Abstract
Organized neuronal firing is critical for cortical processing and is disrupted in schizophrenia. Using 5’ RACE in human brain, we identified a primate-specific isoform (3.1) of the K+-channel KCNH2 that modulates neuronal firing. KCNH2-3.1 mRNA levels are comparable to KCNH2-1A in brain, but 1000-fold lower in heart. In schizophrenic hippocampus, KCNH2-3.1 expression is 2.5-fold greater than KCNH2-1A. A meta-analysis of 5 clinical samples (367 families, 1158 unrelated cases, 1704 controls) shows association of SNPs in KCNH2 with schizophrenia. Risk-associated alleles predict lower IQ scores and speed of cognitive processing, altered memory-linked fMRI signals, and increased KCNH2-3.1 expression in post-mortem hippocampus. KCNH2-3.1 lacks a domain critical for slow channel deactivation. Overexpression of KCNH2-3.1 in primary cortical neurons induces a rapidly deactivating K+ current and a high-frequency, non-adapting firing pattern. These results identify a novel KCNH2 channel involved in cortical physiology, cognition, and psychosis, providing a potential new psychotherapeutic drug target.
A major challenge in modern medicine is to understand cellular and molecular mechanisms underlying common mental illnesses such as schizophrenia, which involve complex genetic and environmental determinants1. Prevailing etiological hypotheses maintain that the interaction of multiple genetic factors with each other and with environmental risk factors results in neurodevelopmental abnormalities predisposing to illness later in life1. Given this complexity, together with imprecise and differing definitions of phenotype, it is not surprising that statistical association studies to identify risk genes for schizophrenia have not enjoyed unanimous replication1,2. Gene identification for complex polygenic disorders, such as schizophrenia, will likely require demonstration that risk variants impact on the biology of the gene in a manner that converges on important aspects of the biology of the illness3. Indeed, this approach has proven important in a number of complex genetic disorders, such as adult onset diabetes, in which multiple genes each account for only a very small share of risk but show stronger effects on related intermediate phenotypes even in normal subjects– e.g body mass index4 or glucose induced insulin release5. Potential biologic intermediate phenotypes related to risk for schizophrenia include abnormalities related to hippocampal (HF) and prefrontal cortices (PFC), which are consistently reported in patients with schizophrenia and in their healthy relatives6–11. Thus, it follows that genes weakly associated with susceptibility for schizophrenia might show relatively robust effects on PFC and HF function in risk-allele carrying populations.
Here, we identify a novel potential schizophrenia susceptibility gene, KCNH2 (a hERG-family potassium channel), using meta-analyses of five independent clinical association samples. Moving beyond statistical association with clinical diagnosis, we describe association with several schizophrenia-linked biological intermediate phenotypes in large samples of normal individuals who carry risk-associated alleles. In searching for a molecular mechanism, we discovered a novel, primate and brain-specific KCNH2 isoform (3.1), encoded in close proximity to the risk-associated SNPs. Expression of KCNH2-3.1 is specifically increased within the HF of schizophrenia patients and in normal individuals who carry risk-associated alleles. Expression of KCNH2-3.1 in rodent cortical neurons causes a dramatic alteration in KCNH2 channel physiology resulting in high-frequency, non-adapting neuronal firing patterns. Such changes in spike frequency may underlie abnormalities in neuronal firing thought to be a fundamental aspect of cortical dysfunction in psychosis12. These convergent results implicate a novel potassium channel in primate cortical physiology and in the etiology and pathophysiology of psychosis, making it a potentially new target for antipsychotic therapy.
Identification of the KCNH2 region associated with schizophrenia
It has become increasingly apparent that genetic risk for schizophrenia and many other complex illnesses will not relate to major amino acid changes and highly detrimental protein mutations, though these may still occur in rare forms of the illness13. Instead, the molecular effects of susceptibility alleles are more likely to be subtle, relating to the regulation and splicing of transcripts or proteins. Thus, the gene expression profiles of patient tissue are a potentially informative starting point to search for candidate risk genes14,15. We selected 10 genes previously reported as differentially expressed in schizophrenic brain16 and genotyped haplotype tagging SNPs in 170 Caucasian families having an offspring with schizophrenia. FBAT analysis17 of gene-based haplotypes demonstrated significant global association with diagnosis in the genomic region of NOS3 (7q36.1) (p<.005; p<.05 after Bonferroni correction for ten association tests) (Supplementary Data S1). In the European sample (CEU) of the HapMap Project18, the block tagged by these SNPs spans portions of NOS3 and KCNH2 (Figure 1; Supplementary Figure S2). The most strongly associated SNP in this 7q36.1 region is within KCNH2 and was associated with schizophrenia following correction for the total number of SNP tests performed across all ten genes (M33 [rs1036145], corrected p = 0.042).
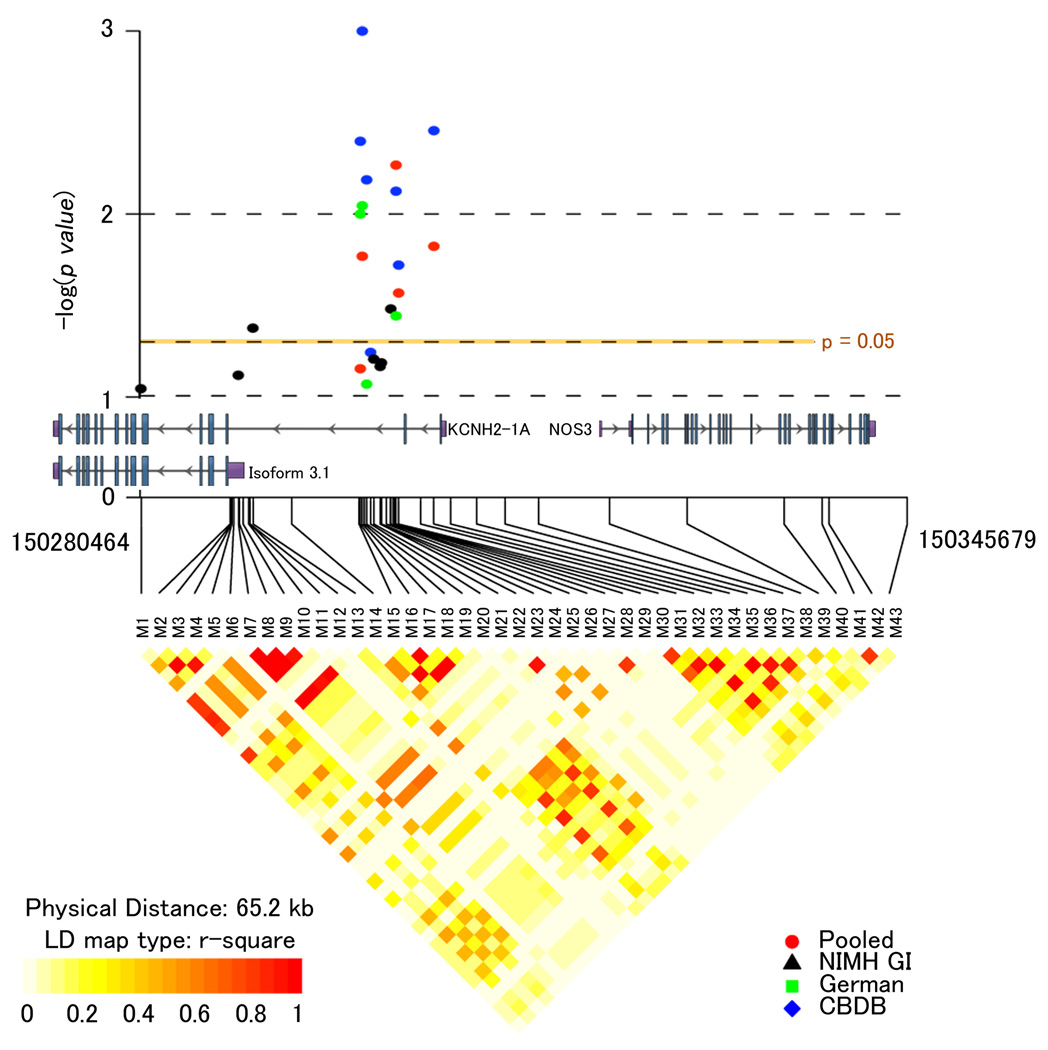
Figure 1. Genetic association of 7q36.1 with risk for schizophrenia.
(Top) Inverse log of the p-value for single SNPs from association results for the CBDB/NIMH Sibling Study (CBDB-blue), NIMHGI (NIMHGI-black), for the German case-control study (German-green) and for the pooled 5 sample meta-analysis (Pooled-red). Only markers with p-values less than 0.1 are shown. A physical map of the region is given and depicts known genes within the region. (Bottom) The LD structure of the genotyped markers is shown for 370 unrelated healthy Caucasian controls and depicted as r2. Graphic created using the R package snp.plotter58.
KCNH2 as a potential risk gene for schizophrenia
Additional SNPs were genotyped (43 in total) in an expanded sample of 296 families (“CBDB family sample”) from the 3’-end of KCNH2 to the 3’-end of NOS3, including 11 SNPs detected through resequencing of the region in schizophrenia patients. Six SNPs were significantly associated with schizophrenia at α < 0.05 (5 with p < 0.01)(Figure 1). Most of the associated markers are in moderate to strong LD, suggesting the region of interest maps to a ∼3 kb segment of intron 2 of KCNH2.
We tested for replication in a large German Caucasian case-control dataset, typing the 6 associated SNPs found in the CBDB sample and one SNP with marginal significance (p=0.057). Significant association at the genotype level was observed for minor allele homozygotes at four of these SNPs, M16, M17, M19 and M30 (Figure 1; Supplementary Data S3). Three additional samples were tested with the same 6 markers: a Caucasian family-based sample from the NIMH Genetics Initiative project19 (NIMHGI), a case-control sample from Armenia and a case-control sample from Italy.. Though the sizes and ascertainment strategies of these samples differed from the larger original samples, significant SNPs were observed in this region and/or the 6 implicated SNPs tended towards the same direction of association though not significantly (Figure 1; Supplementary Data S3).
Given these results and power limitations, a combined family-based and case-control meta-analysis was conducted using the R package catmap20. This meta-analysis showed M17, M30, M31, and M33 to be significantly associated with schizophrenia, with M30 showing the greatest odds ratio across all samples of 1.17 (p = 0.0054) (Supplementary Data S3). “Leave-one-out” sensitivity analysis to determine if the meta-analysis results were driven by the initial family-based result and therefore suggestive of a “winner’s curse” phenomenon21 revealed that removal of any of the included studies still led to significant association using the 4 remaining independent studies for M30 (Supplementary Data S3). The odds ratios in the cumulative analysis are comparable with other genes supported as significantly associated with schizophrenia in larger meta-analyses22,23 and exceed the “Venice interim criteria” for “small summary” findings24. Taken together, these results identify a small intronic region of KCNH2 as a potential susceptibility locus for schizophrenia.
Impact of KCNH2 SNPs on cognition, and cortical structure and function
Given the high expression of KCNH2 within the PFC and HF25, two regions subserving attention and memory processes and consistently implicated in the neuropathology of schizophrenia26,27, we hypothesized that if risk-associated SNPs impact on the biology of these brain regions, cognitive deficits referable to these regions would also be associated with risk genotypes, regardless of disease status. We selected three of the significantly associated SNPs from the meta-analysis (M30, 31, 33, all of which are in strong LD with the fourth positive SNP M17) to test for their effects on seven independent summary measures of cognition28 (Supplementary Data S4) in a sample of healthy, unrelated controls independent from the prior control samples. The use of healthy controls for genetic association at the level of brain function avoids potential confounders related to chronic illness and medical treatment29. Significant association was observed between risk genotypes for each SNP and performance on the IQ/processing speed factor (p = 0.020; Supplementary Figure S4). Thus, even in healthy controls, the same alleles observed more frequently in schizophrenia patients predict significantly lower IQ/processing speed. It is noteworthy that similar cognitive phenotypes have been especially strongly related to genetic risk for schizophrenia in discordant twin samples7.
We also tested whether the same three SNPs would relate to changes in brain structure and physiology. Using MRI-based morphometry, significant volume decreases in HF structures (α = 0.05, FDR corrected), proportional to allelic load, were observed in normal individuals carrying risk-alleles of SNPs M30, M31, and M33 using whole brain and region of interest analyses (Figure 2A; Supplementary Data S6). We next tested whether the same SNPs would influence the physiology of cognitive processing related to HF, again in healthy control risk allele carriers at M30, 31 and 33. fMRI was used to assess regional activation during incidental encoding of a temporal lobe memory task30 in which task performance was controlled. (Supplementary Data S5 for demographic statistics). We observed greater activation (FWE correction α = 0.05) of the HF within healthy control risk allele carriers (Figure 2B; Supplementary Data S5). This pattern of inefficient processing of memory information (i.e. excessive activity for a fixed level of performance) suggests that the tuning of cortical microcircuits critical for information processing within the HF is relatively disordered in individuals with risk-associated alleles. Similar patterns of inefficient processing in HF have been reported for other genes and diseases affecting cognition e.g. 31,32.
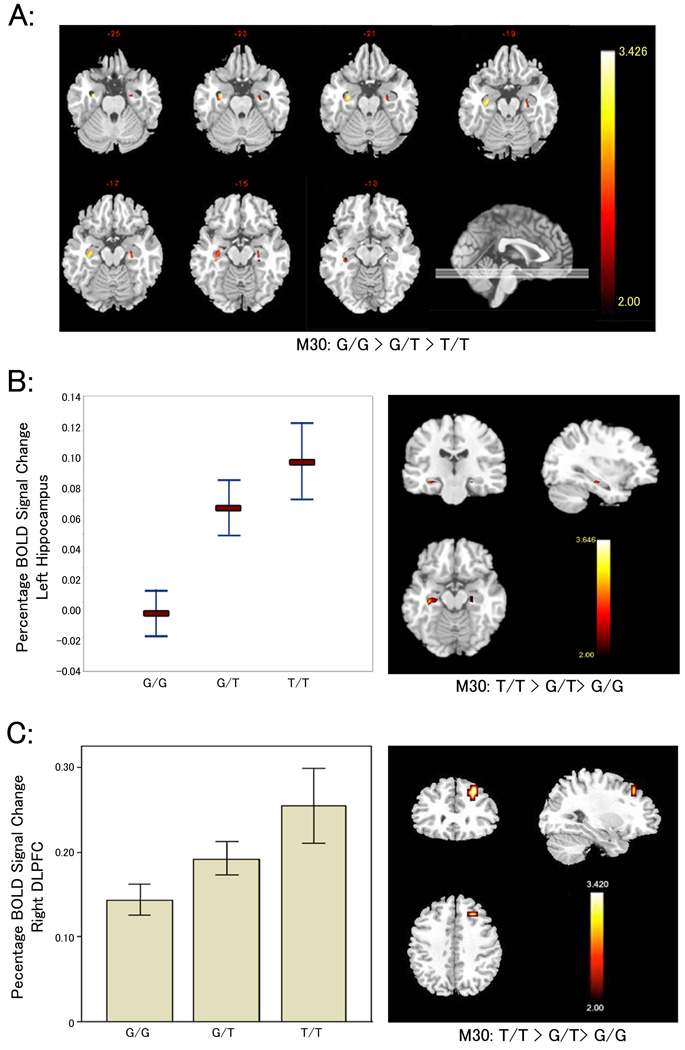
Figure 2. Association of risk SNPs with cognitive measures, brain structure volumes, and regional brain activity during memory-based tasks.
(A) M30 genotype versus hippocampal gray matter volumes in healthy control subjects. Heatmap depicts linear decrease in regional gray matter volume from subjects homozygous for the risk associated allele, A, (n = 16) to heterozygote carriers (n = 61) to non carriers (n= 64). Only voxels corresponding to a p<0.05 FDR corrected threshold are shown. (B) Percent blood oxygen level-dependent (BOLD) signal change in healthy controls during the encoding conditions of a declarative memory task in the left posterior hippocampus (MNI coordinates of peak cluster: −34 −25 −15 mm) showing a significant linear increase of activation in homozygote carriers (n= 14) relative to heterozygote carriers (n=37), and to non carriers (n=28) of the risk associated allele at M30. Plot depicts voxel mean ±1 SEM. Heatmap colors correspond to degree of increase in BOLD signal with each copy of M30 risk allele (T). Only voxels surviving p< 0.05 FWE correction are shown) statistical t-maps and mean (±1 SEM) (D) Thresholded (p< 0.05 FWE corrected) statistical t-maps and mean ((±1 SEM) percent BOLD signal change during the executive working memory task in the right dorsolateral prefrontal cortex (DLPFC) (MNI coordinates of peak cluster: 26 30 42 mm) showing a significant linear increase of activation in homozygote carriers (n=24) relative to heterozygote carriers (n=71), and to non carriers (n=81) of the risk associated allele at M30.
Although hippocampus may be engaged during executive cognition tasks that impact on measures of IQ6,33, recent studies have demonstrated that general intelligence or IQ measures are more closely associated with executive control processes and prefrontal cortex-related structures34–36. Therefore, to more specifically investigate a connection between risk associated alleles and executive-linked physiology, we used fMRI to measure cortical engagement during the N-back working memory task, an executive cognition task well validated as engaging prefrontal cortical circuitry37. Again, normal subjects carrying risk associated alleles of M30, M31, and M33 demonstrated significantly increased activity in an allele load pattern within the DLPFC despite controlled task performance (i.e. inefficient cortical processing) (Figure 2C; Supplementary Data S5). In conjunction with significantly increased HF activity during memory encoding tasks, these data consistently suggest association between risk alleles and impairments in the efficiency of information processing within two areas of the brain implicated in the pathophysiology of schizophrenia.
Effects of schizophrenia and risk genotypes on KCNH2 mRNA expression
The association of KCNH2 with schizophrenia and with related brain phenotypes in multiple independent samples lends statistical and biological support to the involvement of this genomic region in risk of illness. However, these findings do not identify the underlying molecular mechanism. We investigated gene expression in human brain and its relationship to our evidence for genetic association. Using quantitative RT-qPCR, we found expression of KCNH2-1A, to be significantly lower in both the DLPFC (p = 0.016; Figure 3-A) and hippocampus (p = 0.003; Figure 3-A) of schizophrenia patients. Since most patients with schizophrenia receive antipsychotic drugs, which bind to KCNH238, expression differences may be medication artifacts. We, therefore, measured KCNH2 expression within the frontal cortex of rats following 28 days of treatment with clozapine or haloperidol across a range of doses and did not find significant medication effects (Supplementary Figure S6).
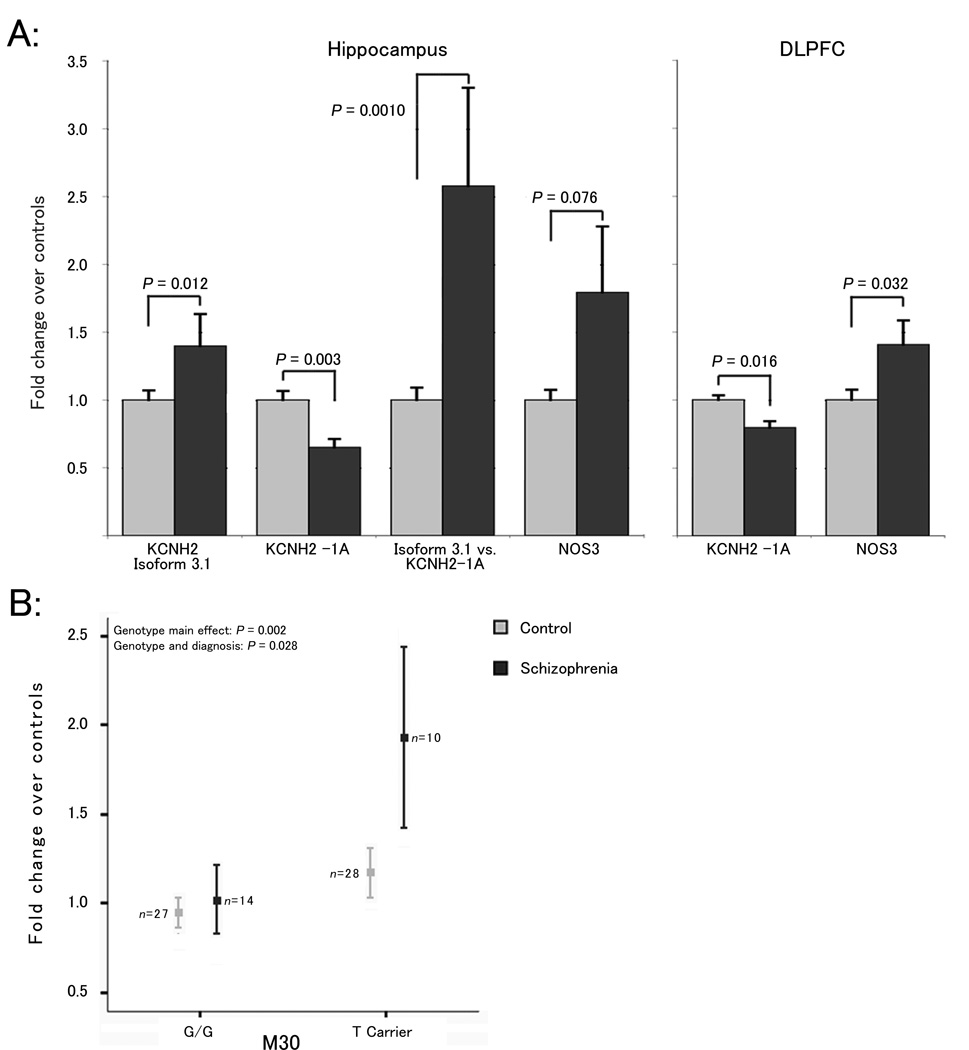
Figure 3. Regional gene expression and association with risk genotype.
(A) Differences in mRNA expression within the hippocampus of 29 schizophrenia patients/59 healthy control subjects, and within the DLPFC of a largely overlapping set of 31 schizophrenia patients/69 healthy control subjects. Expression values are normalized fold differences versus the mean of healthy controls. P-values represent main effect of diagnosis in the final regression model. (B) Association of M30 risk-genotype (T carriers) with isoforms 3.1 expression within the hippocampus (data from A) with respect to diagnosis. All error bars represent one standard error of the mean.
To explore the possibility that the molecular mechanism of the clinical associations involves transcriptional regulation, we tested for genotype effects on expression of KCNH2-1A. No significant effects were observed (e.g. M30 p=0.516; Supplementary Data S7-B). KCNH2-1B, a minor isoform expressed within the brain25, did not show significant differences between controls and schizophrenia patients, nor was its expression significantly associated with risk genotypes (data not shown). NOS3 expression also showed no association with these risk SNPs (M30 p=0.90; Supplementary Data S7-B, Supplementary Note 1). These data suggest that the molecular effects of genetic risk do not impact on the expression of KCNH2-1A or KCNH2-1B; and may instead relate to more complex aspects of gene processing, such as the splicing of transcripts or the expression of other isoforms.
Identification of a novel KCNH2 isoform in human brain
Using 5’-RACE on RNA pooled from the DLPFC of 10 schizophrenia patients and beginning from exon 4 of the full-length transcript (KCNH2-1A; NM_00023) we identified a novel 5’ extension of exon 3 producing an isoform that originates upstream of exon 3 (Figure 1; Supplementary Data S2 and S10). Further cloning from a human cDNA library (Stratagene) revealed that the isoform is expressed endogenously and contains all of the downstream exons of the full-length gene (through exon 15 of KCNH2-1A; Supplementary Data S9). In silico prediction (www.ncbi.nlm.nih.gov) of the longest ORF of KCNH2-3.1 suggests that the majority of the 5’ extension of exon 3 would be untranslated and that the first methionine is in-frame with the conserved portion of KCNH2-1A. As such, KCNH2-3.1 is expected to be missing the first 102 amino acids of KCNH2-1A, replacing them with 6 amino acids unique to this isoform. The translation of KCNH2-3.1 and the predicted difference in protein size was confirmed using western blot in transfected HEK cells (Figure 4B). Western blot in human hippocampus and frontal cortex revealed a protein band of similar molecular weight to that in transfected HEK cells (Figure 4B). Furthermore, neuroblastoma cells transfected with constructs encoding either or both KCNH2-1A or KCNH2-3.1 show overlapping expression of the protein isoforms on the plasma membrane (Figure 4C).
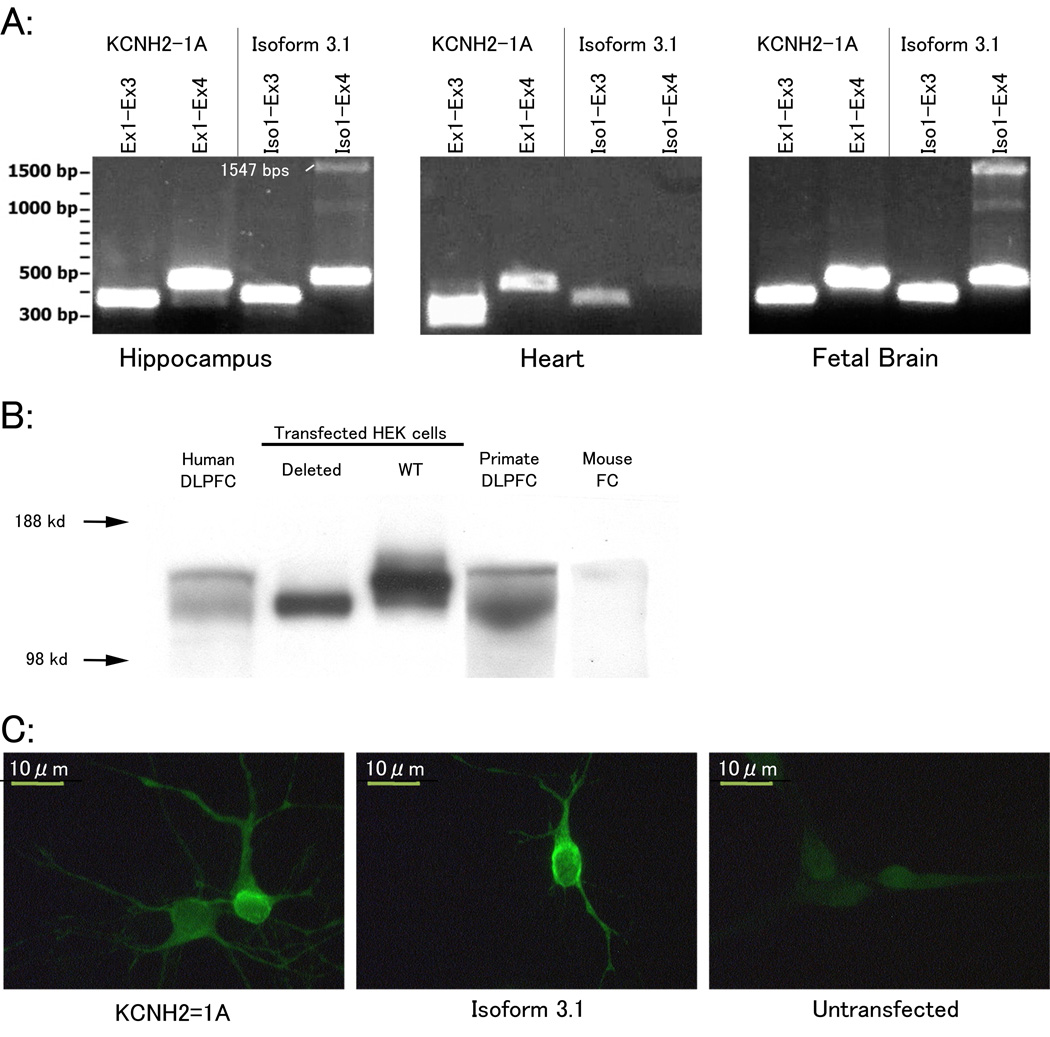
Figure 4. Detection and quantification of Isoform 3.1 mRNA and protein.
(A) PCR of KCNH2-1A and Isoform 3.1 using isoform specific primer pairs in human heart, hippocampus, and fetal brain tissue extracts. PCR products for KCNH2-1A correspond to 373 and 463 bps, respectively, and for Isoform 3.1 at 388 and 478 bp, respectively. The band ∼1.5 kb in Iso1-Ex4 lanes corresponds to small amounts of unspliced RNA or genomic DNA contaminant present in the samples. (B) Protein expression of KCH2-1A and Isoform 3.1 in human, primate, and mouse frontal cortex. Positive control lanes include HEK cells transfected with full-length KCH2-1A (WT) and Isoform 3.1. Proteins extracted from human brain regions show two distinct bands, one equivalent in size to isoform 3.1 transfected HEK cells. However, the larger protein band observed in human brain did not correspond to the size of KCNH2-1A transfected HEK proteins. Instead, the larger band occurs at ∼160 kDa, which is the reported size of KCNH2-1A from in vivo protein extracts suggesting post-translational modifications. (D) Primary rat neurons transfected with KCNH2-1A or Isoform 3.1 containing vectors.
To assess the potential organ selectivity of KCNH2-3.1, we measured the ratio of KCNH2-3.1 to KCNH2-1A mRNA levels across several tissues, including heart and three different brain regions, using quantitative RT-PCR. We found the two forms comparable in expression within several brain regions (Supplementary S9), but KCNH2-3.1 was over 1000-fold less abundant than KCNH2-1A within the heart, suggesting a brain specific role of KCNH2-3.1‥ As ERG family genes are moderately conserved in sequence across several species39, we expected KCNH2-3.1 to be present within other mammals. Using 5’-RACE, we were unable to detect KCNH2-3.1 homologues in mouse brain. Moreover, in silico analysis revealed that the 1.1 Kb region unique to KCNH2-3.1 was highly degenerate outside of primates (supplementary figure S10). Protein expression of KCNH2-3.1 was undetectable in mouse brain but was abundant within rhesus monkey DLPFC (Figure 4B).
As neuronal gene expression is tightly regulated and carefully orchestrated throughout development, we measured the expression profiles of KCNH2-1A and KCNH2-3.1 throughout brain development in the prefrontal cortices of 283 subjects including 39 prenatal samples (14–20 weeks of gestation) (Supplementary Data S11 for sample information). Isoform expression patterns were similar throughout postnatal life but dramatic differences in their relative expression was observed prenatally (Supplementary Data S11). KCNH2-3.1 transcripts are markedly increased prenatally relative to adult levels but then decrease and stabilize shortly after birth. In contrast, KCNH2-1A expression increases throughout prenatal development until it reaches a maximum level that is sustained throughout postnatal life. Though these data offer only a qualitative snapshot of developmental expression patterns, they suggest differential regulation of these isoforms during early brain development and a specific role for KCNH2-3.1 during these early stages.
Higher expression of KCNH2-3.1 in schizophrenia and in subjects with risk-associated genotypes
KCNH2-3.1 mRNA expression was significantly increased within the HF of patients versus healthy control subjects (p = 0.012, Figure 3-A). Moreover, the ratio of KCNH2-3.1 to KCNH2-1A showed a dramatic, 2.5 fold increase in schizophrenia patients (p = 0.0010). This ratio may reflect how isoforms and homologues of KCNH2 co-assemble to create heteromeric potassium channels with unique electrophysiological properties25,40. Additionally, higher levels of KCNH2-3.1 were significantly associated with risk alleles at M30, 31 and 33 even within healthy control subjects. Specifically, diagnosis and M30 genotype each had independent significant main effects on KCNH2-3.1 expression (p = 0.006 and 0.002, respectively) as well as a significant interaction (p = 0.028; Figure 3-B), suggesting that though risk alleles are associated with increased expression of KCNH2-3.1 regardless of subject group, the effect is more pronounced in patients (Figure 3-B). This trend was observed irrespective of race, reducing the potential confounder of genetic background (Supplementary Data S7). Similar results were found with M31 and M33, though interactions with diagnosis were nonsignificant (data not shown). These data support the hypothesis that the mechanism by which the associated SNPs contribute to risk for schizophrenia and related phenotypes involves the regulation of KCNH2-3.1 transcription and further suggest that over-expression of this isoform would result in physiologic effects related to the pathogenesis of the disorder.
Functional role of KCNH2-3.1 revealed by electrophysiology
KCNH2, (hERG1), a member of the tetrameric ether-a-go-go related family of potassium channels39, is best known for its role in slow repolarization during the myocardial action potential, thereby regulating the QT interval41. Its distinct physiological features, namely slow activation, fast inactivation, and slow and voltage-dependent deactivation39,40,42, make KCNH2 an excellent candidate for controlling neuronal firing patterns and cortical network oscillation43–45. Modeling work predicts that KCNH2 is critical for spike-frequency adaptation, or a gradual termination of a train of evoked action potentials, a common firing pattern seen in excitatory neurons in the brain46. Several studies have shown that inhibition of KCNH2 converts low-frequency, adapting to high-frequency, non-adapting neuronal discharges46,47, a firing pattern necessary for sustained neuronal activity subserving numerous complex cognitive functions48.
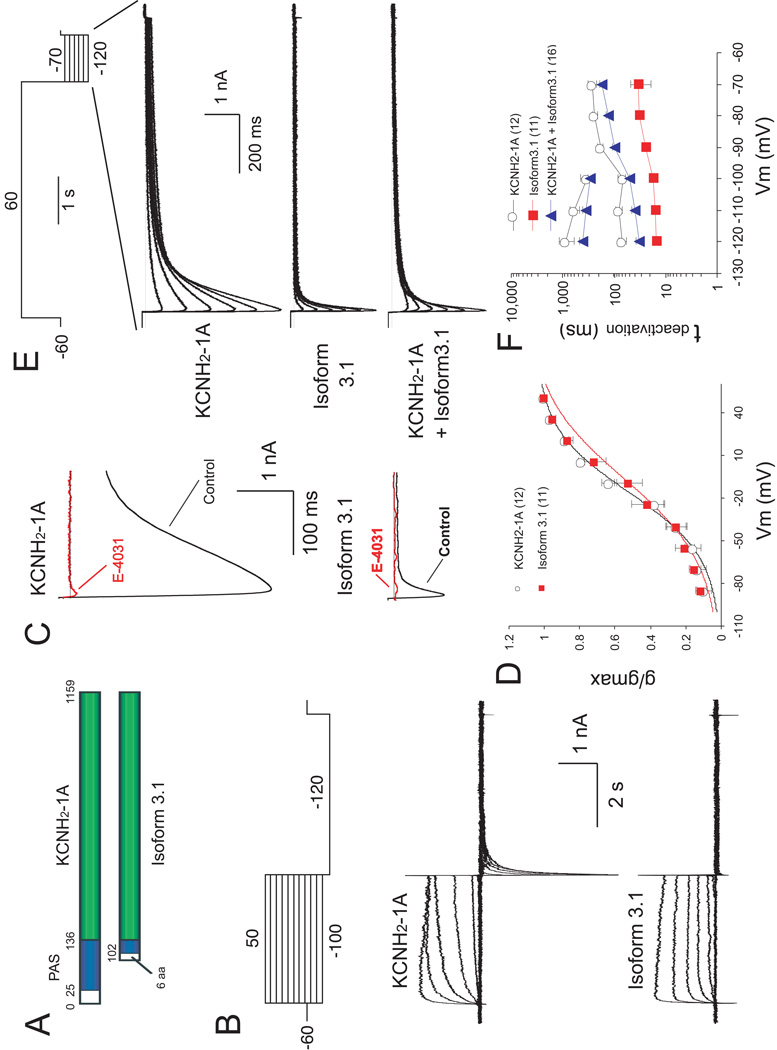
The slow deactivation of the KCNH2 channel is mediated by a PAS domain (aa 25–136) of the KCNH2-1A N-terminus42, most of which is missing in KCNH2-3.1 (Fig. 5A). We performed whole-cell recording on HEK cells transfected with either KCNH2-1A or KCNH2-3.1. When depolarized from −60 mV to various membrane potentials (Fig. 5B, top), KCNH2-3.1 was activated faster than KCNH2-1A (Fig. 5B, compare middle and bottom, Supplementary Fig. 12B).
Figure 5. Characterization of KCNH2 currents in HEK293T cells expressing KCNH2-1A and Isoform 3.1.
(A) Schematic diagram of KCNH2-1A and Isoform 3.1 domain structures. Blue box: PAS domain. Green box: conserved amino acid sequences l between isoforms. Numbers correspond to amino acid positions. (B) Currents evoked by voltage steps (4 sec) from VH of −60 mV to potentials from −100 to +50 mV in 10-mV increments, followed by a voltage pulse to −120 mV (5 sec). Upper panel: voltage protocol. Middle and lower panels: traces (corrected for leak currents) recorded from cells transfected with KCNH2-1A and Isoform 3.1 cDNAs, respectively. (C) Effects of E-4031on tail currents evoked by a test pulse to −120 mV from holding potential of +50 mV, using the same protocol as in B. Traces recorded from the same cells before (black) and after (red) treatment with 10 µM E-4031 are superimposed. Upper and lower panels show KCNH2 currents recorded from cells transfected with KCNH2-1A and Isoform 3.1 cDNAs, respectively. (D) Steady-state activation curves of KCNH2-1A and Isoform 3.1 tail currents, which were induced by the protocol shown in B. The reversal membrane potential (Vrev) under the recording conditions was −96 mV. The V1/2 for KCNH2-1A and Isoform 3.1 are −16.37 ± 2.55 mV and −6.54 ± 4.04 mV, respectively (p<0.05). In this and subsequent figures, the data are presented as mean ±1 SEM. Numbers in parentheses indicate the number of cells recorded. (E) Tail currents evoked by voltage steps from +60 mV to potentials between −120 mV and −70 mV in 10 mV increments. Traces in upper, middle and lower panels represent tail currents of KCNH2-1A, Isoform 3.1 and co-transfection KCNH2-1A with Isoform3.1, respectively. (F) Semilogarithmic plot of deactivation time constants of KCNH2-1A and Isoform3.1 currents at different repolarizing voltages. Upper plot (between −210 and −100 mV) corresponds to τ2 for HEK cells transfected with KCNH2-1A alone or cotransfected KCNH2-1A and Isoform 3.1, whereas the lower plots correspond to τ1 for all transfection combinations.
When the membrane potential was hyperpolarized to −120mV after depolarization, the KCNH2-1A-expressing cells exhibited a large, slowly deactivating “tail current”, which is completely blocked by the hERG specific inhibitor E-403149 (Fig. 5C, top), suggesting that this current is mediated solely by hERG channels. In contrast, cells transfected with KCNH2-3.1 exhibited a dramatically reduced tail current (Fig. 5C, bottom), which was also blocked by E-4031. The I-V relationship showed a dramatic reduction in the peak amplitudes, particularly when cells were more substantially depolarized initially (Supplementary Fig. 12A). In a conductance-voltage plot, KCNH2- 3.1 exhibited a slight, but significantly reduced steady state tail current activation (Fig. 5D; V1/2: KCNH2-1A=−16.37 ± 2.55mV, KCNH2-3.1=−6.54 ± 4.04mV, p<0.05).
When the cells were first depolarized and held at +60mV, and then hyperpolarized at various potentials (Fig. 5E, top), the inward currents mediated by KCNH2-3.1 decayed towards baseline much faster than those by KCNH2-1A (Fig. 5E, middle). The deactivation phase of the KCNH2-1A currents was best fitted by a double-exponential curve, generating a slow and a fast deactivation constant (τ1 and τ2), whereas that of KCNH2-3.1 could be fitted only by a single-exponential curve, with a much slower τ1 at all repolarizing voltages tested (Fig. 5F). Thus, KCNH2-3.1 mediates an inward rectified K+ current with dramatically faster deactivation kinetics. Further, when KCNH2-1A was co-expressed with KCNH2-3.1, the deactivation phase of the tail currents could be fitted by a double-exponential curve, with τ1 and τ2 that were both significantly lower than those in cells expressing KCNH2-1A alone (Figure 5F; ANOVA, p < .05), suggesting codominant channel behavior, intermediate to each channel.
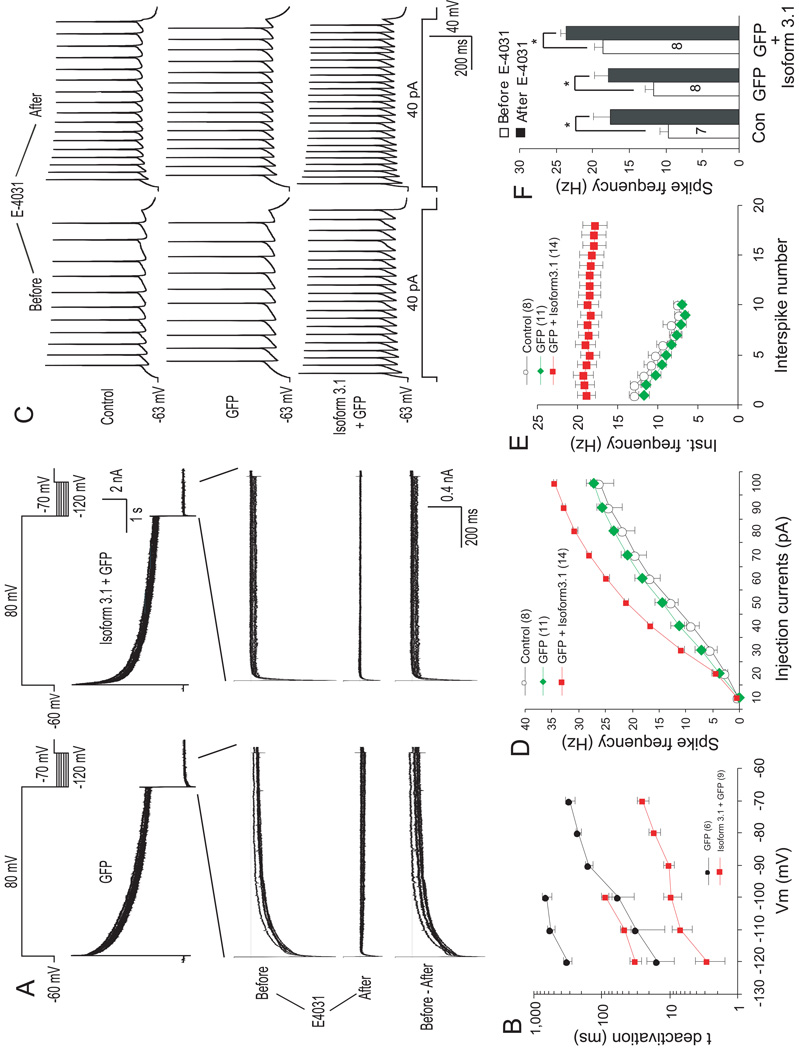
We next examined the functional role of KCNH2-3.1 in cortical neurons. Given that rodent CNS neurons express KCNH2-1A, but not KCNH2-3.1, KCNH2-3.1 introduced into these neurons would presumably complex with the endogenous KCNH2-1A, thereby changing the deactivation kinetics of the K+ current. In rat primary cortical neurons transfected with a GFP marker alone, application of a voltage protocol similar to that in Fig. 5E elicited a series of tail currents that were blocked by E-4031 (Fig. 6A). Subtraction of the currents after E-4031 treatment (middle) from control currents (upper) generated pure “KCNH2-1A-mediated” current (lower) that exhibited typical, slow deactivation kinetics (Fig. 6A, left). In contrast, application of the same protocol to neurons transfected with KCNH2-3.1 (plus GFP marker) elicited a dramatically reduced tail current with significantly faster deactivation (Fig. 6A, right). At −100mV, the introduction of KCNH2-3.1 reduced τ2 (initial fast component) and τ1 (slow component) of tail currents by 5.88 fold and 7.43 fold, respectively (Fig. 6B). Transfection of human KCNH2-1A into rat cortical neurons resulted in tail currents very similar to that of untransfected or GFP-transfected neurons (Compare Fig. 5A left, Supplementary Fig. 12C), further supporting that KCNH2-1A and KCNH2-3.1 have very different deactivation kinetics.
Figure 6. Effect of Isoform 3.1 on repolarization-induced tail currents and firing patterns in cortical neurons.
(A) KCNH2-mediated tail currents in GFP-(left) and Isoform 3.1-transfected (right) neurons before and after application of E4031. Upper plot diagrams the volatage protocol. Lower plot represents E-4031-sensitive current (i.e. hERG current) generated by subtracting pre and post-inhibitor currents. (B) Semilogarithmic plot of deactivation time constants of E-4031-sensitive currents at different re-polarizing voltages in transfected primary cortical neurons. Between −120 and −100 mV decay of E-4031-sensitive currents was fitted by double-exponential functions (lower curves: τ1, upper curves: τ2), whereas between −90 and −70 mV time course followed single-exponential functions (τ). (C) Effect of Isoform 3.1 over-expression in rat cortical neurons on action potential discharge evoked by long depolarizing pulse (40 pA, 1 sec) before (left) and after (right) application of E-4031. (D) Spike frequencies (number of spikes per second) versus applied depolarizing currents in transfected primary cortical neurons. (E) Effect of Isoform 3.1 on spike frequency adaptation depicted as instantaneous frequency (inverse of interpulse interval) versus the corresponding spikes interval evoked by a 4-pA depolarizing pulse. (F) Effect of E-4031 on repetitive action potential discharge of transfected cortical neurons. Error bars ±1 SEM. *: significantly different, p<0.01.
To test whether endogenous KCNH2-1A and exogenous KCNH2-3.1 form a functional complex, we examined action potentials induced by a step depolarization under the current-clamp conditions. In neurons transfected with KCNH2-3.1, a 1-sec depolarization induced significantly more spikes compared with control (non-transfecte) and GFP-transfected neurons (Fig. 6C). A systematic analysis showed a general increase in spike frequencies in KCNH2-3.1-expressing neurons injected with more than 30 pA depolarizing current (Fig. 6D). E-4031 significantly increased firing frequency, even in neurons expressing KCNH2-3.1 (Fig. 6C). Again, the firing behavior of rat neurons expressing human KCNH2-1A was very similar to that of control neurons (Supplementary Fig. 12C and D).
In addition to increasing firing frequency, expression of KCNH2-3.1 appeared to change the firing pattern from a typical, adapting to a non-adapting pattern (Fig. 6C). To better quantify this change, we measured “instantaneous frequency (IF, inverse of the first inter-pulse interval)” versus “spike number”. Indeed, both control and GFP- expressing neurons exhibited a gradual decline of IF, whereas IF remained stable during the entire course of depolarization in KCNH2-3.1-expressing neurons (Fig. 6E). Taken together, these results suggest that expression of KCNH2-3.1 in human PFC significantly increases neuronal excitability and offers a potential mechanism for persistent or overactive neuronal discharges observed in neurons of the PFC during working memory tasks48.
Discussion
We report a novel genetic association with schizophrenia within a small region of intron two of KCNH2, which was observed in a meta-analysis of five independent clinical samples. Our results also suggest that the molecular mechanism of genetic susceptibility relates to changes in gene expression and regulation of a novel isoform of KCNH2 with unique physiologic properties. The sequence of hypothesis-driven experiments and tests leading to this convergent evidence, from initial genetic screening to genetic association with aspects of human hippocampal structure and hippocampal and DLPFC function and gene expression to confirmatory experiments in tissue culture is summarized in Supplementary Figure S13. Though we report here significant association of several SNPs across independent samples and by meta-analysis, the exact risk structure may vary between populations while the regional location and overall effect on KCNH2 gene processing may remain consistent with what is described here. The splicing mechanism related to variation in this region of KCNH2 is yet to be determined (see Supplementary Note 2).
The region of interest on 7q36.1 has not, to date, been reported as a significant locus in the few genome wide association (GWA) studies of psychiatric illnesses50–52. These negative results may reflect the fact that these psychiatric GWA studies have relied largely on Affymetrix 5.0 SNP arrays, whose coverage is notably lacking over this genomic region‥ Conversely, p-values reported here would not be significant if we corrected for all SNPs in the genome. Scanning the genome with hundreds of thousands of SNPs and employing the necessary rigid statistical correction because of no prior probability of any SNP being positive is a popular strategy at the moment, as it makes no assumptions about biology or function. This strategy has the appeal of a level of statistical significance being clear and incontrovertible. However, statistical significance does not, in and of itself, imply biological significance nor does it necessarily identify the genes most likely to be important in unraveling new strategies for prevention and treatment.
In order to move beyond statistical association with clinical diagnosis and to obtain convergent evidence for association between KCNH2 and schizophrenia-related biology, we have performed a series of convergent experiments testing risk-associated SNPs on several intermediate biological phenotypes (Supplementary Figure S13). While we believe our deductive, hypothesis-driven strategy minimizes serendipity, it does involve a number of tests. Thus, the potential for spurious association because of multiple testing is important to consider. Three of the four SNPs showing association in a meta-analysis of 5 independent clinical samples were tested for association with biologic phenotypes related to risk for schizophrenia and to molecular processing of KCNH2 transcripts. A consistent pattern of allelic association was found in normal subjects, involving cognition and structural and functional imaging, and in expression of the 3.1 isoform in brain tissue from patients and controls of various ethnic backgrounds. The likelihood that by chance the same risk-associated alleles would predict variation in each of these independent phenotypes across these diverse samples and always in the direction of abnormality associated with illness is exceedingly remote.
Exploration of the potential electrophysiologic effects of elevated KCNH2-3.1 levels on neuronal repolarization and spike frequency has intriguing implications for both normal cortical function and the pathophysiology of psychosis. Cortical information processing is critically dependent on well-orchestrated, persistent neuronal firing within cortical circuits45. For example, neurons in the PFC exhibit sustained firing of high-frequency activity during the delay period that is thought to be critical for working memory48. The cellular mechanism for this “sustained” firing is not clear. We found that expression of KCNH2-3.1 in cultured cortical neurons results in a much faster deactivation of the hERG channels, leading to a marked increase in spike frequency and a conversion from adapting to predominantly non-adapting firing patterns. Such sustained firing patterns may be important for higher-order cognition and suggests a role for the primate-specific KCNH2-3.1 in normal human cognitive processing. However, this critical evolutionary change in activation kinetics may require specific titration of isoform abundances to achieve optimum signal-to-noise relationships. The 2.5-fold relative increase in KCNH2-3.1 in the schizophrenic brain might imbalance this titration, resulting in abnormally increased neuronal excitability, and disruption of neuronal firing patterns and regulation of signal-to-noise12.
The discovery of KCNH2-3.1 may also have therapeutic implications. Some typical and atypical antipsychotic drugs bind and inhibit KCNH2 with affinities comparable to their affinities for dopamine D2 receptors38. While D2 receptor affinity is thought to account for the therapeutic effects of antipsychotics, KCNH2 binding is responsible at least for side effects such as altered QT interval38 or even sudden cardiac failure53. Given that KCNH2 controls neuronal excitability and firing patterns, could the therapeutic effects of antipsychotic drugs also be related to their affinities for the brain-specific isoforms of KCNH2? Although the binding properties of antipsychotic drugs to KCNH2-3.1 remain to be tested, the unique structure of KCNH2-3.1, its role in non-adaptive firing, its low expression in heart, and elevated expression in brains of schizophrenia patients and genetic risk carriers, makes it reasonable to speculate that selective inhibition of KCNH2-3.1, but not KCNH2-1A, could improve the disorganized firing in schizophrenia brain without eliciting cardiac side effects.
In conclusion, we have identified and characterized a novel, primate-specific, brain-enriched potassium channel, and a potential genetic association of KCNH2 with risk for schizophrenia. . Healthy control carriers of risk alleles demonstrate a schizophrenia-like shift in cognitive traits (IQ/processing speed), in hippocampal volume and physiologic engagement during memory processing, and in prefrontal physiologic engagement during executive cognition. The mechanism of these associations appears to be related to genetic regulation of this novel isoform of KCNH2, which has unique electrophysiological properties and expressional dependence on risk-associated genotypes. Together, these results may provide a novel insight about the etiology of schizophrenia and a new direction for therapeutic discovery.
Methods
Genetic Association Cohorts
Five independent clinical samples were included in this study; two family based Caucasian samples of European ancestry (268 families) and three case controls samples also of European Ancestry (1158 cases, 1704 controls) (see supplementary methods for details). Postmortem brain tissue was taken from the CBDB Postmortem Brain Collection and Life Span Series consisting of tissue from: 31 schizophrenia patients, 69 control subjects, 39 fetal samples, and an additional 244 control subjects covering a range of ages (see Supplementary Methods and Data S8).
Candidate gene screening
10 candidate genes were selected from those previously reported as differentially expressed between schizophrenia patients and healthy controls16. SNPs tagging haplotype blocks determined from HapMap CEU population (release #15) and overlapping with part or all of the gene of interest were genotyped on a screening sample of 175 families (698 Caucasian subjects) (Supplementary Methods and Data S3). All SNPs were genotyped using 5’-exonuclease TaqMan assays as previously described54. Haplotype association testing was performed using family-based association test (FBAT)17, evaluated at α = 0.05 using simulation with 1000 iterations. and subjected to Bonferroni correction.
KCNH2 genotyping and resequencing
43 SNPs within a 65.2 Kb region of KCNH2 and NOS3 (chr7: 150280464 - 150345679) were typed. A total of 13.5Kb was resequenced in 48 schizophrenia patients, including 10.4Kb flanking rs1036145 (chr7:150,299,575-150,309,948) and 3.1Kb upstream of exon 3 (chr7:150,287,750-150,290) with high cross-species conservation in the UCSC genomes database (genome.ucsc.edu). Resequencing found 11 novel SNPs and no coding polymorphisms (supplementary methods for details).
Cognitive Testing
Depending on the cognitive test, data were available for between 230 and 330 normal individuals not part of the clinical association datasets. The tests included tasks aimed at assessing a wide range of cognitive features significantly affected in patients with schizophrenia and in their unaffected siblings.12–15. Factor analysis of 24 performance scores identified seven factors that explained 68% of the variance on these measures28. These 7 factors were used to assess genotype effects on cognitive task performance (see below) and are labeled as follows: Verbal Memory, Nback, Visual Memory, Processing Speed/IQ, Card Sorting, Attention, and Digit Span. Further information regarding the test measures included in each factor is provided in Supplementary Table S5.
Statistical genetics analysis
SNP association within families was analyzed by FBAT17 and by logistic regression in unrelated cases and controls (see supplementary methods). Meta-analyses were performed using the R package catmap20 based on fixed-effects estimates where the heterogeneity (Q-statistic) of genetic effects was not significant at α = 0.05 (markers 17, 19, 30, 31 and 33) and random-effects estimates (markers 16 and 20) where significant differences in genetic effects were found across samples (see supplementary methods).
Genotype effects on cognitive intermediate phenotypes were tested in control subjects using linear regression, adjusting for age and gender. Permutation testing based on 1000 replicates was used to examine the pattern of associations of SNPs M30, 31 and 33 with the cognitive factor scores to assess the likelihood that for each SNP the schizophrenia risk associated allele would be associated with relatively poorer performance on the same factor.
Quantitative real-time PCR (rt-PCR)
Details about the collection, screening, and dissection processes for human brain tissue have been described55 and demographic statistics about the brain specimens used here are provided in supplemental methods. Gene expression levels were measured by quantitative real-time RT-PCR. Probes were designed to differentiate KCNH2 isoforms through hybridizing to isoform-specific exons (Supplementary Methods).
Custom Illumina microarray analysis of fetal tissues
Microarray chips were generated in the NHGRI Microarray Core Facility from 44,544 70mer probes obtained from the Illumina Oligoset HEEBO (http://www.microarray.org/sfgf/heebo.do). RNA samples from the life span DLPFC collection (500 ng) were amplified and labeled using fluorescent dye (Cy5). Samples were each hybridized to microarrays simultaneously with a reference standard (labeled with Cy3) consisting of a pool of RNA from many brain tissue samples. Analysis was done using limma R package56.
Imaging Methods
All imaging subjects were Caucasian healthy volunteers (not part of the clinical association data) without significant differences in age, gender, IQ score, or education across genotype groups (Supplementary Data S5 for demographics). All analyses were done in SPM with SNPs effects assessed with random effects analyses. For gray matter volumes, we performed optimized Voxel Based Morphometry (VBM) following standardized methods with appropriate covariates as described previously22–24 (see supplementary methods) (n=141)
During 3T BOLD fMRI scanning described previously30, subjects performed a declarative memory (n=79) and an executive working memory task (N-back, n=178) to robustly engage the hippocampal formation30 and dorsolateral prefrontal cortex37, respectively (see supplementary methods). Random-effects analyses were performed on a priori defined regions of interest (ROIs) with genotype as a predictor (see Supplementary Methods). False discovery rate (FDR) corrections (α=0.05) were used to control for the expected proportion of false positives among suprathreshold voxels.
Isoform 3.1 Cloning and Expression
5’-RACE was performed using a 5’-RACE System (v2.0) as recommended by the manufacturer (Invitrogen). Template cDNA was made from commercially available total RNA from 10 human cell lines and tissues (Stratagene) (see supplementrary methods). Isoform3.1 cDNA was cloned by long PCR with specific primers and high fidelity DNA polymerase into vector pZero-Blunt (Invitrogen), and then subcloned into vector pcDNA3 (Invitrogen). HEK293 cells were transfected with the KCNH2, Isoform 3.1, and pCDNA3 vector, respectively, using standard methods. Protein expression was measured with Western Blot and immunocytochemistry using standard techniques as described in supplementary methods.
Electrophysiology
HEK293T cells and primary cortical neurons were cultured and transfected by nucleofection, as previously described57. Transfected cells were identified as GFP-positive under fluorescence microscopy, and whole-cell recordings performed, detailed in supplementary methods. Tail currents were obtained during 4 sec voltage steps shown in the upper panels of Fig. 5B, 5E and Fig. 6A, and were fitted by exponential functions and analyzed in detail, as detailed in Supplementary Methods. Transfected cortical neurons were also assessed for repetitive firing properties using a long depolarizing pulse (40 pA, 1 sec), as described in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Drs. John Hardy, Jamie Duckworth, and Parastoo Momeni for technical assistance with high G/C content sequencing. We also thank Drs. John Hardy, David Goldman, Amanda Law, and Wen Chen for their very helpful review of the manuscript. We thank Dr. Richard Straub and Michael Mayhew for their input on statistical genetics analysis, Max Barenboim for help with bioinformatics, and Drs. Mary Herman and Shruti Mitkus for their help with postmortem tissue. We are extremely grateful for the assistance of Guangping Liu and Sue Chen in the cloning and sequencing of Isoform 3.1. We also would like to thank Hans-Jürgen Möller and co-workers at the Dept. of Psychiatry, LMU Munich for their help with subject recruitment and evaluation. S.J. Huffaker was partially supported by the NIH/Cambridge University Health Science Scholars and MSTP Programs. Recruitment of the schizophrenic patients at LMU Munich was supported by GlaxoSmithKline. Human fetal tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD.
Reference
- 1.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 2.Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG, Ioannidis JP. Establishment of genetic associations for complex diseases is independent of early study findings. Eur J Hum Genet. 2004;12:762–769. doi: 10.1038/sj.ejhg.5201227. [DOI] [PubMed] [Google Scholar]
- 3.Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nat Genet. 2000;26:151–157. doi: 10.1038/79866. [DOI] [PubMed] [Google Scholar]
- 4.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grarup N, et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–3111. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 6.Callicott JH, et al. Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry. 2003;160:709–719. doi: 10.1176/appi.ajp.160.4.709. [DOI] [PubMed] [Google Scholar]
- 7.Cannon TD, et al. The inheritance of neuropsychological dysfunction in twins discordant for schizophrenia. Am J Hum Genet. 2000;67:369–382. doi: 10.1086/303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan MF, et al. Relative risk of attention deficits in siblings of patients with schizophrenia. Am J Psychiatry. 2000;157:1309–1316. doi: 10.1176/appi.ajp.157.8.1309. [DOI] [PubMed] [Google Scholar]
- 9.Egan MF, et al. Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry. 2001;50:98–107. doi: 10.1016/s0006-3223(01)01133-7. [DOI] [PubMed] [Google Scholar]
- 10.Honea RA, et al. Is gray matter volume an intermediate phenotype for schizophrenia? A voxel-based morphometry study of patients with schizophrenia and their healthy siblings. Biol Psychiatry. 2008;63:465–474. doi: 10.1016/j.biopsych.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winterer G, et al. Prefrontal broadband noise, working memory, and genetic risk for schizophrenia. Am J Psychiatry. 2004;161:490–500. doi: 10.1176/appi.ajp.161.3.490. [DOI] [PubMed] [Google Scholar]
- 12.Winterer G, Weinberger DR. Genes, dopamine and cortical signal-to-noise ratio in schizophrenia. Trends Neurosci. 2004;27:683–690. doi: 10.1016/j.tins.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Straub RE, Weinberger DR. Schizophrenia genes - famine to feast. Biol Psychiatry. 2006;60:81–83. doi: 10.1016/j.biopsych.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Peirce TR, et al. Convergent evidence for 2',3'-cyclic nucleotide 3'-phosphodiesterase as a possible susceptibility gene for schizophrenia. Arch Gen Psychiatry. 2006;63:18–24. doi: 10.1001/archpsyc.63.1.18. [DOI] [PubMed] [Google Scholar]
- 15.Talkowski ME, Chowdari K, Lewis DA, Nimgaonkar VL. Can RGS4 polymorphisms be viewed as credible risk factors for schizophrenia? A critical review of the evidence. Schizophr Bull. 2006;32:203–208. doi: 10.1093/schbul/sbj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabakaran S, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 17.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 18.The International HapMap Project. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 19.Cloninger CR, et al. Genome-wide search for schizophrenia susceptibility loci: the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet. 1998;81:275–281. [PubMed] [Google Scholar]
- 20.Nicodemus KK. Catmap: case-control and TDT meta-analysis package. BMC bioinformatics. 2008;9:130. doi: 10.1186/1471-2105-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 22.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen NC, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 24.Ioannidis JP, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International journal of epidemiology. 2008;37:120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 25.Guasti L, et al. Expression pattern of the ether-a-go-go-related (ERG) family proteins in the adult mouse central nervous system: evidence for coassembly of different subunits. J Comp Neurol. 2005;491:157–174. doi: 10.1002/cne.20721. [DOI] [PubMed] [Google Scholar]
- 26.Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122(Pt 4):593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 27.Weinberger DR, et al. Prefrontal neurons and the genetics of schizophrenia. Biol Psychiatry. 2001;50:825–844. doi: 10.1016/s0006-3223(01)01252-5. [DOI] [PubMed] [Google Scholar]
- 28.Genderson MR, et al. Factor analysis of neurocognitive tests in a large sample of schizophrenic probands, their siblings, and healthy controls. Schizophr Res. 2007;94:231–239. doi: 10.1016/j.schres.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Hariri AR, Weinberger DR. Imaging genomics. Br Med Bull. 2003;65:259–270. doi: 10.1093/bmb/65.1.259. [DOI] [PubMed] [Google Scholar]
- 30.Hariri AR, et al. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bookheimer SY, et al. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan G, et al. Neural correlates of the emergence of consciousness of thirst. Proc Natl Acad Sci U S A. 2003;100:15241–15246. doi: 10.1073/pnas.2136650100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davachi L, Goldman-Rakic PS. Primate rhinal cortex participates in both visual recognition and working memory tasks: functional mapping with 2-DG. Journal of neurophysiology. 2001;85:2590–2601. doi: 10.1152/jn.2001.85.6.2590. [DOI] [PubMed] [Google Scholar]
- 34.Royall DR, et al. Executive control function: a review of its promise and challenges for clinical research. A report from the Committee on Research of the American Neuropsychiatric Association. The Journal of neuropsychiatry and clinical neurosciences. 2002;14:377–405. doi: 10.1176/jnp.14.4.377. [DOI] [PubMed] [Google Scholar]
- 35.Gray JR, Chabris CF, Braver TS. Neural mechanisms of general fluid intelligence. Nat Neurosci. 2003;6:316–322. doi: 10.1038/nn1014. [DOI] [PubMed] [Google Scholar]
- 36.Duncan J, et al. A neural basis for general intelligence. Science. 2000;289:457–460. doi: 10.1126/science.289.5478.457. [DOI] [PubMed] [Google Scholar]
- 37.Callicott JH, et al. Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex. 2000;10:1078–1092. doi: 10.1093/cercor/10.11.1078. [DOI] [PubMed] [Google Scholar]
- 38.Kongsamut S, Kang J, Chen XL, Roehr J, Rampe D. A comparison of the receptor binding and HERG channel affinities for a series of antipsychotic drugs. Eur J Pharmacol. 2002;450:37–41. doi: 10.1016/s0014-2999(02)02074-5. [DOI] [PubMed] [Google Scholar]
- 39.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wimmers S, Bauer CK, Schwarz JR. Biophysical properties of heteromultimeric erg K+ channels. Pflugers Arch. 2002;445:423–430. doi: 10.1007/s00424-002-0936-4. [DOI] [PubMed] [Google Scholar]
- 41.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 42.Morais Cabral JH, et al. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 43.Bracci E, Vreugdenhil M, Hack SP, Jefferys JG. On the synchronizing mechanisms of tetanically induced hippocampal oscillations. J Neurosci. 1999;19:8104–8113. doi: 10.1523/JNEUROSCI.19-18-08104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazhenov M, Timofeev I, Steriade M, Sejnowski TJ. Potassium model for slow (2–3 Hz) in vivo neocortical paroxysmal oscillations. Journal of neurophysiology. 2004;92:1116–1132. doi: 10.1152/jn.00529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canolty RT, et al. High gamma power is phase-locked to theta oscillations in human neocortex. Science. 2006;313:1626–1628. doi: 10.1126/science.1128115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiesa N, Rosati B, Arcangeli A, Olivotto M, Wanke E. A novel role for HERG K+ channels: spike-frequency adaptation. J Physiol. 1997;501(Pt 2):313–318. doi: 10.1111/j.1469-7793.1997.313bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sacco T, Bruno A, Wanke E, Tempia F. Functional roles of an ERG current isolated in cerebellar Purkinje neurons. Journal of neurophysiology. 2003;90:1817–1828. doi: 10.1152/jn.00104.2003. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Heterogeneity in the pyramidal network of the medial prefrontal cortex. Nat Neurosci. 2006;9:534–542. doi: 10.1038/nn1670. [DOI] [PubMed] [Google Scholar]
- 49.Spector PS, Curran ME, Keating MT, Sanguinetti MC. Class III antiarrhythmic drugs block HERG, a human cardiac delayed rectifier K+ channel. Open-channel block by methanesulfonanilides. Circulation research. 1996;78:499–503. doi: 10.1161/01.res.78.3.499. [DOI] [PubMed] [Google Scholar]
- 50.O'Donovan MC, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 51.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan PF, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Witchel HJ, Hancox JC, Nutt DJ, Wilson S. Antipsychotics, HERG and sudden death. Br J Psychiatry. 2003;182:171–172. doi: 10.1192/bjp.182.2.171-a. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 55.Lipska BK, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Human molecular genetics. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 56.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 57.Nagappan G, et al. Control of extracellular cleavage of ProBDNF by high frequency neuronal activity. Proc Natl Acad Sci U S A. 2009;106:1267–1272. doi: 10.1073/pnas.0807322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23:774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.