 and eri-7 produce separate pre-messenger RNAs (pre-mRNAs)are trans-spliced to form a functional mRNA, eri-6/7. Trans-splicing of eri-6/7 is mediated by a direct repeat that flanks the eri-6 gene. Adenosine to inosine editing within untranslated regions (UTRs) of eri-6 and eri-7 pre-mRNAs reveals a double-stranded pre-mRNA intermediate, forming in the nucleus before splicing occurs. The ERI-6/7 protein is a superfamily I helicase that both negatively regulates the exogenous RNAi pathway and functions in an endogenous RNAi pathway.
RNAi pathways act throughout phylogeny as both an experimental gene-silencing tool and a regulator of endogenous gene expression1,2. The endonuclease Dicer3 and various Argonaute proteins act in specialized roles in most silencing pathways4. C. elegans Dicer interacts with negative regulators of exogenous RNAi5, such as the RNA-dependent RNA polymerase (RdRP) RRF-3 (ref. 6) and the exonuclease ERI-1 (ref. 7). These proteins are required for the production or stability of a subset of endogenous short interfering RNA (siRNAs) suggesting a competition with the exogenous RNAi pathway for shared, rate-limiting factors5,8.
RdRPs amplify siRNAs on mRNA templates in nematodes9-11, fungi and plants1. The feed-forward nature of RNAi and the still unexplained resistance of neurons to RNAi12-14 suggest that there are undiscovered negative regulators of RNAi. To identify such regulators, we conducted a genetic screen for mutations that confer an enhanced response to dsRNA. Here we describe the identification and characterization of adjacent genes, eri-6 and eri-7, which are assembled by a dsRNA-mediated trans-splicing mechanism to regulate RNAi negatively. The dsRNA-dependent production of an RNAi factor suggests that there may be an autoregulatory feedback mechanism in RNAi.
Characterization of eri mutants
To identify negative regulators of C. elegans RNAi, we genetically screened for mutants having an enhanced RNAi (Eri) phenotype. These mutants, unlike wild type, exhibited an enhanced response to green fluorescent protein (gfp) RNAi, downregulating expression of a neuronally expressed gfp transgene (unc-47::gfp)15, and an enhanced response to dsRNA targeting several non-neuronally expressed endogenous genes, including the collagen dpy-13 (ref. 16), the cadherin hmr-1 (ref. 17) and the transcription factor lin-1 (ref. 18). Thirteen of the 44 eri(-) mutants were also thermosensitive sterile and showed X-chromosome non-disjunction phenotypes that define genes that encode the DCR-1-interacting proteins RRF-3, ERI-1, ERI-3 and ERI-5 (refs 5-7). Among the mutations not in this class, mg379 and the independently isolated spontaneous eri allele mg411 enhance RNAi and silence a transgene (mgIs30) that confers a rolling locomotion (Rol) phenotype (Supplementary Fig. 2, and Supplementary Tables 1 and 2). Transgene silencing and exogenous RNAi are mediated by several common factors19, and all known eri(-) mutants silence transgenes6,7. mg379 and mg411, as well as other allele combinations, fail to complement (Supplementary Table 1 and Supplementary Information). mg411 and mg379 were mapped to a 1.7-megabase interval. The 330 genes in the interval were assayed for a loss-of-function phenotype of transgene silencing by using dsRNAs that target each gene20. Two dsRNA clones induced silencing of the mgIs30 transgene, phenocopying the mg411 and mg379 mutations. These dsRNAs target two adjacent, antiparallel open reading frames: C41D11.7, termed eri-7, and C41D11.1, termed eri-6 (Fig. 1a). These RNAi clones are specific for each gene and do not target the adjacent gene or any other gene in the genome. DNA sequencing identified mg411 as a missense mutation in C41D11.7 and mg379 as a 5′ splice donor site mutation in C41D11.1. DNA sequencing of mutants isolated in the genetic screen revealed five more mutations in C41D11.7: mg369, mg380, mg390, mg440 and mg442. An independently isolated deletion allele (mg441) removes eri-6 (Supplementary Fig. 3). The transgene-silencing phenotype of mutants with either eri-7(mg411) or eri-6(mg379) alleles was rescued by transformation of a 9.1-kilobase (kb) fragment that contains only C41D11.1 and C41D11.7 (Fig. 2a, b). The hypersensitivity to RNAi by feeding conferred by the mg411 allele was also rescued by the 9.1-kb fragment (data not shown).
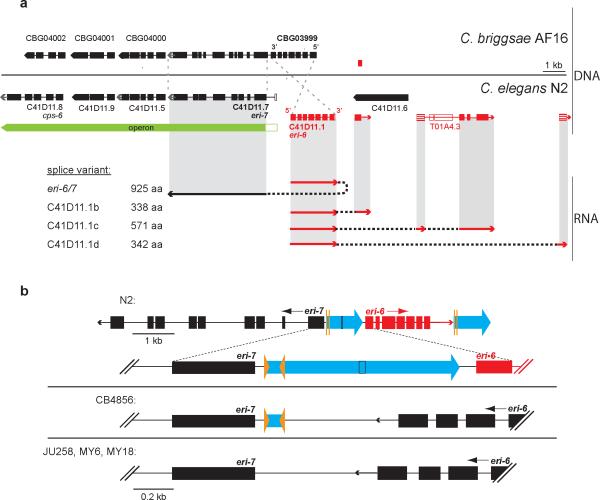
Figure 1. Theeri-6/7 locus in various strains and its gene products in C. elegans N2.
a, Structure of eri-6/7 in C. elegans N2 and the syntenic region in C. briggsae AF16, based on RT-PCR, 3″-RACE and WormBase21. The eri-7 operon is conserved. C. briggsae eri-6 is encoded on the opposite strand from C. elegans and is one gene with C. briggsae eri-7 (CBG03999). Exons are black or red rectangles, depending on the strand on which they are encoded. Open rectangles: predicted exons21, but unconfirmed. Striped exons: discovered experimentally. eri-6 splices to eri-7 24 nucleotidesupstream of the predicted21 eri-6 stop codon. b, Relative orientation of eri-6 and eri-7 in four C. elegans isolates. Blue arrows: the approximate 930-bp direct repeat flanking eri-6. Orange arrowheads: 25-bp inverted repeats.
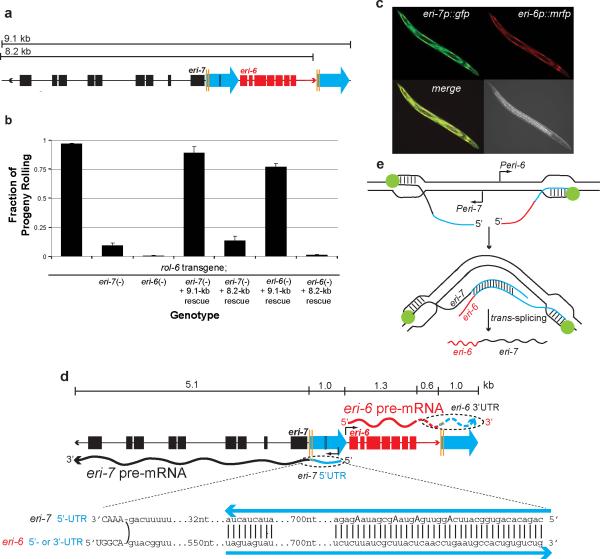
Figure 2. The eri-6/7 mRNA is formed by local trans-splicing.
a, Schematic of eri-6/7 locus used for rescue: 9.1-kb fragment; and 8.2-kb fragment missing the second repeat. b, Rescue of eri-6/7(mg411)-induced silencing of the mgIs30 transgene-conferred rolling phenotype. Number of broods scored = 28, 28, 18, 19, 38, 29 and 18, respectively. Multiple transgenic lines assayed. Error bars: s.e.m. c, The eri-6 and eri-7 promoters express in overlapping tissues (hypodermis and head neurons). eri-6 and eri-7 promoters were fused to rfp and gfp, respectively. d, 5′-RACE experiments show that eri-6 and eri-7 are expressed as separate pre-mRNAs with sufficient nucleotide homology to base-pair, thus facilitating trans-splicing. eri-7 pre-mRNA starts 775 nucleotides (nt) upstream of exon 1, whereas eri-6 pre-mRNA starts 100 nt upstream of eri-6 exon 1 (black arrows). Edited nucleotides (A to G indicative of A to I) in eri-7 pre-mRNA are in bold capital letters. Blue arrows: direct repeats. Capital letters: exon sequence. Curved line: trans-splicing juncture. e, Model of eri-6/7 trans-splicing. eri-6 and eri-7 pre-mRNAs are locally co-transcribed in the same cells. The pre-mRNAs may form a dsRNA intermediate that facilitates co-transcriptional trans-splicing.
Anomalous eri-6/7 gene structure
Although the lack of complementation suggests that eri-6(mg379) and eri-7(mg411) affect a single gene, the mutations were found in ostensibly separate genes on antiparallel strands21. However, the syntenic region in the nematode species C. briggsae surprisingly revealed that eri-6 and eri-7 constitute a single, contiguous gene in this species (Fig. 1a). The 5′ coding region of the C. briggsae gene CBG03999 is homologous to eri-6, whereas the 3′ coding region is homologous to eri-7. We verified the C. elegans genome structure at the eri-6 and eri-7 loci by Southern blotting (Supplementary Fig. 4), and by PCR and sequencing analyses (data not shown). We investigated closer relatives of the C. elegans wild-type N2 strain—27 other natural isolates of C. elegans. Four of these, including an isolate from Hawaii, C. elegans CB4856 (ref. 22), show a fused gene structure similar to that of C. briggsae (Fig. 1b).
eri-6 in the C. elegans strain N2 is flanked by a nearly perfect, approximate 930-base-pair (bp) direct repeat (Fig. 1b). Within this direct repeat is a 25-bp inverted repeat (Fig. 1b and Supplementary Fig. 5). The inverted repeat may have mediated the rearrangement responsible for the difference in eri-6 orientation relative to eri-7 in C. elegans N2 compared with C. elegans CB4856 (Fig. 1b). This hypothesis is supported by the spontaneous allele mg441 (Supplementary Fig. 3 and Supplementary Discussion), which deletes eri-6 in a fashion consistent with a rearrangement having occurred through the approximate 930-bp direct repeats that flank eri-6.
eri-6 and eri-7 are trans-spliced
Sequencing of the expressed sequence tag (EST) yk224h9, and reverse transcription-PCR with (RT- PCR), showed that the divergently transcribed RNAs for eri-6 and eri-7 are assembled into one mRNA in C. elegans strain N2 (Fig. 1a), encoding a protein orthologous to the C. briggsae eri-6/7 gene product. These proteins are members of the superfamily I DNA and RNA helicases (Supplementary Figs 6 and 7). Several eri-6/7 mutations encode amino-acid substitutions in conserved motifs in the ATPase and helicase domains (Supplementary Fig. 7). A transgene containing the eri-6 promoter, the full-length eri-6/7 complementary DNA (cDNA) and the eri-7 3′ UTR rescues both the eri-6(mg379) and eri-7(mg411) phenotypes (Supplementary Table 3).
In addition to the helicase mRNA, three other eri-6 splice variants were identified by EST analysis and 3′ rapid amplification of cDNA ends (RACE), in which eri-6 splices to different downstream exons (Fig. 1a), encoding proteins with no homology to known proteins. Neither the eri-6 mRNA nor the eri-7 mRNA could be detected as the separate mRNAs predicted from the gene structure.
We considered the following two mechanisms of chimaeric RNA formation: (1) trans-splicing of independently transcribed pre-mRNAs; and (2) a low level of DNA inversion in particular cells that would reposition the eri-6 and eri-7 genes in cis. The genomic rearrangement model was alluring because of the observation that such inversions at the eri-6/7 locus are apparent between and within related Caenorhabditis species. However, the rearrangement model is neither supported by Southern blotting (Supplementary Fig. 4) nor extensive PCR analyses designed to detect a cis orientation of eri-6 and eri-7 as a minority component of the genomic DNA of C. elegans N2 (Supplementary Discussion and Supplementary Fig. 8).
The generation of the eri-6/7 mRNA by trans-splicing is, however, supported by multiple experimental data. The trans-splicing juncture of the fused eri-6/7 mRNA uses canonical cis splice sites (Supplementary Fig. 9). Other eri-6 splice variants were detected 3′ of eri-6 and all include exons linked within 12 kb, in cis (Fig. 1a). Bioinformatic searches (Supplementary Discussion) of EST databases and 5′-RACE experiments did not detect distant trans-splicing of eri-6 to other mRNAs. Therefore, it is likely that for trans-splicing to occur, the eri-6 and eri-7 genes need to be in close proximity. Genes can also be juxtaposed through homologous chromosome pairing during meiosis23. Interchromosomal trans-splicing of eri-6 and eri-7 is unlikely to efficiently occur given (1) the lack of complementation observed in eri-7(+)eri-6(mg379)/eri-7(mg369)eri-6(+) animals and (2) sequence analysis of cDNAs cloned from these trans-heterozygotes (Supplementary Discussion).
A prerequisite for co-transcriptional trans-splicing is expression of both pre-mRNAs in the same cells at the same time. eri-7 promoter::gfp and eri-6 promoter::rfp transgenes are expressed in overlapping patterns, with co-expression in hypodermal cells and two pairs of sensory head neurons (ASK and ASI) (Fig. 2c). The eri-7 promoter also expresses in the somatic gonad.
The direct repeat flanking eri-6 (Fig. 1b) could constitute the 5′-UTR of the eri-7 pre-mRNA and, as a reverse complement copy, the eri-6 pre-mRNA 3′- or 5′-UTR. These complementary sequences could bring the pre-mRNAs into physical contact to facilitate trans-splicing. To determine the pre-mRNA sequences of eri-6 and eri-7, we stabilized them by attenuating splicing through RNAi inactivation of rnp-5, which encodes an orthologue of human RNPS1 (ref. 21), a member of the exon-junction complex. 5′-RACE analysis using intron-specific primers identified an eri-7 pre-mRNA 5′ end extending 725 bp into the eri-6/7 intergenic region (685 bp into the first direct repeat). An eri-6 transcription start site was found at up to 100 bp 5′ of the eri-6 start codon (Fig. 2d). We were unable to amplify the eri-6 3′-UTR, possibly because of repetitive, low complexity sequence immediately 3′ of the splice site (5′ of the second direct repeat). The splicing of eri-6 to independent exons located 1-11 kb 3′ of the second direct repeat strongly supports an eri-7 trans-splicing model whereby the eri-6 nascent transcript bears this repeat.
The significance of the second 930-bp direct repeat was tested in rescue experiments using an eri-6/7 (9.1-kb) genomic fragment that contained this repeat, compared with an (8.2-kb) fragment that did not (Fig. 2a, b). The 9.1-kb fragment completely rescued the transgene-silencing phenotypes of both the eri-6(mg379) and eri-7(mg411) alleles, whereas the 8.2-kb fragment only weakly rescued the eri-7 mutation. This nominal eri-6/7 gene activity could be the result of overexpression of eri-6 and eri-7 from the transgenic array bypassing the need for their base pairing, or of a low level of eri-6 transcription from an upstream start site, to include repeat sequence in the 5′-UTR that could mediate base-pairing.
Analysis of cDNA sequences derived from the eri-7 pre-mRNA stabilized in rnp-5 RNAi-treated animals revealed adenosine to guanosine transitions at four positions located within the direct repeat. These transitions are indicative of adenosine to inosine editing of the eri-7 5′-UTR by an adenosine deaminase (ADAR)24, supporting the model that the direct repeat element flanking eri-6 is bidirectionally transcribed to form an RNA duplex intermediate (Fig. 2e) before the two mRNAs are trans-spliced (Fig. 2d). Though ADARs can also edit secondary structures within single RNA strands, no such structure is predicted for the eri-7 5′-UTR.
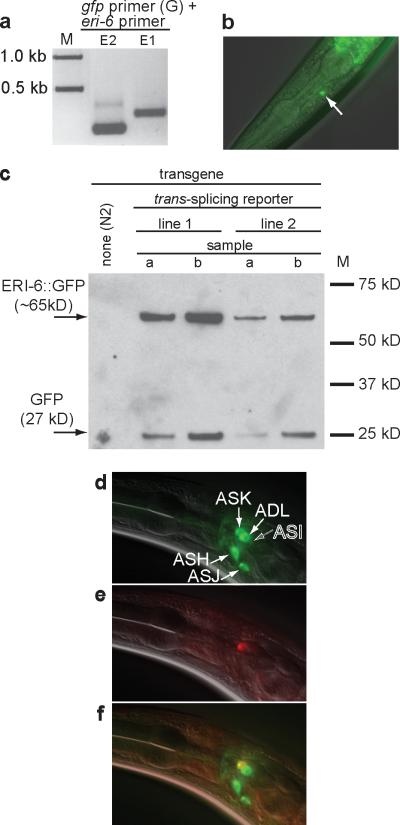
To show that the trans-spliced mRNA is translated into a chimaeric protein, we constructed a trans-splicing reporter in which all but the initial four nucleotides of eri-7 sequence are replaced by a gfp gene lacking a start codon (Supplementary Fig. 10). Sequencing of RT-PCR products (Fig. 3a) from GFP-expressing worms (Fig. 3b) showed that trans-splicing occurred from the transgene to yield an mRNA that consists of eri-6, the first two codons of eri-7, and gfp sequence. Western blotting showed that a protein of the size expected for the ERI-6::GFP fusion can be detected using a GFP antibody, indicating that the eri-6 and eri-7 trans-spliced mRNA is likely translated into a chimaeric ERI-6/7 helicase protein (Fig. 3c).
Figure 3. Chimeric ERI-6/7 protein is expressed through trans-splicing.
a, RT-PCR on gfp-expressing worms using primers in gfp (G) and eri-6 (E1 and E2). Sequencing of the RT-PCR products confirmed trans-splicing. b, Expression of the trans-splicing reporter in ASK. c, Western blot on animals expressing the trans-splicing reporter. A GFP antibody was used. The slower-migrating product (of the expected weight of ERI-6::GFP) is presumably ERI-6::GFP. The faster-migrating product may be GFP expressed using an alternative start codon. d, DiO fills five pairs of sensory neurons. e, ERI-6/7::mCherry is expressed in one amphid neuron. f, Merged image identifies ASK as the ERI-6/7-expressing neuron. d, e, f,
This reporter for eri-6/7 trans-splicing is expressed in the amphid neuron ASK (Fig. 3b) and in the somatic gonad, but not in the hypodermis where the promoter fusions are strongly expressed. A second transgene, bearing the complete eri-6/7 genomic region, fuses mCherry25 to the carboxy terminus of ERI-7 (Fig. 3d-f and Supplementary Fig. 11), and shows expression in ASK and the somatic gonad. One explanation for the lack of hypodermal expression from these trans-splicing-dependent fusion genes is that trans-splicing is inefficient or inhibited in the hypodermis. Alternatively, transcription or translation in the hypodermis does not occur because of sequence in the eri-6/7 gene or mRNA that is not present in the promoter fusion constructs. Interestingly, the eri-6/7trans-splicing reporter is more highly and broadly expressed when RNAi is attenuated by inactivation of the Argonaute gene rde-1 (Supplementary Fig. 12), suggesting that the eri-6/7 is a target of RNAi. A caveat to this result is that transgenes are generally expressed at higher levels when RNAi is defective. Quantitative RT-PCR revealed that eri-6/7 mRNA levels are not increased in animals lacking RDE-1 (data not shown). One explanation for these data is that the dsRNA element of eri-6/7 acts tissue-specifically as a sensor, and the changes in expression are too subtle to detect by quantitative RT-PCR on whole worms.
C. elegans has two ADAR genes: adr-1 and adr-2. The adr-1(-); adr-2(-) double mutant enhances transgene silencing, suggesting that editing of dsRNAs normally inhibits their sensitivity to RNAi26. Perhaps in the absence of ADARs, the unedited eri-6/7 dsRNA intermediate is a better target for the RNAi pathway, thus decreasing eri-6/7 gene activity. However, the level of trans-spliced eri-6/7 mRNA is unchanged in the adr-1(-); adr-2(-) double mutant as analysed by quantitative RT-PCR and by GFP fluorescence of the trans-splicing reporter after adr RNAi (data not shown), suggesting that editing of the double-stranded intermediate does not affect eri-6/7 trans-splicing.
Role of ERI-6/7 in RNAi
eri-6/7 alleles define a new class of Eri mutants. They lack the pleiotropic phenotypes (for example, sterility) typical of when function of the Dicer-interactors ERI-1 to ERI-5 is lost. In addition, eri-6/7(-) mutants do not show the characteristic synthetic multivulva phenotype that is associated with enhanced RNAi mutants of the retinoblastoma pathway27. We analysed the role of eri-6/7 in small RNA pathways. Like eri-1 alleles7, eri-7(mg369) confers an enhanced response (over wild type) to injected double-stranded siRNAs (unc-22) (Supplementary Table 1); however, we did not observe a significantly increased level of siRNAs corresponding to a target gene after exposure to dsRNAs as analysed by northern blotting (data not shown). These data suggest that eri-6/7 acts downstream of processing of the exogenous trigger-dsRNA into siRNAs. Genetic epistasis analyses (Supplementary Table 1 and data not shown) with mutants defective in RNAi (rde-1 (ref. 19), rde-4 (ref. 28), rrf-1 (ref. 11) and mut-7 (ref. 29)) show that, with the exception of the somatic RdRP gene rrf-1, normal function of these genes is required for enhanced and normal RNAi sensitivity. RRF-1 functions in the de novo production of secondary siRNAs after the initial cleavage of the exogenous dsRNA into primary siRNAs by Dicer, RDE-1 and RDE-4 (ref. 11). Unlike rrf-1(-) mutants, rrf-1(-) eri-7(-) double mutants are partly proficient in somatic RNAi targeting hmr-1 and unc-22, suggesting that eri-6/7 negatively acts both in rrf-1-independent RNAi (through primary siRNAs) and in rrf-1-dependent RNAi. These data are consistent with a role of ERI-6/7 downstream of, and dependent on, the siRNA production mediated by RDE-1 and RDE-4. Null alleles of eri-1 or rrf-3, when combined with eri-6/7 null alleles, did not enhance the RNAi sensitivity compared with the single mutants, suggesting that eri-6/7 may act in the same pathway as eri-1 and rrf-3 to enhance RNAi (data not shown).
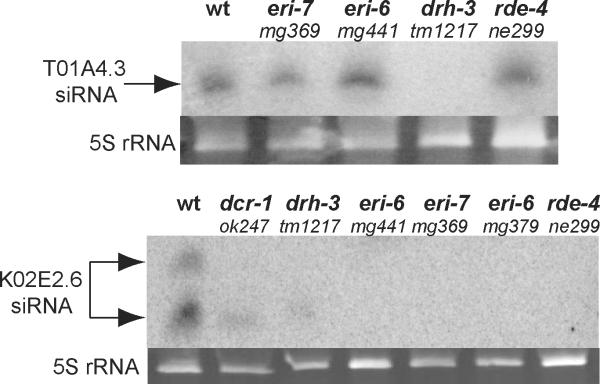
Silencing in response to exogenous dsRNA is increased when certain endogenous RNAi pathways, such as the pathway dependent on eri-1, are defective5. We investigated a role for ERI-6/7 in endogenous RNAi by analysing endogenous siRNA (endo-siRNA) abundance in eri-6/7 mutant worms by northern blotting (Fig. 4). eri-6/7 is required for endo-siRNAs derived from the gene K02E2.6 but not for a Dicer-independent endo-siRNA matching the gene T01A4.3 (Fig. 4), which is thought to be germline-expressed5. ERI-6/7 protein shares these attributes with the DCR-1-interacting proteins ERI-1, RRF-3 and RDE-4 (refs 5, 8) (Fig. 4), as well as with the Argonaute ERGO-1 (ref. 30).
Figure 4. ERI-6/7 is required for endogenous RNAi.
Shown are northern blots probed for two different endogenous siRNAs, targeting the genes T01A4.3 and K02E2.6. Mutants of additional genes known to be required for endogenous RNAi (dcr-1, drh-3 and rde-4) are shown as controls. 5S rRNA is shown as a loading control.
Conclusions
eri-6/7 encodes an RNAi factor that is assembled in C. elegans N2 through trans-splicing via a dsRNA intermediate. The trans-splicing of eri-6 and eri-7 is only observed when the two genes are on the same chromosome, suggesting that it occurs on nascent transcripts. The eri-6/7 double-stranded, premRNA intermediate is edited by ADARs when splicing is inhibited, and may be itself regulated by RNAi.
ERI-6/7 protein is a member of the superfamily I helicases, like the RNAi factors Mov10 in mammals31, SDE3 in Arabidopsis32 and Armitage in Drosophila33. Superfamily I helicases act on either RNA or DNA. Because ERI-6/7 is required for endo-siRNA production or stability, it could function as an RNA helicase in siRNA generation. Our expression data showing exclusive or predominant cytoplasmic expression of a full-length ERI-6/7 protein fused to a fluorescent marker (Supplementary Fig. 11) suggest that it acts as an RNA helicase.
Loss of eri-6/7 causes enhanced exogenous RNAi. Lee et al. (2006) and Duchaine et al. (2006) have proposed that other ERI proteins bridle exogenous RNAi indirectly by competing for factors common to both the endogenous and exogenous RNAi pathways, suggesting that the normal role of ERI-6/7 is either to promote endogenous small RNA pathways or to attenuate exogenous RNAi directly. The loss of some endogenous siRNAs in eri-6/7 mutants is consistent with the former function.
This is the first example of exon-to-exon trans-splicing in C. elegans, and only the third finding of a locus that requires such trans-splicing for the production of a functional protein. The other two examples were reported in Drosophila: mod(mdg4)34,35 and lola36. Each of these genes encodes BTB- zinc-finger transcription factors that achieve extensive protein diversity through alternative trans-splicing. The requirement of proximity between the trans-spliced exons in these cases is not clear35,36.
The polymorphic variation of the eri-6/7 locus between related Caenorhabditis species, and especially between different isolates of the C. elegans species, is remarkable. The inserted repetitive sequence has likely segmented the eri-6/7 gene into two in C. elegans N2 through recombinational events. The ability of the repeat to mediate homology-based trans-splicing, and its bidirectional promoter activity (allowing independent eri-6 and eri-7 expression in N2), have provided a compensatory means of bringing the two fragments together again to encode a composite eri-6/7 mRNA. Beyond the structure of the eri-6/7 locus, the ERI-6/7 proteins seem to have also diverged substantially between C. elegans and C. briggsae. Whereas the mean amino-acid identity between orthologues is 75% (ref. 37), ERI-6/7 shows only 44% identity to its fused orthologue in the related nematode C. briggsae. This is consistent with the rapid evolution that is seen in antiviral RNAi genes38, and with the finding that other C. elegans eri genes are anti-viral39,40.
METHODS SUMMARY
C. elegans was cultured using standard techniques41 and fed on Escherichia coli OP50 or on E. coli HT115 harbouring a dsRNA expressing plasmid (RNAi by feeding).
C. elegans animals were mutagenized using ethylmethanesulphonate. Mutants showing an enhanced RNAi phenotype were selected after gfp RNAi in the F2 and F3 progeny of mutagenized animals carrying a transgene expressing GFP in a subset of neurons. The enhanced RNAi phenotype was further analysed using RNAi to the endogenous genes hmr-1, lin-1 and dpy-13. In addition, several mutants were assayed for their ability to silence the transgene mgIs30 (lim-6::gfp, col-10::lacZ::lin-14, rol-6(su1006)::lin-14 3′-UTR), scoring the Rol phenotype conferred by this transgene. unc-22 siRNA injections were done as described previously7,42.
Mutations were single nucleotide polymorphism-mapped using the Eri and transgene silencing phenotypes. Subsequently, all genes in the mapping interval were inactivated by RNAi and tested for transgene silencing. Inactivation of two genes silenced the transgene mgIs30; these genes were sequenced in the mutants.
Transgenic worms for rescue experiments, expression analysis as well as animals carrying the reporter for trans-splicing were made by microinjection of PCR fragments, cosmids or plasmids. Promoter and full-length fusions to fluorescent proteins were made by splicing by overlapping extension PCR of gfp, mrfp or mCherry to the promoter or in frame with the open reading frame. Western blotting of the eri-6::gfp reporter was done using an anti-GFP antibody.
The gene structure or eri-6/7 in wild-type C. elegans N2, other C. elegans isolates and C. briggsae was confirmed by PCR, sequencing and/or Southern blotting.
eri-6/7 transcripts were analysed by EST sequencing analysis, RT-PCR, 5′- and 3′-RACE. cDNA was cloned and sequenced. Pre-mRNAs and transcriptional start sites were analysed in animals that were deficient in splicing by RNAi of rnp-5.
Endogenous siRNA abundance in mutant animals was assayed by northern blotting42 using StarFire probes5.
METHODS
Isolation of enhanced RNAi mutants
unc-47::gfp worms were mutagenized with ethyl methanesulphonate, and both the F2 and F3 generations were cultured on lawns of E. coli expressing dsRNA that targeted gfp. Mutagenized populations were then screened for rare individuals showing a marked decrease in the number of γ-aminobutyric acid (GABA)-ergic neurons visibly expressing GFP under the dissecting microscope, screening through approximately 50,000 haploid genomes. We identified 44 mutants that show an enhanced exogenous RNAi phenotype. To ensure isolated mutants were Eri and to test for generally enhanced RNAi, we cultured them on E. coli targeting endogenous genes that were expressed in a variety of tissues, asking that they be hypersensitive compared with unc-47::gfp worms to at least two of the following three testers: dpy-13, hmr-1 and lin-1, which result in dumpy, embryonic lethal and sterile, and multivulva phenotypes, respectively. Thermosensitive sterility was assayed by placing L1 worms at 25 °C and scoring for unfertilized eggs in the next generation. The 31 non-sterile mutants were in at least five complementation groups.
Mapping, genetic and expression analyses
eri-6(mg379) and eri-7(mg411) were mapped using a highly polymorphic C. elegans isolate, CB4856. Recombinants generated with either outcrossed strains carrying the mg379 or the mg411 alleles and CB4856 were assayed for their RNAi hypersensitivity phenotype or their ability to silence a rol-6 transgene (mgIs30 [rol-6(su1006)::lin-14 3′-UTR, col-10::lacZ, lim-6::gfp]), respectively, and subsequently genotyped for single nucleotide polymorphisms that exist between the wild-type and CB4856 strains. mg411 was placed into a 3.8-map-unit interval on chromosome I using the transgene silencing phenotype, whereas mg379 was placed into a 4.6-map-unit interval using Eri phenotype lin-1(RNAi). Next, mgIs30 was used to identify genes that cause transgene silencing using feeding RNAi targeting the open reading frames in the mapped mg411 interval. Two open reading frames tested positive: C41D11.1 and C41D11.7. Sequencing of mg411 and mg379 identified the following mutations: mg411 A→T at position 33105 of C41D11, and mg379 G→A at position 34755 of cosmid C41D11. Five additional alleles were obtained by sequencing the eri-6/7 locus in mutants from the ethyl methanesulphonate Eri screen. An eighth, presumably spontaneous, allele (see Supplementary Discussion), eri-6/7(mg441), was isolated by mating a serendipitously obtained Eri strain in the lab with eri-6/7 mutants to ask for non-complementation, followed by sequencing of the eri-6/7 gene.
Rescue experiments were done with the C41D11 cosmid at 5 μg ml-1, and PCR fragments at 5 μg ml-1. Promoter::gfp or mrfp fusions, and the trans-splicing reporter construct, were injected at 10 μg ml-1. Full-length ERI-6/7::mCherry fusion was injected at 5 μg ml-1. DiO was used to identify dye-filling amphid sensory neurons.
Enhanced RNAi assays
unc-22, 23-bp ds-siRNA injections were done as previously described43. The ds-siRNA was injected at 5 mg ml-1. The percentage of F1 twitchers was scored in 330 μM levamisole. Feeding RNAi assays were done at 20 °C. For lin-1(RNAi) and unc-73(RNAi), starved L1 s were placed on E. coli expressing the dsRNA. The next generation was scored for percentage displaying Muv or Pvl (lin-1), or coiler (unc-73) phenotypes. For cel-1(RNAi) and hmr-1(RNAi), worms that were exposed to RNAi starting at the L1 stage were scored as adults for the production of viable progeny. In complementation assays, heterozygotes of mg369/mg379 and mg411/tm1887 were tested for an Eri phenotype based on hypersensitivity to dsRNA targeting hmr-1.
cDNA analysis, 3′- and 5′-RACE
RNA was isolated from mutant worms or worms exposed to RNAi for several generations, using TRI Reagent (Molecular Research Center). RNA was treated with TurboDNase (Ambion) and cDNA was made using the RETROscript kit (Ambion). For 3″-RACE, the First Choice RLM-RACE kit (Ambion) was used; for 5″-RACE the SMART RACE kit (Clontech) was used. 3″- and 5″-RACE products were cloned using Qiagen PCR cloning kit. RT-PCR on worms expressing the trans-splicing reporter was done by hand-picking GFP-expressing worms for RNA isolation. RT-PCR was done using a primer in gfp combined with primers in eri-6. No PCR products were obtained with these primers using genomic DNA of GFP-expressing worms.
Western blotting
To enhance transgene expression, nematodes carrying the trans-splicing reporter were subjected to rde-1 or mut-16 RNAi by feeding. This resulted in brighter and broader expression of GFP. GFP-expressing worms from two independent transgenic lines were hand-picked into sample buffer. Nontransgenic N2 worms served as a negative control. Electrophoresis and western blotting were done according to standard methods. A monoclonal antibody to GFP (Clontech) was used at 1:10,000. A horseradish peroxidase-conjugated goat anti-mouse antibody was used as a secondary antibody.
Northern blotting
Northern blotting for endo-siRNAs was done as previously described42 (http://chronic.dartmouth.edu/VRA/ambroslab.html) using 30 μg of RNA in 12% denaturing polyacrylamide gels. Prehybridization and hybridization were done in 7% SDS, 0.2 M Na-phosphate pH 7.0 at 35 °C. StarFire probe sequences were as described5.
Supplementary Material
Acknowledgements
We thank: S. Kennedy for advice and initiating the enhanced RNAi screen; M. Finney for advice on advanced PCR; C. Zhang for mg441 Eri characterization; Y. Kohara, D. Thierry-Mieg and J. Thierry-Mieg for EST sequences and clones; the Mitani laboratory for deletion strains; the Caenorhabditis Genetics Center for strains; Ruvkun laboratory members for reading the manuscript and many discussions; the laboratories of J. Kaplan and F. Ausubel for discussions; and the Leukemia and Lymphoma Society, and EMBO for funding to S.E.J.F.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Mello CC, Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Parker JS, Barford D. Argonaute: a scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 6.Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alder MN, Dames S, Gaudet J, Mango SE. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 11.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 12.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 13.Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nature Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 14.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 15.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 16.von Mende N, Bird DM, Albert PS, Riddle DL. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988;55:567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- 17.Costa M, et al. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beitel GJ, Tuck S, Greenwald I, Horvitz HR. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 1995;9:3149–3162. doi: 10.1101/gad.9.24.3149. [DOI] [PubMed] [Google Scholar]
- 19.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 21.WormBase. web site. < http://ws170.wormbase.org/> WS170 (9 February 2007)
- 22.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Muse T, Boulton SJ. Meiotic recombination in Caenorhabditis elegans. Chromosome Res. 2007;15:607–621. doi: 10.1007/s10577-007-1146-x. [DOI] [PubMed] [Google Scholar]
- 24.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 26.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 28.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 29.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 30.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 31.Meister G, et al. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 32.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomari Y, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 34.Labrador M, et al. Protein encoding by both DNA strands. Nature. 2001;409:1000. doi: 10.1038/35059000. [DOI] [PubMed] [Google Scholar]
- 35.Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiuchi T, Giniger E, Aigaki T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 2003;17:2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felix MA. Genomes: a helpful cousin for our favourite worm. Curr. Biol. 2004;14:R75–R77. [PubMed] [Google Scholar]
- 38.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 39.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 40.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiernagle T. the C. elegans research community, editor. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. doi/10.1895/wormbook.1.101.1 < http://www.wormbook.org>. [DOI] [PMC free article] [PubMed]
- 42.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- 43.Caplen NJ, et al. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA mediated RNA interference. Hum. Mol. Genet. 2002;11:175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.