Abstract
Endogenous retroviruses have shaped the evolution of mammalian genomes. Host genes that control the effects of retrovirus insertions are therefore of great interest. The Modifier-of-vibrator-1 locus controls level of correctly processed mRNA from genes mutated by endogenous retrovirus insertions into introns, including the pitpnvb tremor mutation and the Eya1BOR model of human branchiootorenal syndrome. Positional complementation cloning identifies Mvb1 as the nuclear export factor Nxf1, providing an unexpected link between mRNA export receptor and pre-mRNA processing. Population structure of the suppressing allele in wild M. m. castaneus suggests selective advantage. A congenic Mvb1CAST allele is a useful tool for modifying gene expression from existing mutations and could be used to manipulate engineered mutations containing retroviral elements.
Mus musculus is a complex species group with subpopulations that have diverged and hybridized since becoming commensal with humans some 10,000 years ago. Allopatric divergence and re-hybridization of mouse lineages are thought to contribute to the diversity and activity of retroviral elements in the current mouse genomes 1,2. Host genes that can influence the expression of newly introduced (or newly mobilized) viral elements might be selected for variants that blunt these effects. Our results suggest that Mvb1 is such a locus.
Mvb1 was originally identified as a strain-derived locus that modifies the neurological mutant, vibrator 3. The vibrator (vb) mutation is a hypomorphic allele of the phosphatidylinositol transfer protein α (PITPα) gene (pitpn) caused by the insertion of an endogenous retrovirus (intracisternal A particle, or IAP) into the fourth intron of the gene, resulting in 5 to 10–fold loss of PITPα expression. Homozygous vibrator mice show severe action tremor, progressive degeneration of interneurons in the brain stem and spinal cord, and uniform juvenile lethality. However, vibrator mice carrying Mvb1 alleles from the wild-derived CAST/Ei inbred strain show reduced tremor severity and survive to adulthood. In principle, this could be due to a change in physiological requirement for PITPα function or to a change in the steady-state expression level of PITPα derived from the mutant allele.
Modifier genes that act on retroviral insertions might be especially useful, as such insertions comprise approximately 15% of spontaneous mutations in laboratory mice 2,4. Here we examine the mechanism of Mvb1-mediated suppression by demonstrating its effect on vibrator RNA expression, by examining its effects on other retrovirus-associated mutations (selected as examples of different retrovirus families and different classes of insertion sites irrespective of family or class frequency), and by positional identification the Mvb1 gene. We show that suppressing Mvb1 alleles elevate steady-state level of correctly processed pitpn mRNA derived from vibrator mutant alleles, creating an in vivo titration of this gene product. We show that Mvb1 also modifies Eya1BOR, a model of human branchiootorenal dystrophy caused by insertion of an IAP element (the class most frequently associated with spontaneous mutations in mice) inserted into introns in the sense orientation 5. We identify Mvb1 by a positional complementation strategy as an allele of the mRNA nuclear export factor Nxf1. Nxf1 protein is known to bind and mediate export of constitutive transport elements (CTEs) in the unspliced genomes of several retroviruses, including rodent IAPs 6, and other retroelements, including human LINEs 7. Interestingly, we find a strong bias against CTE-containing insertions in introns and against sense orientation for elements that are found in introns in the public draft mouse genome. This suggests an orientation-sensitive selective pressure, which could also promote genetic variants that suppress the effects of such insertions, particularly in zones of hybridization among wild mouse populations. Consistent with this selection bias, we show that the IAP-suppressing CAST/Ei allele is the major allele in wild Mus musculus castaneus mice in Southeast Asia and a frequent minor allele in the related Japanese subspecies M. m. molossinus, thought to have arisen by hybridization between M. m. castaneus and M. m. musculus approximately 2000–3000 years ago 8.
RESULTS
Mvb1 is a dosage-sensitive modifier of vibrator RNA accumulation
Mvb1 modifies the severity of vibrator tremor and the associated juvenile lethality 3. To examine this effect in more detail, we monitored the longevity of vb mutant mice congenic on a C57BL/6J (B6) strain background with zero, one or two CAST/Ei alleles of Mvb1 (Figure 1a). Consistent with our behavioral observations, the longevity data indicate a semi-dominant mode of action and high penetrance.
Figure 1. Mvb1 is a semi-dominant modifier of vibrator mRNA level.

a, Dosage-sensitive impact of Mvb1 alleles on lifespan of vb homozygotes. Product-limit analysis of censored survival data over two years is shown for each Mvb1 genotype: B6/B6 in blue, B6/CAST green, and CAST/CAST red. Some animals from the two longer-lived cohorts were sacrificed after 2–24 months; initial and terminal cohort sizes are indicated, censored animals are indicated by a hash mark at the approximate time of removal. b, Suppression of vibrator neuropathology. Sections from red nucleus stained with luxol fast blue and cresyl violet show diminished lipophilic staining of internal membrane structures and eccentric nuclei in a large fraction of cells from vb/vb; Mvb1B6/Mvb1B6 animals (arrows). This pathology is notably absent in suppressed animals. c, Mvb1 alleles control level of RNA derived from vb mutant alleles of pitpn. vb/vb; Mvb1B6/Mvb1B6 tissues express ~18% and vb/vb; Mvb1CAST/Mvb1CAST tissues ~36% of normal pitpn poly(A)+ RNA levels after normalization to Gapd, or Plcb3 or Adrbk1, two genes flanking the Mvb1 interval on chromosome 19. H, heterozygote. d, Aggregate data from several blots similar to C show quantitatively consistent effects of vb and Mvb1 genotypes. For comparisons between blots, all bands were normalized to the average of non-mutant control samples within a blot. e, An aberrant RNA containing the 5′ exons of pitpn is present in total RNA extracts from vibrator, but not littermate controls. f, Chemiluminescent detection illustrates Mvb1 control of PITPα protein level in vb mutant tissue. g, Ratio of PITP proteins from triplicate samples on a single blot detected by 125I-labeled protein A and quantified by phosphorimage analysis.
To extend these observations to the cellular level, we examined histology of vibrator animals homozygous for either the B6 or CAST allele of Mvb1 (Figure 1b). Mutant animals on the B6 background show a high frequency of neuropathology in characteristic nuclei of the hindbrain and spinal cord. Vacuolated cells and cells with reduced staining of internal membrane structures are found in red nucleus, deep cerebellar nuclei, lateral vestibular nucleus, within the reticular formation of the pons, and in the spinal cord. By contrast, mutants homozygous for the CAST allele of Mvb1 show essentially no vacuolated neurons and fewer (and less extreme) examples of neurons with other pathology.
To ask whether Mvb1 acts by altering RNA levels from the mutant allele rather than through bypassing the requirement for this RNA, we examined RNA levels from vibrator mutant and wild-type littermates homozygous for either B6 or CAST alleles (Figure 1c–e). Mvb1 CAST homozygotes accumulate approximately twice as much correctly processed pitpn RNA from the vb allele as their B6 littermates. In contrast, we see no significant difference between Mvb1 genotypes in RNA levels from wild-type alleles of pitpn (data not shown). Similar results were obtained using RNA from liver, demonstrating that Mvb1 activity is not restricted to brain. In addition to increasing the level of normal pitpn RNA derived from vibrator mutant alleles, CAST alleles of Mvb1 appear to decrease the level of a low-abundance, mutant-specific RNA detected by exons 5′ to the IAP insertion site (Figure 1e). Western blots using antibodies specific for either PITPα or PITPβ demonstrate that the changes in RNA level correlate to protein levels (Figure 1f, g).
Mvb1 modifies Eya1BOR, a model of branchio-oto-renal syndrome
In principle, Mvb1 alleles could act either as a locus-specific modifier of pitpn (for example in a feedback regulatory pathway) or as a more general modifier of a step in RNA biogenesis blocked by the IAP insertion. To test this idea, we crossed the suppressing (CAST) allele of Mvb1 to several mutations that involve endogenous retrovirus insertions. The pattern of Mvb1 interactions with these mutations should help to define the mechanism by which Mvb1 acts because each mutation represents a distinct class of retroviral element, mechanism of interference with host gene function, or affected tissues (Table 1).
Table 1.
Mutations tested for suppression by Mvb1CAST.
| Gene | allele | insertion element | site | orientation | phenotypes | Mechanism | Suppression by Mvb1 | source | ref. |
|---|---|---|---|---|---|---|---|---|---|
| Pitpn | vb | IAP | intron | sense | tremor, lethality | decreased expression | yes | lab colony | 1 |
| Eya1 | BOR | IAP | intron | sense | circling, deafness | decreased expression | yes | K. Johnson | 2 |
| Agouti | iy | IAP | intron | antisense | coat color; obesity | ectopic expression | no | Jax | 3 |
| Axin | Fu | IAP | intron | antisense | tailkink | dominant negative | no | Jax | 4 |
| Dab1 | scm | (IAP) | intron | antisense | ataxia | splicing into parental IAP | no | Jax | 5 |
| Hairless | hr | MuLV | intron | sense | complete hair loss | decreased expression | no | Jax | 6 |
| Myo5a | d | MuLV | intron | sense | coat color | abnormal RNA expression | no | DBA/2J | 7 |
| Agouti | a | VL30 | intron | coat color | decreased expression (isoform specific) | no | C57BL/6J | 8 |
References
Hamilton et al. (1997) Neuron 18: 711–722
Johnson et al. (1999) Human Mol Genetics 8: 645–653
Duhl et al. (1994) Nature Genetics 8: 59–65
Vasicek et al. (1997) Genetics 147: 777–786
Ware et al. (1997) Neuron 19: 239–249; Sheldon et al. (1997) Nature 389: 730–733
Cachon-Gonzalez et al. (1994) Proc Natl Acad Sci USA 91: 7717–7721
Mercer et al. (1991) Nature 349: 709–713; Seperack et al. (1995) EMBO J. 14: 2326–2332
Bultman et al. (1994) Genes Dev. 8: 481–490
Eya1BOR is a recessive model of human branchiootorenal syndrome. An IAP insertion into an intron reduces the level of host gene expression ~50% 5. Homozygous animals have a pronounced cochlear malformation and can be recognized behaviorally by characteristic head bobbing, circling, and failure to startle in response to loud noise. In some genetic backgrounds, frequent kidney agenesis and dysgenesis are associated with perinatal and juvenile death.
Mvb1CAST suppresses several Eya1BOR phenotypes. Mutant animals from a digenic cross (F2 from C3H– Eya1BOR × B6. CAST–Mvb1CAST) were assayed for phenotypes dependent on development of the inner ear (Figure 2). Behaviorally, Eya1BOR/Eya1BOR ; Mvb1CAST/Mvb1CAST animals displayed noticeably less head bobbing and circling than Eya1BOR animals with B6 or C3H alleles at Mvb1. More quantitatively, mutant animals were assessed for auditory brainstem responses to a calibrated sound played in one ear. Typical response data are shown in Figure 2a. The difference in average threshold response between Mvb1B6/Mvb1B6 and Mvb1CAST/Mvb1CAST Eya1-mutant animals is highly significant (ANOVA, p<0.001) and the data are consistent with a semi-dominant effect (Figure 2b). Histology was subsequently performed on the inner ear and rated on a scale of 1 to 5 for quality of the cochlear structure by an investigator blind to genotype. Nonmutant and heterozygous mice had normal cochleas irrespective of Mvb1 genotype (Figure 2c). A significant shift in phenotype between Mvb1 genotypes is evident for Eya1BOR mutants and the data suggest a semi-dominant modifying effect. Eya1BOR mice without the Mvb1CAST allele (Figure 2d) had only the basal turn and had a large reduction in the number of spiral ganglion cells (10/12 animals). In contrast, Eya1BOR animals homozygous for Mvb1CAST varied from normal cochlear structure (3/12), through normal structure with a variable loss of spiral ganglion cells in the apical turn (6/12; Figure 2e), to being little different than the non-modified cochlea (3/12). A histogram of scores by genotype is shown in Figure 2f.
Figure 2. Mvb1 modifies Eya1BOR phenotypes and RNA level.
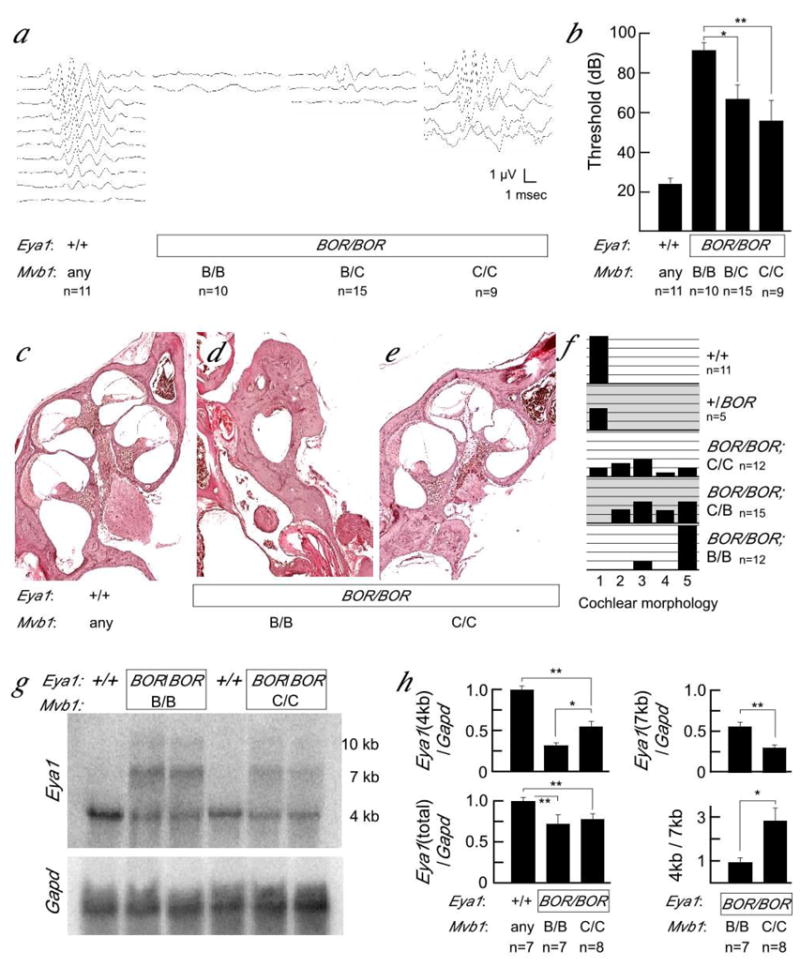
a, Typical auditory brainstem responses to clicks presented at progressively decreasing amplitudes. For each animal, top line represents response to 100dB, followed by 85, 70, 60, 55, 50, 45, 40, 35, and 30 dB or until wave form of the response is lost. b, Distribution of ABR data for each genotype is plotted as decibel level at which response loses waveform. Animals were scored by an experienced investigator blinded to genotype. Group differences between Mvb1B6/Mvb1B6 mutants and the other two genotypes were significant to 0.05 (*) or 0.01 (**) by Student’s t test. c–e Cochlear malformations in Eya1BOR homozygotes are less severe in the presence of Mvb1CAST alleles. Typical histology at the level of the 8th cranial nerve is shown for (C) nonmutant, (D) Eya1BOR/Eya1BOR; Mvb1B6/Mvb1B6, and (E) Eya1BOR/Eya1BOR; Mvb1CAST/Mvb1CAST. f, Histogram of cochlear morphology scores. Each animal was assigned a score from 1 to 5 by an experienced investigator blinded to genotype. 1 = normal cochlea, 3 = nearly normal cochlear turns, but abnormal number of ganglion cells and 5 = cochlea with only the basal turn. Animals shown in c-e were scored 1, 5, and 3, respectively. g, Representative northern blot for quantifying expression of mutant and normal RNAs shows increased level of correctly processed 4 kb message and reduced level of the mutant-specific 7 and 10 kb messages in Eya1BOR animals homozygous for Mvb1CAST compared to Mvb1B6. h, Normalized values from phosphorimage analysis in replicate experiments show significant RNA differences among indicated genotypes using 2-sided t-tests. Level of the wild-type 4 kb RNA is less in mutants than littermate controls (p ≤ 0.001, **) and less in mutants with the B6 allele than those with the CAST allele at Mvb1 (p ≤ 0.015, *). Level of mutant-specific 7 kb RNA is lower in mutants carrying the CAST allele of Mvb1. The total amount of all Eya1 messages is less in mutants than controls, but not significant between Mvb1 genotypes among mutant animals. Differences in the ratio of correct 4 kb product to aberrant 7 kb product significant to 0.011.
Mvb1 also modifies Eya1BOR mortality, caused by kidney dysgenesis/agenesis on the C3H genetic background 5,9. Among 93 F2 Eya1BOR/Eya1BOR animals from a second cross (C3H–Eya1BOR × CAST/Ei), we find a significant absence of C3H homozygotes at Mvb1 (14/93; p>0.01, chi-square test), and a slight excess of CAST homozygotes (35/93) relative to Mvb1 heterozygotes (44/93). By contrast, we see no significant mortality or kidney agenesis in the cross (B6C3–Eya1BOR/+ × B6. CAST–Mvb1) that we used to test suppression of inner ear phenotypes. Taken together, these observations demonstrate activity of Mvb1 on a second IAP-associated mutation, presence of this activity in kidney and developing inner ear, and the existence of additional Eya1 modifiers in B6 and C3H.
To test whether suppression of Eya1BOR by Mvb1CAST also acts through elevation of RNA levels we examined RNA isoform levels on Northern blots (Figure 2g–h). We used adult skeletal muscle as an accessible source of Eya1 RNA from a tissue that remains grossly normal in the mutant. We see both correctly spliced product and two mutant-specific bands of higher molecular weight in samples from mutant animals. Probes corresponding to exons 5′ or 3′ to the insertion site both detect these aberrant bands, suggesting interstitial retention of some inserted sequences. Sequencing RT-PCR products across exons flanking the insertion site confirms the retention of IAP-specific sequences. Importantly, quantification of bands by phosphorimage analysis reveals that suppressing alleles of Mvb1 affect a quantitative shift in RNA isoforms from high molecular weight mutant-specific bands into the correctly processed wild-type product. Together with our results on the pitpnvb mutations, this demonstrates that Mvb1 acts on mutations in distinct pathways in a broad range of tissues and suggests that the mechanism of suppression involves at least one step in RNA processing.
Mvb1 is selective for class of retrovirus and mutagenic mechanism
To further define the mechanism of Mvb1, we tested its effect on other retrovirus-associated mutations, chosen to probe different mechanisms of mutation, irrespective of their frequencies among spontaneous mouse mutations. This group includes mutations with similar structures to vb and Eya1BOR but caused by different classes of insertion element and IAP insertions that act through other mechanisms (Table 1).
Myo5ad 10 and hairless 11–13 are murine leukemia virus (MLV) insertions into introns that inhibit expression of the inserted host gene. Both insertions are in the same transcriptional orientation as the host gene (sense orientation). Mvb1 alleles did not affect the coat color of Myo5ad mutants nor the timing or completeness of hair loss in hairless mutants. Similarly, the nonagouti (a) allele of Agouti is an inactivating VL30 retrotransposon insertion present in the B6 strain 14. Mvb1 alleles do not effect follicle and shaft coloration of hairs taken from either dorsal or ventral locations.
The intermediate yellow allele of Agouti (Aiy) is an antisense-orientation IAP insertion 5′ to the first coding exon. Ectopic expression of Agouti from the LTR promoter of this insertion results in hypo-pigmentation and obesity 15. Mvb1 does not affect the extent or timing of pigmentation or weight gain in Aiy mutant animals. Agouti phenotypes are sensitive to expression level and pattern, so absence of phenotypic difference here strongly suggests that Mvb1 does not alter transcription from the LTR promoter.
Mvb1 shows orientation selectivity for IAP elements. The Fused mutation (AxinFu) is an IAP element insertion in the antisense orientation into the eighth intron of Axin 16, a gene required for axial patterning 17. A proportion of AxinFu transcripts retain a part of the IAP element, introducing a stop codon. This fusion RNA encodes the RGS signaling domain of Axin but lacks the DIX regulatory domain 18,19. AxinFu produces dominant tail kinks with somewhat variable penetrance on several genetic backgrounds we examined. However, we observe no systematic shift in severity or frequency of tail kinks between Mvb1B6 and Mvb1CAST homozygotes carrying one copy of AxinFu.
The Dab1scm mutation causes inappropriate retention of a non-mutagenic, antisense orientation IAP element in an intron, which is spliced out from the premutation normal allele of the parental strain 20,21. No difference in the severity of gait ataxia or frequency of falls was seen between mutant animals homozygous for either allele of Mvb1.
Taken together, these results suggest that Mvb1 acts on sense orientation insertions including at least some class D elements such as IAPs, but not class C insertions such as MLVs, and that the activity might require presence of the retrovirus in the nascent RNA. Interestingly, class C and class D viruses differ in how they balance production of spliced mRNA with export of unspliced genome RNA, a feature relevant to the RNA-level suppression of both pitpnvb and Eya1BOR.
Mvb1 maps to a 210 kb interval containing 11 genes and a recombination hotspot
To identify the Mvb1 gene, we mapped it by recombination in ~7791 informative meioses from an F1 intercross (Figure 3). Mice heterozygous for both vb and Mvb1 alleles were bred and the resulting offspring were genotyped with SSLP or SNP markers flanking the locus. Mice carrying Mvb1-recombinant chromosomes were either scored for vb tremor severity or bred an additional generation to produce homozygous vb/vb mice for scoring. Additional SSLP and SNP markers generated in the course sequencing the interval allowed us to refine the interval to 210 kb. Despite the large number of meioses, we were unable to narrow the region any further by recombination due to a hot spot at the proximal end that accounted for most of the recombination events observed in ~500 kb interval around Mvb1.
Figure 3. Recombination and physical map of the Mvb1 interval.
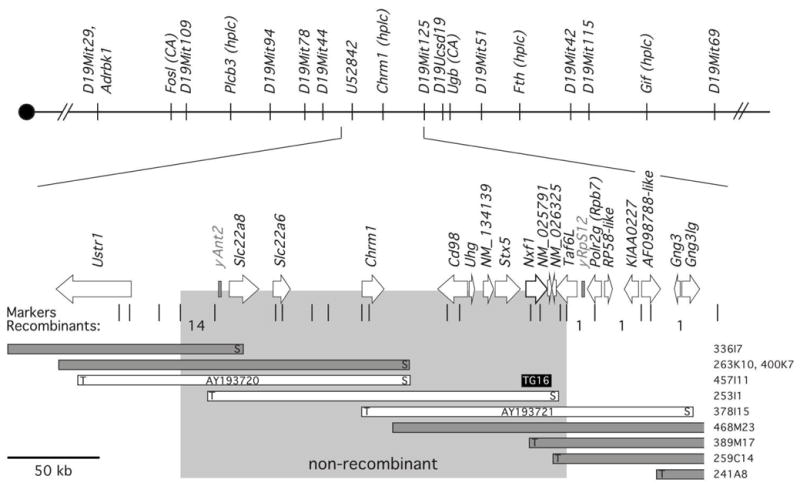
Recombination markers used to position the Mvb1interval in the cross vb/+; Mvb1B6/Mvb1CAST × vb/+; Mvb1B6/Mvb1CAST are indicated. Exclusion mapping resulted in a 210 kb non-recombinant interval. BACs spanning the interval were sequenced to identify positional candidate genes, drawn approximately to scale. Polymorphic microsatellites derived from the genome sequence and SNPs derived from comparative cDNA resequencing used in fine mapping are indicated below the genes, as well as the number of recombinants identified in each interval.
To identify candidate genes in this interval, we sequenced two BACs that span the interval and identified genes using BLAST homology searches and conservation with the draft human genome; these predictions are now replicated in the public mouse genome sequence 4. To identify polymorphic candidate genes, we compared brain RNA expression levels and cDNA sequences from B6 and B6. CAST congenic mice. None of the genes shows dramatic expression differences or changes in RNA size on Northern blots, but the neutral amino acid transporter gene Slc3a2 (also called Cd98, Mdu1, or 4F2), the mRNA export factor Nxf1 (Tap) and the chromatin remodeling complex component, Taf6l (Paf65a) show nonsynonymous substitutions in coding sequence or codon deletions (Taf6l) that suggest them as positional candidate genes. Interestingly, Nxf1 protein is known to mediate nuclear export of unspliced, CTE-containing RNAs 22; in principle this could explain both class-specific and orientation-specific effects of Mvb1, making Nxf1 a strong biological as well as positional candidate gene.
Nxf1 transgenes complement Mvb1
To test whether Nxf1 can complement Mvb1 in vivo, we injected a 16.4 kb genomic fragment containing Nxf1 (B6 allele) into wild-type mouse pronuclei and bred the resulting transgenic mice to our vibrator; Mvb1CAST congenic line (Figure 4a). We assayed transgene expression and complementation in vb/vb; Mvb1CAST/Mvb1CAST; TG/+ mice and non-transgenic controls. Transgenic mice from line 1 express the transgene at levels comparable to the endogenous locus, as shown by RT-PCR sequence assays of polymorphic sites (Figure 4b).
Figure 4. Transgenic complementation of Mvb1.

a, The genomic transgene contains the full length Nxf1 cDNA and flanking sequences as well as the first exon of a predicted gene with EST support. b, Transgenic Nxf1 is expressed at levels comparable to the endogenous copy. RT-PCR product across polymorphic sites was used for DNA sequencing. Peak height at variant sites demonstrates allele-specific expression. Genotypes were established by flanking markers outside the transgenic interval. c, Increased tremor severity in mice carrying the permissive allele transgene in comparison to littermate controls. Mice were scored blinded to genotype, n=11 both groups. The difference is significant, p<0.01 by 1-tailed t-test. A similar sample from mice heterozygous at Mvb1 is not independently significant due to limited sample size, but the combined p-value for both groups is 0.0018. d, Reduced pitpn RNA expression in mice carrying the permissive allele transgene in comparison to littermate controls. e, Quantification of bands indicates that the effect of the transgene is quantitatively similar to heterozygosity at the endogenous locus. For each genotype, the two bars indicate separate measures of the two normal mRNA bands. The number of independent samples is indicated. t-test p-values are significant for effect of the transgene on each band in the Mvb1 C/C background (0.02 and 0.0026) and for both bands combined in the Mvb1 B/C background (0.026), for a combined p-value of 1.4 × 10−6. f, Northern blot from primary cultures of vb/vb; Mvb1B/Mvb1B cells from a single animal untreated (−) or infected with pLentiV (Invitrogen) carrying EGFP, Nxf1 B6 allele, or Nxf1 CAST allele, hybridized to full-length pitpn or Gapd. g, Normalized pitpn/Gapd ratios indicate specific elevation of correctly processed pitpn mRNA in Nxf1 CAST virus infected cultures compared to either control infection or uninfected cells.
Matched pairs of transgenic and non-transgenic mice were assayed for tremor severity and pitpn RNA expression level. Because we placed the permissive B6 allele transgene into the background of the suppressing CAST allele, transgenic mice should exhibit more severe tremor and diminished pitpn RNA level (despite potential over-expression of Nxf1). Phenotypic assessment shows that Nxf1 modifies vibrator: tremor and movement phenotype was assessed by three independent investigators blind to genotype, using a numerical scale of severity based on the distinctive behaviors of vb mutants with different endogenous Mvb1 alleles (Figure 4c). Tremor was scored several times during the life of the animal. Transgenic mice were consistently scored more severely affected than controls by each investigator. The Nxf1 transgene also affects pitpn RNA levels from vb mutant alleles (Figure 4d, e). Northern blot analysis of brain RNA was used to measure pitpn expression from several matched pairs of transgenic and control animals. Transgenic mice averaged 15% less pitpn RNA than the non-transgenic mice after normalization to Gapd and was reduced in 9/9 paired comparisons (p = 0.002). This is roughly comparable to pitpn level expressed by Mvb1 heterozygotes (Figure 1c, d).
To ask whether expression of the CAST allele of Nxf1 can also increase the level of vibrator-derived pitpn RNA in a B6 background, we infected primary skin fibroblast cultures from a single B6-vb/vb mouse with recombinant lentiviral vectors expressing EGFP or Nxf1B6 (control infections) or Nxf1CAST and compared with an uninfected control (Figure 4f, g). Cells infected with virus express approximately 60% more pitpn than the three controls, while cells infected with control viruses express pitpn RNA at levels comparable to uninfected culture.
Nxf1CAST is a natural allele and polymorphic in wild populations
The C57BL6/J and CAST/Ei alleles of Nxf1 differ by two amino acid polymorphisms, S48P and E610G. Genetic variations that distinguish inbred laboratory strains may reflect either natural polymorphisms present in the wild population or new polymorphisms that have been fixed in a lineage after cultivation and under altered selective pressures. To test whether Nxf1CAST represents a natural variation we resequenced both of these polymorphisms and selected other sites from 49 Mus musculus castaneus and 45 M. m. molossinus mice trapped across southeast Asia and Japan as well as several inbred strains (Figure 5a and Table 2). The two amino acid polymorphisms are in strong linkage disequilibrium with each other and with noncoding polymorphisms in this population. Strikingly, the suppressing CAST allele (P48, G610) defines the major haplotype in castaneus, but is a minor allele in molossinus mice. Among inbred strains, the suppressing haplotype is present only in castaneus derivatives.
Figure 5. Nxf1 alleles in inbred strains and wild mice.

a, Geographic distribution of Nxf1 haplotypes by site of collection. Alleles are indicated for four SNPs around exon 2, including the S48P position (underlined) and three SNPs around exon 21, including E610G (underlined). Not shown are 14 additional polymorphic sites in between that increase the diversity of E610 haplotypes, but not the diversity G610 haplotypes (see Table 3). b, Model of the Nxf1-UBA domain from Grant et al24,25, PDB accession 1aoi. Acidic residues are in red, basic in blue and polar in copper. The structure is oriented to place the FG-binding face facing out of the page and a bound FXFG peptide is shown in green. Arrow indicates the position of mouse E610 site. c, The E610G alteration changes a surface charge adjacent to a reported nuclear pore interaction surface. d, Alignment of Nxf1 amino acid sequences around the two variant sites including three mammals, three non-mammal vertebrates, a urochordate and an insect. Low-complexity sequence around S48P (bold type; position relative to the mouse sequence) is less well conserved and comprises less conservative replacements, including insertions (triangles, inserted residues not shown). Sequence around E610G retains most of its structure in that tolerated changes are similar residues. Tree structure indicates only topology among and not distance between species. Nxf1 amino acid sequences around the murine S48P site have low sequence complexity and poor conservation. Alignment of Nxf1 amino acid sequences around the murine E610G site shows stronger conservation. Note E610K in non-mammal vertebrates and fruit flies is accompanied by K608E (circle), conserving an adjacent charge pair in the structure.
The castaneus allele of Nxf1 may have arisen under selective pressure related to retroviral insertions. While one cannot actually prove selection from a retrospective analysis of a single region, several features of the data are suggestive. S48 sits in a poorly conserved region of Nxf1 that is not essential to Nxf1 biochemical function 23. In contrast, E610 is highly conserved among vertebrates and sits in a UBA domain thought to mediate nuclear export through physical interaction with the nuclear pore and exhibits a significant chemical shift upon binding FG peptide 24,25 (Figure 5b–d). The two amino acid polymorphisms (and their haplotypes) are not in Hardy-Weinberg equilibrium across any non-fixed population we examined, consistent with either ongoing natural selection, extensive selectively neutral population admixture, or both. More persuasively, 610G is present on only one 19-marker haplotype across the Nxf1 gene and is the only allelic variant in haplotype to show this pattern. This indicates that 610G is a young allele that has become the majority allele only comparatively recently, a hallmark of positive selection 26.
We further reasoned that if Nxf1 alleles arose under selective pressure due to retroviral insertions, then retained provirus sites in existing genomes should show a pattern consistent with selection against sense strand insertions into introns. We identified 179 sites containing strong matches to a well-characterized IAP-derived CTE 6 in the draft C57BL/6 genome4 and looked for evidence of nearby genes. 18 of the CTE sites occurred within introns of known or predicted genes with experimental evidence (spliced ESTs in at least one organism or conservation of multiple exons with the Fugu genome). Surprisingly, the orientation of these sites with respect to host genes was highly biased, with 15/18 arranged in the antisense orientation (p-value 0.005, 2-tailed t-test). This provides further evidence that sense-oriented retroviral insertions exert a selective pressure, to which the castaneus allele of Nxf1 may be a response.
DISCUSSION
Here we have demonstrated the genetic mechanism, properties, and identity of a novel suppressor of retrovirus insertional mutations. Retrovirus insertions can alter host gene function by altering transcriptional regulation, by interrupting coding sequence, or by reducing the processing efficiency of nascent RNA. Suppressing alleles of Mvb1 derived from CAST/Ei appear to act specifically on hypomorphic mutations caused by insertions into introns that result in reduced expression of a normal mRNA. Mvb1 does not act on IAP insertions known to affect transcriptional initiation. However, we have identified aberrant RNAs in mutant tissues that are reduced in favor of normally processed mRNAs by the presence of Mvb1CAST alleles, strongly implying that Mvb1 plays a role in mRNA processing. This activity is broadly expressed: we observe effects in brain, liver, kidney, muscle, and otic development.
Surprisingly, we find that this altered mRNA processing phenotype results from an allelic alteration at Nxf1, an mRNA export factor homologous to yeast Mex67. Recent work has identified a few key protein factors that link sequential steps in mRNA biogenesis, such as Aly/REF1. Aly forms part of the TREX complex, which links transcription to export in yeast and mammalian cells 27. Aly acts as a transcriptional cofactor 28, and is independently recruited to nascent RNAs with splicing factors 29, and acts in export, in part through interactions with Nxf1 30. Although Nxf1 protein has been shown to interact with other factors in mRNA biogenesis, including Aly, these interactions have been previously interpreted as recruitment of the export machinery to processed mRNA. Our results are the first to show an influence of the canonical export receptor on preceding steps in pre-mRNA processing. This could occur through a kinetic competition among alternate pathways for nascent RNA or through altered RNP configuration on nascent RNAs to which Nxf1 is bound (Figure 6). The location and nature of the E610G polymorphism in the UBA domain suggest a possible alteration in the interaction of Nxf1 with the nuclear pore, which could affect RNP assembly and processing by altering the latency of nascent or partially processed RNA for transport or otherwise moving the transcript away from processing compartments in the nucleus.
Figure 6. Model for Nxf1 modifier activity on retrovirus insertional mutations.
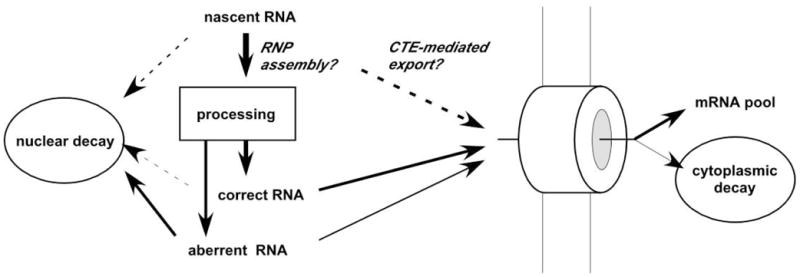
Flow diagram of possible fates for newly synthesized pre-mRNA. Variant alleles of Nxf1 could influence the steady-state ratio of aberrant to correctly processed mRNA by altering a kinetic balance between RNP assembly and CTE-mediated export or by influencing the sites or composition of RNP assembly.
The distribution of Nxf1 alleles in wild mouse populations indicates that this allele has undergone substantial recent fixation among M. m. castaneus mice in southeast Asia, a hallmark of recent selection. Although this could also result from hitchhiking or neutral drift, the fact that this site alters a conserved charge on a functional domain of the protein, yet is the only site for which the major allele occurs on a single extended haplotype argues that this may be a site undergoing adaptive selection. In addition, we show that the class of event on which the CAST Nxf1 allele acts is significantly underrepresented in the C57BL/6 genome (which lacks this compensating mechanism), providing another level of evidence to suggest at least a contextual selective advantage in populations where new retroviral insertion arise with some frequency.
The properties of Mvb1 suggest that it could be used to engineer a binary system for titrating gene expression in vivo. This could be particularly useful for creating mouse models of human disorders where loss of gene expression is a target of therapeutic efforts and dose-responsiveness of functional recovery is unknown (such as Fragile X syndrome and certain cancers) or for titrating the effects of mutations created by insertional elements reported to use an NXF1-binding CTE, such as human L1 LINES 31. Titrating expression from the endogenous promoter by a processing step should allow level-of-expression manipulation independent of timing or cell-type distribution of expression.
METHODS
Mice and Genetic typing
Mutant mice were obtained from the Jackson Laboratory and maintained as stocks locally. The vibrator mutation and the Mvb1 interval from strain CAST/Ei were made simultaneously congenic on C57BL/6J (B6). B6. CAST–Mvb1 mice bred to other mutations were derived from our colony of B6–pitpnvb; Mvb1CAST mice as incipient congenics at N6 or later. B6. CAST–Mvb1 mice bred to transgenics were N15 or later. Genotypes were determined by PCR assays specific for each mutation or for unique microsatellites flanking the mutant locus (Supplementary Table 1)
Molecular biology
RNA was isolated from Trizol (Invitrogen) according to the manufacturer’s protocol. Northern blots were prepared by electrophoresis of formaldehyde or glyoxal treated RNA and capillary transfer to nylon membranes. Cloned probe fragments were verified by DNA sequence, isolated by PCR, and body labeled using random primer DNA synthesis in the presence of 32P-dCTP. Specific binding after hybridization and washing was quantified by phosphorimage analysis on a Molecular Dynamics Storm imager.
Total protein for Western blots was prepared in RIPA buffer, quantified by Bradford assay and subjected to SDS-PAGE.. PITPα and PITPβ were detected with polyclonal antibodies directed against unique carboxyterminal peptides 3 followed by either HRP-coupled secondary antibody and ECL detection (Figure 1E) or 125I-protein A and phosphorimage analysis (Figure 1F).
Recombinant lentiviral genomes were constructed from PCR-amplified fragments cloned into pLenti6/V5-TOPO (Invitrogen). Virions were packaged with VSV-G in the UCSD Gene Therapy Program Vector Development Lab. Primary cells from vibrator mutant mice were infected with viral supernatants and selected for blasticidin resistance prior to harvesting.
Electrophysiology
Hearing threshold was measured using auditory brainstem response (ABR) to click stimuli for the right ear by an investigator blind to Mvb1 genotype. Following anesthesia (ketamine hydrochloride, 50mg/kg, xylazine hydrochloride, 5mg/kg, and acepromazine maleate, 1mg/kg, intraperitonealy), animals were placed in a single-walled acoustic booth (Industrial Acoustics Co, Bronx, NY) on a heating pad. Subdermal electrodes (Astro-Med, Inc. -Grass Instrument Division) were inserted at the vertex (active electrode), the mastoid (reference), and the hind leg (ground). Click stimuli (0.1 msec, 10/sec) were delivered to a Beyer DT 48, 200 Ohm speaker fitted with an ear speculum for placement in the external auditory meatus. The recorded ABR was amplified and digitized by a battery-operated preamplifier and input to an ABR recording system that provides computer control of the stimulus, recording and averaging functions (Tucker Davis Technology, Alachua, FL). Successively decreasing amplitude stimuli were presented in 5 dB steps to the animal and the recorded stimulus-locked activity was averaged (n=512) and displayed. Threshold was defined as the stimulus level between the record with no visibly detectable response and a clearly identifiable response.
Histology
Animals for histological assessment were sacrificed following ABR measures by cardiac perfusion with warm saline and phosphate-buffered, 4% paraformaldehyde. The cochleas were dissected and left in fixative overnight (4° C). They were decalcified in 10% EDTA for 1 week, dehydrated in graded alcohols, and embedded in paraffin. Sections (7 μm) were cut parallel to the modiolus and stained with hematoxylin and eosin.
Transgenic mouse production
An 18 kb Spe I fragment from B6-derived BAC clone 253I1 was subcloned into λBlueStar (Novagen). A 16.4 kb fragment containing 16,406 bp from the Spe I site adjacent to the Stx5 polyadenylation signal through Nxf1 to a Sse8387 I site in the first intron of the non-polymorphic gene encoding cDNA clone AK002249 and approximately 10 bp of pBlueStar cloning site sequence was gel isolated and injected into pronuclei of B6D2 hybrid mice. Pups were screened for presence of the transgene by a competitive three-primer PCR assay using a forward primer corresponding to the cloning site of the vector at the 5′ end of the gene competing with a second forward primer outside the transgenic segment in the endogenous locus and a reverse primer in common to both transgenic and endogenous copies. Neither parental strain nor several derived hybrids has any observed vb-modifying effects in prior reports 3,32 or in non-transgenic littermates tested here.
Table 3.
Nxf1 haplotypes in wild mice.
| Nxf1 Haplotype | Block structure | aa48, 610 | M. m. molossinus | M. m. castaneus | Geographic distribution | Inbred strains |
|---|---|---|---|---|---|---|
| CCGC.ACGTT40ATG3G.GGC | 1.1.1 | P, G | 6 | 69 | Indonesia, Japan, Malaysia, Phillipines, Taiwan | CAST, CASA |
| TCCC.ACGTT40ATG3G.GAC | 3.1.2 | P, E | 1 | Phillipines | ||
| TCCC.ATGTT40ATG3G.GAC | 3.2.2 | P, E | 5 | 6 | Indonesia (Bandar Lanpung), Japan | |
| CCGC.ATGTT44ATG3A.GAC | 1.3.2 | P, E | 11 | Malaysia | ||
| CCGC.ACGTT40ATG3G.GAC | 1.1.2 | P, E | 4 | Japan | ||
| CCGC.ACGTT40ATG3G.AAT | 1.1.3 | P, E | 1 | Indonesia (Bandung ) | ||
| TTCC.ACACC34G0G2A.AAT | 2.4.3 | S, E | 10 | Indonesia (Bandung ) | 129/SvImJ, A/J, AKR/J, BALB/cJ, C3HeB/FeJ, C57BL/6J, DBA/2J, FVB/NJ | |
| TTCT.GCACC34GTA2A.AAT | 4.5.3 | S, E | 66 | Japan | MOLC, MOLD, MOLE, MOLF, MOLG | |
| TTCT.GCACC34GTG2A.AAT | 4.6.3 | S, E | 5 | Japan | ||
| TOTAL haplotypes examined: | 86 | 98 | 15 |
Acknowledgments
We thank Dr. Allen Ryan for advice and assistance with electrophysiology, Drs. X-D. Fu and C. J. Wills for thoughtful discussions and Drs. M. Rosenfeld, R. Kolodner, A. Wynshaw-Boris for helpful comments on draft manuscripts. We thank Dr. Atsushi Miyanohara for assistance with viral packaging, the UCSD Cancer Center Transgenic Mouse Facility for transgenic mouse production and Dr. I. Kalcheva for assistance with BAC sequencing. This work was supported by NIH grants MH59207 (B. A. H.) and DC04268 (E. K.) and the Medical Research Service of the U.S. Department of Veterans Affairs (E. K.). D. G. and J. A. F. were supported in part by a National Research Service Award (2 T32 GM008666-06). B. A. H. is a Pew Scholar in the Biomedical Sciences.
References
- 1.Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 343–436. [PubMed] [Google Scholar]
- 2.Hamilton BA, Frankel WN. Of mice and genome sequence. Cell. 2001;107:13–6. doi: 10.1016/s0092-8674(01)00514-1. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton BA, et al. The vibrator mutation causes neurodegeneration via reduced expression of PITP alpha: positional complementation cloning and extragenic suppression. Neuron. 1997;18:711–22. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 4.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KR, et al. Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum Mol Genet. 1999;8:645–53. doi: 10.1093/hmg/8.4.645. [DOI] [PubMed] [Google Scholar]
- 6.Tabernero C, et al. Identification of an RNA sequence within an intracisternal-A particle element able to replace Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Virol. 1997;71:95–101. doi: 10.1128/jvi.71.1.95-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindtner S, Felber BK, Kjems J. An element in the 3′ untranslated region of human LINE-1 retrotransposon mRNA binds NXF1(TAP) and can function as a nuclear export element. RNA. 2002;8:345–56. doi: 10.1017/s1355838202027759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yonekawa H, et al. Hybrid origin of Japanese mice Mus musculus molossinus: Evidence from restriction analysis of mitochondrial DNA. Mol Biol Evol. 1988;5:63–78. doi: 10.1093/oxfordjournals.molbev.a040476. [DOI] [PubMed] [Google Scholar]
- 9.Xu PX, et al. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nature Genetics. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 10.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–13. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 11.Cachon-Gonzalez MB, et al. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A. 1994;91:7717–21. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter GB, et al. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001;15:2687–701. doi: 10.1101/gad.916701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoye JP, Fenner S, Greenoak GE, Moran C, Coffin JM. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–91. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 14.Bultman SJ, et al. Molecular analysis of reverse mutations from nonagouti (a) to black-and-tan (a(t)) and white-bellied agouti (Aw) reveals alternative forms of agouti transcripts. Genes Dev. 1994;8:481–90. doi: 10.1101/gad.8.4.481. [DOI] [PubMed] [Google Scholar]
- 15.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 16.Vasicek TJ, et al. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–86. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng L, et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–92. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 18.Fagotto F, et al. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol. 1999;145:741–56. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu W, Zeng L, Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–45. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 20.Sheldon M, et al. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–3. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 21.Ware ML, et al. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–49. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 22.Gruter P, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–59. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 23.Bachi A, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–58. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant RP, Hurt E, Neuhaus D, Stewart M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat Struct Biol. 2002;9:247–51. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- 25.Grant RP, Neuhaus D, Stewart M. Structural Basis for the Interaction Between the Tap/NXF1 UBA Domain and FG Nucleoporins at 1 A Resolution. Journal of Molecular Biology. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- 26.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–7. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 27.Strässer K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–8. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 28.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCR-alpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z, et al. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–5. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 30.Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol. 2002;159:579–88. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–60. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 32.Weimar WR, Lane PW, Sidman RL. Vibrator (vb): a spinocerebellar system degeneration with autosomal recessive inheritance in mice. Brain Res. 1982;251:357–64. doi: 10.1016/0006-8993(82)90754-5. [DOI] [PubMed] [Google Scholar]


