Summary
σ28 controls the expression of flagella related genes and is the most widely distributed alternative σ factor, present in motile gram-positive and gram-negative bacteria. The distinguishing feature of σ28 promoters is a long −10 region (GCCGATAA). Despite the fact that the upstream GC is highly conserved, previous studies have not indicated a functional role for this motif. Here we examine the functional relevance of the GCCG motif and determine which residues in σ28 participate in its recognition. We find that the GCCG motif is a functionally important composite element. The upstream GC constitutes an extended −10 motif and is recognized by R91, a residue in Domain 3 of σ28. The downstream CG is the upstream edge of −10 region of the promoter; two residues in Region 2.4, D81 and R84, participate in its recognition. Consistent with their role in base-specific recognition of the promoter, R91, D81 and D84 are universally conserved in σ28 orthologues. σ28 is the second Group 3 σ shown to use an extended −10 region in promoter recognition, raising the possibility that other Group 3 σs will do so as well.
Keywords: Group 3 σ, σ28, extended −10 motif, −10 region, Region 2.4, Region 3.0
Introduction
Bacteria use a family of σ factors to orchestrate transcription. The housekeeping σ, called σ70 in Escherichia coli, directs core RNA polymerase (α2ββ’ω) to the vast bulk of promoters active during exponential phase, whereas alternative σ’s direct core RNA polymerase to mediate the transcription of regulons required for specific tasks such as response to stress, or mediating growth transitions or development (Gross et al., 1998, Paget & Helmann, 2003). The σ70 family of proteins has conserved modular domains with discrete functions. In addition to the N-terminal regulatory domain found only in housekeeping σs, there are three additional domains, each with recognition determinants both for core RNA polymerase and for portions of the promoter (Gruber & Gross, 2003). Thus, Domain 2 recognizes the −10 region of the promoter; Domain 3 recognizes the “extended −10 motif” immediately upstream of the −10 region; and Domain 4 recognizes the −35 region of the promoter (Campbell et al., 2002, Gross et al., 1998). In addition, Region 1.2 recognizes a motif downstream of the −10 region (Haugen et al., 2006, Feklistov et al., 2006). σs have been divided into four groups based on their phylogenetic relationships and modular structure (Paget & Helmann, 2003, Lonetto et al., 1992, Gruber & Gross, 2003). Group 1 σs (housekeeping σs) are essential and have all domains; Group 2 σs are the most related to Group 1 σs but are not essential. Group 3 σs have Domains 2, 3, and 4. Group 4 σs have only Domains 2 and 4 and comprise the largest group of σs.
Promoter recognition has been extensively studied in σ70 and several other Group 1 σs (Siegele et al., 1989, Kenney et al., 1989, Waldburger et al., 1990, Campbell et al., 2002, Sanderson et al., 2003). However, less attention has been devoted 64 to promoter recognition in the alternative σs. σ28, a Group 3 σ, is the most widely distributed alternative σ factor, making it an attractive candidate for study of its promoter recognition. σ28 controls expression of flagella-related genes in all motile Gram-negative and Gram-positive bacteria, and plays a role in development in some non-motile bacteria (e.g. Chlamydia) (Yu et al., 2006b, Yu & Tan, 2003, Shen et al., 2006, Chilcott & Hughes, 2000, Serizawa et al., 2004). The conservation of this σ across millions of years of evolution is indicated by the fact that SigD, the σ28 orthologue in B. subtilis, can substitute for the function of σ28 in E. coli (Chen & Helmann, 1992). Likewise, the σ28 promoter sequence is conserved across organisms (Serizawa et al., 2004, Shen et al., 2006). Bioinformatic analysis of E. coli σ28 promoters suggests that their consensus sequence is TAAAgttt-N11-GCCGATAA (Zhao et al., 2007). Mutational and biochemical analysis of a strong σ28 promoter validated the importance of the TAAA −35 motif and the CGA −10 motif. However, the functional relevance of the highly conserved GC motif of the −10 region remains in question (Yu et al., 2006a, Wozniak & Hughes, 2008).
Additionally, there have been no studies of the amino acid residues in σ28 that mediate recognition of the −10 region. The studies in this report clarify both the functionally relevant bases in the −10 region of the promoter and the amino acid residues in σ28 partially responsible for recognizing this region. Our studies indicate that both the GC and the CGA motifs are functionally important in the −10 region of σ28 promoter, and that this is a composite element. The upstream GC motif is an extended −10 motif recognized by a residue in first helix of Domain 3, whereas the downstream CG is part of the −10 region recognized by residues in Region 2.4 of Domain 2.
Results
Construction of in vivo assay system
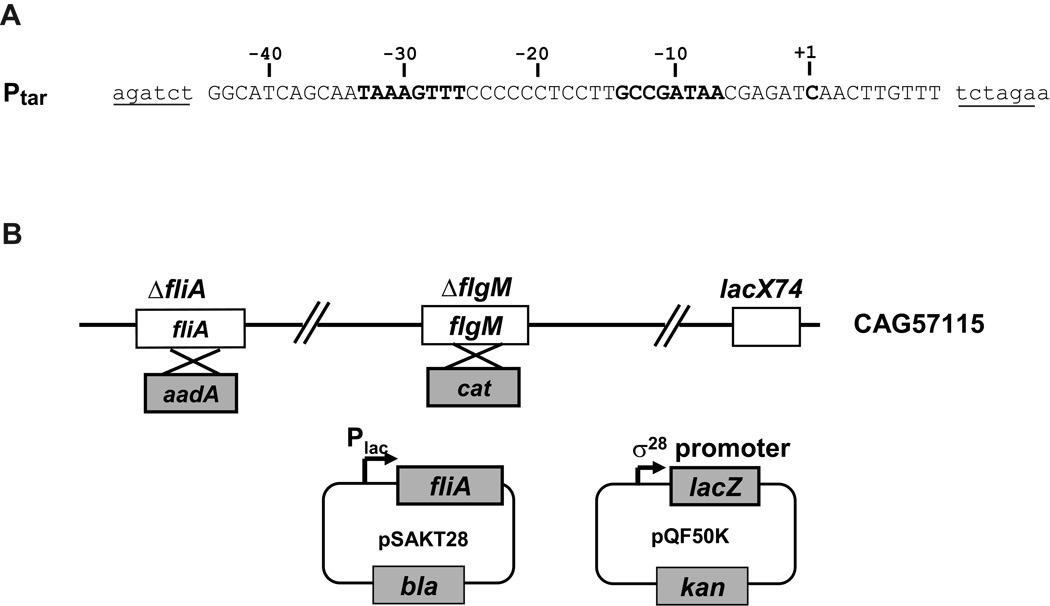
A robust in vivo assay system for identifying the residues in σ that mediate base-specific promoter interactions must be solely dependent on the activity of the particular σ mutant that is being assayed. Such an assay system has two requirements: the cellular regulatory loops that adjust the amount of σ28 to compensate for its activity deficit must be removed; and the promoter utilized must be sensitive to the level and activity of σ. We removed the cellular regulatory loops by using a host strain lacking both endogenous σ28 and its anti-σ, FlgM (Hughes et al., 1993, Chadsey et al., 1998). We based our promoter library on the core region (−44 to +10) of the tar promoter, which has consensus −35 (TAAAGTTT) and −10 (GCCGATAA) sequences (Fig. 1A). Using this promoter ensures that deviations at every position of the promoter can be assayed and that α binding to the UP element will not obscure defects of the mutant σ28 in binding to the promoter. Our assay system utilizes two plasmids, one supplying mutant σ28 alleles, and the other using variants of the σ28 promoter to drive expression of the lacZ reporter (Fig. 1B). Expression from the wild-type (wt) tar promoter was <1 Miller unit without induction and ~ 900 Miller units following induction of wt σ28 (data not shown). Thus, this assay system has an excellent signal to noise ratio.
Figure 1. Components of in vivo assay systems used in this study.
A. Sequence of the Ptar σ28-dependent promoter used in this study. Only the wt sequence is shown: single- or double- base changed promoter mutants were constructed from this parent. The native sequence of the tar promoter region is shown in capital letters; vector sequences are shown in lowercase; BglII and XbaI cloning sites are underlined; −35 and −10 regions, and the transcription start site are shown in bold.
B. Activity of σ28-dependent promoters in the presence of σ28 variants was determined by β-galactosidase assay from the ΔfliA and flgM strain, CAG57115, carrying the plasmids pSAKT28 and pQF50K. σ28expression was induced from pSAKT28 and derivatives (pBK201-pBK212), which has a p15A replication origin and fliA under control of the lac promoter. The reporter plasmid, pQF50K and derivatives (pBK601-pBK618) has a pMB1 replication origin and lacZ under control of the σ28-dependent tar promoter and promoter derivatives.
Determination of functional −10 promoter sequence of σ28
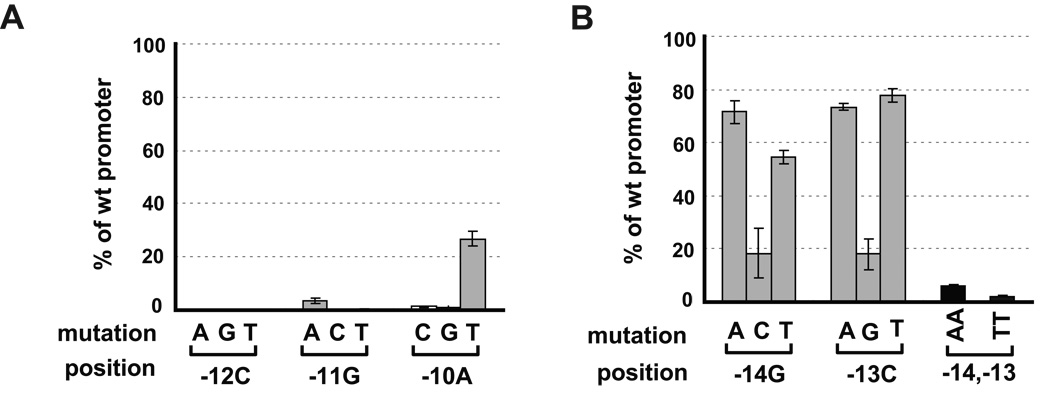
We examined the effect of base changes at positions −14 to −10 in the tar promoter. Every base mutation at the CGA motif resulted in severe defects in transcription, consistent with previous results (Fig. 2A) (Yu et al., 2006a, Wozniak & Hughes, 2008). Additionally, single mutations at the upstream GC motif did not result in a significant decrease in transcription, with exception of −14G to C and −13C to G (Fig. 2B), as has previously been seen by Wozniak and Hughes on the flgK promoter (Wozniak & Hughes, 2008). We considered the possibility that only one of the conserved upstream GC bases was required for activity. Indeed, the two doubly mutated promoters that we tested, −14 −13 GC to TT or AA, were both severely defective in expression, exhibiting only 2~5% of the activity of the wt promoter (Fig. 2B). This defect is far greater than that expected from the behavior of the single mutants, each of which exhibited 60–80% of the activity of the wt promoter. The synergistic effect of the double mutants is consistent with the idea that either −14G or −13C but not both is required for high promoter activity.
Figure 2. Effects of nucleotide changes in the −10 region on activities of tar promoter.
A. Effects of single nucleotide changes at −12C, −11G and −10A positions on activity of the tar promoter.
B. Effects of single or double nucleotides changes at −14G and −13C positions on activity of the tar promoter.
The activity of each single or double nucleotide mutant tar promoter with wt σ28 is shown as a percentage of the measured β-galactosidase activity of the wt promoter. Assay strains are as described in Fig 1B. The promoter mutations and positions are shown on the x-axis. All values are averages of three independent experiments; error bars indicate 1 standard deviation.
Identification of residues in σ28 potentially important for recognition of the −10 region of the promoter
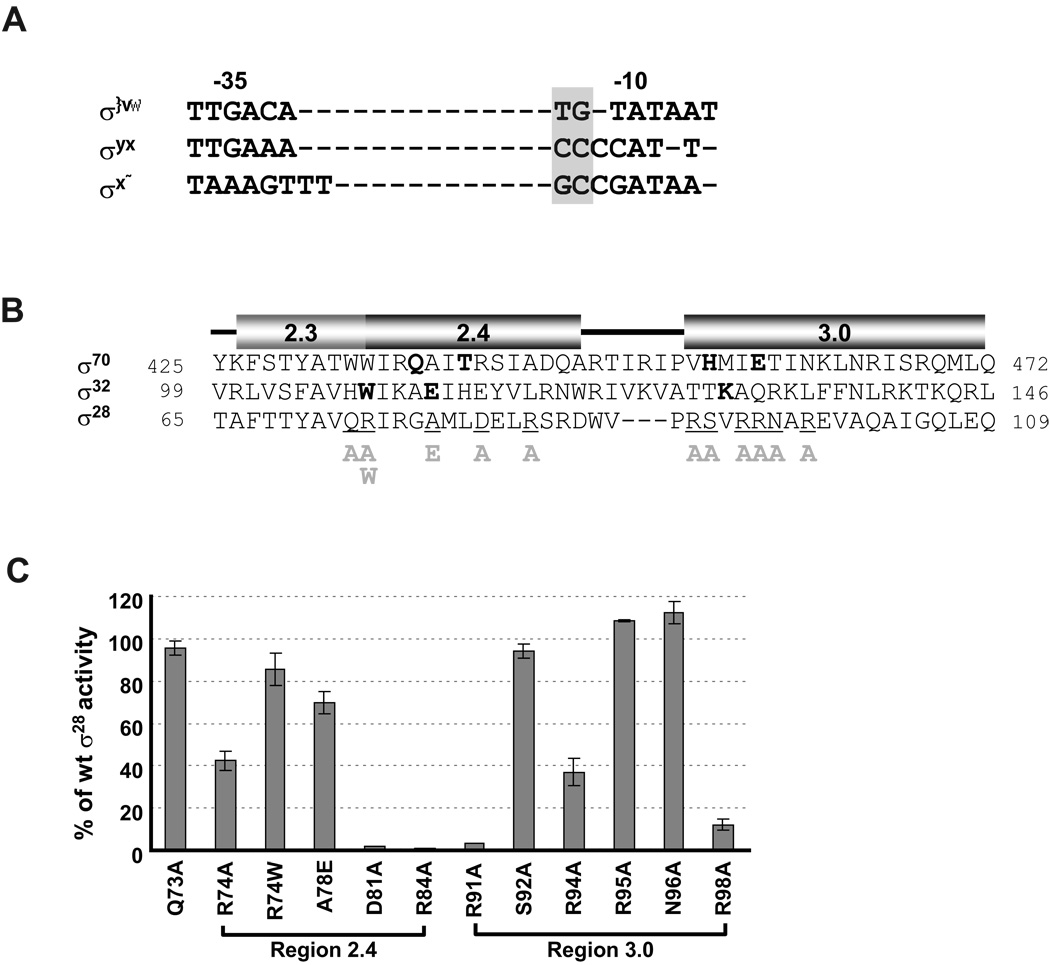
The studies above demonstrated that both the upstream GC motif and the CGA motif of the −10 region are functionally important. We recently found that the upstream CC motif in the promoters of another Group 3 σ, σ32, constitutes an extended −10 motif (Koo et al., 2009). Moreover, the GC motif in σ28 promoters aligns with other extended −10 motifs (Fig. 3A). Together, this led us to direct alanine substitution mutagenesis to σ regions implicated both in extended −10 recognition (Region 3.0 in Domain 3) and in −10 region recognition (Region 2.4 in Domain 2). Selection of amino acid residues for substitution in Region 2.4 was based on sequence conservation among σ28 orthologues and a comparison of σ70, σ32, and σ28 sequences. For Domain 3, we extensively substituted resides in the first helix, as this region is implicated in extended −10 recognition (Fig. 3B). When compared to wt σ28, six of the twelve substitutions in σ28 resulted in a significant decrease in transcriptional activity on wt tar promoter ( ≤ 40% of wt σ28 , Fig. 3C). Three of these were located in Region 3.0 (R91A, R94A and R98A) and three in Region 2.4 (R74A, D81A and R84A). Interestingly, only R74 and D81 corresponded exactly to positions known to be involved in recognition of their cognate promoters of σ70, σ32 and B. subtilis σH (Fig. 3B and Daniels et al., 1990). We then tested whether any of the alanine mutants met the genetic criterion for base-specific interaction; namely, suppression of the promoter defects of base changes limited to particular positions in the promoter. This criterion is based on the idea that if the mutant σ no longer recognizes a particular base, then base changes at that position should show a smaller than expected decrease in expression. This criterion has been used successfully both for transcription factors and for other σs (Koo et al., 2009, Hochschild et al., 1986, Hochschild & Ptashne, 1986, Ebright, 1986, Ebright et al., 1984, Siegele et al., 1989).
Figure 3. Effects of single amino acid substitutions in Region 2.4 and 3.0 of σ28 on activity of the wt tar promoter.
A. Alignment of consensus core −10 and −35 sequences of E. coli σ70, σ32 and σ28 promoters. The extended −10 motifs of each promoter are denoted by grey shading.
B. Alignment of amino acid sequence of Regions 2.3, 2.4 and 3.0 of σ70, σ32 and σ28. Numbers at each end of sequence indicate amino acid position. Amino acid residues of σ70 and σ32 involved in base specific recognition are shown in bold (Koo et al., 2009, Kourennaia et al., 2005, Sanderson et al., 2003, Siegele et al., 1989). Positions of single amino acid substitutions of σ28 used in this study are underlined and the substitutions are shown below the main sequence. The predicted helices at each region was derived from structural data of Thermus aquaticus σA and Aquifex aeolicus σ28 (Campbell et al., 2002, Sorenson et al., 2004) and are shown above the sequences.
C. β-galactosidase activity driven by each σ28 variant on the wt tar promoters are shown as a percentage of measured β-galactosidase activity driven by wt σ28. Assay strains are as described in Fig 1B.The different σ28 amino acid substitutions and their locations are shown on the x-axis. All values are averages of three independent experiments; error bars indicate 1 standard deviation.
Identification of base-specific interactions between Region 2.4 of σ28 and the −10 promoter region
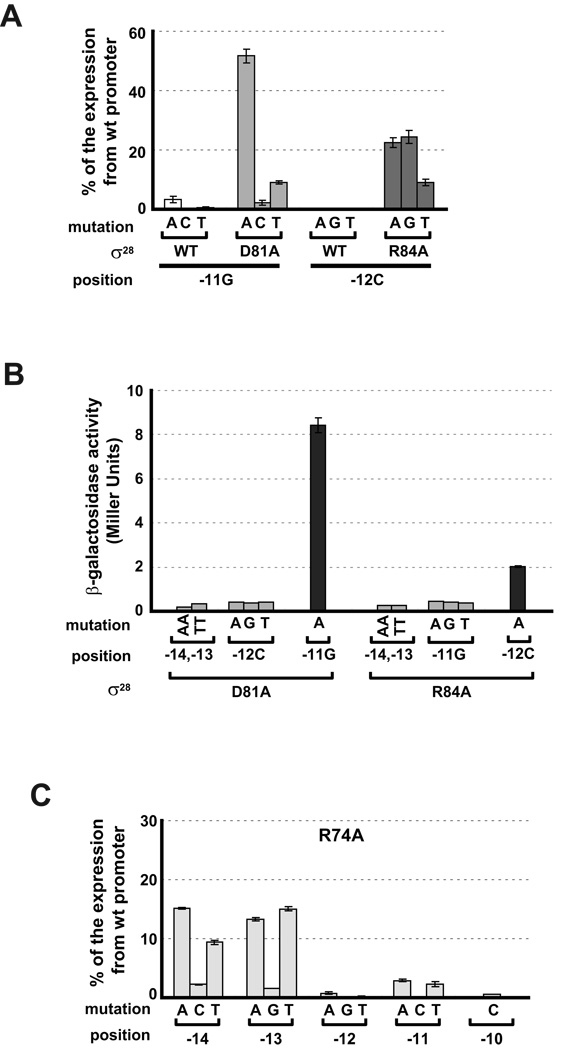
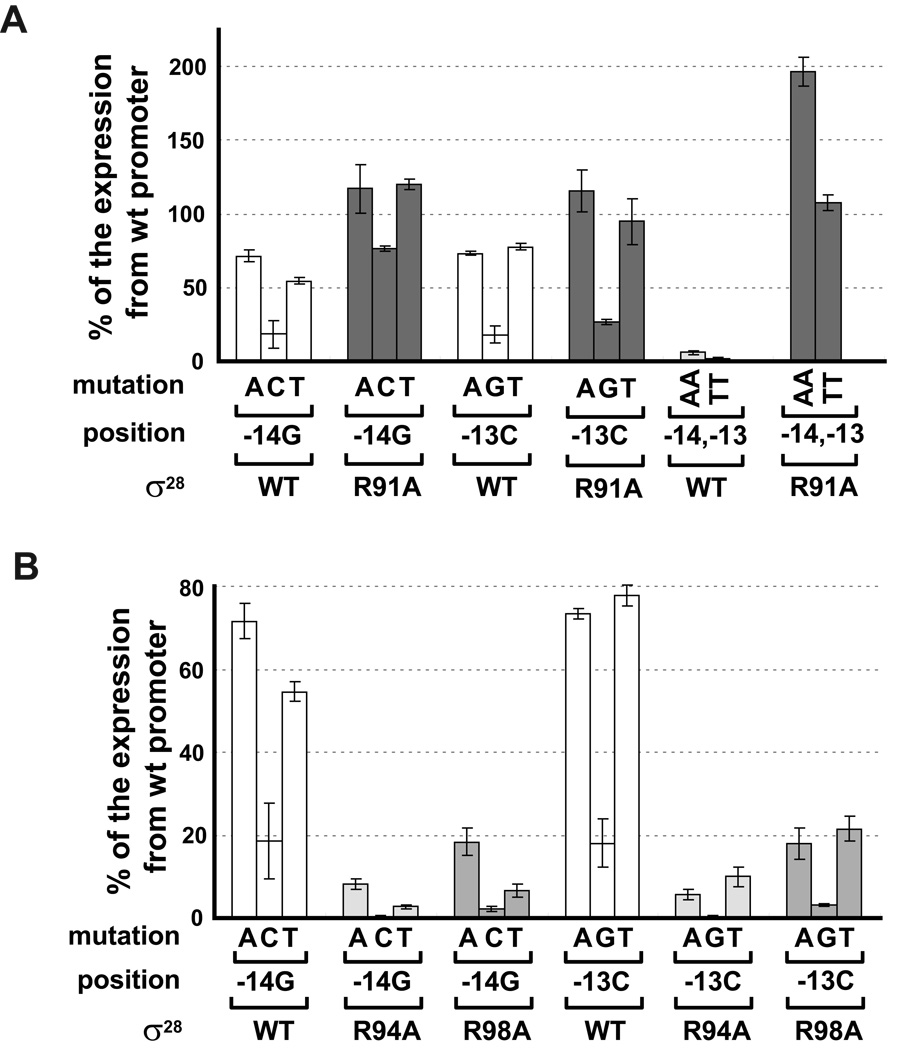
Two of the three Region 2.4 candidates, D81 and R84, met the criteria for mediating a base specific interaction. Both D81A and R84A had significant defects in transcription from wt tar promoter, exhibiting <2% of the transcription mediated by wt σ28 (Fig. 3C) and each suppressed all single base changes at a unique position in the promoter. D81A suppressed all single base changes at −11G at least 10-fold and R84A suppressed all single base changes at −12C at least 20-fold (Fig. 4A). The absolute values of β-galactosidase are very low, because both the mutant promoter and the mutant σ28 independently give severe transcription defects. However, the results are very reproducible and significantly above background. In stark contrast, when D81A is assayed at positions other than −11, β-galactosidase activity is at the background level (compare activity at −14,13 or −12 with that at −11; Fig. 4B). Likewise, when R84A is assayed at positions other than −12, β-galactosidase activity is at the background level (compare activity at −14,13 or −11 with that at −12; Fig. 4B). Taken together, these results support the idea that D81 and R84 participate in base specific recognition of the promoter.
Figure 4. Effect of amino acid substitutions in Region 2.4 of σ28 on mutations at −10 promoter region.
D81A σ2828 and R84A σ28 uniquely suppress single nucleotide changes at positions −11G and −12C of the tar promoter respectively (A and B), whereas R74A σ28 shows less relative activity than wt σ28 on mutant promoters with single nucleotide changes at positions −10 to −14 of the tar promoter (C). For panels A and C, β-galactosidase activity driven by each σ28 variant on mutant tar promoters is shown as a percentage of the β-galactosidase activities of the same σ28 variant on the wt promoter. For panel B, absolute β-galactosidase activity (Miller Units) driven by D81A σ28 and R84A σ28 on mutant tar promoters is indicated. Activities of less than 0.5 Miller Units is at the background level of this assay. Assay strains are as described in Fig 1B. Below the x-axis, mutation and position indicate the base change at the different promoter positions; σ28 indicates the wild type or substituted derivative. All values are averages of three to four independent experiments; error bars indicate 1 standard deviation.
R74 is at the boundary between Regions 2.3 and 2.4 and was tested because the analogous position in σ70 (W434) and in the Group 3 σ, σ32 (W108) were important in −10 recognition and/or melting (Kourennaia et al., 2005, Fenton et al., 2000, Koo et al., 2009, Tomsic et al., 2001). However, R74A did not suppress any mutations in the −10 promoter region. Instead, this ‘loss of function’ mutant had even less relative activity than wt σ28 on promoters that were mutant at the −14G and −13C positions (compare Fig. 4C (R74A) and Fig. 2 (wt)).
Identification of a base-specific interaction between Domain 3 of σ28 and the −10 promoter region
Of the three Domain 3 alanine substitution mutants that reduced expression from the wt promoter, only R91 meets the genetic criteria for mediating a base specific interaction. R91A reduces activity to 5% of wt σ28 (Fig. 3C) and suppressed almost all changes at −13C and −14G to a variable extent (Fig. 5A). Mutations to A or T at both −13 and −14 and to C at −14 were completely or almost completely suppressed. Only the C−13G change was not suppressed. These data suggest that R91 makes base-specific contacts with both −14G and −13C. In support of this idea, we find that R91A suppressed AA and TT changes at −14,−13 completely (Fig. 5A; about 200% for AA and 100% for TT). Interestingly R91 had previously been implicated as important for the transcriptional function of σ28 as it was part of a double mutant (R91C,L207P) isolated to be completely defective in expression from a σ28 promoter (Aldridge et al., 2006). In contrast, R91A did not suppress changes at −11 or −12, exhibiting β-galactosidase activity at the background level of the assay (data not shown).
Figure 5. Effect of alanine substitutions in Region 3.0 of σ28 on nucleotide changes at positions −14G and −13C of tar the promoter.
A. Suppression of single or double nucleotides changes at positions −14G and −13C of the tar promoter by R91A σ28.
B. Effect of R94A σ28 or R98A σ28 on changes at positions −14G and −13G of the tar promoter.
All data are normalized and presented as shown in Fig. 4.
The two other mutants that reduced activity from the wt promoter, R94A and R98A, showed about 40% and 15% of wt σ28 activity on wt tar promoter respectively. Intriguingly, R94A and R98A resulted in even less relative activity than wt σ28 on promoters that were mutant at the −14G and −13C positions (Fig. 5B). R94 and R98 are predicted to be on the same face of an α helix as R91. These residues might interact with promoter DNA nonspecifically to stabilize an interaction between R91 and −14G and −13C positions.
Effects of σ28 variants on transcription in vitro
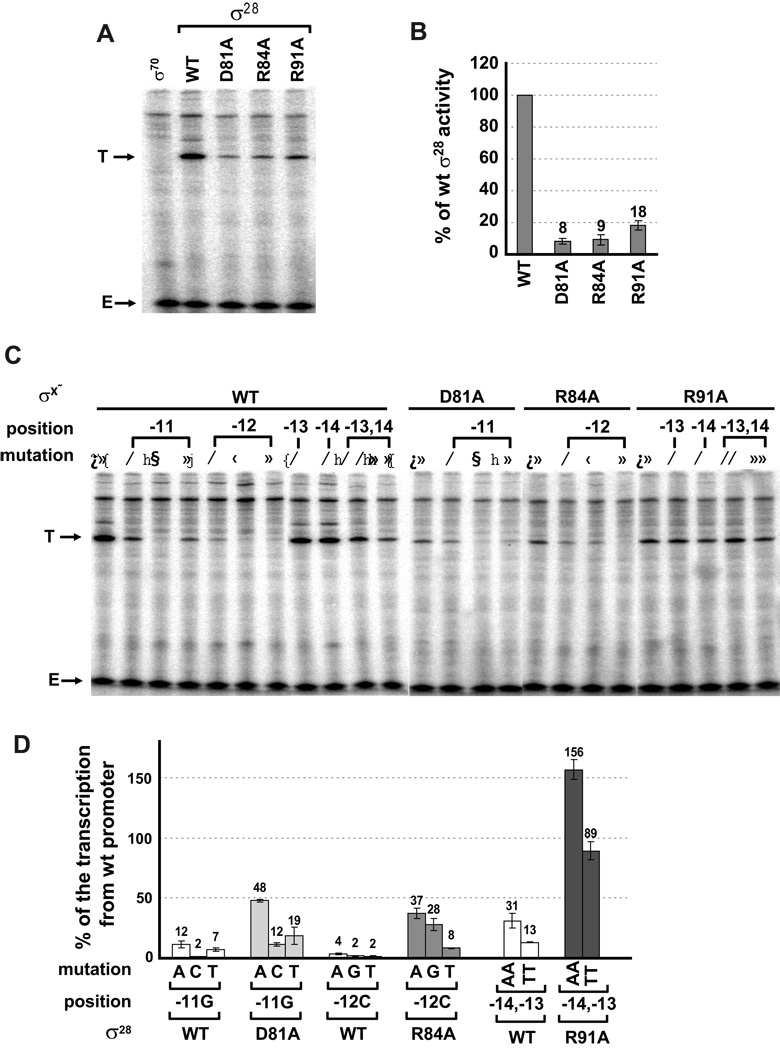
Our in vivo assay identified D81, R84 and R91 as residues in σ28 that might mediate base specific interactions with the promoter. We tested whether we could replicate these results in vitro. We carried out single round in vitro transcription assays on linear templates with the same promoter sequences used for our in vivo assays and compared transcription driven by wt σ28 with that driven by σ28 variants carrying alanine substitutions for each of the three residues implicated in recognition. Results from in vitro studies were consistent with our in vivo assays in all respects. First, the tar promoter we utilized was absolutely σ28 dependent in vitro (Fig. 6A), and the D81A, R84A and R91A σ28 variants exhibited significantly decreased transcription from this promoter (5- to 10- fold; Figs. 6A and B) as was the case in our in vivo assays (Fig. 4A and 5A). Second, transcription of mutated promoters by wt σ28 showed the same rank order of transcription as expression assays in vivo (compare Fig. 6D with 4A and 5A). We note that there was generally more relative transcription from each mutant promoter in vitro than in vivo, especially of the −14, −13 AA and TT mutation (~30 and 13 % of the rate of the wt promoter; Fig. 6D) than in vivo (~5 and 2 %; Fig. 2B), possibly because the in vitro assay does not have wt competitor promoter. Third, and most importantly, we qualitatively reproduced position specific suppression of promoter mutations (Fig. 6C and D). Changes at −11G and −12C significantly decreased transcription with wt σ28 but were significantly suppressed (~3 to 5-fold) by D81A and R84A σ28 respectively.
Figure 6. Effect of mutations in the −10 region of the tar promoter and Regions 2.4 and 3.0 in σ28 on transcription in vitro.
Single round in vitro transcriptions were performed on linear DNA templates by RNA polymerase containing different σ28 variants. DNA templates were prepared as described in the experimental procedures. Each experiment was repeated a minimum of three times; for each experiment a representative gel image is shown and the bar graphs illustrate averaged transcript quantifications from replicate experiments. T; transcript from tar promoter variant driven by different σ28 variants. E; end-labeled 35 nucleotides oligomer.
A. in vitro transcriptions from the wt tar promoter with wt, D81A, R84A and R91A σ28. As a control reaction, transcript driven by σ70 is shown.
B. Quantified and normalized data from A. The bars indicate the relative transcription by the different σ28 variants as a percentage of total transcription with wt σ28.
C. In vitro transcriptions from different tar promoter variants mutated at positions −11 to −14 with wild-type σ28 and suppression of promoter substitutions by D81A, R84A and R91A σ28. Above the gel image, σ28indicates wild-type or the different σ28 variants, position and mutation indicate the base change at the different promoter positions.
D. Quantified and normalized data from C. The bars indicate the relative transcription by the different σ28 variants at the mutant promoters as a percentage of transcription by that variant from the wt promoter. Below the x-axis, mutation and position indicates the base change at the different promoter positions; σ28indicates the wild-type or substituted derivative.
Additionally, R91A completely suppressed the single and double A, T mutations tested at −14G and −13C. That we can reproduce position specific suppression in an in vitro transcription reaction indicates that suppression is a direct effect of the σ28 variant utilized and argues for D81, R84 and R91 mediating base specific interactions with the promoter.
Discussion
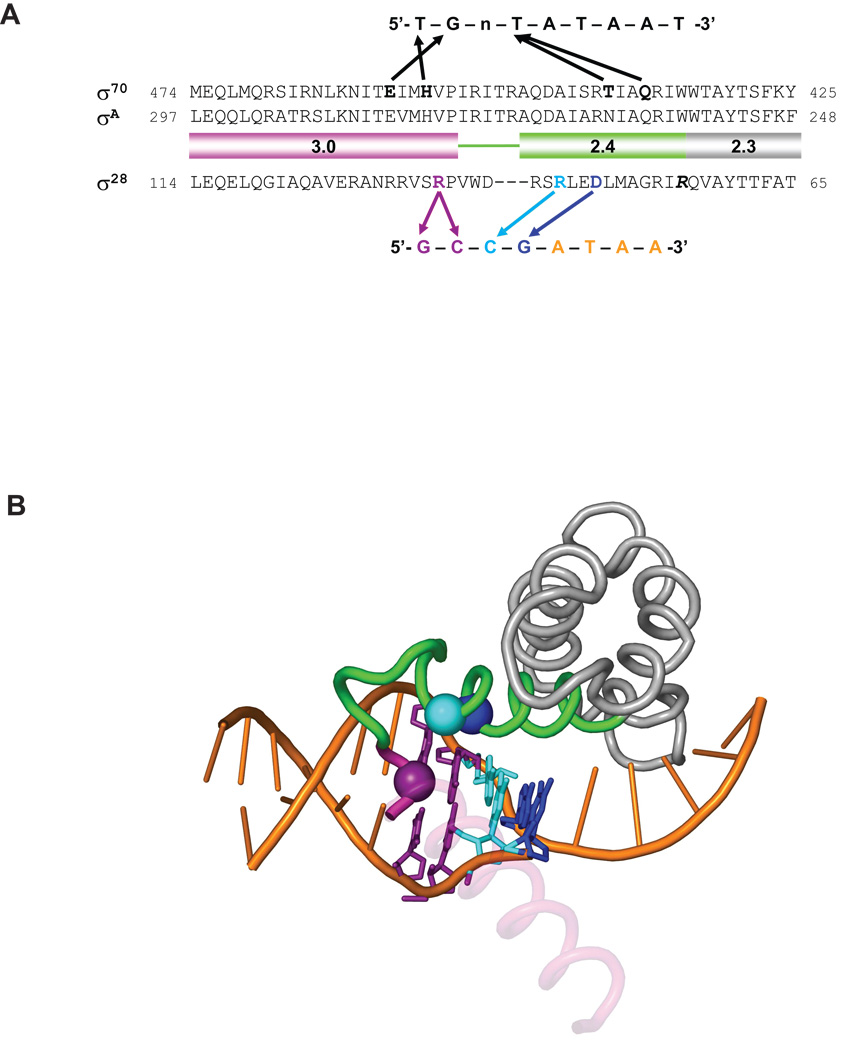
σ28 is the most widely distributed alternative σ factor and its consensus promoter sequence is also well conserved. Thus, information from the study of E. coli σ28 might suggest general mechanisms of transcription initiation by its orthologues in a wide range of bacteria. However, there were major uncertainties about the functionally relevant sequences in the −10 region of its promoter and how they were recognized by σ28. In this study, we focused on the GCCG motif in the −10 region. We examined the importance of each base by using a newly developed sensitive in vivo assay and by reproducing these results in vitro. To identify residues in σ28 contributing to base specific interaction of the GCCG motif, we used a validated genetic criterion: loss of the residue interacting with a particular base should suppress the deleterious effects of promoter mutants only at the interacting position(s). Candidates were chosen using our in vivo assay and confirmed in vitro. Our principal finding was that the GCCG motif is a functionally important composite element: the upstream GC constitutes an extended −10 motif and is recognized by a single residue in Domain 3 of σ28, whereas the downstream CG constitutes the upstream edge of −10 region of the promoter and is recognized by at least two residues in Region 2.4. A schematic comparing the interactions we propose to those in E. coli σ70 and T. aquaticus SigA is presented in Fig. 7A; a model based upon the T. aquaticus holoenzyme/fork-junction promoter DNA complex indicates the positions of these amino acids relative to the DNA (Fig. 7B).
Figure 7. Structural model of recognition of the −10 region of promoters by σ28 and conservation of base-specific promoter recognitions among σ28 orthologues.
A. Comparison of the recognition of the −10 region of promoters by σ70 and σ28. Amino acid sequences of Region 2.3, 2.4 and 3.0 of σ70, σA and σ28 are shown. The −10 consensus promoter sequences for each σ is shown above or below the amino acid sequences. Arrows indicate contacts suggested by the previous studies for σ70 and in this study for σ28.
B. Shown is the structure of σA Regions 2.1–3.0 and the −10 element region of the fork junction promoter DNA from the T. aquaticus holoenzyme/fork-junction promoter DNA complex (Murakami et al., 2002). The rest of the protein and DNA are not shown. The σA Region 2.4 is colored green and Region 3.0 is magenta. Most of the 3.0 helix is transparent for clarity. The positions corresponding to residues of σ28 that make base-specific contacts with the promoter are shown as α-carbon spheres, and color-coded along with the corresponding DNA base-pair: D81 (R264 in T. aquaticus σA)/−11GC, blue; R84 (A267)/−12CG, cyan; and R91 (V277)/−13CG/–14GC, purple. This figure was prepared using PyMol (http://pymol.sourceforge.net/).
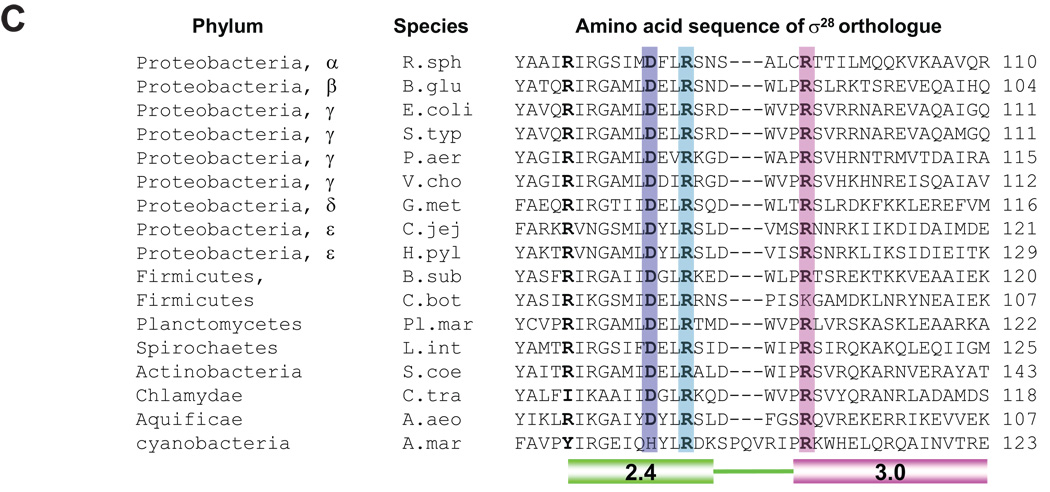
C. Conservation of amino acid residues involved in base-specific recognition of the −10 region of σ28 promoter among σ28 orthologues. Whole amino acid sequences were aligned using ClustalW and sequences from Regions 2.4 to 3.0 are shown. Residues involved in base-specific recognition are denoted by color shading. Residues homologous to R74 of E. coli σ28 are in bold. R. sph, Rhodobacter sphaeroides; B. glu, Burkholderia glumae; E. coli, Escherichia coli; S. typ, Salmonella typhimurium; P. aer, Pseudomonas aeruginosa; V. cho, Vibrio cholerae; G. met, Geobacter metallireducens; C. jej, Campylobacter jejuni; H. pyl, Helicobacter pylori; B. sub, Bacillus subtilis; C. bot, Clostridium botulinum; Pl. mar, Planctomyces maris; L. int, Leptospira interrogans; S. coe, Streptomyces coelicolor; C. tra, Chlamydia trachomatis; A. aeo, Aquifex aeolicus; A. mar, Acaryochloris marina.
The downstream CG is the upstream boundary of the −10 region
We identified D81 as a residue recognizing −11G and R84 as a residue recognizing −12C, based on our finding that an alanine substitution at each position specifically suppressed promoter mutations at the −11 and −12 positions respectively, both in vivo and in vitro. These residues are located in Region 2.4, which mediates recognition of the −10 promoter region, indicating that this CG motif is the upstream boundary of the −10 promoter region. Although suppression is not complete, our conclusion is supported by several additional pieces of data. First, the location of D81 and R84 is consistent with the function proposed, as a structure based alignment indicates that these amino acids are solvent exposed and are in position to contact the DNA (Fig. 7B). Second, D81 and R84 are universally conserved in σ28 orthologues, as expected from residues mediating base specific recognition (Fig. 7C). Additionally, D81 is at the same position in σ28 as residues in other Group 3 σs implicated in interacting directly or indirectly with the upstream portion of the −10 regions of their cognate promoters: M124 of B. subtilis σE (Tatti et al., 1991) and R96 of B. subtilis σH (Daniels et al., 1990). Finally, and provocatively, it has previously been proposed that the DXXR motif is utilized to recognize a CG sequence in the −10 promoter region of Group 4 σs (Wilson & Lamont, 2006). PvdS, a Pseudomonas aeruginosa Group 4 σ has a −10 region that starts with CGT. Alanine substitutions at D77 and R80 of PvdS were defective in binding to this promoter. Conversely, if σs have a DXXR motif in Region 2.4, the −10 region of their promoters are likely to have a CG motif. These σs include the Group 4 σs, CarQ of Myxococcus xanthus, σW and σX of Bacillus subtilis, and σC of Mycobacterium tuberculosis (Wilson & Lamont, 2006). Our finding indicates that using DXXR to recognize a CG motif extends to Group 3 σs.
That R84 mediates −10 recognition has important implications for understanding how σ28 itself is inhibited from promoter binding. Structural and biochemical studies from Darst and collaborators on Aquifex aeolicus σ28 suggested that interdomain interactions inhibit free σ28 from binding to the promoter and that this conformation is stabilized by a salt bridge between R82 and E145, E146 (Sorenson et al., 2004, Sorenson & Darst, 2006). Relevant here is that R84 of E. coli σ28 is the homologous residue of R82 in Aquifex aeolicus. Thus, it seems that the interactions in free σ28 sequester a residue that is critically important in base-specific recognition of the −10 region of the promoter.
All mutations at the −11G and −12C positions of the σ28 promoter were exceptionally deleterious, exhibiting only 1–3% of the activity of the wt promoter. In contrast, mutations at comparable positions of the promoter of another Group 3 σ (σ32 promoter; −13,−14 CC) exhibited between 10~40% of the activity of the wt promoter (Koo et al., 2009, Wang & deHaseth, 2003). What might account for this distinction? This distinction may reflect the fact that σ32 has σRegion 1.2 whereas σ28 does not. Both structural data and sequence alignments suggest that all Group 1 and 2 σs and some Group 3 σs have this region (Sorenson et al., 2004, Lonetto et al., 1992, Campbell et al., 2002, Gruber & Gross, 2003). Region 1.2 appears to have several roles in σ70: it is required for open complex formation (Wilson & Dombroski, 1997), it recognizes the discriminator region, just downstream of −10 region of promoter (Haugen et al., 2006, Feklistov et al., 2006), and it may act as a core RNA polymerase (β’ subsunit coiled-coil)-dependent allosteric switch that modulates recognition of the −10 promoter element by Region 2 (Zenkin et al., 2007). The role of this region in σ32 is unknown, but it may facilitate melting in some way. Because σ28 lacks this region, it may be more dependent on the precise composition of the −10 region for function. If this explanation is true, it begs the question of how a −12,−11 CG motif might facilitate melting. One possibility, originally suggested by Losick for σH of B. subtilis is that nucleotide substitutions at such positions might have an unfavorable effect on the exact conformation of the DNA duplex or its ability to be unwound (Daniels et al., 1990). Alternatively, or in addition, it may serve as a recognition motif for the σ28 residues that facilitate melting.
The upstream GC is an extended −10 motif
Our studies indicate that the highly conserved −14,−13 GC motif is functionally important but contains partially redundant information. Changing either−14G or −13C to A or T results in only a very modest (~20%) reduction in promoter strength in vivo. In stark contrast, a double AA or TT mutation at −14, −13 leads to a dramatic (20 ~ 50- fold) reduction in promoter strength. The synergistic effect of the double mutant on promoter activity suggests that only one of the two bases is required for function. Satisfyingly, the idea that either −14G or −13C, but not both, is required for high promoter activity resolves a discrepancy in the literature. Single changes at the upstream GC in the context of the flgKL promoter, which has both −14G and −13C, caused little or no defect in activity (Wozniak & Hughes, 2008), whereas single changes at −14G in context of the C. trachomatis hctB promoter, which has −14G and −13T caused significant defects in transcription (Yu et al., 2006a). In the latter case, mutation at the −14G position eliminates the upstream GC motif. Interestingly, we found that the −14G to C and the −13C to G single changes each reduce promoter activity ~5-fold. This issue is discussed below.
We identified R91 as the residue recognizing both −14G and −13C, based on our finding that an alanine substitution at R91 suppressed promoter mutations at only −14 and −13. Satisfyingly, the ~20-fold defect of R91A σ28 is approximately equivalent to the defect in expression observed when both −14G and −13C are removed from the wt promoter and substituted with AA or TT. This equivalence lends credence to the idea that R91 recognizes both positions, and also confirmed the importance of the GC motif for promoter function. Importantly, R91 is universally conserved in σ28 orthologues, as expected of a residue that recognizes specific bases in the promoter. Moreover, the hydrogen bond donor properties of arginine are consistent with the phenotype of the substitutions at the −14,−13 positions. Arginine could bond with both the non-template strand G at −14 and the template strand G at −13. Introduction of A or T at either −14 or −13 would still allow binding to the remaining G as well as to the A or T base substitution. There is likely to be sufficient interaction that little or no decrease in expression is observed; hence the requirement for a double mutation. In contrast, a C mutation at either position removes the possibility of hydrogen bonding probably accounting for the fact that single changes of this type exhibit reduced expression (~20% wt) (Luscombe et al., 2001).
R91 is located in the first α-helix in Domain 3 of σ28 and a structure based alignment indicates that R91 is solvent exposed and in position to contact the DNA (Fig. 7B). All residues mediating base-specific interactions with the extended −10 motif identified to date reside in this helix: H455/E458 in σ70 (Fig. 7A) and analogous residues in other Group 1 σs recognizing −15,−14 TG (Fig. 3B, Sanderson et al., 2003); K173 in σs recognizing −13C and possibly −14T (Becker & Hengge-Aronis, 2001); and K130 in σ32 recognizing −16,−15 CC (Fig. 3B, Koo et al. 2009). By analogy to other σs, this upstream −14,−13 GC constitutes an extended −10 motif. Importantly, the residues implicated in extended −10 recognition in different σs are all predicted to be on the same face of the first α-helix in Domain 3, supporting the proposed function of these residues. We note that the precise position of the residues mediating −10 recognition differs in various σ’s, possibly because the length of linker between Domain 2 and 3 in different σs affects the positioning of the first α-helix of Region 3.0 relative to DNA. Alternatively, the somewhat discrepant position of K130 in σ32 could result from the fact that this putative helix is immediately adjacent to an insertion, called the RpoH box, which is the most highly conserved region in σ32 orthologues (Nakahigashi et al., 1995). At least one residue in this region of σ32 interacts with RNA polymerase (Joo et al., 1998) and this interaction might specifically alter the configuration of this helix in σ32.
Group 3 σs have σDomain 3, which encodes the recognition determinants for the extended −10 promoter motif in Group 1 and 2 σs. In addition, these σs utilize promoters that have long −10 regions. These features suggested that the extended −10 motif might be also present in Group 3 σ promoters. However, Domain 3 of the Group 3 σs is the most variable domain in the Group 3 σs, making it dangerous to extrapolate that such recognition occurs. Indeed, two Group 3 σs (σE and σH in B. subtilis) were previously shown not to utilize an extended −10 motif. We demonstrate here that σ28 joins σ32 in utilizing an extended −10 motif. Since the σ28 branch of Group 3 σs is highly divergent from the σ32 branch (Gruber & Gross, 2003, Paget & Helmann, 2003), it is likely that extended −10 recognition will occur in other Group 3 σs In this regard, it is intriguing that promoters recognized by both σB and σF in B. subtilis have GG as their predicted extended −10 motif (Petersohn et al., 2001, Amaya et al., 2001) and also have an arginine at the position comparable to R91 in σ28. Thus, an arginine residue in the first α–helix of Domain 3 of σB and σF might mediate recognition of one or both upstream G’s.
Summary and prospects
We establish that the GCCG motif in the −10 region of σ28 promoters is a composite recognition element: the upstream GC is an extended −10 region recognized by R91, a residue in Domain 3, whereas the downstream CG initiates the −10 recognition region recognized by (at least) D81 and R84 in Region 2.4 of σ28. Thus, σ28 joins σ32 as a Group 3 σ that not only uses extended −10 recognition, but also requires three recognition regions, −35, extended −10 and −10 for successful utilization of the core σ28 promoter. This study raises the possibility that additional Group 3 σs will have promoters exhibiting these properties and poses several provocative avenues of inquiry. We are interested in the features of Group 3 σs that cause them to have such extensive recognition requirements. We also wonder whether such a requirement might have regulatory advantages. For example, extensive recognition requirements prevent the promiscuous promoter recognition characteristic of housekeeping σs. Alternatively, or in addition, the construction of these promoters may discourage their transcription by the housekeeping σs. In this regard, it is interesting that not only many Group 3 σs but also many Group 4 σs have a G and/or C residue(s) at the upstream end of the −10 region. As G/C residues are never found in this position in housekeeping σ’s, this may add to the distinction of promoters recognized by the alternative σs.
Experimental procedures
Strains and plasmids
Strains and plasmids used in this study are listed in Table 1. All strains were grown at 30°C in Luria-Bertani (LB) medium supplemented with appropriate antibiotics if needed.
Table 1.
Strains and plasmids used in this study
| Strains/plasmids | Relevant genotype | Source/reference |
|---|---|---|
| Strains | ||
| DH5α |
fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 |
Invitrogen |
| BL21(DE3)pLysS | dcm ompT hsdS(rB−mB−) gal λ(DE3), pLysS, CmR | Invitrogen |
| DY330 | W3110, ΔlacU169 gal490 λCI857 Δ(cro-bioA) | (Yu et al., 2000) |
| CAG22239 | MG1655 ΔlacX74 | Lab collection |
| CAG57111 | DY330, ΔflgM::cat, CmR | This study |
| CAG57112 | DY330, ΔfliA::aadA, SpR | This study |
| CAG57113 | MG1655 ΔlacX74, ΔflgM::cat, CmR | This study |
| CAG57114 | MG1655 ΔlacX74, ΔfliA::aadA, SpR | This study |
| CAG57115 | MG1655 ΔlacX74, ΔflgM::cat, ΔfliA::aadA, CmR, SpR | This study |
| Plasmids | ||
| pSAKT32 | Vector for σ32 expression. P15A ori, Plac-rpoH, lacIq, ApR | (Wang & deHaseth, 2003) |
| pSAKT28 | Vector for σ28 expression. P15A ori, Plac-fliA, lacIq, ApR | This study |
| pBK201 | σ28-Q73A in pSAKT28, ApR | This study |
| pBK202 | σ28-R74A in pSAKT28, ApR | This study |
| pBK203 | σ28-R74W in pSAKT28, ApR | This study |
| pBK204 | σ28-A78E in pSAKT28, ApR | This study |
| pBK205 | σ28-D81A in pSAKT28, ApR | This study |
| pBK206 | σ28-R84A in pSAKT28, ApR | This study |
| pBK207 | σ28-R91A in pSAKT28, ApR | This study |
| pBK208 | σ28-S92A in pSAKT28, ApR | This study |
| pBK209 | σ28-R94A in pSAKT28, ApR | This study |
| pBK210 | σ28-R95A in pSAKT28, ApR | This study |
| pBK211 | σ28-N96A in pSAKT28, ApR | This study |
| pBK212 | σ28-R98A in pSAKT28, ApR | This study |
| pBK311 | σ28 in pET21a*, ApR | This study |
| pBK312 | σ28-D81A in pET21a*, ApR | This study |
| pBK313 | σ28-R84A in pET21a*, ApR | This study |
| pBK314 | σ28-R91A in pET21a*, ApR | This study |
| pQF50K | Vector, pMB1 ori, 1600 ori, promoterless lacZ, kmR | (Farinha & Kropinski, 1990) |
| pBK601 | Ptar (−44 to +10, WT) in pQF50K, KmR | This study |
| pBK602 | Ptar (G-14A) in pQF50K, KmR | This study |
| pBK603 | Ptar (G-14C) in pQF50K, KmR | This study |
| pBK604 | Ptar (G-14T) in pQF50K, KmR | This study |
| pBK605 | Ptar (C-13A) in pQF50K, KmR | This study |
| pBK606 | Ptar (C-13G) in pQF50K, KmR | This study |
| pBK607 | Ptar (C-13T) in pQF50K, KmR | This study |
| pBK608 | Ptar (C-12A) in pQF50K, KmR | This study |
| pBK609 | Ptar (C-12G) in pQF50K, KmR | This study |
| pBK610 | Ptar (C-12T) in pQF50K, KmR | This study |
| pBK611 | Ptar (G-11A) in pQF50K, KmR | This study |
| pBK612 | Ptar (G-11C) in pQF50K, KmR | This study |
| pBK613 | Ptar (G-11T) in pQF50K, KmR | This study |
| pBK614 | Ptar (A-10C) in pQF50K, KmR | This study |
| pBK615 | Ptar (A-10G) in pQF50K, KmR | This study |
| pBK616 | Ptar (A-10T) in pQF50K, KmR | This study |
| pBK617 | Ptar (G-14A, C-13A) in pQF50K, KmR | This study |
| pBK618 | Ptar (G-14T, C-13T) in pQF50K, KmR | This study |
| pACYC184 | p15A ori, CmR, TetR | (Chang & Cohen, 1978) |
| pDH50 | tac promoter, lacIq, SpR | (Kleerebezem et al., 1995) |
thrombin cleavage site is modified to PreScission protease cleavage site
Construction of in vivo assay strain
The fliA and flgM genes were disrupted using electroporated linear DNA amplified by PCR and E. coli DY330 as described previously (Yu et al., 2000). First, CAG57111 (DY330 ΔflgM::cat) was constructed by replacing open reading frame (ORF) of flgM gene by chloramphenicol resistant cassette (its own promoter and gene). Linear DNA for disruption of flgM was generated by amplification of cat gene in pACYC184 with the following primers; 5’- CCGATAAATAAGCA ACACATGATA AAAGCGCCCTCAATGAGGAATAAACC ATGGAGAAA AAAATCACTGG -3’, and 5’- TAAGCACAGCGGACATCTGGTCGAGGATCTCTGCAAGACGTGTCATACGA TTACGCCCCGCCCTGCCACTC -3’ (underlined sequences are complementary to the upstream and downstream non-coding sequences immediately adjacent to the flgM ORF). CAG57112 (DY330 ΔfliA:aadA) was constructed by replacing the fliA ORF with the spectinomycin resistant gene, aadA. The aadA sequence was amplified from pDH50 using the primers; 5’- CAGAAACGGATAATCATGCCGATAACTCATATAACGCAGGGCTGTTTATC ACCGTGGAAACGGATGAAGGCACG -3’, and 5’- ATCATTAAGAACTCCTGGTAGTCAAAGTTAAAGTGCGGCATTTACTGACG TTATTTGCCGACTACCTTGGTG -3’ (underlined sequences are complementary to the upstream and downstream non-coding sequences immediately adjacent to the fliA ORF).
The assay strain used in this study, CAG57115 (MG1655 ΔlacX74, ΔfliA:aadA, ΔflgM::cat), was constructed by P1 transduction of ΔflgM::cat from CAG57111 and ΔfliA:aadA CAG57112, and selection for chloramphenicol and spectinomycin resistance, repsectively. The presence of each gene was also confirmed by PCR.
Plasmid construction
The plasmid pSAKT28 encoding wt σ28 and pSAKT28 derivatives pBK201-pBK212 encoding σ28 variants were used for expression of σ28 to determine their activities in vivo. The pSAKT28 plasmid derivatives carry the fliA gene under control of the isopropyl-β-D-thiogalactopyranoside inducible Plac promoter as well as the lac repressor gene under the control of a strong mutant promoter (iq) (Wang & deHaseth, 2003). pSAKT28 was constructed from pSAKT32, which encodes rpoH (σ32) by replacing the rpoH ribosome binding site and ORF with the fliA ORF and ribosome binding site. In addition, the start codon of fliA gene (GTG) was changed to ATG. All σ28 variants were constructed by site-directed mutagenesis.
Derivatives of pQF50K (pBK601-pBK618) carrying promoter fragments cloned in the BglII-XbaI sites upstream of lacZ were used for measuring promoter strength and activities of σ’s (Wang & deHaseth, 2003). Promoter mutant derivatives were constructed using PCR by amplifying the promoter regions using mutagenic primers that encompassed either the BglII or XbaI site.
Derivatives of the pET21a vector (Novagen) encoding σ28 and variants were used to overexpress σ28 protein for purification. The vectors were constructed using PCR by amplifying from the pSAKT28 series using primers to create flanking NdeI and HindIII sites and inserted as NdeI-HindIII fragments into pET21a NdeI-HindIII vector. The resultant σs contain an N-terminal His6-tag and PreScission cleavage sit (Patikoglou et al., 2007).
β-galactosidase assay
Assay systems for in vivo transcription were constructed by transforming derivatives of pSAKT28 and pQF50K into CAG57115 sequentially. Overnight cultures in LB media supplemented with 100 µg/ml ampicillin and 30 µg/ml kanamycin at 30°C were diluted 1:100 into fresh media and grown for 90 min with aeration. Selected cultures were then induced with 1 mM IPTG and grown for a further 2 hr before harvesting (final ODs at 600nm was approximately 0.7). β-galactosidase assays were performed as described previously and b-galactosidase activity (in Miller Units) was determined as following equation (Miller, 1972): Units = 1000 × [(OD420 – 1.75 × OD550) / (incubation time in minutes × volume of the culture used in the assay in ml × OD600 of the culture used in the assay)]
Overproduction and purification of σ28’s
E. coli BL21 (DE3)/pLysS cells harboring pET21a encoding wt σ28 or variants were used to overproduce σ28 with an N-terminal His6-tag and PreScission cleavage site for purification. Overnight cultures were diluted 1:100 into fresh media containing 100 µg/ml ampicillin and 30 µg/ml chloramphenicol, and grown aerobically at 30 °C. When the cultures reached OD600 of 0.4, expression of σ28 and variants was induced with 1 mM IPTG and growth continued at 30 °C with aeration for 2 hr. 500 ml of each culture was then harvested by centrifugation and the cell pellets resuspended in buffer A1 (20 mM HEPES, pH 8.0, 300 mM KCl, 1 mM β-mercaptoethanol, 5 mM Imidazole, 1 mM AEBSF). The cells were disrupted by sonication and the insoluble fraction was pelleted by centrifugation at 5,000 × g for 10 min and resuspended in 10 ml of buffer A2 (20 mM HEPES, pH 8.0, 300 mM KCl, 1 mM β-mercaptoethanol, 5 mM Imidazole, 6 M guanidine hydrochloride). The σ28 proteins were purified from the suspension by metal affinity chromatography using Talon metal affinity resin (Clontech) in denaturing condition but proteins were eluted in native condition by omitting guanidine hydrochloride. The N-terminal His6-tag was removed using PreScission protease (GE Healthcare) during dialysis into buffer Q (10 mM Tris, pH 7.5, 50mM NaCl, 1mM DTT) at 4 °C for 16 hr. The sample was further purified by a second, subtractive metal affinity chromatography step to remove uncleaved His6-σ’s and the His6-tag, followed by ion exchange chromatography (HiTrap Q Sepharose; GE Healthcare). As a final step, protein fractions were further purified by gel filtration chromatography (Superdex 75; GE Healthcare) in order to remove residual contaminants and free σ28 fractions were taken and concentrated in buffer B (50 mM Tris, pH 8.0, 200 mM KCl, 1 mM DTT, 0.01% Triton X-100, 50% glycerol). Purified σ28’s were then concentrated in buffer B. Purity of all His6-tag free proteins were >95% judged by SDS-PAGE and native-Protein concentration was determined by the bicinchoninic acid protein assay (Pierce).
In vitro transcription
Run-off single round transcription was performed at 30 °C. Linear DNA templates were generated by amplification of promoter region of pQF50K plasmid series including 220 bp upstream and 200 bp downstream sequence from transcription start site. RNA polymerase core enzyme was purified as described in (Sharp et al., 1999). Holoenzyme was reconstituted by incubation of core RNA polymerase and a 5-fold excess of σ28 on ice for 30 min in binding buffer (20 mM Tris pH 8.0, 100 mM K-acetate, 10 mM Mg-acetate, 0.1 mM EDTA, 1mM DTT, 100 µg/ml BSA, 5% glycerol and 0.05% Tween-20). Holoenzyme was diluted in binding buffer and incubated at 30 °C for 5 min. After incubation of holoenzyme and DNA template at 30 °C for 5 min, transcription was initiated by adding transcription mix and incubated at 30 °C for 6 min. Total reaction mixture was given as a 9 µl of 75 nM holoenzyme, 10 nM template DNA, 200 µM ATP, 200 µM GTP, 200 µM UTP, 10 µM CTP, 100 nM [α-32P]CTP (3000 Ci/mmol), 50 µg/ml heparin in binding buffer. Reactions were stopped by the addition of 7 µl of gel loading solution containing 20 mM EDTA, 80% deionized formamide, 0.1% of bromophenol blue and xylene cyanol FF, and 35-nucleotides 32P end-labeled DNA oligomer. After incubation at 90 °C for 2 min, the samples were subjected to electrophoresis in a 6% acrylamide/7M urea/TBE gel. The transcripts and end-labeled oligo were visualized and analyzed using Molecular Dynamics Storm 560 PhosphoImager scanning system and ImageQuant 5.2 densitometry software.
Acknowledgments
We thank members of the Gross lab for critically reading of the manuscript. We thank Monica Guo for useful suggestions and Seth A. Darst for critically reading this manuscript. This work was supported by National Institutes of Health (NIH) Grants GM36278 (to C.A.G.) and GM53759 (to S.A.D.).
References
- Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. The flagellar-specific transcription factor, sigma28, is the Type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 2006;20:2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya E, Khvorova A, Piggot PJ. Analysis of promoter recognition in vivo directed by sigma(F) of Bacillus subtilis by using random-sequence oligonucleotides. J Bacteriol. 2001;183:3623–3630. doi: 10.1128/JB.183.12.3623-3630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter sigma(S) dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of sigma(S) Mol Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Chadsey MS, Karlinsey JE, Hughes KT. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 1998;12:3123–3136. doi: 10.1101/gad.12.19.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Helmann JD. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc Natl Acad Sci U S A. 1992;89:5123–5127. doi: 10.1073/pnas.89.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D, Zuber P, Losick R. Two amino acids in an RNA polymerase sigma factor involved in the recognition of adjacent base pairs in the −10 region of a cognate promoter. Proc Natl Acad Sci U S A. 1990;87:8075–8079. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RH. Evidence for a contact between glutamine-18 of lac repressor and base pair 7 of lac operator. Proc Natl Acad Sci U S A. 1986;83:303–307. doi: 10.1073/pnas.83.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RH, Cossart P, Gicquel-Sanzey B, Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984;311:232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Farinha MA, Kropinski AM. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, Nikiforov V, Heyduk T, Severinov K, Kulbachinskiy A. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and −10 recognition: mutational analysis of sigma70. Embo J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Douhan J, 3rd, Ptashne M. How lambda repressor and lambda Cro distinguish between OR1 and OR3. Cell. 1986;47:807–816. doi: 10.1016/0092-8674(86)90523-4. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Ptashne M. Homologous interactions of lambda repressor and lambda Cro with the lambda operator. Cell. 1986;44:925–933. doi: 10.1016/0092-8674(86)90015-2. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Joo DM, Nolte A, Calendar R, Zhou YN, Jin DJ. Multiple regions on the Escherichia coli heat shock transcription factor sigma32 determine core RNA polymerase binding specificity. J Bacteriol. 1998;180:1095–1102. doi: 10.1128/jb.180.5.1095-1102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney TJ, York K, Youngman P, Moran CP., Jr Genetic evidence that RNA polymerase associated with sigma A factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci U S A. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Heutink M, Tommassen J. Characterization of an Escherichia coli rotA mutant, affected in periplasmic peptidyl-prolyl cis/trans isomerase. Mol Microbiol. 1995;18:313–320. doi: 10.1111/j.1365-2958.1995.mmi_18020313.x. [DOI] [PubMed] [Google Scholar]
- Koo B-M, Rhodius VA, Campbell E, Gross CA. Dissection of recognition determinants of Escherichia coli σ32 suggests a composite −10 region with an “extended −10” motif and a core −10 element. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06690.x. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourennaia OV, Tsujikawa L, Dehaseth PL. Mutational analysis of Escherichia coli heat shock transcription factor sigma 32 reveals similarities with sigma 70 in recognition of the −35 promoter element and differences in promoter DNA melting and −10 recognition. J Bacteriol. 2005;187:6762–6769. doi: 10.1128/JB.187.19.6762-6769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY; 1972. [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding sigma 32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patikoglou GA, Westblade LF, Campbell EA, Lamour V, Lane WJ, Darst SA. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J Mol Biol. 2007;372:649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M. Global analysis of the general stress response of Bacillus subtilis. J Bacteriol. 2001;183:5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson A, Mitchell JE, Minchin SD, Busby SJ. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- Serizawa M, Yamamoto H, Yamaguchi H, Fujita Y, Kobayashi K, Ogasawara N, Sekiguchi J. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene. 2004;329:125–136. doi: 10.1016/j.gene.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Feng X, Yuan Y, Luo X, Hatch TP, Hughes KT, Liu JS, Zhang YX. Selective promoter recognition by chlamydial sigma28 holoenzyme. J Bacteriol. 2006;188:7364–7377. doi: 10.1128/JB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Sorenson MK, Darst SA. Disulfide cross-linking indicates that FlgM-bound and free sigma28 adopt similar conformations. Proc Natl Acad Sci U S A. 2006;103:16722–16727. doi: 10.1073/pnas.0606482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MK, Ray SS, Darst SA. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- Tatti KM, Jones CH, Moran CP., Jr Genetic evidence for interaction of sigma E with the spoIIID promoter in Bacillus subtilis. J Bacteriol. 1991;173:7828–7833. doi: 10.1128/jb.173.24.7828-7833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli sigma(70) in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J Biol Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- Waldburger C, Gardella T, Wong R, Susskind MM. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, deHaseth PL. Sigma 32-dependent promoter activity in vivo: sequence determinants of the groE promoter. J Bacteriol. 2003;185:5800–5806. doi: 10.1128/JB.185.19.5800-5806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Dombroski AJ. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Lamont IL. Mutational analysis of an extracytoplasmic-function sigma factor to investigate its interactions with RNA polymerase and DNA. J Bacteriol. 2006;188:1935–1942. doi: 10.1128/JB.188.5.1935-1942.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak CE, Hughes KT. Genetic dissection of the consensus sequence for the class 2 and class 3 flagellar promoters. J Mol Biol. 2008;379:936–952. doi: 10.1016/j.jmb.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Di Russo EG, Rounds MA, Tan M. Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli sigma(28) RNA polymerase. J Bacteriol. 2006a;188:5524–5531. doi: 10.1128/JB.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Kibler D, Tan M. In silico prediction and functional validation of sigma28-regulated genes in Chlamydia and Escherichia coli. J Bacteriol. 2006b;188:8206–8212. doi: 10.1128/JB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Tan M. Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the −10 promoter element. Embo J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Liu M, Burgess RR. Adaptation in bacterial flagellar and motility systems: from regulon members to 'foraging'-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]