Abstract
The effects on the structure and function of amicyanin of replacing the axial methionine ligand of the type 1 copper site with leucine have been characterized. The crystal structures of the oxidized and reduced forms of the protein reveal that the copper site is now tri-coordinate with no axial ligand, and that the copper coordination distances for the two ligands provided by histidines are significantly increased. Despite these structural changes, the absorption and EPR spectra of M98L amicyanin are only slightly altered and still consistent with that of a typical type 1 site. The oxidation-reduction midpoint potential (Em) value becomes 127 mV more positive as a consequence of the M98L mutation, most likely due to increased hydrophobicity of the copper site. The most dramatic effect of the mutation was on the electron transfer (ET) reaction from reduced M98L amicyanin to cytochrome c-551i within the protein ET complex. The rate decreased 435-fold, which was much more than expected from the change in Em value. Examination of the temperature dependence of the ET rate (kET) revealed that the mutation caused a 13.6 fold decrease in the electronic coupling (HAB) for the reaction. A similar decrease was predicted from a comparative analysis of the crystal structures of reduced M98L and native amicyanins. The most direct route of ET for this reaction is through the Met98 ligand. Inspection of the structures suggests that the major determinant of the large decrease in the experimentally determined values of HAB and kET is the increased distance from the copper to the protein within the type 1 site of M98L amicyanin.
Copper proteins are common in nature and involved in a variety of biological processes such as respiration, photosynthesis, and redox reactions critical to metabolism (1). The copper centers are classified according to their spectroscopic properties as type 1, type 2 or type 3. Type 1 copper sites are found in a wide range of electron transfer (ET) proteins, including amicyanin and azurin in bacteria, plastocyanin in plants, and multicopper oxidases such as fungal laccase and human ceruloplasmin (1). The type 1 copper site of cupredoxins consists of three relatively strong equatorial ligands, nitrogens of two histidines and sulfur of a cysteine, forming a trigonal plane. A weaker fourth axial ligand is usually, but not always, provided by a sulfur of a methionine (2). Azurin is atypical in that additional coordination to the copper is provided by the backbone carbonyl O of a Gly, giving that copper site a trigonal bipyramidal geometry. In amicyanin from P. denitrificans the three strong equatorial copper ligands are provided by residues His53, His95 and Cys92, and the weak axial ligand is provided by Met98 (3).
Amicyanin (4) serves as a mediator of ET from methylamine dehydrogenase (MADH) (5) to cytochrome c-551i (6). These three proteins are isolated from P. denitrificans as individual soluble proteins but they have to form a ternary complex to catalyze methylamine-dependent cytochrome c-551i reduction (7, 8). The complex of MADH, amicyanin, and cytochrome c-551i is one of the best characterized physiological protein ET systems (9). The proteins have been structurally characterized by x-ray crystallography as the binary complex of MADH and amicyanin and the ternary protein complex, which includes cytochrome c-551i (10). The protein complexes were shown to be catalytically active and able to perform ET in the crystalline state (11). Although it is a thermodynamically favorable reaction, MADH does not reduce cytochrome c-551i in the absence of amicyanin. Furthermore, reduced amicyanin does not reduce oxidized cytochrome c-551i in the absence of MADH at physiologic pH because the oxidation-reduction midpoint potential (Em) value of free amicyanin is much more positive than that of the cytochrome (12). The redox properties of amicyanin are altered on complex formation with MADH so as to facilitate the reaction by lowering the Em value of amicyanin in the complex under physiological conditions (12). ET reactions can be classified as true, gated, or coupled (9, 13). In a true ET reaction, the rate-limiting reaction step is the ET event. Thus, the observed rate is the same as the true rate of ET (kET). ET from Cu(I) of amicyanin to the heme of cytochrome c-551i in the ternary protein complex has been studied in solution by stopped-flow spectroscopy and shown to be a true ET reaction. Analysis of the temperature and ΔGo-dependence of true ET reactions by ET theory (14) yields values for the reorganization energy (λ) and electronic coupling (HAB) which determine the observed ET rates. Using site-directed mutagenesis we have previously altered specific amino acid residues of amicyanin that can modulate its Em value and alter ET rates within the MADH-amicyanin-cytochrome c-551i complex by altering values of ΔGo, HAB, and λ. We have also generated mutations that alter the kinetic mechanisms of these ET reactions and convert true ET reactions to coupled or gated ET reactions (reviewed in (9)).
In previous studies, Met98 which provides the distal axial copper ligand, was converted to Gln and Ala (15, 16). In M98Q amicyanin, the side chain O of the Gln provided the axial copper ligand, but the site exhibited rhombic distortion which was correlated with an increase in λ for the ET from MADH (16). It had been anticipated that the copper site of M98A amicyanin would lack the fourth ligand; however it still possessed a type 1 copper site with a water molecule providing an axial ligand. This M98A amicyanin exhibited similar redox and ET properties to that of native amicyanin (15).
In the present study the axial ligand of amicyanin was converted to leucine by site-directed mutagenesis. It was expected that the leucine cannot provide a ligand as its side-chain does not possess a heteroatom and its side-chain size would prevent water from occupying the site. This mutation is also of interest as there are naturally occurring type 1 copper sites with a leucine in the position typically occupied by methionine. These have been structurally characterized only in multicopper oxidases (17) such as laccase (18) and ceruloplasm (19, 20), although alignment of deduced amino acid sequences indicates that this feature may also be present in certain plantacyanins (21). This study describes the effects of an M98L mutation on amicyanin’s structure, visible and EPR spectra, Em value, and kET, λ and HAB associated with its ET reaction to cytochrome c-551i.
Experimental Procedures
Protein Expression and Isolation
MADH (22), amicyanin (4), and cytochrome c-551i (6) were purified from P. denitrificans as previously described and protein concentrations were calculated using known extinction coefficients for oxidized amicyanin (ε595 = 4610 cm−1 M−1), MADH (ε440 = 26,200 cm−1 M−1), and cytochrome c-551i (ε409 = 102,000 cm−1 M−1).
Site-directed mutagenesis was performed to create M98L amicyanins on double stranded pMEG (23), which contains mauC that encodes amicyanin, using forward and reverse mutagenic primers with the Quikchange® Site-directed Mutagenesis Kit (Strategene). The oligonucleotide sequences used to construct the site-directed mutant were 5′-GACTATCACTGTACCCCGCATCCCTTCCTTCGCGGCAAGGTCG-3′ and its complementary strand of DNA. The underlined bases are those that are changed to create the desired mutation. The entire 555-base mauC-containing fragment was sequenced to ensure that no second site mutations were present and none were found. M98L amicyanin was expressed in BL21 (E. coli) cells and isolated from the periplasmic fraction as described for other recombinant amicyanin mutants (23).
Protein Reconstitution
It was previously observed that the as-isolated M98Q and M98A amicyanins did not have full occupancy of copper in the type 1 site. This was in part due to the presence of zinc (24). An additional procedure (25) was required to unfold the protein, remove any metal ions, and incorporate Cu2+ into the refolded protein. M98L amicyanin was incubated in 10 mM HEPES buffer, pH 8.0, containing 6 M guandinine-HCl, 50 mM EDTA and 2 mM dithiothreitol (DTT). The unfolded apo-protein was diluted 50-fold into 10 mM HEPES buffer, pH 8.0, containing 5 mM DTT. The protein was then dialyzed against 100 mM ammonium acetate, pH 8.0, at 4°C for 4 hr, followed by dialysis against 250 mM ammonium acetate, pH 8.0, overnight. The dialyzed protein solution was then brought to room temperature and titrated against copper sulfate. Copper incorporation was monitored by the increase in visible absorbance and excess copper sulfate was removed by buffer exchange and ultrafiltration. The extinction coefficient of the reconstituted M98L amicyanin was determined by anaerobic reductive titration of the fully oxidized protein with a solution of sodium dithionite which has been standardized by titration with native amicyanin.
Determination of Em values
The Em values of M98L amicyanin free and in complex with MADH in solution were determined by spectrochemical titration as described previously with the native protein (12). The ambient potential was measured directly with a redox electrode which was calibrated using quinhydrone (a 1:1 mixture of hydroquinone and benzoquinone) as a standard with an Em value of +286 mV at pH 7.0. The reaction mixture contained 130 μM amicyanin in 50 mM BisTris propane buffer at the indicated pH, at 25 °C. The mixture was titrated by addition of incremental amounts of a reductant, ascorbate, or an oxidant, ferricyanide. The absorption spectrum of M98L amicyanin was recorded at different potentials and the concentrations of oxidized and reduced M98L amicyanin were determined by comparison with the spectra of the completely oxidized and reduced forms. The data were analyzed according to eq 1 to determine Em values.
| (1) |
For determination of the Em value of M98L amicyanin in complex with MADH, the reaction mixture contained 20 μM M98L amicyanin and 60 μM MADH in 50 mM BisTris propane buffer at pH 7.0, at 25 °C. Potassium ferricyaninde (400 μM), quinhydrone (200 μM), and phenanine ethosulfate (20 μM) were present as mediators. The absorption spectrum of the complex was recorded at different potentials and the concentrations of the oxidized and reduced forms were determined by comparison with the spectra of the completely oxidized complex and a mixture of oxidized MADH with fully reduced amicyanin. Data were then analyzed according to eq 1.
Kinetic Studies of Electron Transfer Reactions
The rates of ET reactions from reduced amicyanin to oxidized cytochrome c-551i in the ternary protein complex were determined using an On-Line Instruments (OLIS, Bogard, GA) RSM stopped-flow rapid scanning spectrophotometer as described previously (8). Experiments were performed in 10 mM potassium phosphate, pH 7.5. Prior to mixing, one stopped flow syringe contained oxidized cytochrome c-551i while the other contained reduced MADH plus reduced amicyanin, so as to ensure that MADH and amicyanin are in complex before the reaction with cytochrome c-551i (8). The concentration of the MADH/amicyanin complex is varied and in excess of a fixed concentration of cytochrome c-551li to maintain pseudo-first-order conditions. As ET from reduced amicyanin to cytochrome c-551i will occur only when the amicyanin is in complex with MADH (7), the concentration of amicyanin in complex [A]bound is calculated according to eq 2, where [M]total is the total concentration of MADH or amicyanin, which are present in equal concentration. Absorbance changes were monitored between 530 and 570 nm to determine the rate of reduction of cytochrome c-551i. The data were fit to the simple kinetic model in eq 3 using eq 4.
| (2) |
| (3) |
| (4) |
Analysis of Electron Transfer Reactions by Electron Transfer Theory
Values for k3 for each ET reaction were obtained at different temperatures and the temperature dependence of k3 was analyzed using eq 5 (14), where λ is the reorganization energy; HAB is the electronic
| (5) |
coupling, h is Plank’s constant, T is temperature, R is the gas constant, and ko is the characteristic frequency of nuclei (1013 s−1), which is the maximum ET rate when donor and acceptor are in van der Waals’ contact and λ=−ΔGo. ΔGo is determined ΔEm value using eq 6 where F is the Faraday constant and n is the number of electrons transferred.
| (6) |
Steady-state kinetics
The assay mixtures for steady-state kinetic experiments of the methylamine-dependent reduction of cytochrome c-551i by the MADH-amicyanin complex contained 18 nM MADH, 21μM cytochrome c-551i and varying concentrations of amicyanin (26). Each reaction was initiated by the addition of 0.1 mM methylamine. Activity was monitored by the change in absorbance due to the reduction of the cytochrome c-551i. Assays were performed in 10 mM potassium phosphate at pH 7.5, and the data were fit to eq 7.
| (7) |
Electron Paramagnetic Resonance Spectroscopy
The copper center of M98L amicyanin (270 μM) in 10 mM potassium phosphate at pH 7.4 containing 5% glycerol was analyzed by EPR spectroscopy on a Bruker (Karlsruhe, Germany) X-band spectrometer equipped with an ER 4119HS high-sensitivity cavity between 4–20 K using an Oxford Instruments low temperature system. For comparison, a sample of native amicyanin was subjected to the identical reconstitution procedure (discussed above) that was applied to M98L amicyanin. These EPR spectra were obtained with 0.2 mW microwave power and 3 G modulation amplitude.
Structure Determinations by X-ray Crystallography
Prior to crystallization trials, the protein was dialyzed against 5 mM sodium monobasic/potassium dibasic phosphate buffer at pH 6.6. Attempts to crystallize M98L amicyanin by following the previously used protocols for native and several other mutant amicyanins (3, 15) failed to yield crystals. Therefore, M98L amicyanin was screened for wide range of conditions using various Hamption screens (Hampton Research, Aliso Vieja, CA, USA). Finally, the crystals of M98L amicyanin were grown by the sitting drop method using 2 μl protein (11 mg/ml) with 2 μl 3.2 M ammonium sulfate in 90 mM Tris at pH 8.0 and 10 mM NaCl. The crystals were grown in 1–2 weeks time. The size of the crystals was ~50× 25×30 microns. Attempts to grow larger crystals using macro-seeding did not succeed. But, these small crystals yielded data to 2.1 Ǻ resolution when tested in the 24ID beamline equipped with Microdiffractometer- MD2 at the NE-CAT, Advance Photon Source using an ADSC Quantum 315 CCD detector. The data for oxidized M98L amicyanin were collected near the absorption edge of the copper. The M98L amicyanin crystallized in a new hexagonal space group P6322, compared to that of native and several other mutant amicyanins solved earlier. The data were integrated, scaled and merged using HKL2000 (27). The reduced M98L amicyanin was prepared by transferring an oxidized crystal to the mother liquor containing an additional component of 8 mM sodium ascorbate and soaking for ~12 min. Reduced M98L amicyanin diffracted to 2.2 Å resolution. Oxidized and reduced crystals were flash-frozen in the liquid nitrogen using frombin as a cryo-protectant. No bleaching of color of the oxidized crystal from the blue Cu(II) to the colorless Cu(I) was observed during data collection indicating that reduction did not occur in the x-ray beam. This is consistent with previous studies of native and other mutants of amicyanin solved previously using data collected with synchrotron radiation which did not show any x-ray induced radiation damage.
The structure of oxidized M98L amicyanin was solved by molecular replacement using the program PHASER (28) of PHENIX (29). A 1.3 Å resolution crystal structure of the native amicyanin (PDB code 1AAC) was used as an initial model, with residue Met98 trimmed at the Cβ atom. A difference Fourier map (Fo-Fc) was then computed using coordinates of the refined native amicyanin model which resulted in clear electron density for leucine at residue 98, and subsequently it was changed to Leu98. For reduced M98L amicyanin, the completely refined oxidized M98L amicyanin was directly submitted to rigid body and positional refinement procedures using the program PHENIX. For cross-validation, about 10% of all reflections were set aside from refinement in order to calculate a free R-factor (30) for the oxidized state (1198 reflections) and reduced state (1068 reflections). No restraints were applied to the copper, to ligand distances, or bond angles. The models were subjected to positional and B-factor refinement alternatively which brought down the R and Rfree to the mid-twenties. Subsequently, water molecules were added to the models using PHENIX. The models were checked and built using COOT (31). A simulated annealing omit map around the copper region was performed for both the oxidized and reduced structures to remove the model bias. A total of 82 and 72 water molecules were added to oxidized and reduced structures, respectively. There was significant negative density at the Cu(II) position of the oxidized structure, and subsequently a copper occupancy of 0.66 was estimated by alternative cycles of B-factor and occupancy refinement (32). No such negative density was observed in the reduced structure even though it was generated by reduction of the oxidized crystal. The final R and Rfree of oxidized and reduced M98L amicyanin are 18.5, 21.4 and 18.4, 20.0, respectively. The final models of oxidized and reduced M98L mutant contain phosphate, chloride, zinc, acetate molecules in addition to copper. The average isotropic temperature factor for the oxidized state is 36.0 Ǻ2 for all protein atoms and 44.0 Ǻ2 for water molecules while 34.0Ǻ2 and 40.4 Ǻ2, respectively, for the reduced state. The data collection, refinement and model statistics are summarized in Table 1. In the Ramachandran plot calculated using PROCHECK (33), all the non-glycine residues for both the oxidized and reduced state are in most-favored/additional allowed regions. The rms deviations were calculated using COOT (31), CCP4MG (34) and CCP4 (35).
Table 1.
Data Collection, Refinement and Model Statistics for M98L Amicyanin Structuresa
| M98L amicyanin oxidized | M98L amicyanin reduced | |
|---|---|---|
| Data Collection | ||
| Wavelength (Å) | 1.3808 | 0.98 |
| Space group | P6322 | P6322 |
| Unit Cell Dimension | ||
| a (Å) | 99.6 | 99.8 |
| b (Å) | 99.6 | 99.8 |
| c (Å) | 70.4 | 70.7 |
| resolution limit | 50-2.1 (2.18-2.1) | 50-2.2 (2.28-2.2) |
| beamline | NE-CAT 24ID-C | NE-CAT 24ID-E |
| I/Sigma(I) | 24.8 (2.9) | 34.8 (6.2) |
| Rmerge (%) | 10.2 (57.9) | 9.1 (58.5) |
| Completeness (%) | 99 (96.2) | 99.9 (99.9) |
| Redundancy | 14.3 | 20.3 |
| Refinement | ||
| resolution range (Å) | 50-2.1 | 37-2.2 |
| R-factor (%) | 18.5 | 18.4 |
| Rfree (%) | 21.4 | 20.0 |
| R-factor (Work + Test sets) (%) | 18.8 | 18.6 |
| No. of Reflections | 11953 | 10632 |
| Model | ||
| No. of amino acids | 105 | 105 |
| No. of water molecules | 82 | 72 |
| No. of Copper | 1 | 1 |
| No. of Zn | 1 | 1 |
| No. of Chloride | 1 | 1 |
| No. Acetate | 1 | 1 |
| No. of phosphate | 1 | 1 |
| Average B-factor (Ǻ2) for protein atoms | 36.0 | 34.0 |
| Residues in generously allowed regions | 0 | 0 |
| Residues in disallowed regions | 0 | 0 |
| No. of atoms with zero occupancy | 6 | 4 |
| Stereo chemical ideality | ||
| bonds (Å) | 0.007 | 0.008 |
| angles (°) | 1.005 | 1.11 |
| dihedral angles (°) | 15.95 | 14.04 |
Values in parentheses are for the outer shell
Analysis of crystal structures for relative values of atomic packing density (ρ), and HAB for ET reactions was performed using the HARLEM computer program (36) to apply the Dutton model for protein ET (37) and the Pathways approach of Beratan, Onuchic and co-workers (38).
Results
Spectroscopic properties
Native amicyanin exhibits a major absorbance maximum centered at 595 nm (4). The ratio of absorbance at 280 nm to 595 nm (ε280/ε595) is approximately 3.5 for pure native amicyanin. For oxidized M98L amicyanin, the absorption maximum is shifted to 601 nm. The A280/A601 for the as-isolated M98L amicyanin is about 5, despite appearing pure as judged by SDS-PAGE. Addition of Cu(II) caused no increase in A601 suggesting that low absorbance was not due to a subpopulation of apoamicyanin, which would be expected to readily incorporate copper. An abnormally low A280/A595 was previously observed for the as-isolated M98Q and M98A amicyanin mutant proteins (24). This was due to partial occupancy of the metal site by zinc rather than copper. As such, the as-isolated M98L amicyanin was subjected to a procedure originally developed by Diederix et al. (25) to remove any metal atoms and then fully reconstitute the metal-binding site with Cu(II). After this procedure was applied to M98L amicyanin, it exhibited an absorption spectrum very similar to that of native amicyanin, except for the shift in absorption maximum (Figure 1, Table 2). The extinction coefficient of the reconstituted M98L amicyanin at 601 nm was determined by reductive titration of the fully oxidized protein with a standard solution of sodium dithionite, and was very similar to that for native amicyanin at 595 nm (Table 2).
Figure 1.
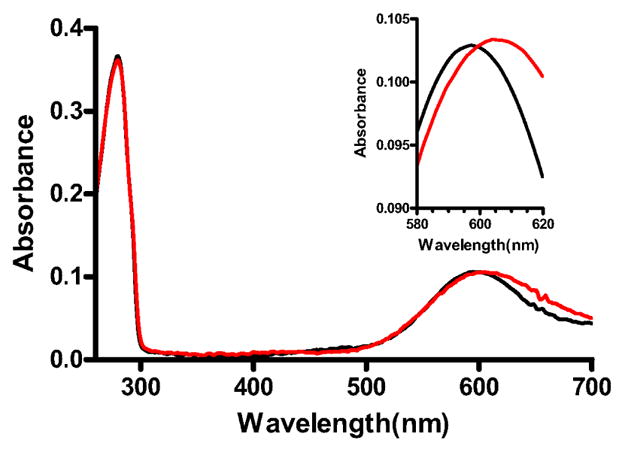
UV-visible absorption spectra of M98L and native amicyanins. Absorption spectra of native (black) and M98L (red) amicyanin were recorded in 10 mM potassium phosphate, pH 7.5. The portion of the spectra of most interest is magnified in the Inset.
Table 2.
Spectroscopic and Redox Properties of Native and M98L Amicyanins
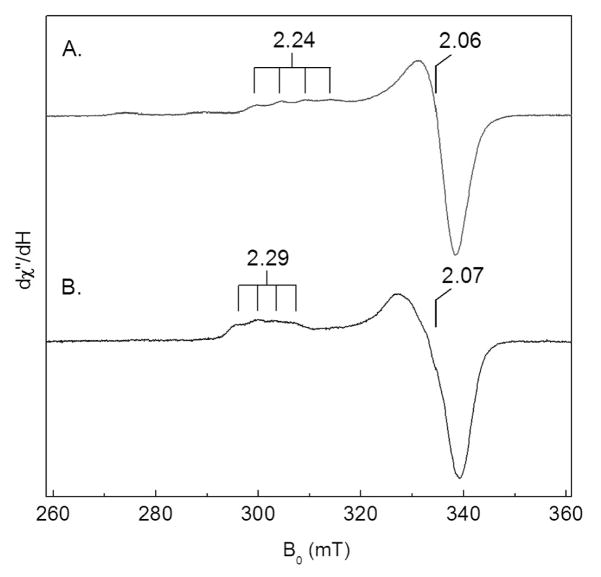
The EPR spectrum of M98L amicyanin exhibits similarities and differences to that of native amicyanin (Figure 2). The EPR spectrum of M98L amicyanin shows a very small hyperfine splitting that is typical of a type I copper center. The g|| values for native and M98L amicyanin are 2.24 and 2.29, respectively, indicating a slightly increased g-anisotropy in the mutant protein. The corresponding A|| values were 53 G and 39 G, respectively. As a control, native amicyanin was also subjected to the protocol to remove and then reconstitute the protein with Cu(II). The absorption and EPR spectra of the reconstituted native amicyanin were essentially unchanged from that of the as-isolated native amicyanin (not shown), confirming that the spectral differences observed with M98L amicyanin were not an artifact of the reconstitution procedure used to fully load the M98L amicyanin site with copper. There is a minor signal in the reconstituted native amicyanin that is likely from adventitious Cu(II) acquired during the reconstitution process.
Figure 2.
X-band EPR spectra of M98L and native amicyanins. Spectra of the reconstituted native (A) and M98L (B) amicyanins were recorded in 10 mM potassium phosphate, pH 7.4, plus 5% glycerol. The spectra were obtained from an average of three scans at 20 K with 0.2 mW microwave power and 3 G values modulation amplitude. The principle g values are indicated.
Crystal Structures of M98L Amicyanin
The oxidized and reduced states of M98L amicyanin were determined at 2.1 Å and 2.2 Å resolution, respectively. This amicyanin mutant was crystallized in the hexagonal space group P6322, which is different from that of native and several mutants of amicyanin solved previously (3, 15). There is a zinc ion present at the surface in both oxidized and reduced structures of M98L amicyanin at a distance of 2.9 Å from the NE1 atom of His95 and 3.6 Å from SD atom of Met51. The zinc was identified on the basis of the anomalous difference Fourier map, and subsequent refinement confirmed its presence. The zinc is located on a crystallographic 3-fold axis and is coordinated by the three copies of His95-NE1 and three copies of Met51-SD. M98L amicyanin was purified initially with some zinc bound instead of copper and subsequently, it was unfolded and reconstituted with CuSO4 to generate the functional protein (discussed under Experimental Procedures). The same was true for M98A and M98Q amicyanins (24) but zinc was not present in the crystal structures of those copper-reconstituted proteins (15). The significance and source of the zinc in the M98L amicyanin structure is unclear. It is possible that the source of zinc could be from impurities of the chemicals used during the crystallization trials. A chloride ion stabilizes the molecule by forming strong hydrogen bonds to Lys68-NZ and Arg99-NH1. A phosphate group is located on the crystallographic two-fold axis and interacts with the residue Lys2-NZ.
The rms deviation between native (PDB code 2OV0) and M98L amicyanin is 0.68 Å with 103 matched Cα atoms based on the secondary structure matching (39) carried out by using CCP4MG (34) (Figure 3). It was previously observed that N-terminal residues 1–21 comprise a very flexible loop segment and adopt variable conformations (40). By omitting this segment, the rms deviation between M98L and native amicyanins is 0.4 Å. By comparing the native and M98L amicyanins, it is noted that the residues in the region 28–31, 49–50, 59–65, 94–97, 99 and 105 deviate by ≥0.5 Å. Glu31 which is predicted to be the point of inter-protein transfer from amicyanin to cytochrome c-551i deviates by 0.6 Å and the region 28–31 of M98L amicyanin deviates by 0.5–0.7 Å with respect to native amicyanin. The side chain atoms CG, CD, OE1 and OE2 of Glu31 did not have electron density in the oxidized state and the temperature factor for the individual atoms this residue varies from 44–58 Å2. In the case of the reduced form, side chain of Glu31 has density for all the atoms at the 0.8σ level and the temperature factor for the individual atoms of the residues varies from 46–69 Å2. The hydrophobic residues of amicyanin 94–97 which interact with MADH deviate by 0.6–0.7 Å with respect to native amicyanin. The analysis of the residue ranges 26–28, 48–52 and 73–74, which are within or close to MADH/amicyanin interface (41) indicate that only Met28, Glu49 and Ala50 show deviation greater than 0.5 Å.
Figure 3.
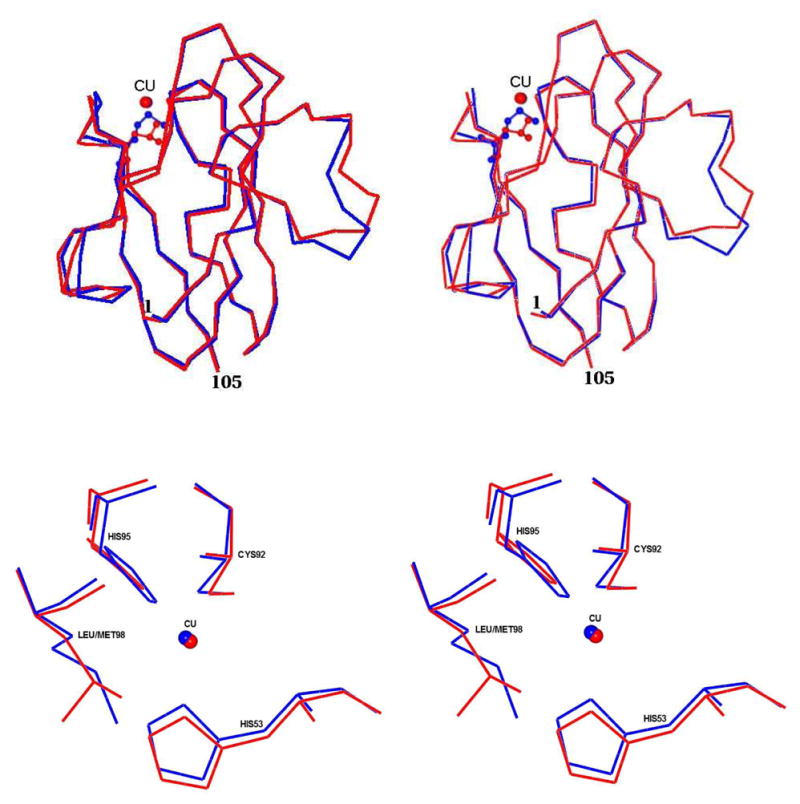
Stereo views of the superposition of oxidized M98L amicyanin with native amicyanin. Top Panel: The entire protein molecules are superimposed with M98L amicyanin in red and native amicyanin (PDB file 2OV0) in blue. Residue 98 (Met/Leu) is shown as a ball and stick in its respective color. The Cu(II) is shown as a red sphere. Lower panel: The type 1 copper sites are superimposed with M98L amicyanin in red and native amicyanin in blue. The Cu(II) in each structure is shown as sphere and in its respective color.
The copper coordination distances in the oxidized and reduced state of M98L and native amicyanins are listed in Table 3. The superposition of the copper binding region of M98L amicyanin with native amicyanin is shown in Figure 3. There is significant negative density at the copper position of the oxidized M98L amicyanin structure and a subsequent occupancy refinement of the copper revealed an occupancy of 66% with B-factor of 40 Ǻ2. There is no such negative density for copper in the reduced M98L amicyanin structure, which was generated by reduction of the oxidized crystal even though the B-factor is 47 Ǻ2. Furthermore, the copper site appears to be fully occupied in the solution state (discussed earlier). It is unlikely that the lower occupancy calculated for the Cu(II) in the oxidized structure is due to multiple orientations for the copper site since the three ligands have very clear density and B-factors comparable to the other protein atoms. The lower calculated occupancy most likely reflects greater thermal motion of Cu(II) in the oxidized structure relative to Cu(I) in the reduced structure.
Table 3.
Copper Coordination Distances in the Type 1 Sites of Native and Met98 Mutant Amicyanins, Laccase and Ceruloplasmin
| Ligandsa | M98L amicyanin (oxidized) | M98L amicyanin (reduced) | Native amicyanin, PDB 2OV0 | M98A amicyanin, PDB 2IDQ | M98Q amicyanin, PDB 2IDT | Laccase, PDB 2Q9O | Ceruloplasmin, PDB 2J5W | M148L Rusticyanin,b PDB 2GY2 |
|---|---|---|---|---|---|---|---|---|
| Cu - SG/Cys92 | 2.21 | 2.19 | 2.17 | 2.14 | 2.16 | 2.18 | 1.94 | 2.20 (2.26) |
| Cu - ND1/His95 | 2.55 | 2.75 | 2.05 | 2.01 | 2.02 | 2.02 | 1.88 | 2.02 (2.09) |
| Cu - ND1/His53 | 2.17 | 2.18 | 1.99 | 1.95 | 1.98 | 1.94 | 2.02 | 2.08 (2.09) |
| Cu-CD2/Leu98 | 3.60 | 3.48 | 3.07c | 2.41d | 2.12/2.13e | 3.60 | 3.52 | 3.08 (2.90) |
Ligands listed below use the residue numbers for M98L amicyanin.
Values in parentheses are for native rusticyanin. Data are from ref (44).
In native amicyanin, this ligand is Met98
In M98A amicyanin this ligand is provided by a water
In M98Q amicyanin the Gln which provides the ligand is present in two alternate conformations.
The rms deviations between Cα atoms calculated after omitting the residues 1–21 of oxidized and reduced M98L, oxidized M98A (PDB code 2IDQ) and oxidized M98Q (PDB code 2IDT) amicyanins are 0.08Ǻ, 0.36 Ǻ and 0.42 Ǻ, respectively. The superposition of copper sites in M98L, M98A and M98Q amicyanins is shown in the Figure 4.
Figure 4.
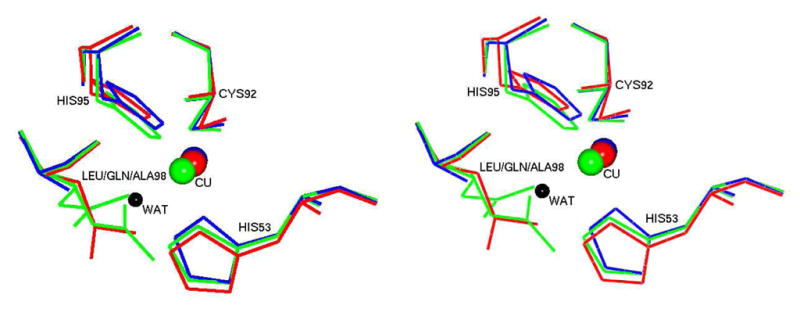
Stereo view of the type 1 copper sites of different Met98 mutants of amicyanin. The structures of M98L (red), M98A (blue, PDB file 2IDQ) and M98Q (green, PDB file 2IDT) are overlayed. The Cu(II) in each structure is shown as sphere in its respective color. The Gln98 in M98Q amicyanin exists in two different conformations and both are shown. The lone active site water which serves as a copper ligand in M98A amicyanin is shown as a black sphere.
Comparison of the Type 1 Copper Sites M98L Amicyanin with those of Laccase and Ceruloplasmin
The multicopper oxidases laccase and ceruloplasmin, each possess a type 1 copper site with leucine in the position normally occupied the axial ligand (19, 42). The superposition of copper sites of M98L amicyanin, laccase and ceruloplasmin is shown in Figure 5. The copper coordination distances for each type 1 copper site are listed in Table 3.
Figure 5.

Stereo view of the type 1 copper sites of M98L amicyanin, laccase and ceruloplasmin. M98L amicyanin is in blue, laccase (PDB file 2Q9O) is in red and ceruloplasmin (PDB file 2J5W) is in green. The Cu(II) in each structure is shown as sphere in its respective color.
The laccase structure which was determined at 1.3 Å resolution contains one type 1 copper, one type 2 copper and two type 3 copper sites (42). The amicyanin superimposes with the type 1 copper site of laccase with a rms deviation of 2.20 Å with 80 matched Cα-atoms based on the secondary structure matching (39) carried out by using CCP4MG (34), while the sequence identity between them is 16.3% (31). When the copper with its four ligands of M98L amicyanin is superimposed with the similar region of laccase, the rms deviation of Cα is 0.60 Å (Figure 5). The difference in position of the copper between M98L amicyanin and laccase is 0.9 Å and the rms deviation between the ligands of copper of M98L amicyanin and laccase varies 0.4–0.8 Å.
The ceruloplasmin structure was determined at 3.8 Å resolution (19). It contains six domains. There are three mononuclear and one trinuclear sites in this structure. The type 1 copper sites are present in the even numbered domains (2, 4, and 6). The copper site in domain 6 is situated ~13 Å from the trinuclear site while the sites in domains 2 and 4 are each at the distance of ~18 Å. The type 1 copper site similar to that of M98L amicyanin is located in domain 2 of ceruloplasmin. The superposition of M98L amicyanin with domain 2 of ceruloplasmin (148 residues) shows a rms deviation of 2.6 Å with 75 matched Cα-atoms based on the secondary structure matching (39) carried out by using CCP4MG (34), despite a sequence identity of only 9.3% between them (31). The superposition of the copper sites yields a rms deviation of 0.70 Å and the rms deviation between ligands of copper varies 0.5–0.9 Å.
Redox Properties
The Em values of M98L amicyanin, both free and in complex with MADH in solution, were determined by spectrochemical titration (Table 2). The Em value of native amicyanin is pH dependent with a pKa value of 7.7 (12). The pKa describes the phenomenon where reduced amicyanin exhibits a pH-dependent conformational change in which the His95 copper ligand rotates out of the coordination sphere of the copper when protonated (12). When amicyanin is in complex with MADH, the redox and pH-dependent conformational change of His95 is restricted. Therefore the Em value of amicyanin in complex with MADH at physiological pH is similar to that of free amicyanin in the pH independent region (i.e., pH 9.0). Thus, complex formation with MADH effectively shifts the pKa value for amicyanin to much lower pH. This is critical for physiologic function, since at physiological pH the Em value of cytochrome c-551i is much less positive than that of free amicyanin (43). Complex formation with MADH lowers the Em value of the bound amicyanin facilitating ET to the cytochrome.
The Em value of M98L amicyanin at pH 7.0 is 127 mV more positive than Em value of native amicyanin (Table 2). Unfortunately, a pH profile for the Em value of M98L amicyanin could not be obtained due to the instability of M98L amicyanin during the redox titrations at higher pH, and so it was not possible to determine the pKa value for this mutant. The Em value of M98L amicyanin in complex with MADH was determined at pH 7.5 (experimental conditions for study of the ET reactions) to be 91 mV more positive than that of native amicyanin in complex with MADH (Table 2). The Em value in complex was used to determine the ΔGo value for the ET reactions described later. The similar increases in Em value for M98L amicyanin in the free state and in complex with MADH, with respect to native amicyanin, suggest that this effect of the mutation is primarily due to a true electronic effect on the intrinsic Em value of the copper rather than simply a change in the pKa of the Em value.
ET from Reduced Amicyanin within the Ternary Protein Complex to Oxidized Cytochrome c-551i
The dependence on complex-bound amicyanin concentration of the observed ET rate from reduced amicyanins to oxidized cytochrome c-551i was determined (Figure 6A, Table 4). From these data, the values of Kd and k3 (see eq 3) were obtained. The results indicate that the M98L mutation does not have any significant effect on the Kd value for formation of the ternary protein complex. However, the observed rate of ET from M98L amicyanin is approximately 435-fold less than that of native amicyanin. The rate at 30 °C of 0.2 s−1 is also much less than the previously reported steady-state kcat of 18 s−1 for methylamine-dependent reduction of cytochrome c-551i by the complex (26). As such, this steady-state assay was performed with M98L amicyanin. The kcat for this reaction was similarly decreased to 0.4 s−1 (data not shown). This indicates that the M98L mutation causes the ET reaction from amicyanin to cytochrome c-551i to become so slow that it is now the rate-limiting step in the overall catalytic and ET reactions catalyzed by the ternary protein complex.
Figure 6.
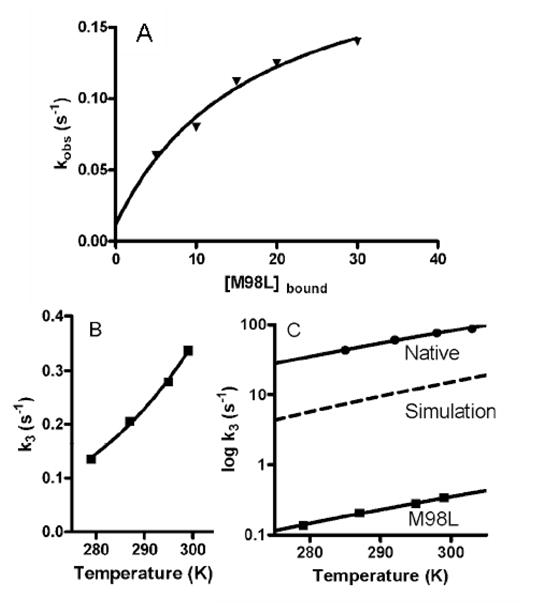
Kinetic and thermodynamic analysis of the ET reaction from Cu(I) of amicyanin to oxidized cytochrome c-551i in the ternary protein complex.
A. Dependence of the observed rate the concentration of amicyanin in complex with MADH. The solid line represents a fit of the data to eq 3.
B. Dependence of the limiting first-order rate constant for the ET reaction on temperature. The data for the reactions of M98L amicyanin are shown. The solid line represents a fit of the data to eq 5.
C. Simulation of the dependence of the rate on temperature due solely to the change in Em value of amicyanin caused by the M98L mutation. The dashed line is a simulation showing the predicted temperature dependence of rate using the ΔGo value for the reaction of M98L amicyanin and the previously determined λ and HAB for the reaction of native amicyanin. This is compared to the experimentally determined rates for the reactions with native and M98L amicyanins.
Table 4.
Kinetic and ET Parameters for the Reactions of Reduced Native and M98L Amicyanins to Oxidized Cytochrome c-551i
| Parameter | Native amicyanina | M98L amicyanin |
|---|---|---|
| k3 (20°C ) s−1 | 87 ± 5 | 0.20 ± 0.03 |
| Kd | 20 ± 6 | 17 ± 13 |
| ΔGo (J/mol) | +3184 | +12061 |
| HAB (cm−1) | 0.3 ± 0.1 | 0.022 ± 0.010 |
| λ (eV) | 1.1 ± 0.1 | 1.1 ± 0.1 |
| λ (kJmol−1)b | 106 ± 10 | 101± 10 |
Taken from ref (8)
λ is sometimes expressed in units of kJ/mol and sometimes as eV; therefore, both values are given.
The ET reaction from M98L amicyanin is thermodynamically less favorable than the reaction from native amicyanin and as such kET is expected to be less. The reaction was studied at different temperatures (Figure 6B). At each temperature the observed decrease in rate for M98L amicyanin is much greater than predicted based solely on the change in Em value (i.e., ΔGo). This means that some parameter other than ΔGo has been altered as a consequence of the mutation. The results of the analysis of these data by eq 5 are shown in Table 5. For the ET reaction from copper to cytochrome c-551i in M98L amicyanin, the value of λ is unaffected by the mutation but the value of HAB is decreased 14-fold as a consequence of the mutation. This is what accounts for the larger than expected decrease in kET. To illustrate the effect of this change in HAB on rate, a simulation of what the rate would be over this temperature range if the only parameter to change was ΔGo is shown and compared to the actual rates determined with native and M98L amicyanin (Figure 6C).
Table 5.
Calculated Electron Transfer Properties from the Structures of Native and M98L amicaynina
| Reduced M98L amicyanin | Reduced native amicyaninb | |
|---|---|---|
| Distance (Cu(I) to Glu31 CA) (Å) | 12.7 | 11.7 |
| Atom packing density (ρ)c | 0.60 | 0.79 |
| Average decay exponential (β)d (Å−1) | 1.66 | 1.30 |
| Electronic coupling (HAB)e | 2.7×10−5 | 4.9×10−4 |
| Calculated HAB M98L/native | 0.055 | |
| Experimental HAB M98L/native | 0.073 ± 0.041 | |
| Maximum ET rate (s−1)f | 7.1×104 | 2.4×107 |
Calculations using HARLEM (36) (http://www.kurnikov.org/harlem_manual/html/index.html)
The structure used is PDB file 2RAC.
Volume fraction between redox cofactors within the united van der Waals radius of intervening atoms (37)
β is a parameter in the term e-βR that represents the exponential fall-off of the electronic tunneling rate with distance.
HAB is the electronic tunneling coupling matrix element between donor and acceptor, units are dimensionless
The predicted free-energy optimized rate of electron transfer when ΔG=−λ
Discussion
The crystal structure of M98L amicyanin confirms the loss of the axial fourth ligand for copper in the type 1 site. The copper ion is in a trigonal configuration. The Cu-Cys92 coordination distance is slightly increased while the other coordination distances are much larger than in native and other Met98 mutant amicyanins (Table 3). The Cu-Leu98 distance is much greater than the axial ligand coordination distance in other amicyanins but similar to the corresponding distances in the laccase and ceruloplasm sites. The truly unique feature of M98L amicyanin is the increased coordination distances for both His ligands. For comparison, the coordination distances of the analogous M148L rusticyanin mutant protein are included in Table 3. It is interesting to note that, in contrast to M98L amicyanin, the M148L rusticyanin mutation causes no increase in coordination distances for the His ligands with respect to native rusticyanin (44). The absorption and EPR spectra of amicyanin are only slightly perturbed by the M98L mutation suggesting that the Cu(II)-Cys interaction, which is least affected by the mutation, has the strongest influence over these features. It is noteworthy that while azurin exhibits a λmax at 626 nm, compared to 595 nm for amicyanin, the M121L azurin mutation resulted in a red shift of 5 nm (45), similar to what was observed with M98L amicyanin (Figure 1).
The small hyperfine splitting at the g|| region of the EPR spectrum is the characteristic signature for a type I copper center and reflects the high covalency of the thiolate-Cu(II) bond in this redox center. In general, the copper hyperfine structure at the g|| region is the sum of the Fermi-contact, spin-dipolar and spin-orbit contributions (46). X-ray absorption spectroscopy and X-alpha scattered wave density function studies at the copper K-edge have shown that the Cu 4px and py orbitals mix into the dx2−y2 ground state of the blue copper site (47, 48). While the crystal structure of M98L amicyanin reveals loss of the axial ligation and lengthening of the two Cu(II)-His coordination distances, the Cu(II)-Cys interaction is relatively unchanged by the mutation (Table 3). The observation of a small hyperfine splitting in the parallel region of the EPR spectrum of M98L amicyanin is consistent with the idea that the Cu(II)-Cys interaction is the major determinant of this spectroscopic feature. Thus, while the axial ligand and distorted tetrahedral geometry do not contribute to the small hyperfine splitting it does influence the exact positioning of the copper within the site. The consequences of this contribution of the axial ligand in amicyanin are discussed later.
The Em value of M98L amicyanin of +421 mV is still much lower than that of the type 1 copper sites with naturally occurring leucine in the position of the axial ligand of laccase and ceruloplasmin which have Em values of 550–800 mV (20, 49). While the axial ligand is important in fine-tuning of Em value of type 1 copper proteins, several other factors may potentially influence Em value including desolvation or hydrophobic effects, hydrogen bonding pattern around the copper site, protein constraint, and intraprotein electrostatic interactions (50).
Typical Em values of type 1 copper proteins with methionine as an axial ligand are around 300 mV, except for rusticyanin which exhibits a significantly more positive Em value (2). Computational studies have attributed the influence of protein permanent dipoles to the atypical high potential of rusticyanin (51). Type 1 copper proteins possessing axial ligands other than methionine show variable Em values. Stellacyanin which has glutamine as an axial ligand (52), exhibits a less positive Em value of 184 mV. Site directed mutagenesis of the axial methionine ligand to glutamine in other type 1 copper proteins, including amicyanin, causes a decrease in the Em value (16, 53, 54). Substitution of leucine for Met98 caused the Em value of amicyanin to become 127 mV more positive (Table 2). Replacement of the methionine axial ligand with leucine in other type 1 copper proteins has caused an increase in Em value similar to what was observed in amicyanin (45, 53, 55, 56). In rusticyanin the Em value increased from 667 to 798 mV (53) and in azurin the Em value increased from 315 to 450 mV (45).
When comparing the Em values of type 1 copper proteins it must be considered that amicyanins exhibit a pH-dependent conformational change in which the His95 copper ligand rotates out of the coordination sphere of the copper when protonated in the reduced state (12, 57). However, in this study the same ΔEm was observed for M98L amicyanin free and in complex with MADH when the conformational change of His95 is restricted (12). Furthermore the magnitude of the ΔEm caused by the M98L mutation of amicyanin is similar to the ΔEm reported for similar mutations of azurin and rusticyanin which do not exhibit this conformational change. Leucine possesses the highest free energy of transfer of any amino acid side chain from cyclohexane to water with 4.92 kcal/mol at pH 7.0 whereas Met and Gln are 2.35 and -5.54 kcal/mol, respectively (58). Removal of the weak axial interaction by mutation of the Met could potentially provide a geometry more favorable for Cu(I) which would increase the Em value. The increase in Em value associated with the M98L mutation likely reflects the change in dielectric constant around the mutated residue and resulting increased hydrophobicity which will preferential stabilize the less charged Cu(I) state. This is also consistent with correlation between Em value and amino acid side-chain hydrophobicity that was determined from studies in which the Met ligand of azurin was replaced with unnatural amino acids (45).
Cupredoxins with a single type 1 copper site function primarily as ET mediators (1). In multicopper oxidases the type 1 copper sites accept electrons from the substrate which is oxidized, and transfer the electrons to a trinuclear copper center where dioxygen binds and is reduced to water. The role of the protein in determining the ET properties of a type 1 copper site has been extensively studied in P. denitrificans amicyanin (9). Its physiologic electron donor and acceptor proteins have been identified and the complex of the three proteins has been characterized structurally and functionally (8–11).
An importance consequence of the M98L mutation is that it significantly alters the rates of ET from copper through amicyanin within the MADH-amicyanin-cytochrome c-551i complex. The influence of the amino acid residue which provides the axial ligand on ET reactivity has been investigated in other blue copper proteins (e.g.,(59, 60)). Such studies have primarily compared changes in reaction rates with changes in Em value and ΔGo and have provided some insight into the effect of these mutations on λ. The current study describes a more detailed analysis of the effect of the axial ligand mutation on kET and includes analysis of HAB as well as λ. The rate of ET from Cu(I) of reduced M98L amicyanin to the cytochrome was 435-fold less than that from native amicyanin. This is in part due to the change in Em value that is caused by the M98L mutation, which makes the ΔGo for the reaction more positive. However, the magnitudes of the changes in ET rate are much greater than can be attributed solely to the change in ΔGo (Figure 6).
The large decrease in kET from reduced amicyanin to oxidized cytochrome c-551i is primarily due to a large decrease in HAB (Table 5). In order to determine the structural basis for the experimentally-determined decrease in HAB, the structures of reduced M98L and native amicyanins were analyzed using the HARLEM program (36) to calculate relative HAB values for ET from Cu(I) through amicyanin. Previous analysis of the crystal structure of the MADH-amicyanin-cytochrome c-551i indicated that the likely point of interprotein ET from amicyanin to cytochrome c-551i is from the protein backbone at residue Glu31 (8). Since a crystal structure of M98L amicyanin in complex with the other proteins is not available, the effects of the M98L mutation on HAB within amicyanin were determined using Cu(I) as the electron donor and the CA of Glu31 in each amicyanin as the electron acceptor. The calculations predict that the HAB for ET through M98L amicyanin will be less than for ET through native amicyanin by a factor of 0.055 (Table 5). This corresponds well with the experimentally determined values of HAB which showed a decrease by a factor of 0.073 ± 0.041. The calculated maximum ET rates from the structures predict a decrease of greater than two orders of magnitude for M98L amicyanin. This also correlates well with the experimental results. It does not correlate exactly because the calculated results assume activationless ET whereas the experimental results include the contribution to kET from ΔGo, which is also affected by the mutation.
Inspection of the structures of M98L and native amicyanins indicates that the predicted change in ρ, and consequently HAB is not because of changes in the overall structure of the protein. With this method of analysis (37) Cu-ligand bonds are given no special treatment and the program calculates the average atom packing distance between donor and acceptor within the united van der Waals’ radii of intervening atoms. The most direct ET route from copper to the site of interprotein ET is through the axial ligand. The calculated change in HAB, which is a consequence of decreased ρ and the corresponding increase in β, is primarily a consequence of the large through space jump which is required for the electron to get from Cu(I) to the protein via Leu98 rather than via Met98 (see Table 3). We previously reported (8) that a Pathways analysis of the MADH-amicyanin-cytochrome c-551i complex analysis using the Greenpath (38) program predicted two sets of ET pathways of comparable efficiency from the type 1 copper to the heme of cytochrome c-551i. In one pathway, the electron exits copper via the Cys92 copper ligand, and in the other, it exits via the Met98 copper ligand. A critical component of the predicted pathway, from which electrons exit via Cys92, is a through-space jump from that Cys to Tyr30. Site-directed mutagenesis of Tyr30 which should have disrupted ET via this pathway did not affect the rate of ET (61). The interpretation of those results was that either ET preferentially occurred via the Met98 pathway or that this ET reaction obeyed a simple distance dependence, independent of specific pathways. The current results support the previous studies and indicate that for amicyanin a critical role for the axial Met98 ligand is for mediation of ET to cytochrome c-551i.
Summary
The structure of M98L amicyanin bears similarities and differences with other natural and mutant type 1 copper proteins with leucine in the analogous position. Comparison of these structures with the physical properties of these proteins has allowed better definition of the role of the axial ligand of the type 1 site. The most common structural feature is the Cu-Cys coordination distance which exerts the strongest influence over the absorption and EPR spectra. The most distinct structural feature of M98L amicyanin is the significantly longer Cu-His coordination distances. This and other studies indicate that in the replacement of methionine with leucine, in the absence of other changes causes an increase of about 130 mV in the Em value. This study also demonstrates that the ET rate from the type 1 copper site may be strongly influenced by the coordination distances. For amicyanin the axial Met ligand, when present, constrains the distorted tetrahedral geometry, but not the spectroscopic features of the type 1 site. It plays a physiologically important role in physically positioning the copper relative to the other ligands, which for amicyanin is critical for its ET function.
Acknowledgments
We wish to thank Professor Scott Mathews, Washington University, St. Louis for helpful discussions.
Abbreviations
- MADH
methylamine dehydrogenase
- ET
electron transfer
- Em
oxidation-reduction midpoint potential
- HAB
electronic coupling
- λ
reorganization energy
- rms
root mean square;
- kET
true electron transfer rate constant;
- ρ
atomic packing density.
Footnotes
This work was supported by NIH Grant GM-41574 (V.L.D.). Use of NE-CAT beamlines and this work is supported by award RR-15301 (NE-CAT facility) from the NCRR at the NIH. Use of the APS is supported by the U.S. DOE, Office of Science, Office of Basic Energy Science, under Contract No. DE-AC02-06CH11357.
Crystallographic coordinates have been deposited in the Protein Data Bank under the file names 3IE9 for Cu(II) M98L and 3IEA for Cu(I) M98L amicyanins.
References
- 1.Adman ET. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 2.Gray HB, Malmstrom BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 3.Cunane LM, Chen Z, Durley RCE, Mathews FS. X-ray crystal structure of the cupredoxin amicyanin from Paracoccus denitrificans, refined at 1.31 Å resolution. Acta Crystallogr D Biol Crystallogr. 1996;D52:676–686. doi: 10.1107/S0907444996001072. [DOI] [PubMed] [Google Scholar]
- 4.Husain M, Davidson VL. An inducible periplasmic blue copper protein from Paracoccus denitrificans. Purification, properties, and physiological role. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 5.Davidson VL. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv Protein Chem. 2001;58:95–140. doi: 10.1016/s0065-3233(01)58003-1. [DOI] [PubMed] [Google Scholar]
- 6.Husain M, Davidson VL. Characterization of two inducible periplasmic c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986;261:8577–8580. [PubMed] [Google Scholar]
- 7.Davidson VL, Jones LH. Complex formation with methylamine dehydrogenase affects the pathway of electron transfer from amicyanin to cytochrome c-551i. J Biol Chem. 1995;270:23941–23943. doi: 10.1074/jbc.270.41.23941. [DOI] [PubMed] [Google Scholar]
- 8.Davidson VL, Jones LH. Electron transfer from copper to heme within the methylamine dehydrogenase-amicyanin-cytochrome c-551i complex. Biochemistry. 1996;35:8120–8125. doi: 10.1021/bi952854f. [DOI] [PubMed] [Google Scholar]
- 9.Davidson VL. Protein control of true, gated and coupled electron transfer reactions. Acc Chem Res. 2008;41:730–738. doi: 10.1021/ar700252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Durley RC, Mathews FS, Davidson VL. Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i. Science. 1994;264:86–90. doi: 10.1126/science.8140419. [DOI] [PubMed] [Google Scholar]
- 11.Merli A, Brodersen DE, Morini B, Chen Z, Durley RC, Mathews FS, Davidson VL, Rossi GL. Enzymatic and electron transfer activities in crystalline protein complexes. J Biol Chem. 1996;271:9177–9180. doi: 10.1074/jbc.271.16.9177. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z, Cunane LM, Chen Z, Durley RC, Mathews FS, Davidson VL. Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin. Biochemistry. 1998;37:17128–17136. doi: 10.1021/bi9817919. [DOI] [PubMed] [Google Scholar]
- 13.Davidson VL. Unraveling the kinetic complexity of interprotein electron transfer reactions. Biochemistry. 1996;35:14035–14039. doi: 10.1021/bi961577p. [DOI] [PubMed] [Google Scholar]
- 14.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 15.Carrell CJ, Ma JK, Antholine WE, Hosler JP, Mathews FS, Davidson VL. Generation of novel copper sites by mutation of the axial ligand of amicyanin. Atomic resolution structures and spectroscopic properties. Biochemistry. 2007;46:1900–1912. doi: 10.1021/bi0619674. [DOI] [PubMed] [Google Scholar]
- 16.Ma JK, Mathews FS, Davidson VL. Correlation of rhombic distortion of the type 1 copper site of M98Q amicyanin with increased electron transfer reorganization energy. Bochemistry. 2007;46:8561–8568. doi: 10.1021/bi700303e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell Mol Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J Biol Chem. 2002;277:37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- 19.Bento I, Peixoto C, Zaitsev VN, Lindley PF. Ceruloplasmin revisited: structural and functional roles of various metal cation-binding sites. Acta Crystallogr D Biol Crystallogr. 2007;63:240–248. doi: 10.1107/S090744490604947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaitseva I, Zaitsev V, Card G, Moshkov K, Bax B, Ralph A, Lindley P. The X-ray structure of human serum ceruloplasmin at 3.1 Å: nature of the copper centres. J Biol Inorg Chem. 1996;1:15–23. [Google Scholar]
- 21.Nersissian AM, Shipp EL. Blue copper-binding domains. Adv Protein Chem. 2002;60:271–340. doi: 10.1016/s0065-3233(02)60056-7. [DOI] [PubMed] [Google Scholar]
- 22.Davidson VL. Methylamine dehydrogenases from methylotrophic bacteria. Methods Enzymol. 1990;188:241–246. doi: 10.1016/0076-6879(90)88040-h. [DOI] [PubMed] [Google Scholar]
- 23.Davidson VL, Jones LH, Graichen ME, Mathews FS, Hosler JP. Factors which stabilize the methylamine dehydrogenase-amicyanin electron transfer protein complex revealed by site-directed mutagenesis. Biochemistry. 1997;36:12733–12738. doi: 10.1021/bi971353m. [DOI] [PubMed] [Google Scholar]
- 24.Ma JK, Lee S, Choi M, Bishop GR, Hosler JP, Davidson VL. The axial ligand and extent of protein folding determine whether Zn or Cu binds to amicyanin. J Inorg Biochem. 2008;102:342–346. doi: 10.1016/j.jinorgbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diederix RE, Canters GW, Dennison C. The Met99Gln mutant of amicyanin from Paracoccus versutus. Biochemistry. 2000;39:9551–9560. doi: 10.1021/bi000648o. [DOI] [PubMed] [Google Scholar]
- 26.Davidson VL, Jones LH. Intermolecular electron transfer from quinoproteins and its relevance to biosensor technology. Anal Chim Acta. 1991;249:235–240. [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected by oscillation methods. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Jensen LH. Solvent model for protein crystals: on occupancy parameters for discrete solvent sites and the solvent continuum. Acta Crystallogr B. 1990;46(Pt 5):650–653. doi: 10.1107/s0108768190005547. [DOI] [PubMed] [Google Scholar]
- 33.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 34.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 35.CCP4. Collaborative Computational Project Number 4. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Kurnikov IV. HARLEM Computer Program. University of Pittsburgh; 2000. [Google Scholar]
- 37.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 38.Regan JJ, Risser SM, Beratan DN, Onuchic JN. Protein electron transport: Single versus multiple pathways. J Phys Chem. 1993;97:13083–13088. [Google Scholar]
- 39.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 40.Carrell CJ, Sun D, Jiang S, Davidson VL, Mathews FS. Structural studies of two mutants of amicyanin from Paracoccus denitrificans that stabilize the reduced state of the copper. Biochemistry. 2004;43:9372–9380. doi: 10.1021/bi049634z. [DOI] [PubMed] [Google Scholar]
- 41.Ma JK, Wang Y, Carrell CJ, Mathews FS, Davidson VL. A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin. Biochemistry. 2007;46:11137–11146. doi: 10.1021/bi7012307. [DOI] [PubMed] [Google Scholar]
- 42.Hakulinen N, Andberg M, Kallio J, Koivula A, Kruus K, Rouvinen J. A near atomic resolution structure of a Melanocarpus albomyces laccase. J Struct Biol. 2008;162:29–39. doi: 10.1016/j.jsb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Gray KA, Knaff DB, Husain M, Davidson VL. Measurement of the oxidation-reduction potentials of amicyanin and c-type cytochromes from Paracoccus denitrificans. FEBS Lett. 1986;207:239–242. doi: 10.1016/0014-5793(86)81496-x. [DOI] [PubMed] [Google Scholar]
- 44.Kanbi LD, Antonyuk S, Hough MA, Hall JF, Dodd FE, Hasnain SS. Crystal structures of the Met148Leu and Ser86Asp mutants of rusticyanin from Thiobacillus ferrooxidans: Insights into the structural relationship with the cupredoxins and the multi copper proteins. J Mol Biol. 2002;320:263–275. doi: 10.1016/S0022-2836(02)00443-6. [DOI] [PubMed] [Google Scholar]
- 45.Berry SM, Ralle M, Low DW, Blackburn NJ, Lu Y. Probing the role of axial methionine in the blue copper center of azurin with unnatural amino acids. J Am Chem Soc. 2003;125:8760–8768. doi: 10.1021/ja029699u. [DOI] [PubMed] [Google Scholar]
- 46.Sinnecker S, Neese F. QM/MM calculations with DFT for taking into account protein effects on the EPR and optical spectra of metalloproteins. Plastocyanin as a case study. Journal of Computational Chemistry. 2006;27:1463–1475. doi: 10.1002/jcc.20426. [DOI] [PubMed] [Google Scholar]
- 47.Shadle SE, Penner-Hahn JE, Schugar HJ, Hedman B, Hodgson KO, Solomon EI. X-ray absorption spectroscopic studies of the blue copper site: metal and ligand K-edge studies to probe the origin of the EPR hyperfine splitting in plastocyanin. J Am Chem Soc. 1993;115:767–776. [Google Scholar]
- 48.Penfield KW, Gewirth AA, Solomon EI. Electronic structure and bonding of the blue copper site in plastocyanin. J Am Chem Soc. 1985;107:4519–4529. [Google Scholar]
- 49.Ducros V, Brzozowski AM, Wilson KS, Brown SH, Ostergaard P, Schneider P, Yaver DS, Pedersen AH, Davies GJ. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 A resolution. Nat Struct Biol. 1998;5:310–316. doi: 10.1038/nsb0498-310. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Webb SP, Ivanic J, Jensen JH. Determinants of the relative reduction potentials of type-1 copper sites in proteins. J Am Chem Soc. 2004;126:8010–8019. doi: 10.1021/ja049345y. [DOI] [PubMed] [Google Scholar]
- 51.Olsson MH, Hong G, Warshel A. Frozen density functional free energy simulations of redox proteins: computational studies of the reduction potential of plastocyanin and rusticyanin. J Am Chem Soc. 2003;125:5025–5039. doi: 10.1021/ja0212157. [DOI] [PubMed] [Google Scholar]
- 52.Koch M, Velarde M, Harrison MD, Echt S, Fischer S, Messerschmidt A, Dennison C. Crystal structures of oxidized and reduced stellacyanin from horseradish roots. J Am Chem Soc. 2005;127:158–166. doi: 10.1021/ja046184p. [DOI] [PubMed] [Google Scholar]
- 53.Hall JF, Kanbi LD, Strange RW, Hasnain SS. Role of the axial ligand in type 1 Cu centers studied by point mutations of met148 in rusticyanin. Biochemistry. 1999;38:12675–12680. doi: 10.1021/bi990983g. [DOI] [PubMed] [Google Scholar]
- 54.Hibino T, Lee BH, Takabe T. Expression and characterization of Met92Gln mutant plastocyanin from Silene pratensis. J Biochem. 1995;117:101–106. doi: 10.1093/oxfordjournals.jbchem.a124693. [DOI] [PubMed] [Google Scholar]
- 55.DeBeer George S, Basumallick L, Szilagyi RK, Randall DW, Hill MG, Nersissian AM, Valentine JS, Hedman B, Hodgson KO, Solomon EI. Spectroscopic investigation of stellacyanin mutants: axial ligand interactions at the blue copper site. J Am Chem Soc. 2003;125:11314–11328. doi: 10.1021/ja035802j. [DOI] [PubMed] [Google Scholar]
- 56.Pascher T, Karlsson BG, Nordling M, Malmstrom BG, Vanngard T. Reduction potentials and their pH dependence in site-directed-mutant forms of azurin from Pseudomonas aeruginosa. Eur J Biochem. 1993;212:289–296. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
- 57.Lommen A, Canters GW. pH-dependent redox activity and fluxionality of the copper site in amicyanin from Thiobacillus versutus as studied by 300- and 600- MHz 1H NMR. J Biol Chem. 1990;265:2768–2774. [PubMed] [Google Scholar]
- 58.Radzicka A, Pedersen L, Wolfenden R. Influences of solvent water on protein folding: free energies of solvation of cis and trans peptides are nearly identical. Biochemistry. 1988;27:4538–4541. doi: 10.1021/bi00412a047. [DOI] [PubMed] [Google Scholar]
- 59.Yanagisawa S, Dennison C. Reduction potential tuning at a type 1 copper site does not compromise electron transfer reactivity. J Am Chem Soc. 2005;127:16453–16459. doi: 10.1021/ja054426v. [DOI] [PubMed] [Google Scholar]
- 60.Di Bilio AJ, Hill MG, Bonander N, Karlsson BG, Villahermosa RM, Malmström BG, Winkler JR, Gray HB. Reorganization energy of blue copper: Effects of temperature and driving force on the rates of electron transfer in ruthenium- and osmium-modified azurins. J Am Chem Soc. 1997;119:9921–9922. [Google Scholar]
- 61.Davidson VL, Jones LH, Graichen ME, Zhu Z. Tyr(30) of amicyanin is not critical for electron transfer to cytochrome c-551i: implications for predicting electron transfer pathways. Biochim Biophys Acta. 2000;1457:27–35. doi: 10.1016/s0005-2728(00)00052-9. [DOI] [PubMed] [Google Scholar]