Abstract
Tissue macrophages comprise a heterogeneous group of cell types differing in location, surface markers and function1. Red pulp macrophages are a distinct splenic subset involved in removing senescent red blood cells2. Transcription factors such as PU.1 and C/EBPα play general roles in myelomonocytic development3,4, but the transcriptional basis for producing tissue macrophage subsets remains unknown. Here we show that Spi-C, a PU.1 related transcription factor, selectively controls the development of red pulp macrophages. Spi-C is highly expressed in red pulp macrophages, but not monocytes, dendritic cells or other tissue macrophages. Spi-C−/− mice exhibit a cell-autonomous defect in the development of red pulp macrophages that is corrected by retroviral Spi-C expression in bone marrow cells, but have normal monocyte and other macrophage subsets. Red pulp macrophages highly express genes involved in capturing circulating hemoglobin and iron regulation. Spi-C−/− mice show normal trapping of red blood cells in the spleen, but fail to phagocytose these red blood cells efficiently, and develop an iron overload localized selectively to splenic red pulp. Thus, Spi-C controls development of red pulp macrophages required for red blood cell recycling and iron homeostasis.
Spi-C belongs to the Spi-subfamily of Ets transcription factors that includes PU.1 and Spi-B5,6 and was initially reported to be expressed in B cells 5,7. We compared expression of PU.1, Spi-B and Spi-C across a panel of immune cells such as B cells, bone marrow (BM) monocytes, dendritic cells and several types of resident tissue macrophages, including red pulp macrophages (RPM) (Fig. 1a, Supplementary Fig. 1). PU.1 was broadly expressed and Spi-B predominantly restricted to B cells8,9. In contrast, Spi-C was highly expressed in RPM, expressed at lower levels in B cells, and essentially absent in other cells. To test for a role of Spi-C in RPM and B cells, we generated mice lacking Spi-C expression by gene targeting (Supplementary Fig. 2). Male and female Spi-C−/− mice are fertile and healthy, with a normal lifespan, but are born at a somewhat lower than expected Mendelian frequency (Supplementary Fig. 2). We also generated Spi-Cnull/null mice, in which the neomycin cassette was deleted from the targeted Spi-C allele (Supplementary Fig. 2).
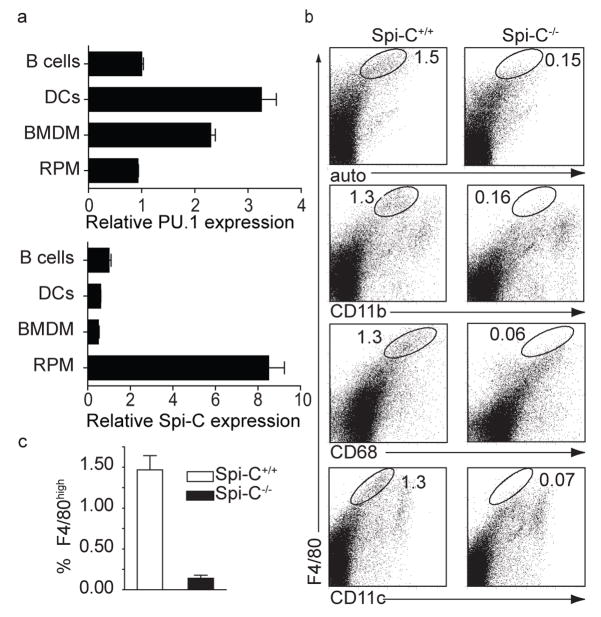
Figure 1. Spi-C−/− mice have a selective loss of red pulp macrophages.
a, PU.1 and Spi-C eexpression was determined by quantitative RT-PCR in purified B cells, dendritic cells (DCs), BM-derived macrophages (BMDM), and red pulp macrophages (RPM). Shown is the normalized mRNA expression relative to expression in B cells. b, Spi-C+/+ and Spi-C−/− spleen cells were stained with antibodies to F4/80, CD11b, CD68, and CD11c and analyzed by flow cytometry. Numbers represent the percentage of cells in the indicated gate. c, Frequency of F4/80hi cells in spleen was determined as a mean (± SD) (n=7) from total splenocytes as shown in b.
RPM have been defined as F4/80hiCD68+CD11blo/− cells with strong autofluorescence10,11. Both Spi-C−/− mice and Spi-Cnull/null mice showed a phenotype characterized by the selective loss of RPM (Fig. 1b and c, Supplementary Fig. 3a), but showed no abnormalities in the development of B cells, conventional dendritic cells, plasmacytoid dendritic cells (Supplementary Fig. 3 and 4, and Supplementary Table 1), or in the development of T cells (Supplementary Fig. 5), and had normal serum immunoglobulin levels (Supplementary Fig. 4f). We confirmed the loss of RPM by immunohistochemical analysis, finding a near total loss of F4/80+ cells in the splenic red pulp (Fig. 2a). By contrast, SIGN-R1-expressing marginal zone macrophages and MOMA-1-expressing metallophilic marginal zone macrophages were present normally in the spleens of Spi-C−/− mice (Fig. 2a). Spi-C−/− mice had normal F4/80+ tissue macrophages in peritoneum and liver, normal macrophage and dendritic cell progenitors (MDP) in BM, and normal monocytes in BM and blood (Supplementary Fig. 6).
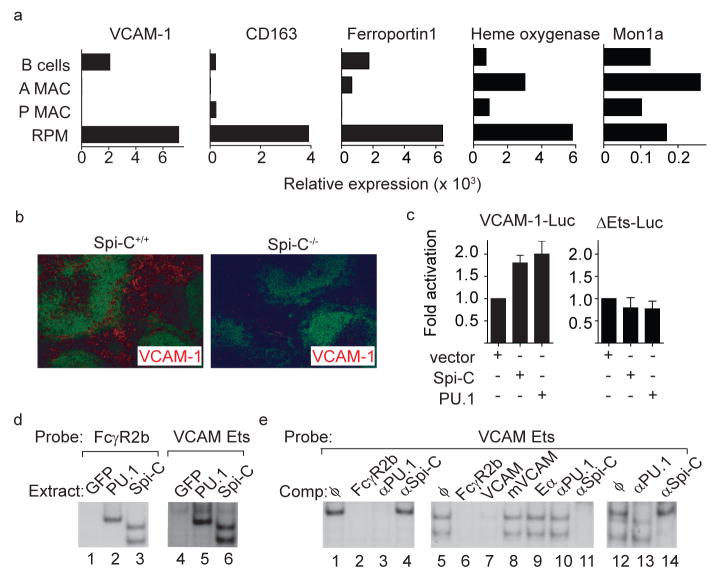
Figure 4. Spi-C regulates VCAM-1 expression.
a, Normalized expression for VCAM-1, CD163, ferroportin I, heme oxygenase, and Mon1a are shown for B cells, alveolar macrophages (A MAC), peritoneal macrophages (P MAC) and red pulp macrophages (RPM) as described in Fig. 1. b, Spi-C+/+ or Spi-C−/− spleen sections were stained for B220 (green) and VCAM-1 (red). c, J774 cells were transfected with the VCAM-1 reporter (VCAM-1-Luc) or the ΔEts-Luc reporter, and with pEF4 (vector), Spi-C- or PU.1-expressing vectors. Cells were analyzed for luciferase activity as described in the Methods. Data is representative of 4 independent experiments (mean ± SD, n=3). d, 293F/T cells were transiently transfected with GFP-RV (GFP), PU1-MIGR1 (PU.1) or Spi-C-RV (Spi-C) and whole cell extracts were analyzed for binding to the FCγR2b or the VCAM Ets probes. e, Extracts from PU.1-expressing cells (lanes 1–4), Spi-C-expressing cells (lanes 5–11) or mixtures of PU.1 and Spi-C extracts (lanes 12–14) were analyzed for binding to the VCAM Ets probe. Shown are competitions (Comp) using unlabelled competitor oligonucleotides or supershifts using anti-sera against PU.1 or Spi-C as indicated.
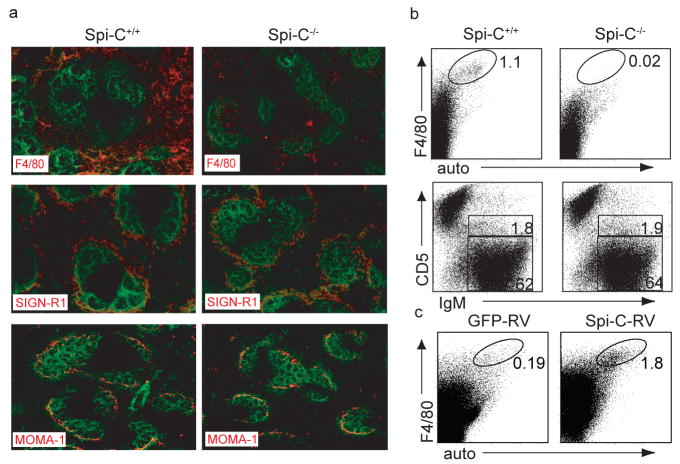
Figure 2. Spi-C−/− mice have a cell-autonomous defect in red pulp macrophages.
a, Spi-C+/+ and Spi-C−/− spleens sections were stained for B220 (green) and F4/80, SIGN-R1 or MOMA-1 (red). b, BM cells from CD45.2+ C57BL/6 Spi-C+/+ or Spi-C−/− mice were transferred into irradiated CD45.1+ B6.SJL mice. Splenocytes were stained for CD45.2, CD45.1, F4/80, CD5 and IgM. After 10 weeks, >97% of spleen cells were donor-derived (CD45.2+CD45.1−). Plots are gated on donor-derived cells. Numbers represent the percentage of donor-derived cells in the indicated gates. c, BM cells from CD45.2+ C57BL/6 Spi-C−/− mice were infected with Spi-C-RV or control RV and transferred into irradiated CD45.1+ B6.SJL mice. After 6 weeks, spleen cells were stained for CD45.2, CD45.1, and F4/80. Plots are gated on donor-derived cells. Results are representative of four mice per group.
Loss of RPM could result from either a cell-autonomous defect or from the loss of a required extrinsic component such as a BM or splenic stromal cell. To distinguish these possibilities, we generated hematopoietic chimeras by transplanting BM cells from CD45.2+ Spi-C+/+ or Spi-C−/− mice into irradiated congenic CD45.1+ recipients (Fig. 2b). In these chimeras, F4/80+ RPM developed only from Spi-C+/+ donor BM, but not from Spi-C−/− donor BM (Fig. 2b). This defect was restricted to RPM, since Spi-C−/− donor BM generated normal B cells (Fig. 2b), and CD11b+ and CD11c+ myeloid lineages, NK cells and T cells (Supplementary Fig. 6e). Development of F4/80+ RPM was restored when Spi-C−/− BM was infected with a Spi-C-expressing retrovirus and allowed to develop in lethally irradiated hosts (Fig. 2c). These findings indicate that the loss of RPM in Spi-C−/− mice results from an intrinsic hematopoietic defect.
Senescent red blood cells (RBCs) are normally phagocytosed by macrophages in spleen and liver, their hemoglobin degraded, and iron transported back into the circulation12–14. To study RBC clearance by splenic macrophages, we used the system of CD47−/− RBCs13. We injected CFSE-labeled CD47−/− RBCs into mice and examined their clearance and uptake by spleen and liver cells (Supplementary Fig. 7). The initial clearance of CD47−/− RBCs from the circulation was similar between Spi-C−/− and Spi-C+/+ mice (Supplementary Fig. 7a). However, we found differences in efficiency of phagocytosis of trapped RBCs by splenic macrophages between Spi-C+/+ and Spi-C−/− mice. In Spi-C+/+ mice, F4/80hi CD68+ RPM showed more than 10-fold higher uptake of CFSE than F4/80loCD68+ cells (Supplementary Fig. 7c, d). In Spi-C−/− mice, which lacked F4/80hiCD68+ macrophages (Supplementary Fig. 7c), the remaining F4/80loCD68+ macrophages showed the low level of phagocytosis as in Spi-C+/+ mice (Supplementary Fig. 7d). Liver cells did not take up labeled CD47−/− RBCs in either wild type or Spi-C−/− mice (Supplementary Fig. 7e). These results suggest that the absence of F4/80hi RPM in Spi-C−/− mice represents the loss of a functional macrophage subset that is normally responsible for efficient phagocytosis of RBCs in the spleen.
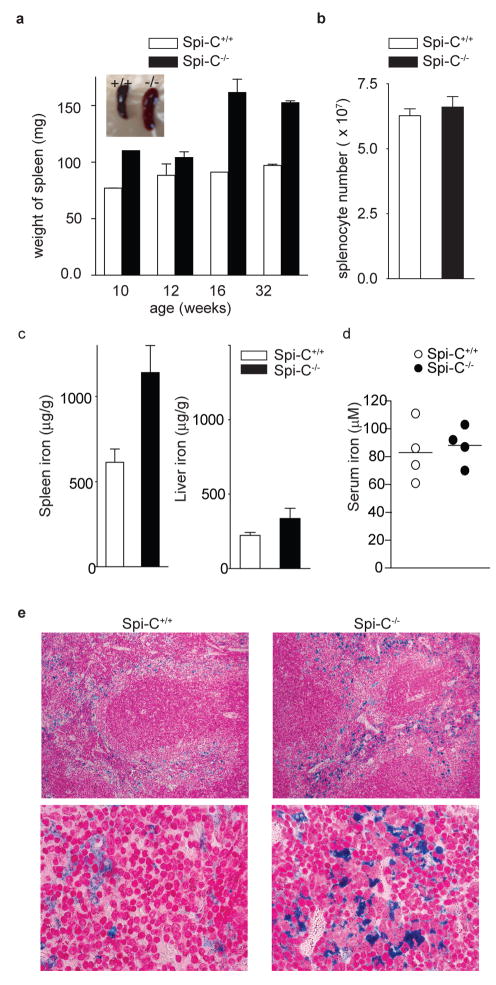
Spi-C−/− mice developed age-dependent increase in the size and weight of the spleen not associated with increased numbers of splenocytes (Fig. 3a, b). Since mice with genetic defects in iron or hemoglobin metabolism can exhibit splenomegaly15,16, we assessed the iron stores of Spi-C−/− mice. Serum and liver iron concentrations were normal in Spi-C−/− mice, but splenic tissue iron concentration was increased (Fig. 3c, d, Supplementary Fig. 8a and b). This iron accumulation was evident histologically in Spi-C−/− mice and largely confined to the red pulp (Fig. 3e, Supplementary Fig. 8c, Fig. 9a), but was not observed in the liver or intestine (Supplementary Fig. 8d, and data not shown). Spi-C−/− mice did not exhibit abnormal erythroid parameters or morphology (Supplementary Table 2, Supplementary Fig. 9c).
Figure 3. Spi-C−/− mice have increased splenic iron stores.
a, Spleens from Spi-C+/+ and Spi-C−/− mice of the indicated age were weighed (n=3 per time point). Representative spleens at 32 weeks (Inset). b, Splenocytes from Spi-C+/+ and Spi-C−/− mice were counted (mean ± SD, n=5). c, Iron levels in spleen (left) and liver (right) of 129 SvEv Spi-C+/+ and Spi-C−/− male mice on a standard diet (mean ± SD, n=4). d, Serum iron levels were determined for 32 weeks old 129 SvEv background Spi-C+/+ and Spi-C−/− male mice on a standard diet. e, Perl’s Prussian blue stain for ferric iron in the spleens of 16-week old 129 SvEv Spi-C+/+ and Spi-C−/− male mice.
To identify potential Spi-C target genes, we compared global gene expression profiles of RPM, alveolar and peritoneal macrophages, monocytes and dendritic cells (Fig. 4a). The hemoglobin scavenger receptor (CD163)17 and the iron transporter ferroportin 1 (Slc40a1)18 were both highly expressed by RPM relative to other macrophage subsets or B cells (Fig. 4a). VCAM-1 was also highly expressed in RPM compared to peritoneal and alveolar macrophages (Fig. 4a), consistent with reported VCAM-1 expression in the red pulp19,20. VCAM-1 is most highly expressed in the red pulp of control spleens, but was virtually absent in Spi-C−/− spleens (Fig. 4b, Supplementary 10a, b). In Spi-C+/+ spleens, VCAM-1 was highly correlated with F4/80 expression (Supplementary Fig. 10a), which were both substantially reduced in Spi-C−/− spleens. In contrast, other tissues did not show this coordinate expression of F4/80 and VCAM-1 (Supplementary Fig. 10a). Peritoneal macrophages expressed high F4/80 without high VCAM-1, and the majority of F4/80+ cells in lymph nodes expressed low levels of VCAM-1 (Supplementary Fig. 10b). However, VCAM-1 expression by B cells was reduced in Spi-C−/− mice compared to Spi-C+/+ mice (Supplementary Fig. 10b). BM monocytes treated with M-CSF21 induced both Spi-C and VCAM-1 expression, whereas treatment with GM-CSF caused much less induction of Spi-C and no induction of VCAM-1 (Supplementary Fig. 10c). Consistently, Spi-C−/− BM cells treated with M-CSF showed reduced levels of VCAM-1 expression compared to wild type cells (Supplementary Fig. 10d). Together, these results suggest the possibility that Spi-C may regulate the expression of VCAM-1.
Reporter analysis and electrophoretic mobility shift assays (EMSA) provide additional evidence that Spi-C regulates VCAM-1 (Fig. 4c–e). PU.1 can augment FcγR2b promoter activity22. PU.1 and Spi-C each augment a VCAM-1 reporter by a similar magnitude22 (Fig. 4c). An Ets consensus element within the proximal VCAM-1 promoter forms complexes with PU.1 and Spi-C in EMSA (Fig. 4d), which are supershifted by anti-PU.1 or anti-Spi-C antisera respectively (Fig. 4e). Formation of the Spi-C EMSA complex was specifically inhibited by an FcγR2b Ets probe and VCAM-1 Ets probe, but not by a mutated VCAM-1 Ets probe. Deletion of the Ets element within the VCAM-1 promoter eliminated reporter augmentation by both PU.1 and Spi-C (Fig. 4c). To test if Spi-C could induce VCAM-1 in cell lines, we transfected the RAW264.7 macrophage cell line with Spi-C-RV or control retrovirus (Supplementary Fig. 10e). Control RAW264.7 cells do not express Spi-C or VCAM-1, and treatment with M-CSF only weakly induced VCAM-1. In Spi-C-expressing RAW264.7 cells, M-CSF strongly induced VCAM-1, but not CD163, ferroportin, Heme oxygenase, or Mon1a. Thus, VCAM-1 appears to be a target of Spi-C in macrophages.
Spi-C was initially identified as a PU.1-related transcription factor expressed in B cells, but its function was unknown5,7. An earlier report based on overexpression studies suggested Spi-C might regulate IgE production23. Differential expression of Spi-C was suggested as evidence for independent ontogeny for splenic B cells and peritoneal B1a B cells24. We find Spi-C−/− mice to have normal B cell development and antibody production, but do not exclude a subtle role for Spi-C in B cell function. However, our evidence supports a critical role for Spi-C in the development of RPM, and shows this cell lineage to be required for normal recycling of red blood cells and iron homeostasis in the spleen.
The known genetic defects of iron metabolism14,25,26 frequently involve proteins that function as iron transporters/exporter (e.g., ferroportin18), as receptors for iron binding proteins (e.g., transferrin receptor27, HFE28), or that function to regulate the activity of these transporters or receptors (e.g., hepcidin29). Such gene defects that directly alter iron transport can produce global changes in iron storage that may affect many tissues. In contrast, the transcription factor Spi-C is the first gene to affect iron metabolism by controlling the development of a particular cell lineage important for recycling red blood cells and their products. Future studies will be directed at analysis of the specific roles of transcriptional targets of Spi-C, such as VCAM-1, in elaborating these functions of red pulp macrophages.
Methods Summary
Mice and reagents
Spi-C−/− mice were generated by deleting the entire protein coding regions exons 2 through 5 of the Spi-C locus. The neomycin resistance cassette was deleted by intercrossing Spi-C+/− mice with CMV-Cre deletor mice to produce Spi-Cnull/null as described in the Methods.
Histochemical analysis
6 μm frozen tissue sections were fixed in cold acetone and blocked with 5% bovine serum albumin (Roche). Monoclonal antibodies used were biotinylated antibody against F4/80 (Caltag), VCAM-1 (eBioscience), MOMA-1 (BMA Biomedicals), SIGN-R1 (ER-TR9, BMA Biomedicals), Alexa488-conjugated antibody against B220 (BD PharMingen), and Streptavadin-Alexa555 (Invitrogen). For Perl’s Prussian blue stain, tussues were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.0), embedded in paraffin, and stained with Perl’s Prussian blue and Pararosaniline (Sigma).
Adoptive transfer of bone marrow cells
Recipient CD45.1+ B6.SJL (Taconic) mice were lethally irradiated with 1100 rads and injected i.v. with 107 BM cells obtained from CD45.2+ F4 or F5 Spi-C+/+ or Spi-C−/− mice. Donor-derived cells in spleen (CD45.2+) were analyzed 10 weeks after transfer by FACS.
Measurement of iron content in spleen, liver and serum
Age and gender matched mice were analyzed for non-heme iron as described30. Livers and spleens were weighed and digested in 3 M hydrochloric acid/10 % trichloroacetic acid, at 65 °C for 20 hr. 10 μl of each acid extract was mixed with 0.5 ml of chromagen reagent. The absorbance at 535 nm was measured by a DU Series 500 Spectrophotometer (Beckman), and compared to an iron standard treated identically. For serum measurements30, 20 μl of serum were incubated with 20 μl of acid reagent for 5 min. Supernatant was mixed with 40 μl chromagen reagent and absorbance at 535 nm measured as above.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (K.M.M.), and the Burroughs Wellcome Fund Career Award for Medical Scientists (B.T.E.)
References
- 1.Taylor PR, et al. Macrophage receptors and immune recognition. Ann Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nature Reviews Immunology. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Ye M, Graf T. Early decisions in lymphoid development. Curr Opin Immunol. 2007;19:123–128. doi: 10.1016/j.coi.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 5.Bemark M, Martensson A, Liberg D, Leanderson T. Spi-C, a novel Ets protein that is temporally regulated during B lymphocyte development. J Biol Chem. 1999;274:10259–10267. doi: 10.1074/jbc.274.15.10259. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson R, Hjalmarsson A, Liberg D, Persson C, Leanderson T. Genomic structure of mouse SPI-C and genomic structure and expression pattern of human SPI-C. Gene. 2002;299:271–278. doi: 10.1016/s0378-1119(02)01078-8. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto S, et al. Prf, a novel Ets family protein that binds to the PU.1 binding motif, is specifically expressed in restricted stages of B cell development. Int Immunol. 1999;11:1423–1429. doi: 10.1093/intimm/11.9.1423. [DOI] [PubMed] [Google Scholar]
- 8.Lloberas J, Soler C, Celada A. The key role of PU.1/SPI-1 in B cells, myeloid cells and macrophages. Immunol Today. 1999;20:184–189. doi: 10.1016/s0167-5699(99)01442-5. [DOI] [PubMed] [Google Scholar]
- 9.Su GH, et al. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor PR, et al. Dectin-2 is predominantly myeloid restricted and exhibits unique activation-dependent expression on maturing inflammatory monocytes elicited in vivo. Eur J Immunol. 2005;35:2163–2174. doi: 10.1002/eji.200425785. [DOI] [PubMed] [Google Scholar]
- 11.Taylor PR, Brown GD, Geldhof AB, Martinez-Pomares L, Gordon S. Pattern recognition receptors and differentiation antigens define murine myeloid cell heterogeneity ex vivo. Eur J Immunol. 2003;33:2090–2097. doi: 10.1002/eji.200324003. [DOI] [PubMed] [Google Scholar]
- 12.Bratosin D, et al. Cellular and molecular mechanisms of senescent erythrocyte phagocytosis by macrophages. A review Biochimie. 1998;80:173–195. doi: 10.1016/s0300-9084(98)80024-2. [DOI] [PubMed] [Google Scholar]
- 13.Oldenborg PA, et al. Role of CD47 as a marker of self on red blood cells. Science. 2000;288:2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 14.De Domenico I, McVey WD, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 15.Biggs TE, et al. Nramp1 modulates iron homoeostasis in vivo and in vitro: evidence for a role in cellular iron release involving de-acidification of intracellular vesicles. Eur J Immunol. 2001;31:2060–2070. doi: 10.1002/1521-4141(200107)31:7<2060::aid-immu2060>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Tolosano E, et al. Enhanced splenomegaly and severe liver inflammation in haptoglobin/hemopexin double-null mice after acute hemolysis. Blood. 2002;100:4201–4208. doi: 10.1182/blood-2002-04-1270. [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen M, et al. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 18.Donovan A, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Gurtner GC, et al. Targeted Disruption of the Murine Vcam1 Gene - Essential Role of Vcam-1 in Chorioallantoic Fusion and Placentation. Genes Dev. 1995;9:1–14. doi: 10.1101/gad.9.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Ulyanova T, et al. VCAM-1 expression in adult hematopoietic and nonhernatopoietic cells is controlled by tissue-inductive signals and reflects their developmental origin. Blood. 2005;106:86–94. doi: 10.1182/blood-2004-09-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leon B, et al. Dendritic cell differentiation potential of mouse monocytes: monocytes represent immediate precursors of CD8− and CD8+ splenic dendritic cells. Blood. 2004;103:2668–2676. doi: 10.1182/blood-2003-01-0286. [DOI] [PubMed] [Google Scholar]
- 22.Schweitzer BL, et al. Spi-C has opposing effects to PU.1 on gene expression in progenitor B cells. J Immunol. 2006;177:2195–2207. doi: 10.4049/jimmunol.177.4.2195. [DOI] [PubMed] [Google Scholar]
- 23.Carlsson R, Thorell K, Liberg D, Leanderson T. SPI-C and STAT6 can cooperate to stimulate IgE germline transcription. Biochem Biophys Res Comm. 2006;344:1155–1160. doi: 10.1016/j.bbrc.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Stoermann B, Kretschmer K, Duber S, Weiss S. B-1a cells are imprinted by the microenvironment in spleen and peritoneum. Eur J Immunol. 2007;37:1613–1620. doi: 10.1002/eji.200636640. [DOI] [PubMed] [Google Scholar]
- 25.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang FD, et al. Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat Genet. 2007;39:1025–1032. doi: 10.1038/ng2059. [DOI] [PubMed] [Google Scholar]
- 27.Fleming RE, et al. Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis. Proc Natl Acad Sci U S A. 2002;99:10653–10658. doi: 10.1073/pnas.162360699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou XY, et al. Knockout of the mouse HFE gene products hemochromatosis. Gastroenterology. 1998;114:A1372. [Google Scholar]
- 29.Roetto A, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 30.Torrance JD, Bothwell TD. Tissue iron stores. In: Cook JD, editor. In Methods in hematology. Vol. 1. 1980. pp. 90–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.