Abstract
Objective
Activating Transcription Factor 6 (ATF6) is a sensor of the endoplasmic reticulum stress response and regulates expression of several key lipogenic genes. We utilized a two-stage design to investigate whether ATF6 polymorphisms are associated with lipids in subjects at increased risk for cardiovascular disease (CVD).
Methods and Results
In stage 1, 13 tag-SNPs were tested for association in Dutch samples ascertained for Familial Combined Hyperlipidemia (FCHL) or increased risk for CVD (CVR). In stage 2, we further investigated the SNP with the strongest association from stage 1, a Methionine/Valine substitution at amino-acid 67, in Finnish FCHL families and in subjects with CVR from METSIM, a Finnish population-based cohort. The combined analysis of both stages reached region-wide significance (P=9×10−4), but this association was not seen in the entire METSIM cohort. Our functional analysis demonstrated that Valine at position 67 augments ATF6 protein and its targets Grp78 and Grp94 as well as increases luciferase expression through Grp78 promoter.
Conclusions
A common nonsynonymous variant in ATF6 increases ATF6 protein levels and is associated with cholesterol levels in subjects at increased risk for CVD, but this association was not seen in a population-based cohort. Further replication is needed to confirm this variant's role in lipids.
Keywords: Activating Transcription Factor 6, cardiovascular risk, cholesterol, association, lipids
High serum total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C) and apolipoprotein B (apoB) are major risk factors for cardiovascular disease (CVD). Cholesterol levels are tightly regulated through a feedback pathway from the endoplasmic reticulum (ER) to the nucleus.1 Sterol regulatory element binding protein 2 (SREBP2), an ER resident transcription-factor, has a central role in cellular cholesterol homeostasis.2 In response to cholesterol demand the active domain of SREBP2 is released to the cytoplasm and migrates to the nucleus where it binds sterol response element (SRE) sequences in the promoter of target genes, such as the LDL receptor gene.2
ATF6 is also an ER membrane-bound transcription-factor and a major regulator of the unfolded protein response (UPR).3 The UPR is an evolutionary conserved stress-signalling pathway from the ER to the nucleus. Various stressful conditions, e.g. nutrient deprivation, oxidative stress, high and low glucose concentrations, viral infections, obesity, and increased synthesis of secretory proteins, lead to accumulation of unfolded proteins and trigger the UPR.4,5 Besides a role for ATF6 in the UPR, lipogeneic properties have been ascribed to ATF6, and crosstalk with the SREBP2 pathway has been demonstrated.6 There is growing evidence that ATF6 mediated attenuation of SREBP2 fine-tunes the cholesterol biosynthetic pathway in cells under conditions of ER stress.6 More specifically, Zeng et al. demonstrated in vitro that changes in glucose levels influences lipogenesis via ATF6-mediated inhibition of SREBP2.6 Additional parallel pathways through which ATF6 could modulate cholesterol homeostasis may exist, suggested by the presence of ATF6-binding elements in the promoter of the apoB gene.7
We investigated whether genetic variation in the ATF6 gene is associated with plasma TC, LDL-C, and apoB levels, and whether it contributes to the complex genetic background of CVD. We employed a two-stage design. In stage 1, we performed genotyping of tag-SNPs in the ATF6 gene region to test for association in Dutch samples ascertained for Familial Combined Hyperlipidemia (FCHL) or increased risk for CVD (CVR). An amino-acid substitution (methionine[67]valine) with the strongest evidence of association was further investigated in stage 2 study samples. We also functionally demonstrated that this variant augments ATF6 protein levels and its downstream targets.
Methods
For complete description of the Methods, please see the online supplementary material available at http://atvb.ahajournals.org.
Study Participants
The study design was approved by the ethics committees of the participating centres and all subjects gave written informed consent.
Stage 1 study samples consisted of Sample 1 (Dutch CVR) with a total of 393 unrelated subjects at increased risk for CVD, i.e. age 40–70 years and either hypertension (HT), or body mass index (BMI) >25 kg/m2 from the Cohort study of Diabetes and Atherosclerosis Maastricht8, and Sample 2 (Dutch FCHL) with a total of 195 unrelated probands and spouses from families with FCHL9.
Stage 2 study samples consisted of Sample 3 (Finnish FCHL) with 715 individuals from 61 Finnish FCHL families9, and Sample 4 (Finnish CVR ) with 1,371 subjects with CVR selected from 5,112 male subjects in the on-going Finnish population-based cohort, METSIM (METabolic Syndrome In Men)9 using the same ascertainment criteria as in Sample 1. All of these study samples are described in detail in the Supplementary Methods.
Statistical Analyses
Association analyses with continuous traits were performed using linear regression for the genotypic model. The genotypic test is a two degrees of freedom test of an additive (β̂ add) and a dominance-deviation (β̂ dev) effect. TThe β̂ dev coefficient reflects a deviation from an additive effect. A recessive character is suggested when the sign of β̂ dev is opposite of β̂ add, and a dominant character is suggested when the signs of both coefficients are in the same direction. A full recessive or dominant model is observed when the magnitude of the effects is equal |β̂dev| = |β̂ add|. To avoid violating the assumptions of the test statistics, we estimated empirical p-values in the combined analysis of Sample 1 and 2 by combining the χ2 statistics from 20,000 random permutations of each sample.9 Association analysis in families (Sample 3) was performed with the genotypic model of the family-based association test (FBAT) software.10 The combined analysis of stage 1 and 2 and the meta-analysis of stage 1, Sample 3 and the METSIM cohort were performed using Fisher’s method11 for combining P-value, as described in the Supplementary Methods.
Cell studies
Primary preadipocytes were isolated from subcutaneous adipose tissue from 11 subjects who underwent fat-biopsies. Hela cells were used for transfection experiments with the following plasmids: Cytomegalovirus-galactosidase; Flag-ATF6-(1–373[67]-Methionine); Flag-ATF6-(1–373[67]-Valine); Flag-ATF6-(1–373[67]-Leucine); Grp78 promotor(−284-+221)-Luciferase, and pGL2p. Reporter assays included luciferase and β-galactosidase assays. Western blot experiments were done using specific antisera against ATF6, FLAG, KDEL (Grp78 and Grp94), beta-actin, and GAPDH.
Results
ATF6 protein levels correlate with plasma lipid levels
We investigated the potential role of the ATF6 gene in hyperlipidemia by first evaluating the relation between basal ATF6 levels in cultured human primary pre-adipocytes and plasma TC, LDL-C, and apoB levels. We observed a significant positive correlation between ATF6 levels in vitro and plasma TC, LDL-C and apoB levels of the corresponding subjects (r=0.65, P=0.032; r=0.72, P=0.018 and r=0.76, P=0.006, respectively) (Supplemental Figure 1).
Stage 1 association analysis
We utilized a two-stage design to investigate whether variants within the ATF6 gene are associated with lipid levels in subjects at increased risk to develop CVD. In stage 1, tag-SNPs selected to capture the common genetic variation in ATF6 were investigated in two independent Dutch samples comprising of 393 individuals with increased cardiovascular risk (CVR) (Sample 1), and 195 unrelated FCHL probands and their spouses (Sample 2). In stage 2, the strongest signal was further investigated in two Finnish studies: 715 subjects from 61 FCHL families (Sample 3) and in 1,371 subjects with CVR (Sample 4) from the METSIM cohort. Finally, a combined analysis of the two stages was performed to reach a region-wide significance. Clinical characteristics of the study samples are shown in Supplementary Table 1.
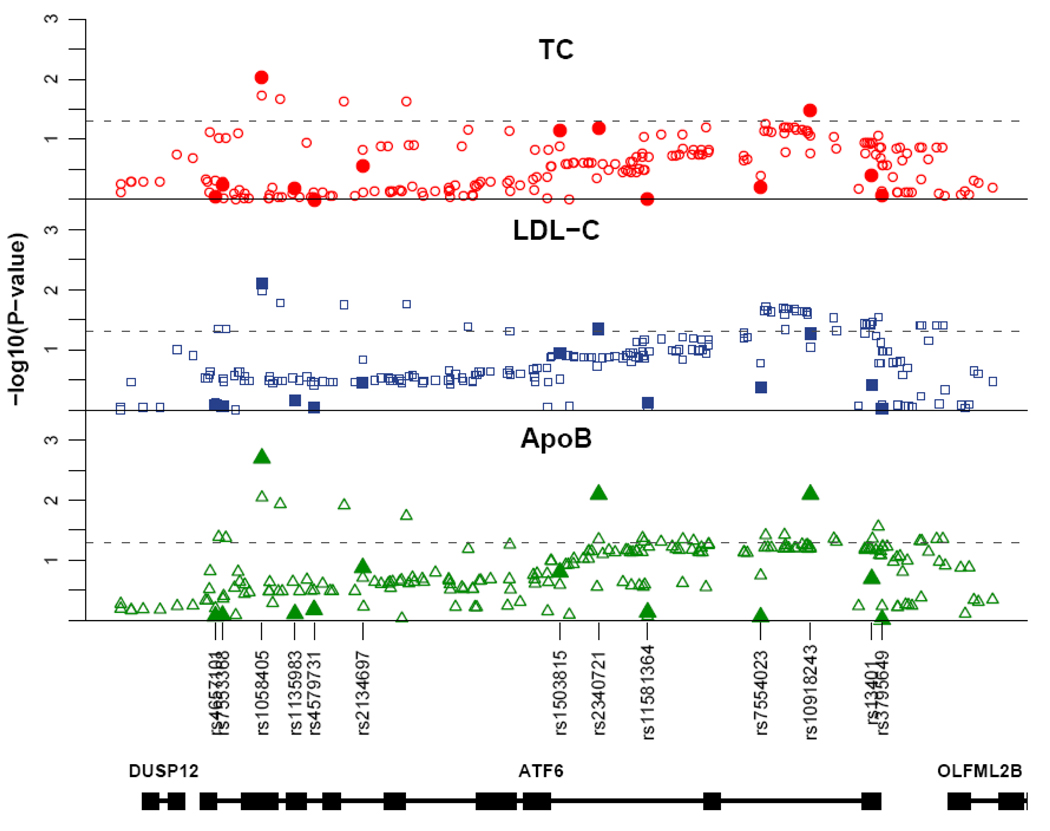
In stage 1, we tested a total of 13 SNPs for association with TC, LDL-C and apoB levels using multivariate linear regression for the genotypic model. The most significant association was observed for SNP3 (rs1058405) with TC (P=0.009, β̂ dev=−(0.2–0.3) and β̂ add=0.2–0.3), LDL-C (P=0.008, β̂ dev=−(0.2– 0.3) and β̂ add=0.2) and apoB (P=0.002, β̂ dev=−0.3 and β̂ add=0.3) (Table 1 and Figure 1). The distributions of TC, LDL-C and apoB between the genotype groups suggest a recessive effect for SNP3 which is also demonstrated by the equal magnitude and opposite sign of the additive and dominance-deviation effects. SNP8 and SNP11 were associated with P-values <0.05 (Supplemental Table 2 and Figure 1) but only SNP3 resulted in P-values <0.05 for all traits. We confirmed that the direction of the association is the same in both Sample 1 and 2 for all SNPs with P-value <0.05, as the χ2 statistics could go in either direction. Furthermore, it should be noted that SNP3 is in high LD with SNP8 (r2=0.7) and in low LD with SNP11 (r2=0.2).
Table 1.
Association of Met[67]Val substitution (SNP3) in Stage I.
| Trait | Met[67]Val | Sample1 Dutch CVR | Sample2 Dutch FCHL | Stage I | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N=384 | R2† | N=181 | R2† | P‡ | ||||||
| β̂ *add(SE) | β̂ *dev (SE) | β̂ *add (SE) | β̂ *dev (SE) | |||||||
| Val/Val | 5.55±0.15 | 6.72±0.29 | ||||||||
| TC | Met/Val | 5.20±0.07 | 0.19(0.9) | −0.21(0.12) | 1.20% | 5.90±0.17 | 0.28(0.11) | −0.33(0.17)§ | 3.64% | 0.009 |
| (mmol/L) | Met/Met | 5.22±0.07 | 5.96±0.13 | |||||||
| Val/Val | 3.72±0.14 | 4.43±0.26 | ||||||||
| LDL-C | Met/Val | 3.28±0.06 | 0.23(0.9) | −0.30(0.12)§ | 2.01% | 3.94±0.15 | 0.21(0.11) | −0.20(0.17)∥ | 2.03% | 0.008 |
| (mmol/L) | Met/Met | 3.34±0.06 | 3.90±0.12 | |||||||
| Val/Val | 1.24±0.04 | 1.31±0.07 | ||||||||
| ApoB | Met/Val | 1.10±0.02 | 0.25(0.9) | −0.33(0.12)§ | 2.45% | 1.11±0.04 | 0.31(0.12) | −0.32(0.17) | 4.16% | 0.002 |
| (g/L) | Met/Met | 1.12±0.02 | 1.11±0.03 | |||||||
Trait values represent the marginal mean evaluated at the average age and sex ±SEM.
β̂ add, indicates the standardized beta coefficients per each copy of the rare allele (additive term) and β̂ dev for the dominance-deviation term.
R2 indicates the proportion of variance explained by the genotypic model.
The p-values represent the results of the combined analysis of Sample 1 and 2, as described in Methods.
P < 0.05 for the significance of deviation from an additive model (^β dev ≠ 0).
P > 0.1 for the significance of deviation from an additive model.
Figure 1.
Association results of stage 1. Filled symbols represent the –log10 of the P-values obtained from linear regression test for the genotypic model. The open symbols represent the –log10 of the P-values obtained from Bayesian regression test with SNP imputation. The dashed lines indicates P=0.05. The location of the SNPs is shown in relation to the gene structure of ATF6 and its adjacent genes.
Next, we used an imputation-based regression method to extend our association analysis to non-tagged SNPs in the ATF6 region (Supplementary Methods). Furthermore, as we test for association with all common SNPs (MAF > 5%) in the region using a Bayesian-regression approach, we can statistically assess which SNP (tagged or non-tagged) most likely affects the traits. We obtained the strongest evidence of association for SNP3 (i.e. the largest estimated Bayes Factor (BF)) for all the traits (Figure 1). The BF we observed for apoB (BF=1.03) can be considered as strong evidence of association, and the ones for LDL-C (BF=0.77) and TC (BF=0.6) as substantial evidence of association. Taken together, these data suggest that in Stage 1, SNP3 is the best regional-candidate for affecting plasma lipid levels.
SNP3 is a coding variant that translates into the Methionine[67]Valine amino-acid substitution (Supplemental Figure 2B). According to the dbSNP-database SNP3 is possibly a tri-allelic variant (A-G-T →Met-Val-Leu). However, the leucine variant is most likely a sequencing error as it was originally identified in a sample size of 4 individuals with validation status unknown. Furthermore, we did not observe the leucine variant in 200 subjects of Sample 1 that we specifically screened using an allele-specific primer for it.
Stage 1 and 2 association analysis
SNP3 that provided a P-value <0.05 for all traits in the stage 1 analysis was tested in a combined analysis of stage 1 and 2. We analyzed the stage 2 samples using the same genotypic model as in stage 1 and performed a combined analysis of the two stages using Fisher’s method11 for combining P-values weighted by the sample size. This analysis included a total of 2,674 subjects at increased risk to develop CVD. We observed a significant association between SNP3 and TC (P=9×10−4), LDL-C (P=0.007) and apoB (P=0.005) levels (Table 2). Similar to the results obtained in the stage 1 samples, higher lipid levels were also observed with the Val allele in the stage 2 samples (Table 2). In the FCHL families (Sample 3), both the lipid genotypic mean values and the Z-scores for the homozygote rare group (all ZV/V >1.7) suggest a recessive effect for SNP3, similar to Samples 1 and 2 (Table 2). However, an additive effect could not be rejected in the CVR sample selected from the METSIM cohort (Sample 4), as none of the β̂ dev terms where significantly different from zero (Table 2). The association between SNP3 and TC levels is region-wide significant, as it surpasses the Bonferroni correction for 13 SNPs tested in stage 1 with 3 traits, as well as 6 additional tests performed in stage 2 (Adjusted P-value 0.0009×45=0.04). However, it should be noted that as we tested three correlated traits (average correlation=0.85) the Bonferroni correction for multiple testing, that presumes that tests are independent, is conservative for our analyses.
Table 2.
Association of Met[67]Val substitution (SNP3) in Stage I and II.
| Trait | Met[67]Val | Sample 3 Finnish FCHL | Sample 4 Finnish CVR | Stage I+II | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N=715 | Z*V/V | Z* M/V | R2† | N=1,371 | R2† | P§ | ||||
| β̂ ‡add(SE) | β̂ ‡dev(SE) | |||||||||
| Val/Val | 6.47±0.23 | 5.61±0.09 | ||||||||
| TC | Met/Val | 5.96±0.11 | 1.78 | −1.68 | 2.37% | 5.56±0.04 | 0.09(0.05) | 0.05(0.06)∥ | 0.54% | 0.0009 |
| (mmol/L) | Met/Met | 6.15±0.12 | 5.43±0.04 | |||||||
| Val/Val | 4.22±0.20 | 3.60±0.08 | ||||||||
| LDL-C | Met/Val | 3.82±0.10 | 2.07 | −1.71 | 2.34% | 3.57±0.04 | 0.07(0.05) | 0.04(0.06)∥ | 0.32% | 0.0072 |
| (mmol/L) | Met/Met | 3.99±0.10 | 3.48±0.03 | |||||||
| Val/Val | 1.32±0.07 | 1.10±0.03 | ||||||||
| ApoB | Met/Val | 1.11±0.03 | 2.32 | −1.32 | 3.03% | 1.12±0.01 | 0.03(0.05) | 0.08(0.07)∥ | 0.28% | 0.0049 |
| (g/L) | Met/Met | 1.13±0.03 | 1.09±0.01 | |||||||
Trait values represent the marginal mean evaluated at the average age and sex ±SEM.
ZV/V, indicates the Z value obtained from the FBAT genotypic model test for the Val/Val genotype and ZM/V for the Met/Val genotype.
R2 indicates the proportion of variance explained by the genotypic model.
β̂ add, indicates the standardized beta coefficients per each copy of the rare allele (additive term) and β̂ dev for the dominance-deviation term.
The p-values represent the results of the combined analysis of stage 1 and 2, as described in Methods.
P > 0.1 for the significance of deviation from an additive model.
Meta-analysis of SNP3 in a population-based cohort
As we obtained a region-wide significant association between SNP3 and plasma TC levels, we also included all available subjects of the METSIM cohort (i.e CVR and non-CVR) in a meta-analysis of all samples weighted by the proportion of the sample size. We did not observe significant associations for TC in meta-analysis of all subjects (n=5,812, χ2 =4.42, P=0.62). However, as we excluded subjects with T2DM from our study (Stage 1 and 2) we also performed the meta-analysis while excluding subjects with T2DM or family history of T2DM from METSIM. The subjects with family history of T2DM also exhibit impaired glucose homeostasis as shown in Supplemental Table 3. We observed a significant association between SNP3 and TC levels (n=3,471, χ2 =18.09, P=0.006), suggesting that the effect of SNP3 may be different between individuals with and without T2DM and history of T2DM.
Furthermore, the effect of SNP3 was more pronounced in the stage 1 and 2 analysis that included subjects from METSIM with CVR (Sample 4) (Table 2). To further investigate this relationship between SNP3, T2DM and CVR, we examined the effect of their multiplicative interaction on serum TC levels (Supplementary Methods). We observed that plasma TC levels are significantly affected by the interaction of SNP3 and T2DM or family history of T2DM (standardized β̂ gr=−0.12 (SE=0.05), P=0.01), whereas an opposite direction of interaction was observed between SNP3 and CVR on serum TC levels (standardized β̂gr=0.10 (SE=0.05), P=0.03). These data suggest opposite effects for the Val variant on TC levels between individuals with T2DM and CVR, with a decreasing effect in subjects with T2DM or family history of T2DM and an increasing effect in subjects with CVR.
Functional characterization of the methionine[67]valine variant
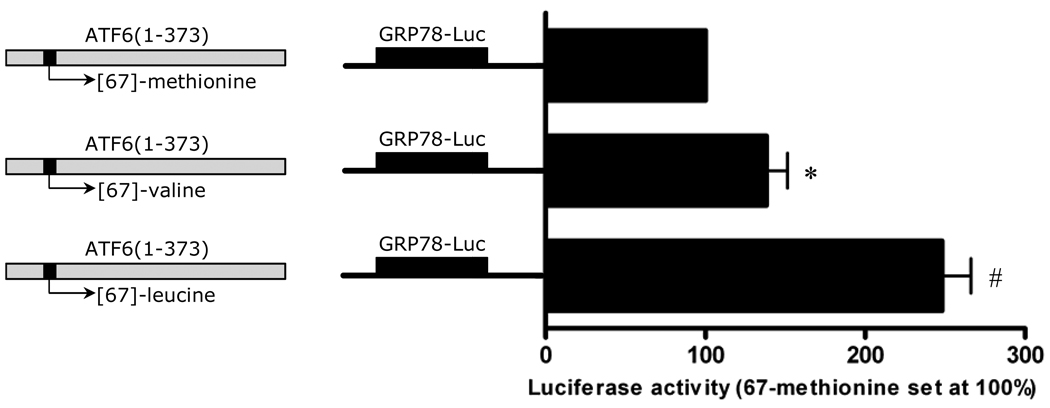
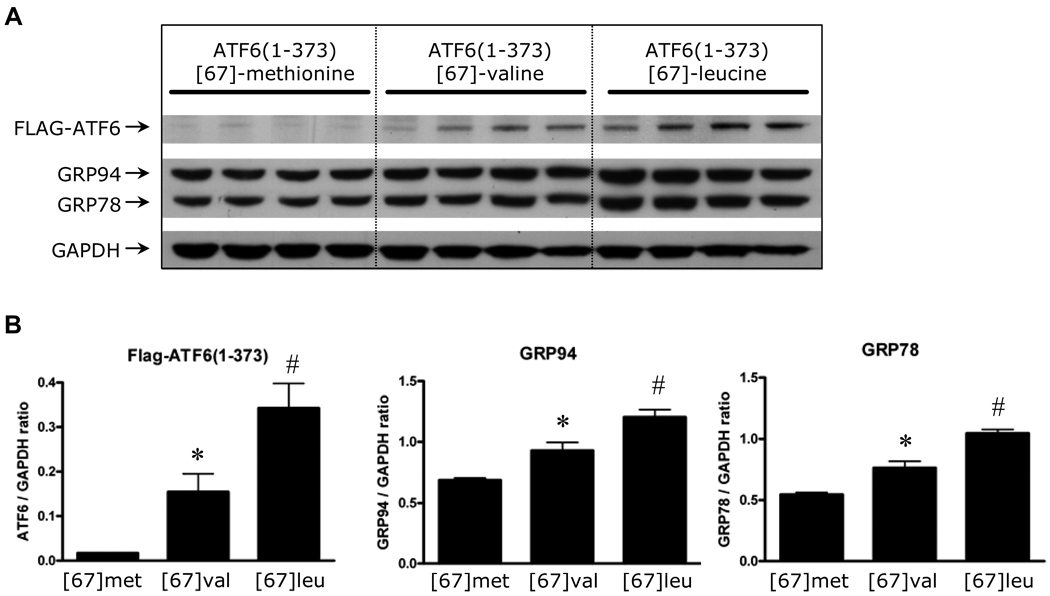
The Met[67]Val substitution is located in the transcription activation domain (TAD) of ATF6, more specifically in the VN8-like region (amino-acid 61–68) which is critical for transcription activity and degradation of ATF6 (Supplemental Figure 2).12 We observed that this amino-acid substitution was computationally predicted to be damaging to the protein (PSIC score=2.08) based on phylogenetic and structural information (PolyPhen server tool).13 To explore the possibility that the Met[67]Val polymorphism alters the protein function, we assessed the effect of amino-acid substitution at position 67 on the protein levels and transcriptional activity of ATF6 in-vitro. Although the Leucine-variant is probably a sequencing error, we also tested it functionally. Accordingly, we prepared three constructs of FLAG-tagged ATF6(1–373), that mimic active ATF6 generated by ER-stress. These three constructs only varied in amino-acid 67. We tested the three constructs in HeLa cells by assaying for luciferase expression through the GRP78 promoter. The constructs, FLAG-ATF6(1–373)-[67]-valine and FLAG-ATF6(1–373)-[67]-leucine had significantly higher luciferase activity than the Met allele construct (1.4 and 2.5 fold change, respectively) (Figure 2). We also compared the protein levels of endogenous Grp78 and Grp94, which are direct targets of ATF6, in cells transfected with FLAG-ATF6 constructs. In agreement with the results from the luciferase assay, the levels of Grp78 and Grp94 were significantly higher in cells transfected with FLAG-ATF6(1–373)-[67]-valine and FLAG-ATF6(1–373)-[67]-leucine, than those transfected with the Met allele construct (Figure 3A and 3B). Furthermore, FLAG-ATF6 levels were also increased, suggesting a gain-of-function substitution. Taken together, these functional data demonstrate that this variant augments ATF6 protein levels and its downstream transcriptional targets.
Figure 2.
Potency of FLAG-ATF6(1–373)-[67]-methionine, FLAG-ATF6(1–373)-[67]-valine, and FLAG-ATF6(1–373)-[67]-leucine to induce luciferase expression in Hela cells through the GRP78 promoter. Hela cells were transfected with one of the FLAG-ATF6(1–373) plasmids (1µg). Cells were cotransfected with either pGL2p, or GRP78-luciferase reporter plasmid (10µg), and CMV-β-gal (1µg). Each value represents the mean and SEM of 4 independent transfections from triplicate cultures. * p<0.05, # p<0.001
Figure 3.
Potency of FLAG-ATF6(1–373)-[67]-methionine, FLAG-ATF6(1–373)-[67]-valine, and FLAG-ATF6(1–373)-[67]-leucine to induce endogenous GRP94 and GRP78 levels. Panel A: Hela cells, transfected with one of the FLAG-ATF6(1–373) plasmids (1µg) were extracted and analyzed for expression of FLAG-ATF6, Grp78, Grp94 and GAPDH by western blot using specific antiserum antibodies for FLAG, KDEL (Grp78 and Grp94), and GAPDH, respectively. Panel B: quantification of the blots by densitometry analysis. Each value represents the mean and SEM of 4 independent transfections). * p<0.05, # p<0.01
Discussion
Our two-stage association analysis of tag-SNPs in the ATF6 gene revealed a region-wide significant association between the Met[67]Val amino-acid substitution and increased TC levels in samples ascertained for FCHL and risk factors for CVD. Furthermore, we also functionally demonstrate that the Met[67]Val substitution augments ATF6 levels and its downstream transcriptional targets. These findings are also supported by a strong positive correlation between the ATF6 protein levels in cultured pre-adipocytes and plasma lipids levels in the corresponding individuals. Obviously human liver would have been the most preferable tissue for these types of protein experiments. However, liver biopsy is an invasive procedure that can cause potentially life-threatening complications. Therefore, we utilized adipose biopsies, also relevant for studies of lipid metabolism.
The Met[67]Val substitution in ATF6 (rs1058405) is a common variation. Its MAF ranges from 7% in African American to 23% and 29% in Asian and European populations, respectively. It is an excellent functional candidate as it resides within the VN8-like domain of ATF6 (Supplemental Figure 2). The VN8 domain was identified in the virion protein 16 (VP16) transcription factor from herpes simplex virus type I, and is required for transcriptional activation and rapid proteasomal degradation of VP16.14,15 More recently, Thuerauf et al. identified a VN8-like domain in ATF6, which is 75% identical to the VN8 domain in VP16.12 Analogous to VP16, the ATF6-VN8 region was found to be critical for transcriptional activity and subsequent degradation of ATF6. Mutating amino-acid 62 and 64 in the ATF6-VN8 region caused a 5-fold reduction in transcriptional activity.12 Our transfection experiments with mutated ATF6 constructs revealed that the valine variant at position 67 increases ATF6 transcriptional activity in cultured cells. Furthermore, ATF6 levels were increased with the Val allele when compared to the Met allele construct. According to the dbSNP-database rs1058405 is possibly a tri-allelic variant (Met-Val-Leu). The leucine variant could be a sequencing error as it was originally identified in a sample size of 4 individuals with validation status unknown, and we did not detect it in 200 subjects that we specifically screened for the leucine variant. Nevertheless, mutating amino acid 67 to leucine further increased ATF6 levels, as well as its targets, Grp78 and Grp94, illustrating the importance of amino-acid 67 for ATF6 transcription activity.
Zeng and colleagues recently elucidated a potential molecular mechanism through which ATF6 can modulate cholesterol homeostasis.6 In response to glucose starvation, activated ATF6 can interact with SREBP2 and bind as a heterodimer to SREs in the promoters of cholesterogenic genes, e.g. the LDL receptor. Bound to SRE, the ATF6-SREBP2 heterodimer recruits HDAC1, and together they exert a major inhibitory effect on SREBP2 transcription activity.6 It is tempting to propose an additional inhibitory effect on SREBP2 activity due to the Met[67]Val gain-of-function-polymorphism, which would further reduce the number of LDL receptors that bind and internalize LDL and VLDL particles, consequently increasing plasma cholesterol levels. Recently, it has also been demonstrated that prolonged high glucose conditions induce ER stress and ATF6 activity.5 These findings both suggest that glucose levels may present a confounding factor in the genetic analysis of ATF6. We observed an interaction between the valine variant and T2DM on plasma TC levels, suggesting that impaired glucose homeostasis may confound the associations with ATF6. However, the potential confounding role of glucose homeostasis and the exact mechanism through which the Met[67]Val substitution in ATF6 affects cholesterol metabolism remains to be addressed in future studies.
Recently, the GWAS data of predominantly population-based cohorts were analyzed for lipid traits.16 SNP3 was not significantly associated with LDL-C in this meta-analysis using an additive model (P>0.05). However, as the types of genes identified in a GWAS naturally depend and reflect the characteristics of the study samples, it is likely that other variants and genes will be identified in studies based on CVR. In our analyses of all samples and the METSIM cohort, the association signal originated specifically from subjects at increased cardiovascular risk without T2DM or family history of T2DM. Notably, although our combined P-value of both stages indicates a significant overall association, the association signal and the proportion of variance explained by SNP3 were less significant in Sample 4, selected from the population based METSIM cohort. Furthermore, a recessive mode of inheritance was suggested throughout the samples except for Sample 4, as evident by the sign and the significance of the dominant-deviation term. These differences may reflect the fact that population-based cohorts are not optimal for detecting modest effects, particularly those that are largely recessive. As CVD risk factors are highly prevalent in the population due to modern lifestyle, sampling cases from a population-based cohort hampers the possibilities to collect cases based on the true genetic background. This environmental “noise” may have resulted in a limited power to detect the effect of SNP3 in the METSIM cohort. Therefore, the genetic model and the significance of SNP3 on cholesterol levels need to be addressed in future replication studies using large samples ascertained for CVR.
The effect of SNP3 suggest that this SNP alone is not a major determinant of cholesterol levels, but rather a modifier variant that influences cholesterol levels. However, the combined effect of SNP3 and additional risk factors is likely to be more pronounced, and thus, more easily detectable in study samples that are enriched for CVD risk factors. This may explain the consistent association observed in the CVR and FCHL samples and the lack of association signal in the entire unascertained METSIM cohort or recent GWAS for lipids.16 As exemplified by our current study and given the complex architecture of common diseases, such as CVD, it may be more important to select replication samples using the specific attributes of the initial study than the size of the replication sample in general.
In conclusion, our data suggest that the Met[67]Val substitution in the ATF6-gene is associated with cholesterol levels in subjects with increased risk to develop CVD. We also demonstrate that the Met[67]Val substitution is a gain-of-function polymorphism that increases ATF6 transcriptional activity.
Supplementary Material
Acknowledgments
We thank the patients and family members for their participation in this study. We also thank E. Nikkola and M. Lupsakko for laboratory technical assistance.
Sources of Funding
This research was supported by an EFSD/Novo Nordisk research grant to CS, by the NIH grants HL-28481 and HL082762 to PP, by the NIH grants HL-075573, NS-25037 and HL-085577 awarded to CCG. M-RT is supported by the Clinical Research Institute HUCH Ltd and Finnish Heart Foundation, and DW-V by the NHGRI grant T32 HG02536. SM was supported by a travel grant from the Netherlands Organization for Scientific Research (NWO) to visit the Glembotski laboratory.
Footnotes
Disclosures
T.dB. has been employed by GlaxoSmithKline. This work was not supported in full or part by GlaxoSmithKline. S.M, M.vG, T.A, R.V, C.vdK and T.dB are inventors on patent WO2006125513: “Genetic association of polymorphisms in the ATF6-alpha gene with insulin resistance phenotypes”.
References
- 1.Sakai J, Rawson RB. The sterol regulatory element-binding protein pathway: control of lipid homeostasis through regulated intracellular transport. Curr Opin Lipidol. 2001;12:261–266. doi: 10.1097/00041433-200106000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 4.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Seo HY, Kim YD, Lee KM, Min AK, Kim MK, Kim HS, Won KC, Park JY, Lee KU, Choi HS, Park KG, Lee IK. Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases insulin gene expression via up-regulation of orphan nuclear receptor small heterodimer partner. Endocrinology. 2008;149:3832–3841. doi: 10.1210/en.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY. ATF6 modulates SREBP2-mediated lipogenesis. Embo J. 2004;23:950–958. doi: 10.1038/sj.emboj.7600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 8.Kruijshoop M, Feskens EJ, Blaak EE, de Bruin TW. Validation of capillary glucose measurements to detect glucose intolerance or type 2 diabetes mellitus in the general population. Clin Chim Acta. 2004;341:33–40. doi: 10.1016/j.cccn.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Lee JC, Weissglas-Volkov D, Kyttälä M, Dastani Z, Cantor RM, Sobel EM, Plaisier CL, Engert JC, van Greevenbroek MM, Kane JP, Malloy MJ, Pullinger CR, Huertas-Vazquez A, Aguilar-Salinas CA, Tusie-Luna T, de Bruin TW, Aouizerat BE, van der Kallen CC, Croce CM, Aqeilan RI, Marcil M, Viikari JS, Lehtimäki T, Raitakari OT, Kuusisto J, Laakso M, Taskinen MR, Genestq J, Pajukanta P. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am J Hum Genet. 2008;83:180–192. doi: 10.1016/j.ajhg.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Fisher RA. Statistical Methods for Research Workers. 4th Ed. Oliver & Boyd: Edinburgh; 1932. [Google Scholar]
- 12.Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–20739. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- 13.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci U S A. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molinari E, Gilman M, Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 1999;18:6439–3447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.