Abstract
The neural ectoderm of vertebrates forms when the BMP signaling pathway is suppressed. Herein we review the molecules that directly antagonize extracellular BMP and the signaling pathways that further contribute to reduce BMP activity in the neural ectoderm. Downstream of neural induction, a large number of “neural fate stabilizing” (NFS) transcription factors are expressed in the presumptive neural ectoderm, developing neural tube, and ultimately in neural stem cells. Herein we review what is known about their activities during normal development to maintain a neural fate and regulate neural differentiation. Further elucidation of how the NFS genes interact to regulate neural specification and differentiation should ultimately prove useful for regulating the expansion and differentiation of neural stem and progenitor cells.
Keywords: BMP, FGF, Wnt, neural stem cells, transcriptional network, neural fate stabilization
Introduction
Nearly 100 years ago, Hilde Mangold and Hans Spemann (1924) excised small bits of tissue from the point at which cells first involute in the amphibian gastrula and grafted these into the ventral belly region of host embryos. Surprisingly, a secondary body axis formed at the transplant site, and it included a neural tube and axial mesoderm (notochord, somites). By using explants and hosts from differently pigmented species so that the donor cells could be identified, they demonstrated that the transplanted piece, now known as Spemann’s “organizer”, did not give rise to neural ectoderm, but instead formed dorsal mesoderm. They reasoned that the transplanted cells signaled adjacent host cells to become the neural tissue of the secondary axis; thus, the phenomenon of neural induction was discovered. Equivalent organizing regions with neural inducing capacity are found in zebrafish (shield), chick (Henson’s node), and mouse (node) embryos (Beddington, 1994; Shih and Fraser, 1996; Smith and Schoenwolf, 1998; Boettger et al., 2001). Hence, neural induction is one of the primary steps in vertebrate neural development, and understanding how it is accomplished is of fundamental importance.
Since this landmark observation, numerous studies focused on identifying the inducing molecule(s) that are necessary to establish neural fate in vertebrate embryos. In addition, a large number of transcription factors whose expression is initiated by neural inductive signaling was identified. How these factors cooperatively stabilize the neural fate of embryonic cells is only beginning to be appreciated. While understanding the acquisition of neural fate during embryogenesis is exciting in its own right, it also appears that these same signaling factors and transcriptional regulators may be expressed in endogenous niches of neural stem cells in the adult brain. Therefore, the developmental program that specifies a neural fate in the embryo may prove to be critically important for coaxing neural stem cells to repopulate damaged tissue. This review will summarize the current state of knowledge of the process of neural induction, the genes involved in neural fate stabilization, and speculate on how this information may play a role in regulating the differentiation of neural stem cells.
Neural Induction
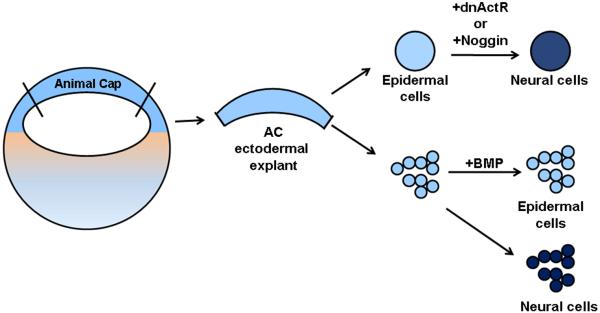
The organizer forms in the dorsal mesoderm in response to secreted factors from a signaling center located in the adjacent presumptive dorsal endoderm, which in amphibians is called the Nieuwkoop Center (Nieuwkoop, 1967; Gerhart et al., 1989; Agius et al., 2000; Vonica and Gumbiner, 2007). In Xenopus, the Nieuwkoop Center forms where the expression domains of VegT, Vg1, Nodal-related (Nr) proteins, and β-Catenin overlap (Fig. 1). The Spemann-Mangold experiment predicted that a neural inducing substance emanated from the organizer. While several studies sought to identify the neural inducer(s), more recent work led to the hypothesis that ectoderm forms neural tissue not by an activating signal, but by default (Sato and Sargent, 1989; Wilson and Hemmati-Brivanlou, 1995; Hawley et al., 1995; Weinstein and Hemmati-Brivanlou, 1997). Pieces of naïve embryonic ectoderm dissected from the animal pole of Xenopus blastulae (called animal cap [AC] ectodermal explants) form non-neural epidermis when cultured without the addition of growth factors (Fig. 2). However, if the AC ectodermal explant is first dissociated into single cells, the cells become neural, indicating that: (1) short range cell-to-cell signaling is required for the formation of epidermis, and (2) in the absence of this signaling, neural tissue forms by default (Sato and Sargent, 1989; Wilson and Hemmati-Brivanlou, 1995). In support of this hypothesis, experiments showed that the inhibition of transforming growth factor-β (TGF-β) signaling through the use of a dominant-negative Activin receptor caused AC ectodermal explants to express neural marker genes (Hemmati-Brivanlou et al., 1992; Hemmati-Brivanlou and Melton, 1994; Hemmati-Brivanlou and Thomsen, 1995; Hawley et al., 1995). However, the dominant-negative Activin receptor interferes with signaling of all of the members of the TGF-β family, including TGF-β, Activin, Nr, and bone morphogenetic proteins (BMPs). Further work demonstrated that BMPs are the important ligands for neural versus epidermal fate induction.
Figure 1.
The dorsal-ventral axis is specified by the Nieuwkoop center and the Spemann organizer in Xenopus. (A) Prior to gastrulation the dorsal axis is specified by the Nieuwkoop center (green circle), formed as a result of overlapping expression of VegT, Vg1, Nodals (Xnr), and β-Catenin (βcat). (B) The Nieuwkoop center secretes dorsalizing molecules that induce the formation of the Spemann organizer (blue oval) in the dorsal mesoderm. The organizer expresses Chordin (Chd), Noggin (Nog), Follistatin (Foll), Cerberus (Cer), and in Xenopus, Nodal related-3 (Xnr3). These molecules actively antagonize BMP, which is secreted from the ventral side (red arrows). V is ventral; D is dorsal. Line intersecting the Nieuwkoop center represents the dorsal blastopore lip.
Figure 2.
The default model of neural induction in Xenopus. An ectodermal explant dissected from the animal cap of a blastula stage embryo forms epidermis (light blue). If BMP signaling is inhibited in those cells by expressing a dominant-negative Activin receptor (dnActR) or the extracellular antagonist Noggin, then neural tissue forms (dark blue). Dissociation of the explant into single cells without the addition of exogenous factors also leads to the formation of neural tissues. However, if exogenous BMP is added to the dispersed cells, they adopt an epidermal fate (light blue).
BMP signaling
Several studies pointed to the BMP signaling pathway as key for neural induction, but as a negative regulator, not a positive inducer. First, using expression screening techniques to identify novel genes that induce a neural fate in AC ectodermal explants, secreted antagonists of BMPs were discovered. These proteins are secreted from the organizer (Fig. 1), and include Noggin (Zimmerman et al., 1996), Chordin (Piccolo et al., 1996), Follistatin (Fainsod et al., 1997), Cerberus (Bouwmeester et al., 1996), and XNr3 (Smith et al., 1995). They bind to BMP in the extracellular space and inhibit activation of its receptor (Fig. 3); when this occurs in the naïve embryonic ectoderm, neural tissue forms instead of epidermis (Meinhardt, 2001; Vonica and Gumbiner, 2007). Second, when expression of one of these secreted proteins in the organizer is blocked, BMP signaling is maintained on the dorsal side of the embryo and the neural plate does not form (Wessely et al., 2004). Third, when dominant-negative BMP4 protein, dominant-negative BMP receptors, or any of the BMP antagonists are expressed in AC ectodermal explants, anterior neural genes are directly induced. Similarly, when these proteins are expressed on the ventral side of embryos, a secondary axis containing neural tissue forms (Hawley et al., 1995).
Figure 3.
Pathways involved in neural induction. BMP2, 4, or 7 dimers bind to the serine-threonine receptor complex, leading to phosphorylation of the type I receptor by the type II receptor, which in turn phosphorylates (P) an appropriate Receptor regulated (R)-Smad (Smad1, 5, or 8). This phosphorylation enables the R-Smad to form a complex with the Common-Smad, Smad4, and this complex translocates to the nucleus to activate (non-neural) or repress (neural) target genes. The binding of secreted factors from the organizer, such as Noggin (Nog), to extracellular BMP prevents activation of its receptor complex. In addition, FGF and Wnt signaling lead to the inhibition of bmp4 transcription. FGF signaling also can prevent the phosphorylation of Smads1/5/8.
Mutational analysis and knock-down experiments in zebrafish, mouse, and frog embryos also demonstrate that BMP inhibition is required to form neural tissue. In zebrafish, double mutants for the BMP antagonists, chordino (a Chordin homolog) and ogon (a secreted Frizzled homolog), have a ventralized phenotype (Miller-Bertoglio et al., 1999; Yabe et al., 2003). Xenopus tropicalis embryos depleted of Chordin, Noggin, and Follistatin protein have nearly a complete loss of the central nervous system (CNS) (Khokha et al., 2005). Mouse mutants lacking the BMP antagonists, Chordin (Bachiller et al., 2000), Noggin (McMahon et al., 1998), or Cerberus (Belo et al., 2000), express only posterior neural genes, and double mutants for Noggin and Chordin fail to develop anterior brain structures (Bachiller et al., 2000). These studies indicate that across vertebrates there is a conserved and necessary role for BMP inhibition in the induction of anterior neural tissue.
There are more than 30 known BMP proteins (Balemans and Van Hul, 2002), but it is BMP4 that is mainly involved in epidermal induction and neural inhibition (Sasai et al., 1995; Wilson and Hemmati-Brivanlou, 1995; Xu et al., 1995). Its expression is restricted to the non-neural ectoderm and ventral mesoderm, and over-expression ventralizes embryos (Fainsod et al., 1994; Hemmati-Brivanlou and Thomsen, 1995). A number of BMP target genes have been identified and these include Msx1 (Foerst-Potts and Sadler, 1997; Tucker et al., 1998; Feledy et al., 1999; Ishimura et al., 2000; Takeda et al., 2000; Yamamoto et al., 2000; Yamamoto et al., 2001; Tribulo et al., 2003), Msx2 (Foerst-Potts and Sadler, 1997), Gata2 (Walmsley et al., 1994; Friedle and Knochel, 2002), Vent1 (Gawantka et al., 1995; Onichtchouk et al., 1998; Friedle et al., 1998; Rastegar et al., 1999), and Vent2 (Ladher et al., 1996; Onichtchouk et al., 1996; Onichtchouk et al., 1998; Rastegar et al., 1999; Trindade et al., 1999; Friedle and Knochel, 2002). Msx2 is not likely to be involved in inhibiting the onset of expression of early neural genes because it is not expressed until the mid-gastrula stage, which is after neural induction occurs. In contrast, Msx1, Gata2, Vent1, and Vent2 are expressed throughout the embryonic ectoderm of the animal pole of blastula embryos, and as gastrulation proceeds their expression domains are restricted to the ventral tissues in a pattern similar to that of BMP4 (Hemmati-Brivanlou and Thomsen, 1995).
Gain-of-function studies indicate that Msx1, Vent1, and Vent2 are involved in restricting the expression of neural genes to the dorsal ectoderm. Injection of Msx1 mRNA inhibited neural tissue formation induced by Noggin (Ishimura et al., 2000), and over-expression of a dominant-activator form of Msx1 prevented ventralization by BMP. However, whereas knock-down of Msx1 function with anti-sense morpholino oligonucleotides (MOs) demonstrated that it is required for the ventralizing activity of BMP4, it is not required for epidermal development, in hibition of neural induction, or axis formation (Suzuki et al., 1997; Yamamoto et al., 2000). Knock-down experiments have not been reported for the Vent proteins, but over-expression of Vent1 (Gawantka et al., 1995) or Vent2 (Ladher et al., 1996; Onichtchouk et al., 1996; Friedle and Knochel, 2002) induces epidermis and inhibits the formation of both dorsal mesoderm and neural tissue. Additionally, both Vent proteins are required to restrict expression of reporter constructs of Geminin (Taylor et al., 2006) and Sox3 (Rogers et al., 2008), two early neural ectoderm transcription factors (discussed below), to the dorsal ectoderm.
Even though inhibition of BMP signaling is sufficient to induce neural tissue in AC ectodermal explants, it is not sufficient to induce it in the ventral, non-neural ectoderm of the embryo (Wilson and Hemmati-Brivanlou, 1995; Hawley et al., 1995; Rogers et al., 2008). For example, inhibition of BMP signaling in ventral ectoderm via over-expression of Smad6, an inhibitor of the BMP effector Smad1, repressed epidermis formation but failed to induce ectopic expression of neural markers (Delaune et al., 2005; Chang and Harland, 2007). It is possible that either a second downstream pathway needs to be silenced. For example, BMP inhibition is sufficient to induce neural tissue in ventral ectoderm if signaling through both Smad1 and Smad2 are inhibited (Chang and Harland 2007). There also is evidence that additional instructive or permissive signals are required. Several experiments in frog and chick demonstrate that neural tissue is induced in non-neural ectoderm when BMP signaling is inhibited in the presence of fibroblast growth factor (FGF) (Delaune, Lemaire et al. 2005; McMahon and Moon, 1989; Christian et al., 1991a; Rodriguez-Gallardo et al., 1997; Alvarez et al., 1998; Wilson et al., 2001).
FGF signaling
In frog, there are four known FGF receptors (FGFR1-4α) implicated in neural development due to their expression in the ectoderm prior to neural induction and the subsequent restriction of their expression to neural tissue. Because FGFR2 and FGFR4α are up-regulated at the onset of neural induction, they may play a specific role in the process (Hongo et al., 1999). To determine if FGF signaling is required for neural induction, signaling was inhibited by incubation in SU5402, a chemical inhibitor of FGF signaling (Mohammadi et al., 1997), or by over-expression of a dominant-negative FGFR1 receptor (XFD) (Amaya et al., 1991) or a dominant-negative FGFR4α receptor (Δ4α) (Hongo et al., 1999). XFD inhibits FGF3 and FGF4 signaling through FGFR1, FGFR2, and FGFR3, whereas Δ4α blocks FGF8 signaling through FGFR4α (Ueno et al., 1992; Hongo et al., 1999; Bainter et al., 2001). XFD and SU5402 inhibited the induction of neural tissue in neuralized AC ectodermal explants as indicated by a loss of the pan-neural markers, Ncam and Sox2 (Launay et al., 1996; Ribisi et al., 2000; Delaune et al., 2005). Over-expression of Δ4α led to a loss of anterior neural tissue (marked by Nrp1, Bf1, and En2) in whole embryos and in neuralized AC ectodermal explants (Hongo et al., 1999). Furthermore, although dissociated AC ectodermal explants are thought to be neuralized by the reduction in levels of BMP protein due to cell dispersal, recent studies showed that BMP signaling still occurs in these cultures (Hurtado and De Robertis, 2007), and that MAP kinase signaling may drive the conversion of cells to neurons (De Robertis and Kuroda, 2004; Kuroda et al., 2005). While these studies suggest that FGF signaling is required for the formation of neural tissue in dissociated AC ectodermal explants, disruption of FGF signaling via the MAP kinase pathway by inhibiting Ras did not affect the induction of anterior neural genes, but inhibited the posteriorization of neural tissue in AC ectodermal explants (Ribisi et al., 2000). This is consistent with several previous studies indicating that FGF is required for the anterior-posterior patterning of the neural plate (reviewed in Gould and Grainger, 1997; Gamse and Sive, 2000). Moreover, treatment of embryos with SU5402 decreased some dorsal mesodermal markers, including Noggin; thus, the decrease in neural tissue may be accomplished indirectly by reducing the amount of dorsal mesoderm available to induce it (Delaune et al., 2005). Taken together, these experiments demonstrate that signaling through FGFR4α may be required for the induction of anterior neural tissue, and that FGFR1, FGFR2, and MAP kinase signaling are required for posteriorizing neural tissue.
Other studies demonstrate a role for FGF in neural induction; however, they do not demonstrate that the function of FGF is independent of its ability to inhibit BMP transcription or the BMP signaling pathway (Baker et al., 1999; Hongo et al., 1999; Ishimura et al., 2000; Wilson et al., 2000; Wilson et al., 2001; Pera et al., 2003; Sheng et al., 2003; Rentzsch et al., 2004; Wittler and Kessel, 2004; Linker and Stern, 2004; Delaune et al., 2005). In chick and zebrafish, FGF signaling inhibits BMP gene expression (Furthauer et al., 1997; Wilson et al., 2000), and in frog and zebrafish, FGF signaling inhibits BMP signaling through the phosphorylation and inactivation of its effector protein, Smad1 (Schier, 2001; Pera et al., 2003). In this manner, FGF may cooperate with or be redundant with the activity of extracellular BMP antagonists. Taken in total, these data support a role for FGF in patterning the anterior-posterior axis of neural tissues and in reinforcing the antagonism of the BMP pathway, but its requirement for neural induction independent of BMP inhibition remains in question (Lamb and Harland, 1995; Wilson et al., 2000; Takemoto et al., 2006; Rogers et al., 2008; Weisinger et al., 2008).
Wnt signaling
Maternal Wnt signaling and the inhibition of zygotic Wnt are important for the formation of neural tissue. The establishment of the dorsal-ventral axis and formation of the Nieuwkoop Center are dependent on the dorsal accumulation of the Wnt effector, β-Catenin (Larabell et al., 1997). However, it is unclear if there is a specific role for Wnt signaling in neural induction. Supporting a requirement for Wnt signaling in axis formation, over-expression of either Wnt1 or Wnt3a, which are expressed in neural tissue, induced a secondary axis (Steinbeisser et al., 1993). However, their ability to induce neural tissue in naïve ectoderm has not been tested. The strongest evidence for a role for Wnt signaling in neural induction were experiments demonstrating the induction of Ncam in AC ectodermal explants in the absence of mesoderm formation in response to mRNA injections of either Wnt8 or an activated form of β-Catenin. Both inhibited BMP4 expression, leading to the proposal that Wnt signaling sensitizes AC ectodermal explants to extracellular BMP inhibition (Baker et al., 1999). However, it is worth noting that Wnt8 normally is expressed in the ventral ectoderm, not the dorsal, presumptive neural ectoderm (Christian et al., 1991b), and in whole embryos it induces dorsal mesoderm (Otte and Moon, 1992), indicating an indirect effect. Also, loss of Wnt signaling by over-expression of a dominant-negative transcriptional effector (TCF3) or a dominant-negative receptor (Frizzled8) did not interfere with the induction of neural markers by Noggin (Rogers et al., 2008). Thus, there is little evidence for a direct role of Wnt signaling in neural induction.
In fact, studies in both frog and chick support a requirement for the inhibition of Wnt signaling for neural induction. In frog, over-expression of a dominant-active form of β-Catenin inhibited the expression of Sox2 and Sox3 in whole embryos and inhibited the induction of Sox3 by Noggin in AC ectodermal explants (Heeg-Truesdell and Labonne, 2006). Similarly, over-expression of Wnt3a in chick embryos inhibited both the neuralization of ectoderm by BMP inhibition and FGF signaling (Wilson et al., 2001). However, like FGF, Wnt signaling does play an important role in the anterior-posterior patterning of the neural plate (Gould and Grainger, 1997; Gamse and Sive, 2000).
Neural Fate Stabilization
As the presumptive neural ectoderm is established via neural inductive signaling, a large number of transcription factors is co-expressed in broad overlapping domains (Fig. 4; Sasai, 1998; Moody and Je, 2002). The mRNAs of several of these genes (Geminin, Sox3, Sox11, SoxD) are found throughout the dorsal ectoderm at the onset of gastrulation. The transcripts of others (FoxD5, Sox2, Zic1, Zic2, Zic3) are concentrated in a broad band near the blastoporal lip. The transcripts of a third group (Xiro1, Xiro2, Xiro3) are detected in two dorso-lateral bands near the blastoporal lip. Although a few are expressed maternally, most are expressed around the onset of gastrulation and continue to be expressed in the neural tube (Fig. 4). Experiments in Xenopus demonstrated that while most of these transcription factors do not induce ectopic neural tissue on their own, when they are over-expressed dorsally by mRNA injection, they all expand the neural plate. Previous studies propose that some of these genes modulate the competence of neural ectoderm to respond to signaling molecules (Sox2, Sox3, Zic1, Zic3), some maintain an immature neural state (Geminin, FoxD5, Zic2), whereas others promote the expression of neural determination/differentiation bHLH genes (SoxD, Sox11, Xiro1, Xiro2, Xiro3). How these transcription factors relate to each other to stabilize a neural fate and regulate the progression from the initially-induced neural ectoderm to neural progenitor cells has yet to be revealed.
Figure 4.
Twelve neural genes have overlapping expression patterns during the establishment of the Xenopus neural plate. (A) At the onset of gastrulation (st10), some NE genes are expressed throughout the dorsal ectoderm (orange: geminin [gem], sox3, sox11, soxD), some are expressed only in a broad dorsal band adjacent to the blastopore lip (bl) (blue: foxD5, sox2, zic1-3), and the Xiro genes (yellow) are expressed weakly in two posterior-lateral bands. Dorsal view with animal pole (AP) to the bottom. (B) The temporal expression patterns of these genes also overlap extensively. Four genes (foxD5, gem, sox3, zic2) are expressed maternally (oocyte) with detectable mRNAs through blastula stages; sox11 is expressed in oocytes but is not detected at blastula stages. The zygotic expression of most of these genes begins at late blastula (st9) to early gastrula (st10). The expression of nearly all is maintained through neural tube (nt) stages; in contrast, foxD5 is lost as the neural plate is established (st14).
Sox genes
Sox genes are named for their similarity to the testis determination factor Sry, and a conserved high-mobility group (HMG) (Sry-related HMG-box) domain that confers their DNA binding ability (Gubbay et al., 1990; Sinclair et al., 1990; Wegner, 1999). They are transcription factors that bind to the minor groove of DNA, inducing a sharp bend and regulating gene transcription (Guth and Wegner, 2008). Of the ten groups of Sox proteins, only SoxD (a Xenopus member of the SoxG group; Wegner, 1999), SoxB (Sox1, 2, 3, 14, 21), SoxC (Sox4 and 11), and SoxE (Sox8, 9, 10) groups are involved in neural induction and development (Guth and Wegner, 2008).
Sox2 and Sox3 are well documented early pan-neural markers (Uwanogho et al., 1995; Collignon et al., 1996; Penzel et al., 1997; Rex et al., 1997; Mizuseki et al., 1998a; Uchikawa et al., 1999; Graham et al., 2003; Linker and Stern, 2004; Takemoto et al., 2006; Rogers et al., 2008), and are part of the SoxB1 (Sox1, Sox2, Sox3) subfamily of transcriptional activators (Penzel et al., 1997; Uchikawa et al., 1999). They are both expressed in neural progenitor cells throughout CNS development and are required for neural progenitor maintenance (Graham et al., 2003; Ellis et al., 2004; Pevny and Placzek, 2005; Rogers et al., 2009). Their expression patterns are similar but not identical (Fig. 4). Sox3 is expressed maternally and is pan-ectodermal until the mid-gastrula stage when expression is restricted to the dorsal ectoderm (Penzel et al., 1997). In contrast, Sox2 expression begins at the onset of neural induction only in the neural ectoderm (Nitta et al., 2006). Other differences include: Sox3 is repressed in the floor plate of the neural tube prior to Sox2, expressed much more strongly in the otic placodes, and expressed in the developing lens, while Sox2 is expressed in the retina (Penzel et al., 1997; Elkins and Henry, 2006; Nitta et al., 2006; Rogers et al., 2008). These differences in their spatio-temporal expression patterns indicate that they are regulated differently, and promoter studies and bioinformatic analysis indicate that this is true (Uchikawa et al., 2003; Rogers et al., 2008). The Sox2 regulatory region has a neural induction module that is conserved across species, and in chick this module responds to both FGF and Wnt signaling for activation in the posterior neural plate (Takemoto et al., 2006). This module is present upstream of Xenopus tropicalis Sox2, but is not in the flanking sequence of Sox3. Thus far, reporter studies in transgenic frog embryos reveal that Xenopus Sox3 expression is induced by the inhibition of BMP signaling and is restricted to the neural plate by the BMP targets, Vent1 and Vent2 (Rogers et al., 2008).
Rescue experiments indicate that the SoxB1 proteins are redundant in tissues in which they are co-expressed. In mouse, loss of Sox2 in the CNS is compensated for by the up-regulation of Sox3 expression. Neurogenesis is decreased in these mice, but neural stem cells retain multipotency (Miyagi et al., 2008). In chick, over-expression of Sox1 rescues the loss of Sox2 (Graham et al., 2003). Although the SoxB1 proteins can compensate for each other, each gene is also expressed in unique tissues during development and loss in that tissue can result in severe phenotypes. Mice mutant for either Sox2 or Sox3 have relatively normal CNS formation (Malas et al., 2003; Taranova et al., 2006). However, loss of Sox2 results in diminished neurogenesis in the mouse retina (Ferri et al., 2004) and anophthalmia in humans (Fantes et al., 2003). Sox3-null mice have craniofacial abnormalities and defective pituitary development (Rizzoti et al., 2004; Rizzoti and Lovell-Badge, 2007), and Sox1-null mice suffer from epilepsy due to a complete loss of neurons in the ventral striatum (Malas et al., 2003; Ekonomou et al., 2005).
In early neural development, vertebrate SoxB1 proteins function as transcriptional activators (Bylund et al., 2003; Graham et al., 2003; Uchikawa et al., 1999). It has been proposed that they counteract neuronal differentiation induced by the SoxB2 subfamily of repressor proteins (Sox14 and Sox21) (Bylund et al., 2003). In this way, the two subgroups of SoxB proteins together may maintain a balance between proliferation and differentiation. In concordance, over-expression of both Sox2 and Sox3 in frog and zebrafish leads to a loss of neurogenesis in the cranial placodes (Dee et al., 2008; Schlosser et al., 2008) and expansion of neural progenitors in the frog and chick neural tube at the expense of epidermal development and neuronal differentiation (Kishi et al., 2000; Graham et al., 2003; Rogers et al., 2009).
The SoxC gene, Sox11, is expressed in the developing nervous system of chick (Uwanogho et al., 1995), mouse (Hargrave et al., 1997), zebrafish (de Martino et al., 2000), and frog (Hyodo-Miura et al., 2002; Brugmann et al., 2004). Sox11 is induced in response to BMP inhibition by Chordin (Hyodo-Miura et al., 2002) and by ectopic expression of the FoxD5 transcription factor (Yan et al., 2009). In Xenopus animal cap assays, Sox11 expression induces pan-neural (Ncam) and anterior neural (Otx2, En2) markers through an interaction with the MAP kinase, NLK (Hyodo-Miura et al., 2002). Sox11 is thought to be functionally redundant with the other SoxC proteins, Sox4 and Sox12, due to their overlapping expression patterns, ability to bind to the same DNA gene sequence, and weak single gene knockout phenotypes in mouse (Hoser et al., 2008; Dy et al., 2008). In chick, while Sox11 is expressed in the neural epithelium, it also is up-regulated in maturing neurons (Uwanogho et al., 1995), and functions in the differentiation of neural progenitors into neurons (Bergsland et al., 2006).
SoxD, a member of the SoxG group, is unique to amphibians (Mizuseki et al., 1998b, Guth and Wegner, 2008,). It is first detected at late blastula stages throughout the embryonic ectoderm, and like Sox3, it is then restricted to the dorsal ectoderm during gastrulation. SoxD is induced in response to the inhibition of BMP signaling, and like Sox2, can be induced in AC ectodermal explants by Zic1. Little is known about the requirement for FGF or Wnt signaling for the expression of SoxD, although the MEK5/ERK5 pathway has been implicated in its neuralizing activity (Nishimoto et al., 2005). Over-expression of SoxD in whole embryos expanded neural progenitors marked by Nrp1, induced the neural determination bHLH gene Ngnr1, and inhibited epidermal gene expression. Inhibition of SoxD by a dominant-negative construct suppressed neural tissues, a phenotype that was not rescued by Sox2, indicating that SoxD acts downstream (Mizuseki et al., 1998b).
Geminin
Geminin is a novel coiled-coil protein whose mRNA is expressed in a similar pattern as Sox3 (Fig. 4); it is maternally expressed and ubiquitous in the animal pole ectoderm until gastrulation when it is first enriched dorsally, and then restricted to the neural ectoderm (Kroll et al., 1998). After gastrulation, Geminin expression is pan-neural with an anterior bias and encompasses a region wider than that of either Sox2 or Sox3 (Kroll et al., 1998; Kroll, 2007). Geminin expression can be induced by inhibiting BMP signaling, and it is maintained in the proliferative regions of the developing nervous system (Kroll, 2007). Geminin maintains a neural progenitor population by inhibiting re-initiation of DNA replication, thereby maintaining chromosomal integrity and preventing cell cycle exit (Seo and Kroll, 2006). The latter is controlled by an antagonistic interaction with the catalytic subunits of the SWI/SNF complex, Brg1 and Brahma (McGarry and Kirschner, 1998; Seo et al., 2005). Over-expression of Geminin inhibits epidermal development and neuronal differentiation, and expands the neural progenitor population (Kroll et al., 1998; Papanayotou et al., 2008). It has been suggested that Geminin maintains cell cycling during neural plate stages and represses various aspects of differentiation, allowing for the expansion of neural ectoderm (Luo and Kessel, 2004; Pitulescu et al., 2005; Seo and Kroll, 2006).
Experiments using a human Geminin regulatory region driving GFP expression indicated that as for Sox3, Vent1 and Vent2 restrict Geminin to the dorsal side of the embryo (Kroll et al., 1998). Dorsal expression of this Geminin reporter construct required positive regulation by the Wnt signaling effector, TCF3 (Taylor et al., 2006). Additionally, recent experiments demonstrated that Geminin expression is activated directly by two other early neural proteins, Sox3 (Rogers et al., 2009) and FoxD5 (Yan et al., 2009).
FoxD5
FoxD5 is a member of the forkhead/winged helix family of transcription factors (Solter et al., 1999; Fetka et al., 2000; Sullivan et al., 2001). Although there are homologous genes in zebrafish (fkd8; Odenthal and Nusslein-Volhard, 1998), mouse (FoxD4/Fkh2; Kaestner et al., 1995), and human (FOXD5 [2q13]; Katoh and Katoh, 2004), no functional information is currently available for any of these. In Xenopus, FoxD5 maternal transcripts are localized to the animal pole, similar to those of Sox3 and Geminin. However, at gastrulation, FoxD5 zygotic transcripts are more restricted than those of Sox3 and Geminin in the presumptive neural ectoderm (Fig. 4). Unlike the other early neural transcription factor genes, FoxD5 expression is not maintained throughout neural development, but is extinguished as the neural folds elevate and fuse, except at the midbrain/hindbrain junction and the tail bud. Also, unlike the other neural genes, FoxD5 is only weakly induced by Noggin, not induced by Chordin, but is induced strongly by the maternally regulated Siamois pathway via Cerberus (Sullivan et al., 2001).
It is proposed that FoxD5, like Geminin and the SoxB1 proteins, functions to maintain a proliferating neural progenitor population. Dorsal injections of FoxD5 mRNA expanded expression of other early neural genes (Sox2, Sox3, Otx2), and repressed neural patterning (En2, Krox20) and neural differentiation (Ngnr1, NeuroD, N-tub) genes (Sullivan et al., 2001). While expression of FoxD5 induced several neural genes in AC ectodermal explants (Sullivan et al., 2001), it only induces a few of these in the ventral ectoderm of embryos (Yan et al., 2009), suggesting that it acts downstream of neural induction. Microinjections of FoxD5 dominant-activator and dominant-repressor constructs revealed that FoxD5 functions as a transcriptional repressor to expand the neural plate and deletion analysis revealed that it is dependent on the C-terminal domain of the protein. These results indicate that FoxD5 contributes to maintaining an undifferentiated neural ectoderm during the early steps of neural plate formation.
Zic genes
Three Zic genes (Zic1, Zic2, Zic3), members of the zinc finger family of transcription factors, are expressed in the early neural ectoderm (Fig. 4). All three are expressed in the presumptive neural ectoderm during early gastrulation, and later become restricted to the lateral margins of the neural plate in mouse, fish, and frog (reviewed in Moody and Je, 2002). In Xenopus, Zic1 and Zic3 are transcribed 30-60 minutes after the Chordin gene, and can be induced in AC ectodermal explants by BMP antagonism (Nakata et al., 1997; Kuo et al., 1998; Mizuseki et al., 1998a; Nakata et al., 1998), indicating that they are likely transcribed in immediate response to neural induction. Expression of Zic1 allows ectoderm to be more sensitive to neural induction by Noggin (Kuo et al., 1998), confirming its role in neural fate stabilization. Over-expression of Zic1 and Zic3 expands the neural plate and neural crest, and concomitantly represses epidermal fate. In AC ectodermal explants, Zic1 activates other early neural markers (Nrp1, SoxD) in the absence of mesoderm induction. Zic1 and Zic3 both expand the expression of neural determination/differentiation bHLH genes (Mizuseki et al., 1998a), suggesting that they promote the transition to neural differentiation.
Zic2 is expressed in a similar pattern to Zic1 and Zic3 (Brewster et al., 1998; Nakata et al., 1998), and it similarly expands the neural plate and neural crest, and reduces epidermal markers (Brewster et al., 1998). In contrast, Zic2 over-expression represses neural determination/differentiation bHLH genes, and can counteract the formation of ectopic neurons produced by Ngnr1 mRNA injection. These studies suggest that during neural plate formation Zic1 and Zic3 promote the onset of neural differentiation, whereas Zic2 maintains cells in a more immature progenitor state.
Iroquois genes
Genes within the Drosophila Iroquois complex (Iro/Irx) encode homeodomain proteins that are required for the activation of proneural bHLH genes (Gomez-Skarmeta et al., 1996; Gomez-Skarmeta and Modollel, 2002). Several vertebrate homologues have been cloned, three of which in Xenopus (Xiro1, Xiro2, Xiro3) are expressed in restricted bands in the neural ectoderm (Fig. 4) just prior to the earliest expressed neural determination bHLH genes (Ngnr1 and NeuroD). Later in development, Xiro1-3 are expressed only in the dorso-lateral neural tube and neural crest. All three Xiro genes are induced only weakly by Noggin, but their expression is greatly enhanced by the addition of posteriorizing factors, such as Wnt and FGF. These data indicate that they are expressed downstream of the afore-described early neural genes, which do not require posteriorizing factors for their induction. In Xenopus, over-expression of each of the three Xiro genes causes the neural plate to expand, and promotes the onset of neural differentiation (Bellefroid et al., 1998; Gomez-Skarmeta et al., 1998), but suppresses terminal differentiation into neurons (de la Calle-Mustienes et al., 2002). Thus, these genes are predicted to function in a position intermediate between the above-described early neural genes and the neural determination/differentiation bHLH genes (Fig. 5).
Figure 5.

A summary of the data discussed in the text predicts a gene regulatory network that results in the stabilization of neural fate. After these 12 NFS genes are induced, FoxD5 up-regulates Geminin, Sox11, and Zic2 expression. These three genes regulate each other and together (bracket) they differentially affect the expression of the other 8 NFS genes. Geminin and Sox2/Sox3 directly regulate each other, and increased Zic1 up-regulates Sox2 and SoxD. FoxD5, Geminin, Sox11, and Zic2 maintain neural ectodermal cells in an immature, stem-like state, whereas Zic1, Zic3, SoxD, and Xiro1-3 promote the onset of neural differentiation.
Do early neural transcription factors interact to regulate neural fate stabilization?
Because of their overlapping expression patterns and their common over-expression phenotype of expanding the neural plate, it seems likely that together these early neural genes coordinately stabilize the newly induced neural fate, regulate neural plate formation, and control the onset of neural differentiation. However, our understanding of how this is accomplished is woefully incomplete. Several studies indicate that one aspect of stabilizing neural fate is by modifying the signaling pathways involved in neural induction. For example: (1) in the presence of Zic1 a lower concentration of Noggin is required to induce neural ectoderm (Kuo et al., 1998); (2) Sox11 induces neural marker genes by antagonizing Wnt signaling (Hyodo-Miura et al., 2002); and (3) Geminin, Sox3, and Xiro1 each antagonize some aspect of the BMP4 signaling pathway (Kroll et al., 1998; Glavic et al., 2001; Gomez-Skarmeta et al., 2001; Rogers et al., 2008), and FoxD5 reduces the nuclear localization of the BMP signaling effectors, phosphorylated SMAD1/5/8, reduces the ventral epidermal expression of BMP4 target genes, and up-regulates the expression of Szl, a secreted BMP antagonist (Yan et al., 2009). Thus, one aspect of neural fate stabilization appears to be the regulation of the strength of signaling pathways, in particular that of BMP4, in the neural ectoderm. However, we know virtually nothing about how these several early neural genes, henceforth called neural fate stabilizing (NFS) genes, may cooperatively cause these effects.
The over-expression studies reviewed above indicate that the NFS genes can be placed into two functional groups: those that promote an immature, undifferentiated neural state (Geminin, Sox2, Sox3, FoxD5, Zic2), and those that promote the onset of neural differentiation (SoxD, Sox11, Zic1, Zic3, Xiro1, Xiro2, Xiro3). Are the genes within each group simply redundant, or do they have distinct roles in the transition from neural induction to neural differentiation? Answers to these questions are only just beginning to be experimentally addressed.
Recently, as a first step in putting the NFS genes into transcriptional order, we found that when endogenous FoxD5 levels in the neural ectoderm were reduced by targeted injection of anti-sense MOs, the expression of each of the 11 other NFS genes was down-regulated (Yan et al., 2009). This suggests that FoxD5 is necessary for either the induction or the maintenance of their expression. As expected from previous studies (Sullivan et al., 2001), over-expression of FoxD5 expanded the neural plate and thus the domains of each of the 11 other NFS genes; this was a cell non-autonomous effect because the neural plate was expanded equally well in regions devoid of the FoxD5-expressing cells, which were marked by a lineage tracer. However, when only the lineage-tagged cells expressing elevated levels of FoxD5 were examined, several responses were observed: (1) some genes that promote an immature neural fate (Geminin, Zic2) were up-regulated; (2) Sox11, which previously had been characterized as promoting neural differentiation (Bergsland et al., 2006), also was up-regulated; (3) some genes that promote an immature neural fate but are required for neural differentiation (Sox2, Sox3) were transiently and weakly repressed; and (4) genes that promote the onset of neural differentiation (Zic1, Zic3, SoxD, Xiro1, Xiro2, Xiro3) were strongly repressed. Thus, FoxD5 appears to maintain an immature neural state by differentially regulating genes that either repress or promote neural differentiation. Further experiments demonstrated that FoxD5 directly regulates Geminin, Sox11, and Zic2 transcription, and that these three genes together phenocopy FoxD5 over-expression and rescue FoxD5 MO-mediated knock-down phenotypes.
These results indicate that the NFS genes coordinately stabilize neural fate and regulate the initiation of neural differentiation, and data from several laboratories predict a NFS gene regulatory network (Fig. 5). FoxD5 acts upstream of the other NFS genes, primarily by regulating Geminin, Sox11, and Zic2. This transcriptional triad cooperatively carries out most of the effects of FoxD5 on the other NFS genes. They also regulate each other’s expression, indicating a network rather than a linear path of transcriptional regulation. There also are mutual interactions between Geminin and SoxB1 genes. Chick Geminin interacts in a complex at the N2 enhancer to promote Sox2 transcription (Papanayotou et al., 2008), and frog Sox2 and Sox3 directly regulate Geminin expression (Rogers et al., 2009). As mentioned above, Zic1 up-regulates both Sox2 and SoxD expression. In addition, those genes that promote the onset of neural differentiation (Zic1, Zic3, SoxD, Xiro1, Xiro2, Xiro3) feedback to repress FoxD5 expression, perhaps to allow the neural ectodermal cells to initiate differentiation (Yan et al., 2009). These studies are just the beginning of defining the regulatory network that promotes the transition from neural induction to differentiation. Much more work is needed to fully understand the gene interactions that comprise this network, and to determine what other factors are involved.
How Might NFS Genes Regulate Neural Stem Cells?
Stem cell therapy is expected to have widespread benefits to patients suffering from neurodegenerative diseases and neuronal loss due to trauma, stroke and congenital disease. Although the mature CNS harbors niches of self-renewing neural stem cells (Doetsch et al., 1999; Alvarez-Buylla et al., 2001), these make a significant contribution to only a few areas of brain (Cameron and McKay, 2001; Pencea et al., 2001). Therefore, many laboratories world-wide are researching how to manipulate different sources of stem cells to produce specific types of neurons for therapeutic treatments. It is most exciting that many of the NFS genes have already been implicated in controlling the differentiation of neural stem and progenitor cells. Sox2 and Sox3 are necessary for neural differentiation (Kishi et al., 2000; Wegner and Stolt, 2005; Wang et al., 2006). Each maintains neural stem/progenitor cells in a proliferative state upstream of neuronal terminal differentiation genes (Li et al., 1998; Zappone et al., 2000; Bylund et al., 2003; Graham et al., 2003; Ellis et al., 2004; Chung et al., 2006; Wang et al., 2006). Sox2 is required for adult neural stem cell maintenance and its loss causes cells to leave the ventricular zone and exit the cell cycle (Graham et al., 2003; Ellis et al., 2004; Episkopou, 2005). In contrast, Sox1 and Sox3 appear to regulate neural progenitor cell states (Kan et al., 2004; Barraud et al., 2005; Chung et al., 2006; Wang et al., 2006). Sox11 is reported to be up-regulated as neural stem cells transition to neural progenitor cells, and later it maintains pan-neural genes in neuronal progenitors downstream of bHLH differentiation factors (Uwanogho et al., 1995; Wegner and Stolt, 2005; Bergsland et al., 2006). Thus, together these Sox genes may function downstream of FoxD5, Geminin, and Zic2 to promote the initial steps from neural stem to neural progenitor cell. FoxD5 transiently represses Sox2, Sox3, and Sox11 (Yan et al., 2009), suggesting that it may act to delay the stem-to-progenitor transition. Zic genes, which promote the expression of neural determination/differentiation bHLH factors, are expressed in proliferative neural progenitor cells and are required for their expansion (Aruga et al., 2002; Ebert et al., 2003; Inoue et al., 2007). Iro/Irx genes also are involved in the initiation of expression of the neural determination/differentiation bHLH factors. Later, they play important roles in patterning the CNS (Rodriguez-Sequel et al., 2009; Stedman et al., 2009), and appear to regulate the number of cranial placode progenitors (Feijoo et al., 2009).
Conclusions
Much has been learned about the endogenous signaling pathways and transcriptional factors that induce the neural ectoderm and stabilize its neural fate. However, new experimental tactics will be needed to sort out how the various signaling pathways intersect to produce a neural fate, and likewise how the several NFS transcription factors regulate one another. Ultimately, producing neurally-committed stem and progenitor cells in sufficient quantities and at the appropriate state of differentiation for clinical applications requires that we understand the molecular mechanisms that regulate the normal process of in vivo neural development in greater detail.
Acknowledgements
Some of the work presented in this review was supported by NIH grant NS23158 (SAM) and NIH grant NS048918 (ESC).
References
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM. Endodermal nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez IS, Araujo M, Nieto MA. Neural induction in whole chick embryo cultures by FGF. Dev Biol. 1998;199:42–54. doi: 10.1006/dbio.1998.8903. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla AJ, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nature Rev. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Aruga J, Tohmonda T, Homma S, Mikoshiba K. Zic1 promotes expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev Biol. 2002;244:329–341. doi: 10.1006/dbio.2002.0598. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Bainter JJ, Boos A, Kroll KL. Neural induction takes a transcriptional twist. Dev Dyn. 2001;222:315–327. doi: 10.1002/dvdy.1210. [DOI] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999;13:3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- Barraud P, Thompson L, Kirik D, Bjorklund A, Parmar M. Isolation and characterization of neural precursor cells from the Sox1-GFP reporter mouse. Eur Neurosci. 2005;22:1555–1569. doi: 10.1111/j.1460-9568.2005.04352.x. [DOI] [PubMed] [Google Scholar]
- Beddington RSP. Induction of a secondary neural axis by the mouse node. Development. 1994;120:613–620. doi: 10.1242/dev.120.3.613. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belo JA, Bachiller D, Agius E, Kemp C, Borges AC, Marques S, Piccolo S, De Robertis EM. Cerberus-like is a secreted BMP and nodal antagonist not essential for mouse development. Genesis. 2000;26:265–270. [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger T, Knoetgen H, Witte L, Kessel M. The avian organizer. Int J Dev Biol. 2001;45:281–287. [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cameron HA, McKay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol. 2001;435:406–417. doi: 10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- Chang C, Harland RM. Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development. 2007;134:3861–3872. doi: 10.1242/dev.007179. [DOI] [PubMed] [Google Scholar]
- Christian JL, Gavin BJ, McMahon AP, Moon RT. Isolation of cDNAs partially encoding four Xenopus Wnt-1/int-1-related proteins and characterization of their transient expression during embryonic development. Dev Biol. 1991a;143:230–234. doi: 10.1016/0012-1606(91)90073-c. [DOI] [PubMed] [Google Scholar]
- Christian JL, McMahon JA, McMahon AP, Moon RT. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development. 1991b;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- Chung S, Shin BS, Hedlund E, Pruszak J, Ferree A, Kang UJ, Isacson O, Kim KS. Genetic selection of sox1 GFP-expressing neural precursors removes residual tumorigenic pluripotent stem cells and attenuates tumor formation after transplantation. J Neurochem. 2006;97:1467–1480. doi: 10.1111/j.1471-4159.2006.03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Glavic A, Modolell J, Gomez-Skarmeta JL. Xiro homeoproteins coordinate cell cycle exit and primary neuron formation by upregulating neuronal-fate repressors and downregulating the cell-cycle inhibitor XGadd45-γ. Mech Dev. 2002;119:69–80. doi: 10.1016/s0925-4773(02)00296-4. [DOI] [PubMed] [Google Scholar]
- de Martino S, Yan YL, Jowett T, Postlethwait JH, Varga ZM, Ashworth A, Austin CA. Expression of sox11 gene duplicates in zebrafish suggests the reciprocal loss of ancestral gene expression patterns in development. Dev Dyn. 2000;217:279–292. doi: 10.1002/(SICI)1097-0177(200003)217:3<279::AID-DVDY6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee CT, Hirst CS, Shih YH, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Doetsche F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, Liu Y, Hunsaker TL, Johnson JE. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 auto-regulation. Development. 2003;130:1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- Ekonomou A, Kazanis I, Malas S, Wood H, Alifragis P, Denaxa M, Karagogeos D, Constanti A, Lovell-Badge R, Episkopou V. Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 2005;3:e186. doi: 10.1371/journal.pbio.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins MB, Henry JJ. Isolation and characterization of a novel gene, xMADML, involved in Xenopus laevis eye development. Dev Dyn. 2006;235:1845–1857. doi: 10.1002/dvdy.20824. [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Episkopou V. SOX2 function in adult neural stem cells. Trends Neurosci. 2005;28:219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- Fainsod A, Steinbeisser H, De Robertis EM. On the function of BMP-4 in patterning the marginal zone of the Xenopus embryo. EMBO J. 1994;13:5015–5025. doi: 10.1002/j.1460-2075.1994.tb06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- Feijoo CG, Saldias MP, De la Paz JF, Gomez-Skarmeta JL, Allende ML. Formation of posterior cranial placode derivatives requires the Iroquois transcription factor irx4a. Mol Cell Neurosci. 2009;40:328–337. doi: 10.1016/j.mcn.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Feledy JA, Beanan MJ, Sandoval JJ, Goodrich JS, Lim JH, Matsuo-Takasaki M, Sato SM, Sargent TD. Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev Biol. 1999;212:455–464. doi: 10.1006/dbio.1999.9374. [DOI] [PubMed] [Google Scholar]
- Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Fetka I, Doederlein G, Bouwmeester T. Neuroectodermal specification and regionalization of the Spemann organizer in Xenopus. Mech Dev. 2000;93:49–58. doi: 10.1016/s0925-4773(00)00265-3. [DOI] [PubMed] [Google Scholar]
- Foerst-Potts L, Sadler TW. Disruption of Msx-1 and Msx-2 reveals roles for these genes in craniofacial, eye, and axial development. Dev Dyn. 1997;209:70–84. doi: 10.1002/(SICI)1097-0177(199705)209:1<70::AID-AJA7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Friedle H, Knochel W. Cooperative interaction of Xvent-2 and GATA-2 in the activation of the ventral homeobox gene Xvent-1B. J Biol Chem. 2002;277:23872–23881. doi: 10.1074/jbc.M201831200. [DOI] [PubMed] [Google Scholar]
- Friedle H, Rastegar S, Paul H, Kaufmann E, Knochel W. Xvent-1 mediates BMP-4-induced suppression of the dorsal-lip-specific early response gene XFD-1′ in Xenopus embryos. EMBO J. 1998;17:2298–2307. doi: 10.1093/emboj/17.8.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furthauer M, Thisse C, Thisse B. A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development. 1997;124:4253–4264. doi: 10.1242/dev.124.21.4253. [DOI] [PubMed] [Google Scholar]
- Gamse J, Sive H. Vertebrate anteroposterior patterning: the Xenopus neurectoderm as a paradigm. Bioessays. 2000;22:976–986. doi: 10.1002/1521-1878(200011)22:11<976::AID-BIES4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. EMBO J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Danilchik M, Doniach T, Roberts S, Rowning B, Stewart R. Cortical rotation of the Xenopus egg: consequences for the anteroposterior pattern of embryonic dorsal development. Development. 1989;107(Suppl):37–51. doi: 10.1242/dev.107.Supplement.37. [DOI] [PubMed] [Google Scholar]
- Glavic A, Gomez-Skarmeta JL, Mayor R. Xiro-1 controls mesoderm patterning by repressing bmp-4 expression in the Spemann organizer. Dev Dyn. 2001;222:368–376. doi: 10.1002/dvdy.1189. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta J, de La Calle-Mustienes E, Modolell J. The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and downregulates Bmp4. Development. 2001;128:551–560. doi: 10.1242/dev.128.4.551. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferre-Marco D, Modolell J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Glavic A, de la Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Modollel J. Iroquois genes:genomic organization and function in vertebrate neural development. Curr Opin Genet Dev. 2002;12:403–408. doi: 10.1016/s0959-437x(02)00317-9. [DOI] [PubMed] [Google Scholar]
- Gould SE, Grainger RM. Neural induction and antero-posterior patterning in the amphibian embryo: past, present and future. Cell Mol Life Sci. 1997;53:319–338. doi: 10.1007/PL00000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guth SI, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave M, Wright E, Kun J, Emery J, Cooper L, Koopman P. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- Heeg-Truesdell E, Labonne C. Neural induction in Xenopus requires inhibition of Wnt-beta-catenin signaling. Dev Biol. 2006;298:71–86. doi: 10.1016/j.ydbio.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Thomsen GH. Ventral mesodermal patterning in Xenopus embryos: expression patterns and activities of BMP-2 and BMP-4. Dev Genet. 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Wright DA, Melton DA. Embryonic expression and functional analysis of a Xenopus activin receptor. Dev Dyn. 1992;194:1–11. doi: 10.1002/aja.1001940102. [DOI] [PubMed] [Google Scholar]
- Hongo I, Kengaku M, Okamoto H. FGF signaling and the anterior neural induction in Xenopus. Dev Biol. 1999;216:561–581. doi: 10.1006/dbio.1999.9515. [DOI] [PubMed] [Google Scholar]
- Hoser M, Potzner MR, Koch JM, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado C, De Robertis EM. Neural induction in the absence of organizer in salamanders is mediated by MAPK. Dev Biol. 2007;307:282–289. doi: 10.1016/j.ydbio.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo-Miura J, Urushiyama S, Nagai S, Nishita M, Ueno N, Shibuya H. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells. 2002;7:487–496. doi: 10.1046/j.1365-2443.2002.00536.x. [DOI] [PubMed] [Google Scholar]
- Inoue T, Ota M, Ogawa M, Mikoshiba M, Aruga J. Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors. J Neurosci. 2007;27:5461–5473. doi: 10.1523/JNEUROSCI.4046-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura A, Maeda R, Takeda M, Kikkawa M, Daar IO, Maeno M. Involvement of BMP-4/msx-1 and FGF pathways in neural induction in the Xenopus embryo. Dev Growth Differ. 2000;42:307–316. doi: 10.1046/j.1440-169x.2000.00514.x. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Monaghan AP, Kern H, Ang SL, Weitz S, Lichter P, Schutz G. The mouse fkh-2 gene. Implications for notochord, foregut, and midbrain regionalization. J Biol Chem. 1995;270:30029–30035. doi: 10.1074/jbc.270.50.30029. [DOI] [PubMed] [Google Scholar]
- Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Human FOX gene family. Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Kroll KL. Geminin in embryonic development: coordinating transcription and the cell cycle during differentiation. Front Biosci. 2007;12:1395–1409. doi: 10.2741/2156. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Kuo JS, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Fuentealba L, Ikeda A, Reversade B, De Robertis EM. Default neural induction: neuralization of dissociated Xenopus cells is mediated by Ras/MAPK activation. Genes Dev. 2005;19:1022–1027. doi: 10.1101/gad.1306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher R, Mohun TJ, Smith JC, Snape AM. Xom: a Xenopus homeobox gene that mediates the early effects of BMP-4. Development. 1996;122:2385–2394. doi: 10.1242/dev.122.8.2385. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay C, Fromentoux V, Shi DL, Boucaut JC. A truncated FGF receptor blocks neural induction by endogenous Xenopus inducers. Development. 1996;122:869–880. doi: 10.1242/dev.122.3.869. [DOI] [PubMed] [Google Scholar]
- Li M, Pevny L, Lovell-Badge R, Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131:5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- Malas S, Postlethwaite M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, Meldrum B, Constanti A, Episkopou V. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience. 2003;119:421–432. doi: 10.1016/s0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Moon RT. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H. Organizer and axes formation as a self-organizing process. Int J Dev Biol. 2001;45:177–188. [PubMed] [Google Scholar]
- Miller-Bertoglio V, Carmany-Rampey A, Furthauer M, Gonzalez EM, Thisse C, Thisse B, Halpern ME, Solnica-Krezel L. Maternal and zygotic activity of the zebrafish ogon locus antagonizes BMP signaling. Dev Biol. 1999;214:72–86. doi: 10.1006/dbio.1999.9384. [DOI] [PubMed] [Google Scholar]
- Miyagi S, Masui S, Niwa H, Saito T, Shimazaki T, Okano H, Nishimoto M, Muramatsu M, Iwama A, Okuda A. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998a;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998b;21:77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Moody SA, Je HS. Neural induction, neural fate stabilization, and neural stem cells. ScientificWorldJournal. 2002;2:1147–1166. doi: 10.1100/tsw.2002.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc Natl Acad Sci U S A. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD. The “organization centre”. 3. Segregation and pattern formation in morphogenetic fields. Acta Biotheor. 1967;17:178–194. doi: 10.1007/BF01601987. [DOI] [PubMed] [Google Scholar]
- Nishimoto S, Kusakabe M, Nishida E. Requirement of the MEK5-ERK5 pathway for neural differentiation in Xenopus embryonic development. EMBO Rep. 2005;6:1064–1069. doi: 10.1038/sj.embor.7400515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, Takahashi S, Haramoto Y, Fukuda M, Onuma Y, Asashima M. Expression of Sox1 during Xenopus early embryogenesis. Biochem Biophys Res Commun. 2006;351:287–293. doi: 10.1016/j.bbrc.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. Fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–1456. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- Otte AP, Moon RT. Ectopic induction of dorsal mesoderm by overexpression of Xwnt-8 elevates the neural competence of Xenopus ectoderm. Dev Biol. 1992;152:184–187. doi: 10.1016/0012-1606(92)90168-g. [DOI] [PubMed] [Google Scholar]
- Papanayotou C, Mey A, Birot AM, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencea V, Bingaman KD, Freedman LJ, Luskin MB. Neurogenesis in the subventricular zone and rostral migratory stream of the neonatal and adult primate forebrain. Exp Neurol. 2001;172:1–16. doi: 10.1006/exnr.2001.7768. [DOI] [PubMed] [Google Scholar]
- Penzel R, Oschwald R, Chen Y, Tacke L, Grunz H. Characterization and early embryonic expression of a neural specific transcription factor xSOX3 in Xenopus laevis. Int J Dev Biol. 1997;41:667–677. [PubMed] [Google Scholar]
- Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu M, Kessel M, Luo L. The regulation of embryonic patterning and DNA replication by geminin. Cell Mol Life Sci. 2005;62:1425–1433. doi: 10.1007/s00018-005-4553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar S, Friedle H, Frommer G, Knochel W. Transcriptional regulation of Xvent homeobox genes. Mech Dev. 1999;81:139–49. doi: 10.1016/s0925-4773(98)00239-1. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Bakkers J, Kramer C, Hammerschmidt M. Fgf signaling induces posterior neuroectoderm independently of Bmp signaling inhibition. Dev Dyn. 2004;231:750–757. doi: 10.1002/dvdy.20244. [DOI] [PubMed] [Google Scholar]
- Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Ribisi S, Jr, Mariani FV, Aamar E, Lamb TM, Frank D, Harland RM. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev Biol. 2000;227:183–196. doi: 10.1006/dbio.2000.9889. [DOI] [PubMed] [Google Scholar]
- Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- Rizzoti K, Lovell-Badge R. SOX3 activity during pharyngeal segmentation is required for craniofacial morphogenesis. Development. 2007;134:3437–3448. doi: 10.1242/dev.007906. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gallardo L, Climent V, Garcia-Martinez V, Schoenwolf GC, Alvarez IS. Targeted over-expression of FGF in chick embryos induces formation of ectopic neural cells. Int J Dev Biol. 1997;41:715–723. [PubMed] [Google Scholar]
- Rodriguez-Seguel E, Alarcon P, Gomez-Skarmeta The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev Biol. 2009;329:258–268. doi: 10.1016/j.ydbio.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Archer TC, Cunningham DD, Grammer TC, Casey EM. Sox3 expression is maintained by FGF signaling and restricted to the neural plate by Vent proteins in the Xenopus embryo. Dev Biol. 2008;313:307–319. doi: 10.1016/j.ydbio.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Harafuji N, Archer T, Cunningham DD, Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Identifying the missing links: genes that connect neural induction and primary neurogenesis in vertebrate embryos. Neuron. 1998;21:455–458. doi: 10.1016/s0896-6273(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- Sato SM, Sargent TD. Development of neural inducing capacity in dissociated Xenopus embryos. Dev Biol. 1989;134:263–266. doi: 10.1016/0012-1606(89)90096-1. [DOI] [PubMed] [Google Scholar]
- Schier AF. Axis formation and patterning in zebrafish. Curr Opin Genet Dev. 2001;11:393–404. doi: 10.1016/s0959-437x(00)00209-4. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Kroll KL. Geminin’s double life: chromatin connections that regulate transcription at the transition from proliferation to differentiation. Cell Cycle. 2006;5:374–379. doi: 10.4161/cc.5.4.2438. [DOI] [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Shih J, Fraser SE. Characterizing the zebrafish organizer: microsurgical analysis at the early-shield stage. Development. 1996;122:1313–1322. doi: 10.1242/dev.122.4.1313. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Smith WC, McKendry R, Ribisi S, Jr, Harland RM. A nodal-related gene defines a physical and functional domain within the Spemann organizer. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- Smith JL, Schoenwolf GC. Getting organized: new insights into the organizer of higher vertebrates. Curr Top Dev Biol. 1998;40:79–110. doi: 10.1016/s0070-2153(08)60365-8. [DOI] [PubMed] [Google Scholar]
- Solter M, Koster M, Hollemann T, Brey A, Pieler T, Knochel W. Characterization of a subfamily of related winged helix genes, XFD-12/12′/12” (XFLIP), during Xenopus embryogenesis. Mech Dev. 1999;89:161–165. doi: 10.1016/s0925-4773(99)00195-1. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from different species. In: Willier BH, Oppenheimer JM, editors. Foundations of Experimental Embryology. Hafner, NY: 1924. pp. 144–184. [Google Scholar]
- Stedman A, Lecaudey V, Havis E, Anselme I, Wassef M, Gilardi-Hebenstreit P, Schneiderr-Maunoury S. A functional interaction between Irx and Meis patterns the anterior hindbrain and activates krox20 expression in rhombomere 3. Dev Biol. 2009;327:566–577. doi: 10.1016/j.ydbio.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Steinbeisser H, De Robertis EM, Ku M, Kessler DS, Melton DA. Xenopus axis formation: induction of goosecoid by injected Xwnt-8 and activin mRNAs. Development. 1993;118:499–507. doi: 10.1242/dev.118.2.499. [DOI] [PubMed] [Google Scholar]
- Sullivan SA, Akers L, Moody SA. foxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transcriptional repression that is dependent on the C-terminal domain. Dev Biol. 2001;232:439–457. doi: 10.1006/dbio.2001.0191. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development. 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- Takeda M, Saito Y, Sekine R, Onitsuka I, Maeda R, Maeno M. Xenopus msx-1 regulates dorso-ventral axis formation by suppressing the expression of organizer genes. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:157–168. doi: 10.1016/s0305-0491(00)00194-2. [DOI] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Kamachi Y, Kondoh H. Convergence of Wnt and FGF signals in the genesis of posterior neural plate through activation of the Sox2 enhancer N-1. Development. 2006;133:297–306. doi: 10.1242/dev.02196. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Wang T, Kroll KL. Tcf- and Vent-binding sites regulate neural-specific geminin expression in the gastrula embryo. Dev Biol. 2006;289:494–506. doi: 10.1016/j.ydbio.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Trindade M, Tada M, Smith JC. DNA-binding specificity and embryological function of Xom (Xvent-2) Dev Biol. 1999;216:442–456. doi: 10.1006/dbio.1999.9507. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev Dyn. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Ueno H, Gunn M, Dell K, Tseng A, Jr, Williams L. A truncated form of fibroblast growth factor receptor 1 inhibits signal transduction by multiple types of fibroblast growth factor receptor. J Biol Chem. 1992;267:1470–1476. [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Vonica A, Gumbiner BM. The Xenopus Nieuwkoop center and Spemann-Mangold organizer share molecular components and a requirement for maternal Wnt activity. Dev Biol. 2007;312:90–102. doi: 10.1016/j.ydbio.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley ME, Guille MJ, Bertwistle D, Smith JC, Pizzey JA, Patient RK. Negative control of Xenopus GATA-2 by activin and noggin with eventual expression in precursors of the ventral blood islands. Development. 1994;120:2519–2529. doi: 10.1242/dev.120.9.2519. [DOI] [PubMed] [Google Scholar]
- Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J Comp Neurol. 2006;497:88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Hemmati-Brivanlou A. Neural induction in Xenopus laevis: evidence for the default model. Curr Opin Neurobiol. 1997;7:7–12. doi: 10.1016/s0959-4388(97)80114-6. [DOI] [PubMed] [Google Scholar]
- Weisinger K, Wilkinson DG, Sela-Donenfeld D. Inhibition of BMPs by follistatin is required for FGF3 expression and segmental patterning of the hindbrain. Dev Biol. 2008;324:213–225. doi: 10.1016/j.ydbio.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Wessely O, Kim JI, Geissert D, Tran U, De Robertis EM. Analysis of Spemann organizer formation in Xenopus embryos by cDNA macroarrays. Dev Biol. 2004;269:552–566. doi: 10.1016/j.ydbio.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Graziano E, Harland R, Jessell TM, Edlund T. An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr Biol. 2000;10:421–429. doi: 10.1016/s0960-9822(00)00431-0. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature. 2001;411:325–330. doi: 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- Wittler L, Kessel M. The acquisition of neural fate in the chick. Mech Dev. 2004;121:1031–1042. doi: 10.1016/j.mod.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Xu RH, Kim J, Taira M, Zhan S, Sredni D, Kung HF. A dominant negative bone morphogenetic protein 4 receptor causes neuralization in Xenopus ectoderm. Biochem Biophys Res Commun. 1995;212:212–219. doi: 10.1006/bbrc.1995.1958. [DOI] [PubMed] [Google Scholar]
- Yabe T, Shimizu T, Muraoka O, Bae YK, Hirata T, Nojima H, Kawakami A, Hirano T, Hibi M. Ogon/Secreted Frizzled functions as a negative feedback regulator of Bmp signaling. Development. 2003;130:2705–2716. doi: 10.1242/dev.00506. [DOI] [PubMed] [Google Scholar]
- Yamamoto TS, Takagi C, Hyodo AC, Ueno N. Suppression of head formation by Xmsx-1 through the inhibition of intracellular nodal signaling. Development. 2001;128:2769–2779. doi: 10.1242/dev.128.14.2769. [DOI] [PubMed] [Google Scholar]
- Yamamoto TS, Takagi C, Ueno N. Requirement of Xmsx-1 in the BMP-triggered ventralization of Xenopus embryos. Mech Dev. 2000;91:131–141. doi: 10.1016/s0925-4773(99)00290-7. [DOI] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol. 2009;329:80–95. doi: 10.1016/j.ydbio.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]