Abstract
Background
Cytochrome P4502C9 (CYP2C9) plays a vital role in drug metabolism. There has been an increased effort to identify polymorphisms within the gene and determine their clinical consequences. However, most of these efforts have focused on populations of European descent. Herein we report the influence of CYP2C9 genotype on warfarin dose among European American and African American patients. We also identify two new mutations; one in the coding region and one in the non-coding region of the CYP2C9 gene.
Methods
Patients (≥20 years of age) are enrolled after obtaining medical, lifestyle and concomitant medication history. Changes in International Normalized Ratio (INR), warfarin dose, co-medications, diet, physical activity and the occurrence of complications are documented. CYP2C9 genotype was determined using PCR-RFLP and pyrosequencing. Differences in genotype frequencies and HWE assumptions were assessed using χ2 statistics and exact tests. The genotype dose association was evaluated using multivariable linear regression.
Results
This report includes 490 patients (mean age 60.6 ± 15.6, 51.3% men). African American patients comprise 48.9% of the cohort with mean follow-up of 13.5 (±10.6) months. Both the CYP2C9 *2 and *3 allele were more frequent in European Americans (11.24%, 5.1%) compared to African Americans (1.1% and 1.8%). CYP2C9 *5 (0.9%), *6 (0.4%), and *11 (1.1%) variants were only observed in African Americans. The variant genotype is more frequent among European Americans compared to African Americans (29.8% vs. 9.73%, p<0.0001). Warfarin dose was significantly related to CYP2C9 genotype (p<0.0001) both in univariate and multivariate analyses. Multivariable race-specific analyses highlight the contribution of CYP2C9 genotype among European American but not among African American patients.
Conclusion
The variant CYP2C9 genotype is more frequent among European Americans compared to African Americans. Among African Americans the variant genotype frequency is higher than previously reported. CYP2C9 genotype predicts warfarin dose in European Americans, but not in African Americans.
Keywords: CYP2C9, Pyrosequencing, African American, European American, Warfarin Dose
Introduction
Cytochrome P450s are a superfamily of oxygen-activating enzymes involved in the metabolism of a gamut of endogenous and exogenous substrates.[1] Although the annotation of the human genome has revealed the presence of some 57 human P450 genes, less than a dozen play a critical role in the hepatic clearance of drugs. [2, 3]Cytochrome P4502C9 (CYP2C9) has drawn considerable attention for several reasons. [4] First, within the CYP2C subfamily CYP2C9 is the largest contributor to the hepatic microsomal P450 content. [5] Second, it is involved in the metabolism of up to 15% of all drugs that undergo phase I metabolism, second only to Cytochrome P4503A4. [2] Third, it plays a vital role in drug metabolism serving as the principal metabolic pathway for clinically important drugs such as warfarin, tolbutamide, phenytoin, etc. [6] It has been implicated in causing adverse drug reactions and drug-drug interactions that pose unique therapeutic management problems, especially for CYP2C9 substrates which have low therapeutic index (e.g. warfarin and phenytoin). [7, 8]Fourth, the gene coding for CYP2C9 exhibits significant polymorphism. [9]Some data suggest that up to 40% of Caucasian populations are carriers of alleles that encode for a partially defective enzyme which exacerbates adverse drug reactions and drug-drug interactions involving CYP2C9 substrates, especially those with intrinsically low margin of safety. [4, 10]
Recognition of polymorphisms with CYP2C9 spearheaded efforts to determine their clinical consequences. However, most of these efforts have focused on populations of European descent. [11–18]Herein we report CYP2C9 allele frequencies among European American and African American patients and evaluate the influence of CYP2C9 on warfarin dose stratified by race. We detail genotyping methodology and describe study design and cohort characteristics at enrollment. We also describe two new mutations in the CYP2C9 gene (one coding change and one noncoding change).
Methods
Study Setting
The Pharmacogenetic Optimization of Anticoagulation Therapy (POAT) is an ongoing prospective cohort study aiming at defining the influence of CYP2C9 polymorphisms on the dose of warfarin and risk of warfarin related complications during a 2-year follow-up period. As part of the study protocol CYP2C9 genotype was assessed in all participants.
The study is being conducted at the University of Alabama at Birmingham (UAB) enrolling patients from the anticoagulation clinic at The Kirklin Clinics (TKC-AC) and the Jefferson Clinic P.C., Jefferson County Health System (CGH-JC) under the approval of the respective Institutional Review Boards. At both clinics patient care is managed via a physician approved prescriptive authority protocol that provides a standardized approach to warfarin dose adjustments based on International Normalized Ratio (INR) results, management of over-anticoagulation and under-anticoagulation and frequency of follow-up.
Inclusion and Exclusion
Patients ≥20 years of age are identified at the initiation of chronic warfarin therapy. Patients are considered eligible if the intended duration of anticoagulation therapy was at least 2 years, the target INR range was 2–3 and are to be managed at one of the two anticoagulation clinics.
Data Collection
A structured interview form is used at the time of enrollment to obtain a detailed medical history. Information on self-reported race, indication for therapy, demographics, height and weight, concomitant medications and co-morbid conditions is documented. Lifestyle and socioeconomic data included smoking, alcohol use (number of alcoholic drinks per week)[19–21], education, annual household income, medical insurance, compliance, level of physical activity[22, 23], and dietary vitamin K intake (number of servings of foods rich in vitamin K per week). [24] All patients are followed for up to two years from initiation of therapy at monthly intervals. At each visit changes in factors influencing warfarin response are documented.
Blood collection, DNA extraction and genotyping methodology
Approximately 8 ml of blood is collected in a PAXgene tube (Qaigen Inc., Valencia, CA) at the time of patient consent. DNA is extracted using the PAX gene blood DNA extraction kits and stored at 2–8 °C.
Cytochrome P450 2C9 (CYP2C9) genotype determination
Although the initial aim was to determine *1, *2 (C430T, R144C), *3 (A1075C, I359L), *5 (C1080G, D360E) and *6 (818delA (frameshift) variants of CYP2C9, evolving evidence indicates the *11 (C1003T, R335W) variant has reduced metabolic efficiency. [25, 26]Therefore, this variant was also included in this study. The *10 allele (A815G, E272G) is a rare allele reported in European Americans. [26] Although CYP2C9.10 protein in vitro did not indicate altered catalytic activity, the genotype was readily determined in the pyrosequencing reaction of the *6 allele, and therefore included. All of these alleles are documented on the Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee (http://www.cypalleles.ki.se). These pyrosequencing methods allow for the determination of six alleles with four PCR reactions and four sequencing reactions.
Pyrosequencing
Genomic DNA (10–30 ng) is amplified with I unit Ampli Taq Gold (Applied Biosystems, Foster City, CA) in 40 μl reaction containing 1X PCR buffer, 0.2 pm/μl biotinylated forward primer, 0.2 pm/μl reverse primer (Eurogentec, San Diego, CA) (Table 1), 2 mM final MgCl2, and 0.5 mM final dNTPs. An initial denaturation of 95 °C for 8 minutes is used followed by 95 °C for 15 sec, 56 °C for 30 sec and 72 °C for 15 sec for 45 cycles. This reaction is followed by a final extension at 72 °C for 5 min (GeneAmp PCR 9700 System, Applied Biosystems, Foster City, CA).
Table 1.
PCR sequencing primers for Pyrosequencing CYP2C9 alleles
| PCR primersa | bpb size | 2C9 allele | Sequencing primer |
|---|---|---|---|
|
| |||
| Exon 3 | |||
| F: B-5′-AAACAGAGACTTACAGAGCTC-3′ R: 5′-CTAACAACCAGACTCATAATG-3′ |
381 | *2 | 5′-GGGCTTCCTCTTGAAC-3′ |
| Exon 5 | |||
| F: B-5′-CAGAGCTTGGTATATGGTATG-3′ R: 5′-TCGTAAACACAGAACTAGTCAAC-3′ |
323 | *6,*10 | 5′-AAGCTTTTGTTTACATTTT-3′ |
| Exon 7 | |||
| F: B-5′-CTGAATTGCTACAACAAATGTG-3′ R: 5′-GATACTATGAATTTGGGACTTC-3′ |
314 | *11 | 5′-TTGCATGCAGGGGCT-3′ |
| F: B-5′-TGCACGAGGTCCAGAGAT-3′ Reverse primer same as above |
155 | *3,*5 | 5′-GCTGGTGGGGAGAAG-3′ |
F: forward primer; R: reverse primer, B: biotin labeled primer
bp indicates PCR fragment base pair size
2C9 allele: cytochrome P4502C9 single nucleotide polymorphism
The entire biotinylated PCR product is mixed with 40 μl of 2× Binding-Washing buffer II (10 mM Tris-HCL, 2 M NaCl, I mM EDTA and 0.1% Tween 20, pH 7.6) and immobilized with 3 μl (10 ug/ul) streptavidin-coated polystyrene beads (Amersham Biosciences, Piscataway, NJ). Samples are mixed at room temperature for 10 minutes. To achieve DNA strand separation, a vacuum prep tool (Biotage, Foxboro, MA) is used and PCR products are isolated through alkaline denaturation and wash steps (Biotage, Foxboro, MA). Beads are released into wells of a PSQ™ sequencing plate (Biotage, Foxboro, MA) containing 20 pmoles of the appropriate sequencing primer (Table 1). The reactions annealed at 90°C for 5 min and cooled at RT for 10 min. Substrates and enzymes as well as dNTP from the manufacturer’s kit (Biotage, Foxboro, MA) were placed into a cartridge (supplied by Biotage) and placed into the PSQ 96 MA Pyrosequencer (Biotage, Foxboro, MA) for sequence determination. These methods allow the determination of the six alleles with four PCR reactions (Table 1).
CYP2C9*2 was determined with one PCR reaction amplifying exon 3 and a single sequencing primer. The CYP2C9*6 and CYP2C9*10 alleles were determined with a second PCR reaction amplifying exon 5 and one sequencing primer. The CYP2C9*3, CYP2C9*5, and CYP2C9*11 alleles were determined with two PCR reactions for exon 7, each with a separate sequencing primer.
Polymerase chain reaction with restriction fragment length polymorphism (PCR-RFLP) was also used to assess the CYP2C9*2 variant in 293 patients as described by Sullivan–Klose et al [27] with some modifications. Each 25 μL reaction contained 1X Thermopol Buffer II (New England Biolabs, Beverly, MA), 3 mM magnesium sulfate, 50 μM dNTPs, 0.02 U/μl standard Taq DNA polymerase (New England Biolabs, Beverly, MA), 100ng genomic DNA, and a 0.25 μM of forward (5′-TACAAATACAATGAAAATATCATG-3′) and reverse 5′-CTAACAACCAGACTCATAATG-3′) primer for CYP2C9*2. Thermocylcing conditions (touch down protocol) consisted of 94°C for 5 minutes followed by 10 cycles of 94°C for 30seconds, 55°C → 50°C (touch down protocol, −0.5°C per cycle) for 30 seconds and 72°C for 30 seconds. This is followed by 20 cycles of 94°C for 30 seconds, 50°C for 30 seconds and 72°C for 15 seconds with a 5 minute extension step at 72°C. Ten μls of the CYP2C9*2 amplicon were digested overnight at 37°C using the restriction endonuclease Ava II under manufacturer recommended directions (New England Biolabs, Beverly, MA). The product digests are then separated by electrophoresis through a 2.5% agarose gel and visualized by staining with ethidium bromide.
Statistical Methods
Dose was defined in two ways: the average maintenance dose required to maintain anticoagulation for the duration of therapy and stable dose at which therapy was considered stabilized (3 consecutive INRs in target range, with these INR measurements encompassing a period of at least 2 weeks, with a maximum difference between the mean daily dosages of 10%). The distribution of dose was marginally skewed to the right therefore log transformation was done to attain normality. The association of dose and CYP2C9 was evaluated using linear regression after accounting for the effects of covariates. Genotypes are categorized into three groups: homozygous wild type [*1/*1], heterozygotes (*1/Variant [*V] or homozygotes [*V/*V]) for the variant allele. The homozygous wild type is considered the referent group for all analyses. Student’s t-tests are used to test differences for continuous variables and the χ2 test of independence to assess the differences for categorical variables between racial groups. All analyses are performed using SAS version 9.1 (SAS Institute, Cary NC) at a non-directional α level of 0.05.
The assumption of Hardy Weinberg Equilibrium (HWE) is tested using the χ2 test of independence. We also tested the HWE assumption using the “HARDY” genetic analysis software (http://linkage.rockefeller.edu/soft/list2.html#h) to obtain exact test statistics. This program uses the Markov Chain Monte Carlo (MCMC) algorithm for association in two-dimensional contingency tables, and for testing Hardy-Weinberg equilibrium. [28]
Results
All patients meeting eligibility criteria (n=526) were approached to participate in the study after consent of the treating physician. Thirty-six (6. 9%) patients (17 African Americans, 19 European Americans, 15 men, 21 women) declined participation for various reasons. The age, gender and racial makeup of patients declining participation did not differ from that of those who agreed to participate. The age, gender and racial makeup of patients enrolled in the study was similar to that of patients initiated on warfarin therapy at UAB, residing in the five-county Birmingham metropolitan area. A majority (94.4%) of patients resided within the five-county Birmingham metropolitan area, with 81% residing in the Jefferson County. The gender and racial makeup of participants from Jefferson County is similar to that reported of its residents in the 2000 US census. [29]
This report includes 490 patients (mean age 60.6 ±15.6, range 26 to 93 years) followed from initiation of chronic warfarin therapy. Patients were recruited between August 2003 and August 2006. African American patients comprise 48.9% of the cohort and men 51.3% with an average of 13.5 (±10.6) months of follow-up accrued.
Table 2 displays the socio-demographic and selected lifestyle characteristics of study participants by race. The mean age was significantly higher in European American compared to African American patients (63.5 vs. 57.5 years, p<0.0001). There were no significant differences in gender distribution, height, weight and body mass index (BMI; kg/M2). More European American patients were light drinkers (alcohol intake: 1–7 drinks/week) (p<0.0001) and were more physically active (p=0.002) compared to African American patients. European American patients were more likely to have medical insurance (90.8% vs. 73.6%, p<0.0001), higher education (p<0.0001) and higher income (p<0.0001). European American patients were less likely to have smoked or to engage in smoking at the time of enrollment (p=0.005).
Table 2.
Socio-demographic and lifestyle characteristics of the POAT study participants.
| Characteristic | African American (n=239) | European American (n=249) | p-value |
|---|---|---|---|
| Age, mean (SD) | 57.5 (15.8) | 63.5 (14.8) | <0.0001 |
|
| |||
| Height (inches) | 67.6 (4.1) | 68.2 (6.4) | 0.20 |
|
| |||
| Weight (lbs) | 196.0 (51.2) | 193.2 (51.7) | 0.55 |
|
| |||
| Body Mass Index, mean (SD) | 30.2 (7.4) | 29.3 (7.5) | 0.25 |
|
| |||
| N (%) | N (%) | ||
|
| |||
| Gender | |||
| Female | 126 (52.7) | 111 (44.6) | 0.07 |
| Male | 113 (47.3) | 138 (55.4) | |
|
| |||
| Alcohol (drinks per week)1 | |||
| 0 | 185 (77.4) | 150 (60.2) | <0.0001 |
| 1–7 | 34 (14.2) | 83 (33.3) | |
| >8 | 19 (8.0) | 16 (6.4) | |
|
| |||
| Smoking Status1 | |||
| Current | 48 (20.0) | 23 (9.2) | 0.005 |
| Past | 83 (34.7) | 95 (38.2) | |
| Never | 104 (43.5) | 123 (49.4) | |
|
| |||
| Level of Physical Activity1 | |||
| Wheelchair bound | 14 (5.9) | 7 (2.8) | 0.003 |
| Uses Walker/Cane | 39 (16.3) | 24 (9.6) | |
| Ambulates without assistance | 46 (19.2) | 43 (17.3) | |
| Physically active | 137 (57.3) | 163 (65.5) | |
| Consistent/Intensive exercise | 1 (0.42) | 11 (4.4) | |
|
| |||
| Education | |||
| < High School | 73 (30.5) | 21 (8.4) | <0.0001 |
| High School | 110 (46.1) | 90 (36.1) | |
| College | 55 (23.0) | 106 (42.6) | |
| Graduate School | 1 (0.4) | 32 (12.9) | |
|
| |||
| Annual Household Income1 | |||
| <15,000 | 96 (40.2) | 31 (12.5) | <0.0001 |
| 15,000–25,000 | 73 (30.5) | 29 (11.6) | |
| 25,000–50,000 | 65 (27.2) | 84 (33.7) | |
| 50,000–100,000 | 3 (1.2) | 84 (33.7) | |
| >100,000 | 1 (0.4) | 21 (8.4) | |
|
| |||
| Clinic | |||
| The Kirklin Clinic | 151 (63.2) | 231 (92.8) | <0.0001 |
| Cooper Green Hospital* | 88 (36.8) | 18 (7.2) | |
|
| |||
| Medical Insurance | |||
| Medicare | 66 (27.6) | 103 (41.4) | <0.0001 |
| Medicare Medicaid | 5 (2.1) | 0 (0.0) | |
| Medicaid | 14 (5.9) | 9 (3.6) | |
| Private | 91 (38.1) | 114 (45.8) | |
| None | 63 (26.3) | 23 (9.2) | |
Mean (SD) displayed for continuous variables and frequency counts (column percent) for categorical variables
Information on missing for smoking (n=13), level of physical activity (n=3), alcohol (n=2), income (n=1)
2 Hispanic patients excluded, all significant p-values bolded
Private insurance includes various private insurance plans such as Blue Cross Clue Shield, Aetna, Travelers, etc.
The clinical characteristics of patients enrolled (n=488) by race are presented in Table 3. As of August 2006 there is no difference in the duration of follow-up accrued by race (p=0.89). The prescribed target INR for all patients was 2–3. Stroke (p=0.007) and venous thromboembolism (both venous and pulmonary, p= 0.002 and 0.0004 respectively) were more common indications for warfarin therapy in African American patients, while atrial fibrillation was a more common indication in European Americans. There were no significant differences by site of thromboembolism except venous thromboembolism, which was more prevalent in African American compared to European American patients (p< 0.0001). This finding was consistent with the higher prevalence of prior history of venous thromboembolic event in the African American patients (p=0.013). Multivariable race-specific analyses highlighting the contribution of FVL mutation to the risk of venous thromboembolic events in European American (p = 0.03) but not in African American patients (p = 0.95) have recently been reported. [30]
Table 3.
Clinical characteristics of the POAT study participants.
| Characteristic | African American (n=239) | European American (n=249) | p-value |
|---|---|---|---|
| Follow-up accrued (months, Mean ± SD) | 13.5 (10.5) | 13.6 (10.7) | 0.89 |
|
| |||
| Indication for warfarin therapy* | |||
| Deep Vein Thrombosis | 84 (35.1) | 56 (22.5) | 0.002 |
| Pulmonary Thromboembolism | 41 (17.1) | 17 (6.8) | 0.0004 |
| Recurrent Venous Thromboembolism | 20 (8.4) | 14 (5.6) | 0.23 |
| Atrial Fibrillation | 76 (31.8) | 147 (59.0) | <0.0001 |
| Valvular Heart Disease | 26 (10.9) | 40 (16.1) | 0.09 |
| Low Left Ventricular Ejection Fraction | 40 (16.7) | 29 (11.6) | 0.11 |
| Cardiac Thrombus | 10 (4.2) | 12 (4.8) | 0.73 |
| Myocardial Infarction | 62 (25.9) | 80 (32.1) | 0.13 |
| Transient Ischemic Attack | 15 (6.3) | 20 (8.0) | 0.45 |
| Stroke | 53 (22.2) | 32 (12.8) | 0.007 |
| Peripheral Vascular Disease | 27 (11.3) | 38 (15.3) | 0.20 |
|
| |||
| Site of thromboembolism** | |||
| Arterial | 99 (41.1) | 112 (45.0) | 0.43 |
| Venous | 118 (49.4) | 76 (30.5) | <0.0001 |
| Both | 25 (10.5) | 19 (7.6) | 0.27 |
| None | 34 (14.2) | 44 (17.7) | 0.30 |
|
| |||
| Comorbidity | |||
| History of Myocardial Infarction | 8 (3.3) | 19 (7.6) | 0.04 |
| History of CABG/PTCA | 25 (10.5) | 53 (21.3) | 0.001 |
| Cardiomyopathy | 14 (5.9) | 16 (6.4) | 0.79 |
| Coronary Artery Disease | 74 (31.0) | 90 (36.1) | 0.22 |
| Congestive Heart Failure | 60 (25.1) | 51 (20.5) | 0.22 |
| Hypertension | 103 (43.1) | 99 (39.8) | 0.45 |
| Hyperlipidemia | 49 (20.5) | 84 (33.7) | 0.001 |
| Diabetes Mellitus | 83 (34.7) | 75 (30.1) | 0.28 |
| Malignancy | 23 (9.6) | 49 (19.7) | 0.002 |
| Prior Hemorrhage | 21 (8.8) | 11 (4.4) | 0.051 |
| Renal insufficiency | 43 (18.0) | 30 (12.0) | 0.07 |
| End Stage Renal Disease | 28 (11.7) | 7 (2.8) | <0.0001 |
|
| |||
| Number of Comorbid Conditions | |||
| Low (0 or 1) | 81 (33.9) | 64 (25.7) | 0.046 |
| Medium (2 to 4) | 112 (46.9) | 117 (47.0) | |
| High (5 or more) | 46 (19.2) | 68 (27.3) | |
2 Hispanic patients excluded, significant p-values bolded
patients with orthopedic surgery excluded due to short (3–6 month) treatment duration, patients with mechanical heart valve and hypercoagulable state excluded due to higher intensity of anticoagulation required
Arterial thromboembolism includes patients with MI, Stroke & TIA. Venous thromboembolism includes patients with DVT & PE. Both include patients with venous and arterial events. None includes patients with no thromboembolic events (e.g. Atrial Fibrillation),
The prevalence of individual comorbid conditions differed across racial groups. More European American patients had a prior history of myocardial infarction (p=0.04), undergone coronary artery bypass grafting or percutaneous coronary angioplasty (p=0.001), hyperlipidemia (p=0.001), and malignancy (p=0.002) compared to African American patients. The prevalence of end stage renal disease (p<0.0001) was significantly higher and renal insufficiency marginally higher (p=0.07) in African American patients compared to European American patients. European American patients had higher number of comorbid conditions at baseline compared to African American patients (p=0.046).
Five patients refused to submit a blood sample after enrollment and CYP2C9 genotype has not been determined for 39 patients. CYP2C9*2 variants were assessed using both PCR-RFLP and pyrosequencing. Initial pyrosequencing results for CYP2C9*2 were indeterminate for six samples due to low signal strength (n=2), failed dispensation (n=1) and potentially new single nucleotide polymorphisms (n=3). Repeat pyrosequencing confirmed CYP2C9 genotype as *1/*1 for three samples. Pyrosequencing called the CYP2C9 genotype as *2/*2 for eleven samples. However, eight samples were graded as “check” or “fail” by the pyrosequencing software. Resequencing of these samples for exon 3 [27] confirmed the genotype as *1/*2 heterozygote in all eight samples. Three samples were genotyped unequivocally as homozygous for CYP2C9*2 and were considered “pass” by the software. All three pyrograms were identical and PCR-RFLP confirmed the genotype as *2/*2 homozygote, as did resequencing of one of the samples.
For the CYP2C9*2 variant the concordance between pyrosequencing and PCR-RFLP methodologies was 99.3%. The discordant results involved two European Americans with a previously unreported non-coding change (C429T) in the region of the *2 mutation (C430T). The CYP2C9 genotype for these two patients was determined to be *1/*2 by PCR-RFLP (Figure 1, sample #81) but a new mutation at base pair 429 was detected by pyrosequencing. C429T was noted as an extra peak on the pyrogram (the incorporation of an A instead of the expected negative dispensation on the antisense strand) and a decrease in the height of the preceding G peak. This change was confirmed as C429T on the sense strand by genomic sequencing. Figure 2 shows the predicted histograms and resulting pyrograms for CYP2C9 exon 3 in individuals genotyped as *1/*1 (Panel 1), *1/*2 (Panel 2) and *1/C429T (Panel 3). Sequencing of genomic DNA confirmed this C429T non-coding change as a silent mutation (D143D); therefore the CYP2C9 genotype in these two patients was designated *1/*1 for all further analyses.
Figure 1.
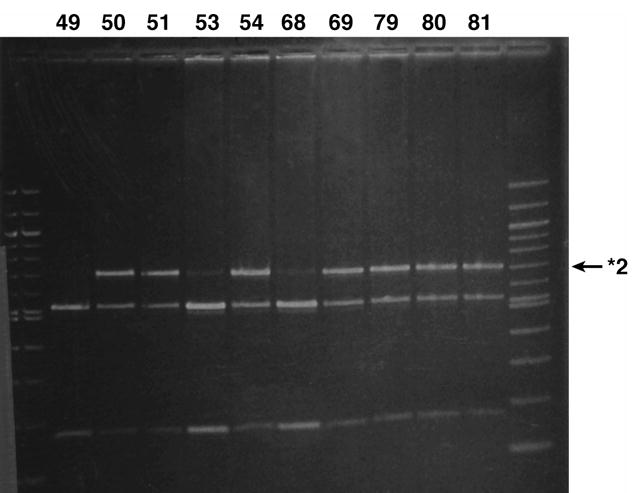
CYP2C9 *2 variant as assessed by PCR-RFLP as described by Sullivan Klose et al. [27]The first and last lane contains molecular weight markers (100 base pair ladder). The remaining 10 lanes correspond to the Ava II digested PCR products from patients 49, 50, 51, 53, 54, 68, 69, 79, 80 and 81 separated by electrophoresis through a 2.5% agarose gel and visualized by staining with ethidium bromide. The CYP2C9 genotype for patients 49, 53 and 68 is CYP2C9*1/*1 and CYP2C9*1/*2 for patients 50, 51, 54, 69, 79, 80 and 81. The genotype for patient #81 (second last lane) is falsely determined to be *1/*2 due to a previously unreported non-coding change at base pair 429. The genotype for patient 81 was CYP2C9*1/*1 by pyrosequencing (Figure 2)
Figure 2.
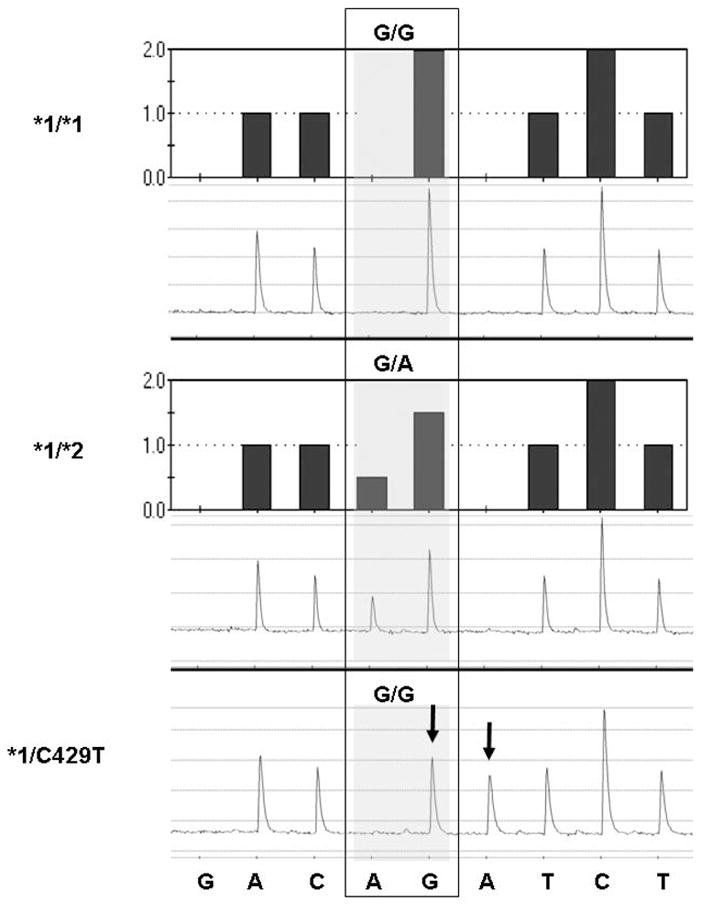
Predicted histograms and actual pyrograms showing the CYP2C9*2 genotyping test. The reverse strand was sequenced; the sequence at the bottom refers to the nucleotide dispensation. While the dispensation order is G(negative)AC[AG]A(negative)TCT, the actual sequence analyzed (reverse strand) is AC[A/G]GTCCT (variant nucleotides in brackets with the mutant nucleotide in bold). The predicted histogram is shown at the top, and the actual pyrogram is shown below. The shaded boxes indicate the regions in which the two variable nucleotides were dispensed, with the base designation of the sample at base pair 430 at the top. The actual genotype of the sample is indicated at the left. Samples are from individuals genotyped as CYP2C9*1/*1 (top panel), CYP2C9*1/*2 (middle panel) and the bottom panel shows a pyrogram from patient #81 with an abnormal pyrogram (see arrows) with the disappearance of a G and the appearance of an A at bp 429 on the antisense strand (confirmed by sequencing as heterozygous for a noncoding mutation 429C>T on the sense strand).
Figure 3 shows the predicted histograms and resulting pyrograms for CYP2C9 exon 7 in individuals presenting with *1/*1 (Panel 1), *1/*3 (Panel 2), 1/*5 (Panel 3), and *1/C1078T (Panel 4). Panel 4 depicts the pyrogram from an African American man who was found to carry a previously unreported polymorphism, confirmed by DNA sequencing, to represent a G1078A mutation in the same region as the CYP2C9*3 mutation. The resulting codon change ‘GAC to AAC’ would lead to the substitution of D360N in the CYP2C9 protein. This patient, in addition to two Hispanic patients was excluded from the analysis assessing the HWE.
Figure 3.
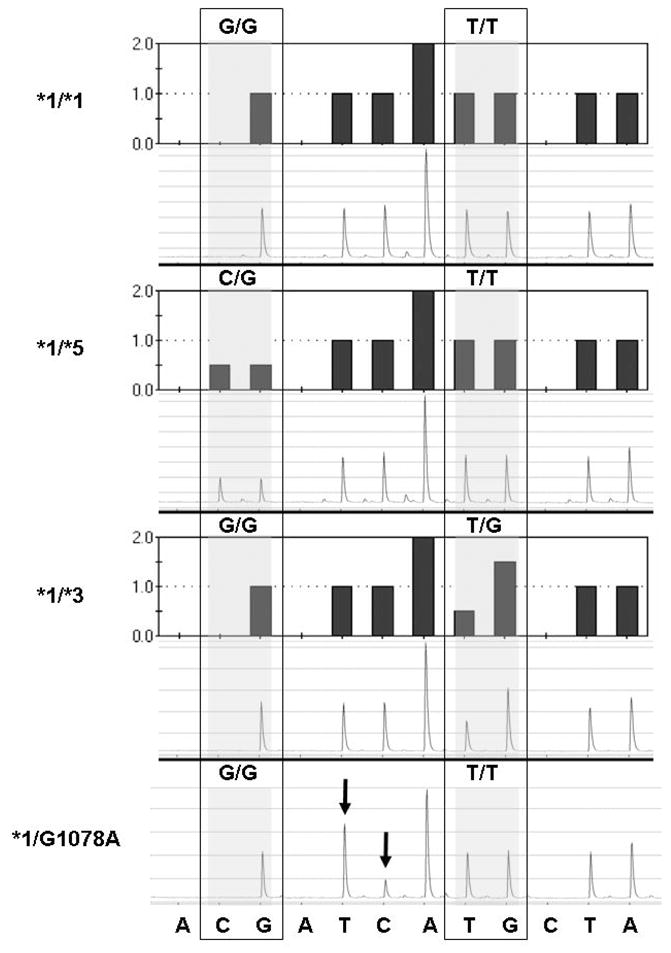
Histograms and pyrograms showing CYP2C9*5 and CYP2C9*3 genotyping tests. The sequence at the bottom refers to the nucleotide dispensation. While the dispensation order is A(negative)[CG]A(negative)TCA[TG]C(negative)TA, the actual sequence (reverse strand) analyzed is [C/G]TCAA[T/G]GTA (variant nucleotides in brackets with mutant nucleotides in bold). The shaded boxes indicate the variable region in which two nucleotides were dispensed, with the base designation of each allele. Pyrograms shown are examples of individuals homozygous for CYP2C9*1/*1 or heterozygous for CYP2C9*1/*5 and CYP2C9*1/*3. The final pyrogram is from individual # 43 who shows an abnormal pyrogram (see arrows) with the appearance of an extra T at bp 1078 and the disappearance of a C (antisense strand), confirmed by sequencing of the genomic DNA as a G1078A mutation on the sense strand producing a new coding change D360N).
Of the variant alleles tested only the CYP2C9*2 (11.24%), and CYP2C9*3 (5.1%) alleles were observed among European Americans. Therefore the inferred frequency of CYP2C9*1 allele is 83.7%. Of the variants tested, CYP2C9*2 (1.1%), CYP2C9*3 (1.8%), CYP2C9*5 (0.9%), CYP2C9*6 (0.4%), and CYP2C9*11 (1.1%) were observed among African Americans. Therefore the inferred frequency of CYP2C9*1 allele is 94.7%. The prevalence of variant CYP2C9 genotypes (Table 4) is significantly higher in European Americans compared to African Americans (29.82%% vs. 9.73%, p <0. 0001). The distribution of CYP2C9 genotype differed by race (p <0.0001) but not by gender (p = 0.13, data not shown). CYP2C9 genotype frequencies were found to be in HWE among European Americans (all p-values >0.45) and African Americans (all p-values >0.75).
Table 4.
CYP2C9 genotype and average warfarin dose (mg/day ± SD) distribution among participants of the POAT cohort.
| Genotype | All Participants | African Americans | European Americans | |||
|---|---|---|---|---|---|---|
| N=444 | N=226 | N=218 | ||||
| N (%) | Dose | N (%) | Dose | N (%) | Dose | |
| *1/*1 | 357 (80.4) | 5.7 (± 2.5) | 204 (90.3) | 6.1 (± 2.6) | 153 (70.2) | 5.3 (± 2.4) |
| *1/*2 | 45 (10.1) | 4.7 (± 2.0) | 5 (2.2) | 6.6 (± 1.59) | 40 (18.3) | 4.5 (± 2.0) |
| *1/*3 | 26 (5.8) | 4.1 (± 1.4) | 7 (3.1) | 5.0 (± 1.0) | 19 (8.7) | 3.8 (± 1.4) |
| *1/*5 | 3 (0.7) | 6.3 (± 1.9) | 3 (1.3) | 6.3 (± 1.9) | 0 (0.0) | - |
| *1/*11 | 5 (1.1) | 5.8(± 2.0) | 5 (2.2) | 5.8(± 2.0) | 0 (0.0) | - |
| *2/*2 | 3 (0.7) | 2.9 (± 1.6) | 0 (0.0) | - | 3 (1.4) | 2.9 (± 1.6) |
| *2/*3 | 3 (0.7) | 1.8 (± 0.6) | 0 (0.0) | - | 3 (1.4) | 1.8 (± 0.6) |
| *3/*6 | 1 (0.2) | 2.0 | 1 (0.4) | 2.0 | 0 (0.0) | - |
| *5/*6 | 1 (0.2) | 4.0 | 1 (0.4) | 3.0 | 0 (0.0) | - |
Subjects recruited during the intervalof August 2003 – August 2006
Analysis excludes Hispanic patients (n=2)
Genotype frequencies present as number of participants with genotype and (percent)
Average daily warfarin dose (mg/day ± SD) analysis included 437 patients
Table 4 summarizes the average maintenance dose by CYP2C9 genotype. CYP2C9 significantly influenced warfarin dose among European Americans (p=0.0007) but not African Americans (p=0.59). Dose difference among patients with CYP2C9*5, *6, and *11 alleles could not be determined due to the rarity of these variants. Restricting analysis to CYP2C9*2 and *3 alleles did not change the magnitude of the dose-genotype associations. Combining genotype into groups according to the presence of variant alleles did not alter the dose-genotype associations among European Americans or African Americans (Table 5).
Table 5.
Warfarin daily dose stratified by CYP2C9 genotype
| Dose (mg/day) | CYP2C9*1/*1 | CYP2C9*1/* V | CYP2C9* V/* V | p-value |
|---|---|---|---|---|
| Average Maintenance | 5.3 [4.0–6.8] | 4.4 [3.0–5.9] | 2.12 [1.8–3.2] | <0.0001 |
| African American | 5.8 [4.2–7.5] | 5.3 [4.7–6.3] | 3.0 [2.0–4.0] | 0.16 |
| European American | 4.9 [3.7–6.3] | 4.0 [2.8–5.7] | 2.1 [1.5–2.4] | <0.0001 |
| Stable Dose | 5.0 [3.9–6.4] | 4.3 [2.9–5.7] | 2.1 [1.5–2.8] | <0.0001 |
| African American | 5.0 [3.9–6.6] | 5.0 [4.3–6.2] | -1 | 0.71 |
| European American | 5.0 [3.9–6.4] | 1.0 [2.9–5.3] | 2.1 [1.5–2.8] | 0.0003 |
V denotes ≥ variant allele (2, *3, *5, *6, *10, or *11)
Median daily dose (interquartile range) presented. All p-values are based on Kruskal-Wallis Test
two patients were not stabilized at the time of data analysis
Analysis excludes Hispanic patients (n=2)
Although the variability in dose explained by CYP2C9 genotype decreased slightly from the univariate to the multivariable model, the significance of the dose-genotype association remained unchanged. In the multivariable analysis CYP2C9 genotype accounted for 3.5% variability in average warfarin dose (p<0.0001) and 5.6% for stable warfarin dose (p<0.0001) which was largely due the influence of CYP2C9 among European Americans. The variability in warfarin dose explained by CYP2C9 genotype was 6.4% (average dose) and 7.9% (stable dose) among European Americans and 1% African American (Table 6). European American, but not African American patients with a variant CYP2C9 genotype required significantly lower warfarin dose compared to their wild-type genotype comparators.
Table 6.
Percent variation in warfarin log-dose explained by variant CYP2C9 genotype
| Average Maintenance Dose |
Stable Dose |
|||||
|---|---|---|---|---|---|---|
| Model | CYP2C9 | p | Model | CYP2C9 | p | |
| All patients | 27.0% | 3.5% | <0.0001 | 25.7% | 5.6% | <0.0001 |
| Daily Dose (mg) | 5.3 vs. 4.0 | <0.0001 | 5.3 vs. 4.1 | <0.0001 | ||
| European Americans | 19.4% | 6.4% | <0.0001 | 21.6% | 7.9% | 0.0003 |
| Daily Dose (mg) | 4.9 vs. 3.7 | <0.0001 | 5.2 vs. 3.8 | <0.0001 | ||
| African Americans | 28.0% | 1.0% | 0.092 | 33.6% | 1.0% | 0.25 |
| Daily Dose (mg) | 5.6 vs. 5.3 | 0.22 | 5.4 vs. 5.0 | 0.16 | ||
Warfarin daily dose is predicted dose for wild type versus variant genotype. Analysis for stable dose included 232 patients
Adjusted for age, gender, BMI, drug interactions, average alcohol intake, average vitamin K intake, number of comorbid conditions
CYP2C9 genotype effects are present for variant genotype (*1/*V and *V/*V) versus *1/*1* genotype. V denotes variant allele (2, *3, *5, *6, *10, or *11).
Discussion
Although the efficacy of warfarin is proven, it is vastly underused with difficulty in management of therapy and fear of complications being the main deterrents. CYP2C9, the primary enzyme involved in the metabolism of warfarin, is known to be polymorphic. Although the influence of these polymorphisms has been documented in Caucasians, their influence in African Americans is not extensively documented. To our knowledge, the 226 African Americans who comprise 50% of the POAT cohort represent the largest population of African Americans genotyped for CYP2C9. Inclusion of the recently discovered variants [26, 31, 32] in the genotyping provides a robust estimate of the CYP2C9 allele frequencies in this racially underrepresented group.
As previously reported, the variant CYP2C9 genotype is more common in European Americans compared to African Americans (29.8% vs. 9.73%). Inclusion of rare alleles such as CYP2C9*5, CYP2C9*6 and CYP2C9*11 resulted in the variant genotype frequencies to be higher among African Americans (5.3%) than previously reported (3.7%) for this population. [9, 33] As previously reported CYP2C9*2 and CYP2C9*3 were the most common poor-metabolizer CYP2C9 alleles, among both European American and African American patients, although the frequency of these alleles was higher in the former group. The allele frequencies for CYP2C9*2 and CYP2C9*3 are consistent with those reported previously.[9, 34]
African American populations contain known or putative poor-metabolizer alleles (CYP2C9*5, CYP2C9*6 and CYP2C9*11) which are rarely found in European Americans. As reported by Dickmann et al [31] the CYP2C9*5 allele was observed only in African American patients. However the frequency of the CYP2C9*5 allele was lower in our study, 0.9% versus 1.7% reported previously. This difference is probably due to differences in racial admixture which is known to vary among African Americans residing within different geographic regions of the US. [35–37] Our study is the first to examine the frequencies of the CYP2C9*6 and CYP2C9*11 alleles in a large African American population. CYP2C9*6 (818delA, frameshift) contains a premature stop codon, resulting in a null allele that dramatically decreases the metabolism of phenytoin and warfarin. [31, 32]Based on studies with recombinant CYP2C9 [25, 26] CYP2C9*11 is predicted to have greatly diminished catalytic activity, but has not received extensive clinical study. One study [38] reported significantly lower warfarin dose requirements in patients with the CYP2C9*1/*11 genotypes compared to those with CYP2C9*1/*1 genotype. The rarity CYP2C9*6 (0.4%) and CYP2C9*11 (1.1%) allele did not facilitate evaluation of the dose-genotype association.
We identified two new mutations, one new non-coding change (C429T, Figure 2) in the same region as CYP2C9*2 mutation. This C429T change, a silent noncoding mutation (D143D) was noted in 2 (of 218 participants, 0.92%). This noncoding change is important because it destroys the AvaII site commonly used in RFLP analysis of the CYP2C9*2 allele, invalidating this test. This example discrepancy between the two genotyping methodologies demonstrates the superiority of pyrosequencing over RFLP genotyping tests.
Amino acids 359 and 360 lie within one of the predicted substrate recognition sites of the CYP2 family of enzymes.[39] This area seems to be a “hot spot” for mutations in CYP2C. The new D360N coding change found in one African American in this study represents a new mutation within this substrate recognition site. Several defective alleles which arise from mutations in this region greatly affect the affinity and catalytic activity of the CYP2C9 enzyme resulting in large variability in dosage requirements of CYP2C9 substrates. [40]These alleles include CYP2C9*3 (I359L)[27], CYP2C9*4 (I359T) (found in Japanese), [41]CYP2C9*5 (D360E) in African-Americans. [31] A Y358C mutation has also been reported to the NCIdbSNP homepage (rs1057909). Newer crystal structures show that the hydrophobic substrate pocket includes the adjacent amino acids L362 and L366 [42] which limit access for warfarin to the heme group. [43] The proximity of I359 and D360 to these amino acids can potentially result in defective alleles. Although rare, these alleles can produce clinically meaningful changes in warfarin dose and toxicity.
This study confirms the association of CYP2C9*2 and CYP2C9*3 polymorphisms on warfarin dose among European Americans after accounting for the effects of multiple potential covariates. It also confirms recently findings by Kealey et al [33] on the lack of significant influence of CYP2C9*2 and CYP2C9*3 on warfarin dose in African Americans. Our findings further extend this lack of association after inclusion of CYP2C9*5, *6 and *11 among African American patients. Although African American patients with 2 variant alleles required lower doses (Table 5), trend tests were not statistically significant, probably due to the small sample size in this subgroup.
We also recognize limitations of the present study. Documentation of vitamin K intake was based on patient report using vitamin K inventory [24] not quantified by assay measurements. However, all measurements were used consistently among all participants; therefore, bias if any should be non-differential. Although the focus of the current study was to define the association of CYP2C9 genotype on warfarin dose and risk of complications we recognize that drug response is influenced by multiple genes.[44] [45, 46] At least one other gene; Vitamin K Epoxide Reductase (VKORC1) has been consistently shown to significantly influence warfarin dose in European Americans [14, 15, 47, 48] and recently among African Americans.[49] Expansion of genotyping efforts, within this prospective cohort, to include other genes along the warfarin pharmacodynamic and pharmacokinetic pathways will facilitate gene-gene interaction studies and help tailor warfarin therapy based on genetic, clinical and demographic characteristics. Inclusion of underrepresented racial groups such as African Americans will help identify genetic and non-genetic determinants of drug response and quantify their influence in this subgroup where the risk of thromboembolic diseases and the resultant mortality is disproportionately high. [50]
Conclusion
The variant CYP2C9 genotype is more frequent among European Americans compared to African Americans. Among African Americans the variant genotype frequency is higher than previously reported. CYP2C9 genotype predicts warfarin dose in European Americans, but not in African Americans.
Executive Summary
▪This prospective study documents the frequencies of CYP2C9 polymorphisms (*2, *3, *5, *6, *11) in European Americans and African Americans.
▪The variant genotype is more frequent among European Americans (29.8%) compared to African Americans (9.7%).
▪CYP2C9 polymorphisms influenced warfarin dose requirements in European Americans but not African Americans in univariate and multivariable analyses.
▪CYP2C9 genotype predicts warfarin dose in European Americans, but not in African Americans.
▪Further studies are needed to identify other genetic and non-genetic predictors of warfarin dose in African Americans.
Acknowledgments
Supported in part by a grant from the National Institute of Neurological Disorders and Stroke (Grant Number: K23NS45598-01) and in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
The first author wishes to acknowledge Dr. Edward Faught for his support and mentorship and Ellen Funkhouser for her critical review of this manuscript. We are grateful to all the patients that participated in the study. We thank Janice Ware for her untiring efforts with patient recruitment and to the staff of the Anticoagulation Clinic at The Kirklin Clinic, the Cooper Green Hospital and Jefferson Clinic P.C for their help with identification of potential participants. We also thank the physicians, especially Drs. Mark Wilson, and Melissa Baird; at the University of Alabama at Birmingham and the Health Service Foundation for their support of this research. Thanks to Steve Duncan and Darlene Green of the University of Alabama at Birmingham, Office of Data resources for their work with the POAT database and quality assurance.
Abbreviations
- CYP2C9
Cytochrome P4502C9
References
- 1.Rendic S, Di Carlo FJ. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev. 1997;29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 3.Nelson DR. Introductory remarks on human CYPs. Drug Metab Rev. 2002;34:1–5. doi: 10.1081/dmr-120001385. [DOI] [PubMed] [Google Scholar]
- 4.Rettie AE, Jones JP. Clinical and toxicological relevance of CYP2C9: drug-drug interactions and pharmacogenetics. Annual Review of Pharmacology & Toxicology. 2005;45:477–94. doi: 10.1146/annurev.pharmtox.45.120403.095821. [DOI] [PubMed] [Google Scholar]
- 5.Lasker JM, Wester MR, Aramsombatdee E, Raucy JL. Characterization of CYP2C19 and CYP2C9 from human liver: respective roles in microsomal tolbutamide, S-mephenytoin, and omeprazole hydroxylations. Arch Biochem Biophys. 1998;353:16–28. doi: 10.1006/abbi.1998.0615. [DOI] [PubMed] [Google Scholar]
- 6.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–38. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirmohamed M, Park BK. Cytochrome P450 enzyme polymorphisms and adverse drug reactions. Toxicology. 2003;192:23–32. doi: 10.1016/s0300-483x(03)00247-6. [DOI] [PubMed] [Google Scholar]
- 8.Pirmohamed M, Park BK. Genetic susceptibility to adverse drug reactions. Trends Pharmacol Sci. 2001;22:298–305. doi: 10.1016/s0165-6147(00)01717-x. [DOI] [PubMed] [Google Scholar]
- 9.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002;54:1257–70. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 10.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Herman D, Locatelli I, Grabnar I, et al. Influence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J. 2005;5:193–202. doi: 10.1038/sj.tpj.6500308. [DOI] [PubMed] [Google Scholar]
- 12.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 13.Margaglione M, Colaizzo D, D’Andrea G, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000;84:775–8. [PubMed] [Google Scholar]
- 14.Schalekamp T, Brasse BP, Roijers JF, et al. VKORC1 and CYP2C9 genotypes and acenocoumarol anticoagulation status: interaction between both genotypes affects overanticoagulation. Clin Pharmacol Ther. 2006;80:13–22. doi: 10.1016/j.clpt.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Schalekamp T, Brasse BP, Roijers JF, et al. VKORC1 and CYP2C9 Genotypes and Phenprocoumon Anticoagulation Status: Interaction Between both Genotypes Affects Dose Requirement. Clin Pharmacol Ther. 2007;81:185–93. doi: 10.1038/sj.clpt.6100036. [DOI] [PubMed] [Google Scholar]
- 16.Tassies D, Freire C, Pijoan J, et al. Pharmacogenetics of acenocoumarol: cytochrome P450 CYP2C9 polymorphisms influence dose requirements and stability of anticoagulation. Haematologica. 2002;87:1185–91. [PubMed] [Google Scholar]
- 17.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96:1816–9. [PubMed] [Google Scholar]
- 18.Veenstra DL, Blough DK, Higashi MK, et al. CYP2C9 haplotype structure in European American warfarin patients and association with clinical outcomes. Clin Pharmacol Ther. 2005;77:353–64. doi: 10.1016/j.clpt.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Havrda DE, Mai T, Chonlahan J. Enhanced antithrombotic effect of warfarin associated with low-dose alcohol consumption. Pharmacotherapy. 2005;25:303–7. doi: 10.1592/phco.25.2.303.56955. [DOI] [PubMed] [Google Scholar]
- 20.McGriff-Lee NJ, Csako G, Chen JT, et al. Search for predictors of nontherapeutic INR results with warfarin therapy. Ann Pharmacother. 2005;39:1996–2002. doi: 10.1345/aph.1E381. [DOI] [PubMed] [Google Scholar]
- 21.Wittkowsky AK, Devine EB. Frequency and causes of overanticoagulation and underanticoagulation in patients treated with warfarin. Pharmacotherapy. 2004;24:1311–6. doi: 10.1592/phco.24.14.1311.43144. [DOI] [PubMed] [Google Scholar]
- 22.Lenz TL, Lenz NJ, Faulkner MA. Potential interactions between exercise and drug therapy. Sports Med. 2004;34:293–306. doi: 10.2165/00007256-200434050-00002. [DOI] [PubMed] [Google Scholar]
- 23.Shibata Y, Hashimoto H, Kurata C, Ohno R, Kazui T, Takinami M. Influence of physical activity on warfarin therapy. Thromb Haemost. 1998;80:203–4. [PubMed] [Google Scholar]
- 24.Booth SLSJ, Weihrauch JL, Ferland G. Vitamin K (phylloquinone) content of foods. J Food Consump Anla. 1993;6:109–20. [Google Scholar]
- 25.Allabi AC, Gala JL, Horsmans Y, et al. Functional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among black Africans. 2004:113–8. doi: 10.1016/j.clpt.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 26*.Blaisdell J, Jorge-Nebert LF, Coulter S, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–37. doi: 10.1097/01.fpc.0000114759.08559.51. Identification of CYP2C9*11 allele. [DOI] [PubMed] [Google Scholar]
- 27*.Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. Identification of CYP2C9*3 allele. [DOI] [PubMed] [Google Scholar]
- 28*.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–72. Exact test for assessing HWE assumptions. [PubMed] [Google Scholar]
- 29.U.S., Census, Bureau. State and County QuickFacts. 2000 [Google Scholar]
- 30.Limdi NA, Beasley TM, Allison DB, Rivers CA, Acton RT. Racial differences in the prevalence of Factor V Leiden mutation among patients on chronic warfarin therapy. Blood Cells Mol Dis. 2006;37:100–6. doi: 10.1016/j.bcmd.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Dickmann LJ, Rettie AE, Kneller MB, et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60:382–7. doi: 10.1124/mol.60.2.382. Identification of CYP2C9*5 allele. [DOI] [PubMed] [Google Scholar]
- 32.Redman AR, Dickmann LJ, Kidd RS, Goldstein JA, Ritchie DM, Hon YY. CYP2C9 genetic polymorphisms and warfarin. Clin Appl Thromb Hemost. 2004;10:149–54. doi: 10.1177/107602960401000205. [DOI] [PubMed] [Google Scholar]
- 33*.Kealey C, Chen Z, Christie J, et al. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2007;8:217–25. doi: 10.2217/14622416.8.3.217. Influence of CYP2C9*2 and *3 alleles on warfarin dose in African Americans. [DOI] [PubMed] [Google Scholar]
- 34.Xie HG, Kim RB, Wood AJ, Stein CM. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol. 2001;41:815–50. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 35.Kuffner T, Whitworth W, Jairam M, McNicholl J. HLA class II and TNF genes in African Americans from the Southeastern United States: regional differences in allele frequencies. Hum Immunol. 2003;64:639–47. doi: 10.1016/s0198-8859(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 36.Reed TE. Caucasian genes in American Negroes. Science. 1969;165:762–8. doi: 10.1126/science.165.3895.762. [DOI] [PubMed] [Google Scholar]
- 37.Reitnauer PJ, Go RC, Acton RT, et al. Evidence for genetic admixture as a determinant in the occurrence of insulin-dependent diabetes mellitus in U.S. blacks. Diabetes. 1982;31:532–7. doi: 10.2337/diab.31.6.532. [DOI] [PubMed] [Google Scholar]
- 38.Tai G, Farin F, Rieder MJ, et al. In-vitro and in-vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenet Genomics. 2005;15:475–81. doi: 10.1097/01.fpc.0000162005.80857.98. [DOI] [PubMed] [Google Scholar]
- 39.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 40.Shintani M, Ieiri I, Inoue K, et al. Genetic polymorphisms and functional characterization of the 5′-flanking region of the human CYP2C9 gene: in vitro and in vivo studies. Clin Pharmacol Ther. 2001;70:175–82. doi: 10.1067/mcp.2001.117367. [DOI] [PubMed] [Google Scholar]
- 41.Imai J, Ieiri I, Mamiya K, et al. Polymorphism of the cytochrome P450 (CYP) 2C9 gene in Japanese epileptic patients: genetic analysis of the CYP2C9 locus. Pharmacogenetics. 2000;10:85–9. doi: 10.1097/00008571-200002000-00011. [DOI] [PubMed] [Google Scholar]
- 42.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–8. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 43.Seifert A, Tatzel S, Schmid RD, Pleiss J. Multiple molecular dynamics simulations of human p450 monooxygenase CYP2C9: the molecular basis of substrate binding and regioselectivity toward warfarin. Proteins. 2006;64:147–55. doi: 10.1002/prot.20951. [DOI] [PubMed] [Google Scholar]
- 44*.Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. Influence of multiple genes on warfarin dose. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadelius M, Sorlin K, Wallerman O, et al. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 2004;4:40–8. doi: 10.1038/sj.tpj.6500220. [DOI] [PubMed] [Google Scholar]
- 46.Kimmel SE, Christie J, Kealey C, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2007 doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- 47.Bodin L, Verstuyft C, Tregouet DA, et al. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood. 2005;106:135–40. doi: 10.1182/blood-2005-01-0341. [DOI] [PubMed] [Google Scholar]
- 48.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–33. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 49*.Schelleman H, Chen Z, Kealey C, et al. Warfarin Response and Vitamin K Epoxide Reductase Complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007 doi: 10.1038/sj.clpt.6100144. Influence of VKORC1 on warfarin dose in African Americans. [DOI] [PubMed] [Google Scholar]
- 50.Thom T, Haase N, Rosamond W, et al. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:e85–151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI] [PubMed] [Google Scholar]


