Abstract
The S. pombe protection of telomeres 1 (SpPot1) protein recognizes the 3′ single-stranded ends of telomeres and provides essential protective and regulatory functions. The ssDNA-binding activity of SpPot1 is conferred by its ssDNA-binding domain, Pot1-DBD (residues 1-389), which can be further separated into two distinct domains, Pot1pN (residues 1-187) and Pot1pC (residues 188-389). Here we show that Pot1pC, like Pot1pN, can function independently of Pot1-DBD and binds specifically to a minimal nonameric oligonucleotide, d(GGTTACGGT), with a KD of 400 +/- 70 nM (specifically recognized nucleotides in bold). NMR chemical shift perturbation analysis indicates that the overall structures of the isolated Pot1pN and Pot1pC domains remain intact in Pot1-DBD. Furthermore, alanine scanning reveals modest differences in the ssDNA-binding contacts provided by isolated Pot1pN and within Pot1-DBD. Although the global character of both Pot1pN and Pot1pC is maintained in Pot1-DBD, chemical shift perturbation analysis highlights localized structural differences within the G1/G2 and T3/T4 binding pockets of Pot1pN in Pot1-DBD, which correlate with its distinct ssDNA-binding activity. Furthermore, we find evidence for a putative interdomain interface on Pot1pN that mediates interactions with Pot1pC that ultimately result in the altered ssDNA-binding activity of Pot1-DBD. Together, these data provide insight into the mechanisms underlying the activity and regulation of SpPot1 at the telomere.
Keywords: telomeres, ssDNA-binding domain, end-protection, OB fold, Pot1
Eukaryotic chromosomes terminate in a conserved 3′ single-stranded DNA overhang. If left unattended, this overhang triggers the activation of the DNA damage response leading to chromosomal abnormalities that halt cellular proliferation (1). This outcome is circumvented by the protective functions of specialized telomere-associated proteins collectively referred to as the telomere end-protection (TEP) family. In addition to their protective functions, TEP proteins also participate in numerous regulatory functions at telomeres, including controlling the nucleotide addition activity of telomerase (2-7), coordinating lagging (3′ → 5′) strand synthesis (8), fixing the termination point of lagging strand resection (9) and controlling the formation of higher order telomere structure (i.e., t-loops and G-quartet structures) (10, 11). As a result of the critical nature of these regulatory activities, TEP proteins are essential for normal cellular proliferation and long-term survival.
A universally shared feature of TEP proteins is their ability to specifically bind to the 3′ ssDNA ends of telomeres through a conserved ssDNA-binding domain (DBD). Structural and bioinformatics (12) studies of TEP DBDs from TEBPα/β (13-15) in ciliates, Cdc13 in Saccharyomyces cerevisiae (16), and the Pot1 proteins from Schizosaccharyomyces pombe (17) and humans (18) reveals that TEP DBDs are structural homologs, containing the conserved OB (Oligonucleotide/Oligosaccharide-Binding) superfold (19, 20). The characterization of a wide array of proteins and individual domains reveals a trend in which ssDNA-binding activity of TEP proteins is mediated by multiple OB-folds, a feature also shared by other ssDNA-binding proteins, such as E. coli SSB (21) and human replication protein A (RPA) (22).
The DBD in S. pombe protection of telomeres 1, Pot11-389 (hereafter referred to as Pot1-DBD) is formed from one structurally characterized N-terminal OB-fold domain, Pot1pN (17, 23), and a second uncharacterized domain, Pot1pC (24, 25). Pot1-DBD binds specifically to a minimal dodecameric oligonucleotide, d(GGTTACGGTTAC), with a tight binding affinity of 46 pM at 50 mM NaCl (specifically recognized nucleotides in bold) (24). The Pot1pN domain of Pot1-DBD can be independently expressed and exhibits altered ssDNA-binding activity relative to Pot1-DBD. Pot1pN binds a minimal oligonucleotide consisting of a single repeat of the conserved hexameric S. pombe telomere sequence (25), d(GGTTAC) with an affinity of 190 nM at 50 mM NaCl (specifically recognized nucleotides shown in bold) (23). Structural characterization of this domain reveals a ssDNA-recognition interface composed of a single face of the OB-fold β-barrel (consisting of β3 and β5) and two structured loops (Loop12 and Loop45), that together form a well-defined binding groove (Supplemental Figure 1). This ssDNA-binding interface contains three distinct binding pockets that constrain the bound oligonucleotide in a highly compacted orientation: the G1/G2, T3/T4 and A5/C6 binding pockets (Supplemental Figure 1). The bound oligonucleotide interacts with the protein via internucleotide stacking interactions between the protein and G1/G2, T3/T4 and A5/C6 as well as a complex array of hydrogen bonds formed primarily between the protein and the nucleotide bases. These interactions are augmented by the presence of non-canonical hydrogen bonding interactions between G1/T3 and G2/T4 (17). The extensive biochemical and structural information available on isolated Pot1pN is contrasted by the absence of corresponding data regarding the ssDNA-binding activity of Pot1pC. Comparing and contrasting the individual ssDNA-binding characteristics of these independent domains with that of Pot1-DBD will provide a means towards understanding how these two domains cooperatively function within Pot1-DBD to facilitate telomere recognition.
To further our biochemical understanding of Pot1pC, we have cloned, expressed and assessed its ability to bind ssDNA independently of Pot1-DBD. Pot1pC (residues 178-389), like Pot1pN, is a well-folded domain that can be readily expressed and purified from E. coli. Furthermore, this domain is biochemically active and specifically binds a minimal nonamer oligonucleotide, d(GGTTACGGT), with a KD of 400 nM at 50 mM NaCl (specifically recognized nucleotides are indicated in bold). These results indicate, that like Pot1pN, the ssDNA-binding activity of Pot1pC is distinct from that exhibited by Pot1-DBD. To understand how Pot1pN and Pot1pC synergize within Pot1-DBD to facilitate the recognition of telomeric ssDNA, we performed a structural and biochemical comparison of Pot1-DBD with the isolated domains using alanine scanning mutagenesis and NMR minimal chemical shift perturbation analysis. Alanine scanning of ssDNA-contacting residues within Pot1pN and Pot1-DBD indicate that the overall ssDNA-binding interface defined by the Pot1pN/d(GGTTAC) complex is generally preserved in Pot1-DBD. Furthermore, MCSP studies indicate that the global backbone amide conformations of isolated Pot1pN and Pot1pC are retained in Pot1-DBD. In spite of these conserved characteristics, a detailed MCSP analysis of Pot1-DBD detects the localized perturbation of residues within the G1/G2 and T3/T4 binding pockets of the Pot1pN domain of Pot1-DBD that appear to contribute to the different ssDNA-binding activities of these two proteins. Finally, we identify a putative domain/domain interface mediating interactions between Pot1pN and Pot1pC within Pot1-DBD. Mutagenesis experiments support a role for this interface in dictating the specific nucleotide recognition demonstrated by Pot1-DBD and indicate that the coupling of these two domains leads to the non-additive binding affinity observed for Pot1-DBD. Taken together, these results suggest that the ssDNA-binding activities of Pot1pN and Pot1pC in isolation are not fully retained in Pot1-DBD, but are subtly altered to produce the ssDNA-binding activity exhibited by Pot1-DBD.
Experimental Procedures
Chemicals, reagents and proteins
All chemicals and reagents were obtained from Fisher Scientific unless otherwise indicated. Chromatography materials were purchased from GE Healthcare. Oligonucleotides were commercially synthesized and reversed-phase HPLC purified (Integrated DNA technologies). The plasmid encoding Pot1pN was graciously provided by Professor Thomas Cech (23). All mutagenesis performed on Pot1pN and Pot1-DBD was performed using the reagents and protocols provided in the Quikchange Mutagenesis Kit (Invitrogen). NMR studies were performed with complexes of 15N-labeled protein and their respective cognate ssDNA oligonucleotides at 1.25 mM (Pot1pN), 350 μM (Pot1pC) and 400 μM (Pot1-DBD). All samples contained an excess of 0.5 molar equivalents of ssDNA to ensure that all available protein was present in a homogeneous, ssDNA-bound complex. Pot1pN studies were conducted in NMR sample buffer 1 (50 mM potassium phosphate, pH 6.13, 50 mM sodium chloride, 1 mM deuterated DTT, 10% deuterium oxide), while Pot1pC and Pot1-DBD studies were performed in NMR sample buffer 2 (100 mM triethanolamine, pH 8.5, 100 mM potassium chloride, 10 mM DTT, 10% deuterium oxide).
Pot1pC expression and purification
The 26 kDa, C-terminally hexahistidine tagged Pot1pC domain (residues 178-389) from the S. pombe Pot1 protein was cloned into pET21b (Novagen) and expressed in BL21(DE3)C41 E. coli (26). E. coli were grown at 37°C in 2×YT media containing 50 μg/ml ampicillin to an OD600 of 0.5-0.6. Cells were then placed on ice for one hour after which protein production was induced by addition of 0.5 mM IPTG and continued shaking at 220 RPM for 18 hours at 15°C. Cells were harvested by centrifugation following the induction period.
For purification, cells were thawed on ice, resuspended in lysis buffer (25 mM Tris, pH 8.0, 50 mM potassium phosphate, 300 mM sodium chloride, 2 mM imidazole, 200 μg/ml lysozyme, 10% glycerol, 3 mM BME and 10 mM magnesium chloride), lysed by sonication and treated with 40 μg/ml DNase I (Roche) for 30 minutes at 4°C. Cell debris were removed by centrifugation and the soluble fraction was batch incubated with Ni-charged Fast Flow Chelating Sepaharose (GE Healthcare) at 4°C for ∼18 hours. Following elution with 600 mM imidazole, Pot1pC-containing fractions were extensively dialyzed at 4°C against 50 mM potassium phosphate, pH 8.0, 150 mM sodium chloride and 3 mM BME. Dialyzed Pot1pC was then further purified by Superdex S200 16/60 (GE Healthcare) size exclusion chromatography (SEC) in the same buffer containing 5 mM BME. Monomeric Pot1pC was obtained in ∼2-3 mg/L purified yields and was judged > 95% pure by Coomassie brilliant blue stained SDS-PAGE (Supplemental Figure 2, inset). Pot1pC was routinely concentrated to 200 μM, flash frozen in liquid nitrogen, and stored at -70° C in gel filtration buffer.
Expression, purification and preparation of uniformly 15N-labeled Pot1pC for NMR studies were performed as described above with the following modifications. Uniform 15N-labelling was performed by expressing Pot1pC in modified minimal M9 media containing 1.5 g/L of 99% 15(NH4)2SO4 with purified yields of ∼1 mg/L (27).
Expression and purification of Pot1pN and alanine mutant constructs
The 22 kDa, N-terminally hexahistidine tagged Pot1pN domain from the S. pombe Pot1 protein and various alanine mutants were expressed in BL21(DE3) E. coli (26), purified and stored according to the protocols established by Lei et al. (23) with final purified yields of ∼25 mg/L. Expression, purification and preparation of uniformly 15N-labeled Pot1pN for NMR studies were performed as described previously (28).
Expression and purification of Pot1-DBD and alanine mutant constructs
The 47 kDa, C-terminally hexahistidine tagged Pot1-DBD domain from the S. pombe Pot1 protein and various alanine mutants was expressed in BL21(DE3)C41 E. coli (26), purified and stored according to the protocols established by Croy et al. (24) with final purified yields of ∼3-4 mg/L. Expression, purification and preparation of uniformly 15N-labeled Pot1-DBD for NMR studies were performed as for unlabeled Pot1-DBD with the following modifications. Due to complications arising from the misfolding and aggregation of Pot1-DBD when expressed in minimal media, it was necessary to co-express Pot1-DBD with the folding chaperones GroEL and GroES (data not shown). BL21(DE3)C41 E coli (26) were prepared by transformation with pGRO7 (Takara Bio Inc.) which contains a chloramphenicol selection marker with GroEL and GroES under control of an L-arabinose inducible promoter. BL21(DE3)C41_pGRO7 cells were then transformed with pET21b which contains an ampicillin selection marker with Pot1-DBD under an IPTG inducible promoter. E. coli were grown at 37°C in M9 minimal media containing 50 μg/ml ampicillin, 10 μg/ml chloramphenicol, 0.5 mg/ml L-arabinose to an OD600 of 0.5-0.6. Cells were immediately placed on ice for one hour after which Pot1-DBD expression was induced by addition of 0.5 mM IPTG and continued shaking at 220 RPM for 6 hours at 37°C. Uniformly 15N-labelled Pot1-DBD was extracted and purified in yields of ∼2-3 mg/L as previously described (24).
Electrophoretic mobility shift assay (EMSA) of the ssDNA binding activity of Pot1pC
All oligonucleotides (Table 1) were 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase according to standard protocols (Invitrogen). Free [γ-32P]ATP not incorporated onto the oligonucleotide was removed by passage through a G25 spin column (GE Healthcare). Labeled oligonucleotides were stored at 4°C and typically used within one week of labeling. Active concentration of Pot1pC was determined using an EMSA assay with concentrations of ssDNA (6.7 μM) which were approximately 15-fold above the estimated KD typically measured for Pot1pC. Pot1pC was serially diluted over a broad concentration range (7 nM – 100 μM) in binding buffer (20 mM potassium phosphate, pH 8.0, 50 mM sodium chloride, 3 mM BME, 10% glycerol) and complexes were formed with 32P-radiolabeled ssDNA at 4°C for one hour. Pot1pC/ssDNA complexes were loaded onto 5% non-denaturing polyacrylamide gels containing 1× TBE (89 mM Trizma Base, 89 mM boric acid and 2 mM ethylenediaminetetraacetic acid) and 5% glycerol and run in 1× TBE buffer at 100 volts for 30 minutes at 4°C. Following electrophoresis, gels were dried on filter paper (Whatman), exposed to phosphorimaging screens and visualized on a Typhoon Phosphorimager (GE Healthcare). Data were quantified using ImageQuant version 5.1 (GE Healthcare) and KaleidaGraph version 4.0 (Synergy Software). EMSA experiments used to determine the KD were performed in an identical fashion with the following differences: The concentration of ssDNA (typically ∼100 pM) was kept ∼100-fold below the measured KD and Pot1pC was diluted over a lower concentration range (8 nM - 100 μM). EMSA data was collected using triplicate independent experiments, each performed on separate days with different batches of protein. Raw data from each trial was quantified by phosphorimaging, converted to normalized fraction bound (Fbound) and globally fit using non-linear curve regression to a standard two-state binding model to calculate the KD. For the weaker Pot1pC/ssDNA interactions complete shifting of the labeled oligonucleotide could not be achieved due to the limitations in the maximum stable concentration of Pot1pC (200 μM). Therefore, a lower KD limit for these binding reactions was determined by the inclusion of a synthetic limiting data point for saturation at high Pot1pC concentration (10 mM) if binding greater than 20% was observed at the highest concentrations. Errors in the reported KD values are the standard error of the mean extracted from the global fits and are a measure of the reproducibility of the experiment. However, due to the limitations of EMSA, we have conservatively considered changes of less than two fold insignificant.
Table 1.
KD and free energy changes for Pot1pC bound to telomeric ssDNA oligonucleotides of varying length.
| Oligonucleotide Name | Oligonucleotide Sequence | KD (nM)a |
ΔΔG (kcal/mol)b |
Fold Changec |
|---|---|---|---|---|
| 12mer | GGTTACGGTTAC | 700±200 | 0.5±0.1 | 2 |
| 11mer-3′ | GGTTACGGTTA | 600±100 | 0.4±0.01 | 2 |
| 10mer-3′ | GGTTACGGTT | 300±40 | -0.3±0.01 | 1 |
| 9mer-3′ | GGTTACGGT | 400±70 | --- | --- |
| 8mer-3′ | GGTTACGG | 1900±300 | 1 ±2 | 5 |
| 7mer-3′ d | GGTTACG | 10500±1900 | >3 | 26 |
| 6mer | GGTTAC | NDB | --- | --- |
| 11mer-5′ d | GTTACGGTTAC | 3200±400 | >2 | 8 |
| 10mer-5′ d | TTACGGTTAC | 11000±1500 | >3 | 30 |
| 9mer-5′ d | TACGGTTAC | 29000±4600 | >3 | 70 |
| 8mer-5′ d | ACGGTTAC | 15000±2,000 | >3 | 40 |
| 7mer-5′ d | CGGTTAC | 35000±8,000 | >4 | 90 |
NDB = no detectable binding was observed for this oligonucleotide
KD values were obtained from three independent EMSA experiments conducted at 50 mM sodium chloride, 20 mM potassium phosphate, 3 mM BME, pH 8.0 at 4° C. Errors represent the standard error of equilibrium binding constants obtained from three independent binding experiments.
ΔΔG = RTln(KD/KD9mer3′), R = 1.9872 cal mol-1 K-1, T = 277.15 K
Fold difference is relative to the 9mer-3′ sequence.
Saturation was not observed for these weak Pot1pC/ssDNA interactions. KD values were calculated using a synthetic data point as described in the materials and methods and represent a lower limit for binding.
Filter binding analysis of the ssDNA binding activity of Pot1pN, Pot1-DBD and alanine mutant constructs
Filter binding assays to determine active concentration and apparent binding affinity for Pot1pN and Pot1-DBD were carried out essentially as described in Lei et al. (23) and Croy et al. (24) using a dual layer filter system. Filter binding assays for Pot1pN were conducted with binding buffer containing 15 mM NaCl. However, due to sensitivity limitations arising from the extremely tight affinity of Pot1-DBD for d(GGTTACGGTTAC) (the binding is not measurable at 15 mM and is 46 pM at 50 mM NaCl), filter binding experiments were conducted at increased NaCl concentrations (400 mM). These conditions attenuate the apparent binding affinity to reasonably detectable levels. Briefly, filter binding assays used to determine KD values were performed with concentrations of ssDNA kept at least 10-fold below the respective non-activity corrected KD values of Pot1pN and Pot1-DBD (∼100 pM for both Pot1pN and Pot1-DBD). Raw data from each binding experiment was quantified by phosphorimaging and converted to fraction bound (Fbound) and globally fit using non-linear curve regression to a standard two-state binding model to calculate the KD. Saturation of the nitrocellulose filters occurred at protein concentrations of 10 μM; as such, complete saturation of binding for the weaker Pot1pN/ssDNA and Pot1-DBD/ssDNA interactions could not be achieved. Therefore, a lower KD limit was determined by inclusion of a synthetic limiting data point at high protein concentration. Errors in the KD values represent the standard error of the mean extracted from the global fits and are a measure of the reproducibility of the experiment. However, due to the limitations of the filter binding experiment, we have conservatively considered changes of less than two fold insignificant.
1H/15N minimal chemical shift perturbation (MCSP) analysis of the Pot1/oligonucleotide complexes
Varian Biopack gradient-selected, sensitivity-enhanced 15N-1H HSQC spectra for the Pot1pN/d(GGTTAC) complex were collected on a Varian Inova 600 equipped with a cold probe. Varian Biopack gradient-selected, sensitivity-enhanced TROSY 15N-HSQC spectra for the Pot1pC/d(GGTTACGGT) and Pot1-DBD/d(GGTTACGGTTAC) complexes were collected on a Varian Inova 800 or 900 MHz spectrometer equipped with a cold or room temperature probe, respectively. 15N-HSQC pulse sequences were derived from Varian BioPack pulse sequences with minor modifications. Acquired spectra were analyzed using CCPNMR analysis v2.0.6 (29). Values for the minimal chemical shift perturbations were calculated according to the following equation (30):
| (1) |
Results
Pot1pC represents an autonomous domain present in Pot1-DBD
Full telomeric ssDNA-binding activity of S. pombe Pot1 is conferred by residues 1-389 which form an independent, biochemically active ssDNA-binding domain, Pot1-DBD (Figure 1A) (24). Pot1-DBD contains one structurally characterized OB-fold domain, Pot1pN (17) followed by a second, uncharacterized domain, Pot1pC (Figure 1A). Although this domain contains no significant sequence similarity to other members of the TEP family, secondary structure analysis suggests it may also adopt an OB fold structure (24, 25). Building upon the notion that Pot1pN can be expressed independently of Pot1-DBD and specifically binds GT-rich ssDNA, we set forth to determine whether or not Pot1pC is autonomous or requires association with Pot1pN for activity. We identified two potential constructs of Pot1pC for cloning and protein expression tests: residues 178-389 and residues 189-389. Expression tests revealed that the construct spanning residues 178-389 yielded more soluble protein when compared with the one spanning residues 189-389. Therefore, the construct spanning residues 178-389 was further characterized as Pot1pC. A C-terminal hexahistidine tag was added to Pot1pC to facilitate purification via affinity chromatography followed by gel filtration. Monomeric Pot1pC could be concentrated to 5-10 mg/mL without signs of oligomerization or aggregation/precipitation (data not shown), suggesting that this construct is well behaved in solution and represents a stable, independent domain of Pot1-DBD.
Figure 1.
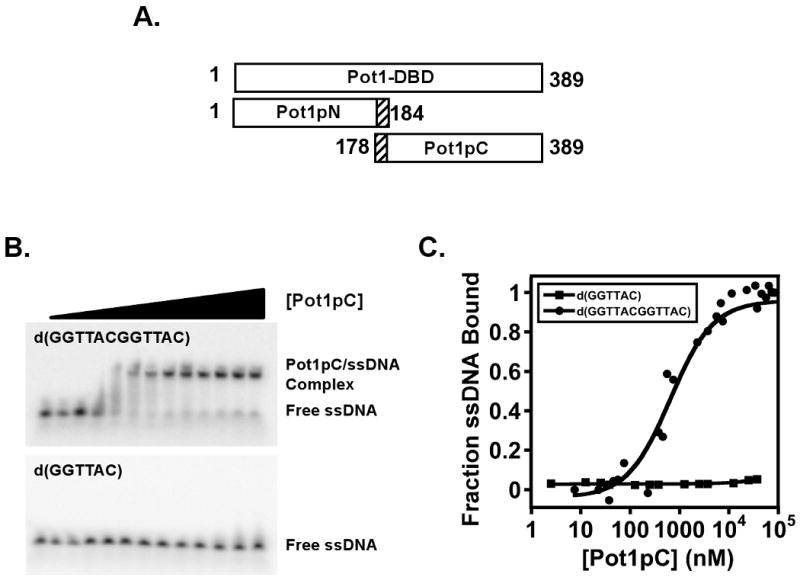
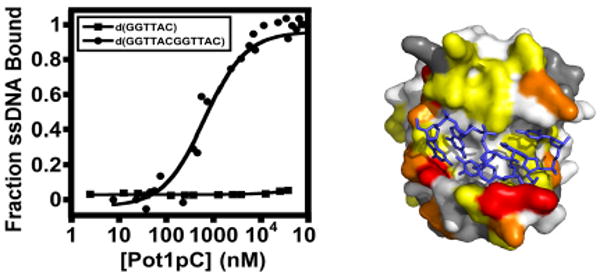
Constructs used and characterization of the ssDNA-binding activity of Pot1pC using EMSA/gel shift assays. A) Residue boundary limits for the ssDNA-binding domain of S. pombe Pot1, Pot1-DBD, and the independently expressed Pot1pN and Pot1pC domains are given. B) Representative data from an EMSA assay showing the binding of Pot1pC to d(GGTTACGGTTAC) (top panel), but not to d(GGTTAC) (lower panel). C) Normalized data collected from triplicate independent EMSA studies of Pot1pC binding to d(GGTTACGGTTAC) (filled circles) and binding to d(GGTTAC). Data are plotted as a function of fraction of each respective oligonucleotide bound versus Pot1pC concentration (nM).
A minimum of nine nucleotides is required for high affinity Pot1pC-ssDNA binding
Pot1-DBD and Pot1pN recognize the conserved hexameric d(GGTTAC) repeats within S. pombe telomeres, either singly (Pot1pN/d(GGTTAC)) or in tandem (Pot1-DBD/d(GGTTACGGTTAC)) (23, 24). These observations suggest that, if the ssDNA-binding properties of Pot1-DBD are an additive function of its respective domains, then the Pot1pC domain of Pot1-DBD should recognize the hexameric d(GGTTAC) oligonucleotide. Surprisingly, we find that Pot1pC exhibits very weak affinity for d(GGTTAC) (KD ≫ 40 μM). However, when we tested the binding of the Pot1-DBD minimal sequence, d(GGTTACGGTTAC), to Pot1pC, we found a significant increase (> 57-fold) in binding affinity for this longer oligonucleotide (700 +/- 200 nM) (Figure 1BC and Table 1). This binding is specific for a GT-rich telomeric sequence, as no affinity was observed to the complementary C-rich sequence, d(CCAATGCCCAATGC) (data not shown).
The minimal binding sequence for Pot1pC was determined using a series of truncated oligonucleotides based on d(GGTTACGGTTAC) oligonucleotide. Table 1 shows the effects on binding affinity resulting from progressive individual truncations made from either the 5′ or 3′ end of d(GGTTACGGTTAC). Removal of the terminal 3′ nucleotides (TAC-3′) from d(GGTTACGGTTAC) had a negligible impact (< 2-fold) on the apparent binding affinity. However, continued 3′ truncation resulted in significantly weakened binding affinities (5-fold and 25-fold, respectively, for further one- and two-nucleotide truncations) (Table 1). In contrast, removal of 5′ nucleotides from d(GGTTACGGTTAC) was not tolerated as removal of the 5′ guanosine resulted in an 8-fold decrease in binding affinity (Table 1). These results indicate that Pot1pC recognizes a minimal nonameric oligonucleotide sequence of d(GGTTACGGT) with an apparent KD of 400 +/- 70 nM (Table 1). In light of these results we examined whether the non-additive characteristics of Pot1-DBD binding to d(GGTTACGGTTAC) results from the truncation of a preferred oligonucleotide containing a complete Pot1pN and Pot1pC minimal binding sequence. These studies revealed minimal changes in binding affinity with either the 15 nucleotide sequence d(GGTTACGGTTACGGT) or other longer sequences (Supplementary Table 1). Taken together these data support a model in which the individual biochemical functions of Pot1pN and Pot1pC are non-additive in the context of Pot1-DBD.
Positions 2 and 7 are specifically recognized by Pot1pC in d(GGTTACGGT)
To determine which nucleotides in d(GGTTACGGT) are specifically recognized by Pot1pC, individual bases were substituted with their corresponding complement, (G⇔C) and (A⇔T). Table 2 and Figure 2 show the effects on apparent binding affinity of Pot1pC for d(GGTTACGGT) resulting from the series of individual complementary nucleotide substitutions. In order to maintain consistency with the Pot1-DBD studies (24) we set the thermodynamic threshold for specific nucleotide recognition at ΔΔG of > 1 kcal/mol. Using this criterion, we found that substitutions made at positions 2 and 7 resulted in significant changes in apparent binding affinity, while the remaining nucleotides were minimally affected (ΔΔG < 1 kcal/mol) (Figure 2 and Table 2). These results indicate that the biochemically derived specificity profile for Pot1pC bound to the minimal nonamer oligonucleotide can be represented as d(GGTTACGGT) (specifically recognized nucleotides in bold). Combined with binding affinity data for Pot1pC in isolation, the nucleotide specificity results indicate that the ssDNA-binding characteristics observed for Pot1-DBD do not simply result from the additive functions of its constitutive domains, Pot1pC and Pot1pN.
Table 2.
KD and free energy changes for Pot1pC bound d(GGTTACGGT) complement mutant oligonucleotides
| Oligonucleotide Name | Oligonucleotide Sequence | KD (nM)a | Fold Changeb |
|---|---|---|---|
| 9mer-3′ | GGTTACGGT | 400±60 | ---- |
| 9mer_G1C | CGTTACGGT | 1000±100 | 3 |
| 9mer_G2C | GCTTACGGT | 3000±600 | 8 |
| 9mer_T3A | GGATACGGT | 600±100 | 2 |
| 9mer_T4A | GGTAACGGT | 700±300 | 2 |
| 9mer_A5T | GGTTGCGGT | 400±70 | 0 |
| 9mer_C6G | GGTTAGGGT | 300±50 | 1 |
| 9mer_G7C | GGTTACCGT | 3000±500 | 8 |
| 9mer_G8C | GGTTACGCT | 800±200 | 2 |
| 9mer_T9A | GGTTACGGA | 2000±400 | 5 |
KD values were obtained from three independent EMSA experiments conducted at 50 mM sodium chloride, 20 mM potassium phosphate, 3 mM BME, pH 8.0 at 4°C. Errors represent the standard error of apparent equilibrium binding constants obtained from three independent experiments.
Fold difference is relative to the 9mer-3′ sequence.
Figure 2.
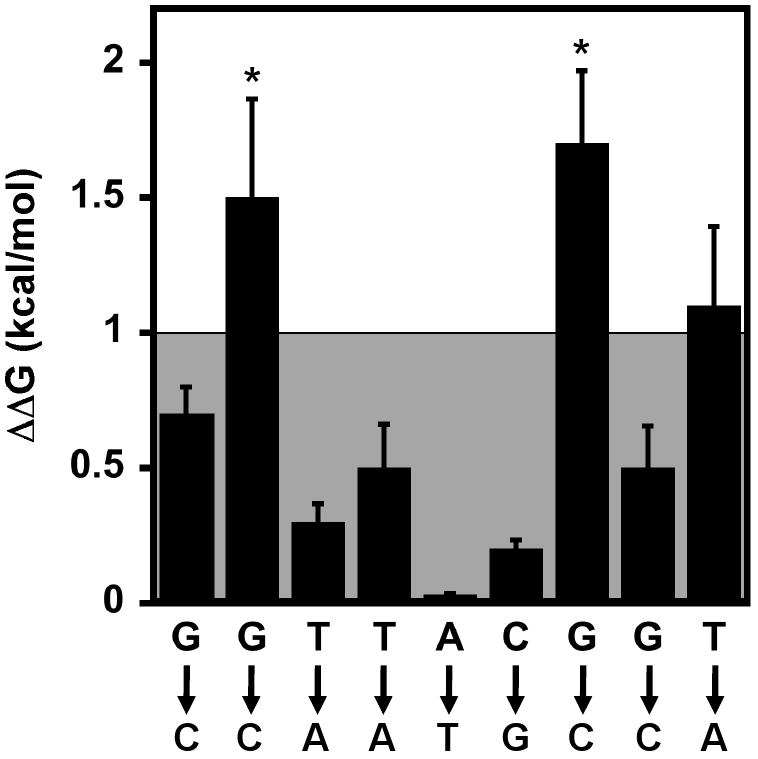
The effect of complementary base substitutions on the binding of Pot1pC to d(GGTTACGGT). ΔΔG values for each nucleotide substitution were calculated as follows: ΔΔG=RTln(KDsub//KDd(GGTTACGGT)), where R = 1.9872 cal/mol*K, T = 277.15 K and KDd(GGTTACGGT) = 400 nM. Asterisks represent binding reactions for which binding was observed, but saturation was not reached. To maintain consistency with the Pot1-DBD studies {Croy, 2006 #647}, any nucleotide substitution displaying a change in ΔΔG > 1 kcal/mol (outside the shaded box) was designated as specifically recognized.
The overall structure of Pot1-DBD is well represented by the isolated Pot1pN and Pot1pC domains
The observed differences between the biochemical behaviors of Pot1-DBD, Pot1pN and Pot1pC may be due to structural perturbations of Pot1pN and Pot1pC relative to their independent structures. We used NMR 15N-1H chemical shift mapping to probe for changes in the local chemical environments of individual backbone amides in Pot1pN and Pot1pC in the context of Pot1-DBD. The instabilities of Pot1-DBD and Pot1pC in the absence of ssDNA precluded a comparative assessment between the ssDNA-free structures of Pot1-DBD, Pot1pN and Pot1pC by NMR. The addition of cognate ssDNA to Pot1-DBD and Pot1pC significantly improved their solution behaviors, allowing for an assessment of their respective backbone structures. An overlay of the individual spectra of the Pot1pN/d(GGTTAC) and Pot1pC/d(GGTTACGGT) complexes provides a very good approximation of the spectrum observed for Pot1-DBD/d(GGTTACGGTTAC) (Figure 3 and Supplemental Figure 3). Such a large degree of spectral overlap indicates that the structures of isolated Pot1pN and Pot1pC are largely conserved in the context of Pot1-DBD. Our minimal chemical shift perturbation (MCSP) analysis indicates that isolated Pot1pN is not significantly perturbed relative to Pot1-DBD (Figure 3 and Supplemental Figures 3 and 4). Of the 154 peaks present in the 15N-1H HSQC spectrum of Pot1pN/d(GGTTAC), we find that 10 peaks (6 %) are strongly perturbed (> 0.1 ppm), 15 peaks (10 %) are moderately perturbed (0.071-0.1 ppm) and 23 peaks (15%) are weakly perturbed (0.051-0.070 ppm), while the remaining 106 peaks (68%) exhibit no detectable perturbation when compared with Pot1-DBD (Supplemental Figure 4). Similar to Pot1pN, MCSP analysis indicates that the majority of peaks present in isolated Pot1pC are minimally perturbed in the context of Pot1-DBD. Of the 172 peaks present in the 15N-1H HSQC spectrum of isolated Pot1pC, we find that 8 peaks (5%) are strongly perturbed (MCSP > 0.1 ppm), 8 peaks (5%) are moderately perturbed (0.071-0.1 ppm) and 6 peaks (4%) are weakly perturbed (0.051-0.070 ppm), while the remaining 150 peaks (86%) exhibit no detectable perturbation when compared with Pot1-DBD. Taken together these results indicate that the overall backbone structures of isolated Pot1pN and Pot1pC are well conserved in Pot1-DBD.
Figure 3.
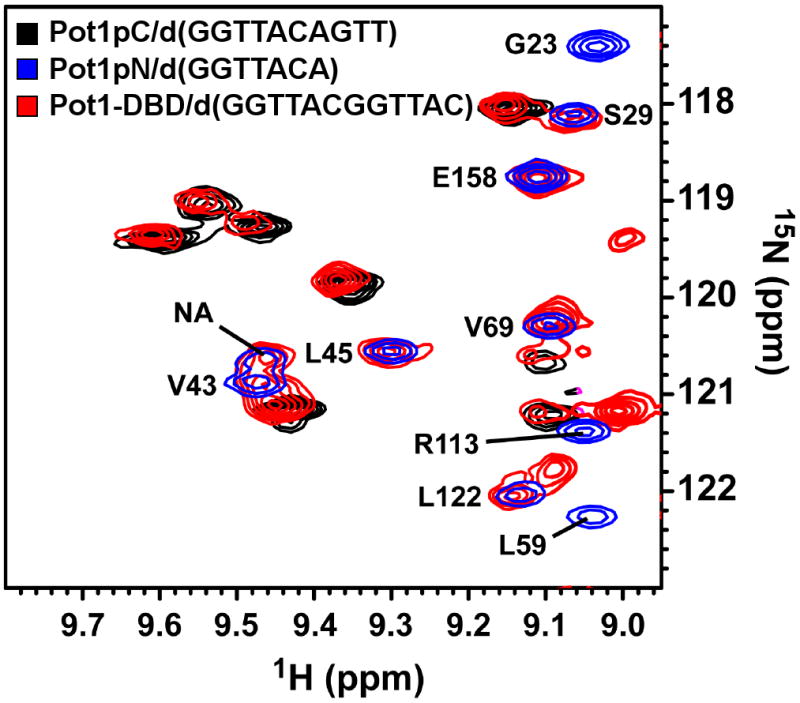
Select region of the 15N-1H HSQC spectrum for the Pot1pN/d(GGTTAC) complex (blue) and the 15N-1H HSQC-TROSY spectra for the Pot1pC/d(GGTTACGGT) (black) and Pot1-DBD/d(GGTTACGGTTAC) (red) complexes. Spectra were collected on Varian 900 (Pot1-DBD), 800 (Pot1pC) and 600 (Pot1pN) MHz spectrometers equipped with either cold (600 and 800 MHz) or room temperature (900 MHz) probes at 30°C for Pot1pN and Pot1-DBD and 25°C for Pot1pC. Peaks present in the Pot1pN/d(GGTTAC) complex spectra have been annotated with their respective one letter amino acid designations and respective positions (peaks corresponding to Pot1pC and Pot1-DBD have not been assigned and are unlabeled).
Figure 4.
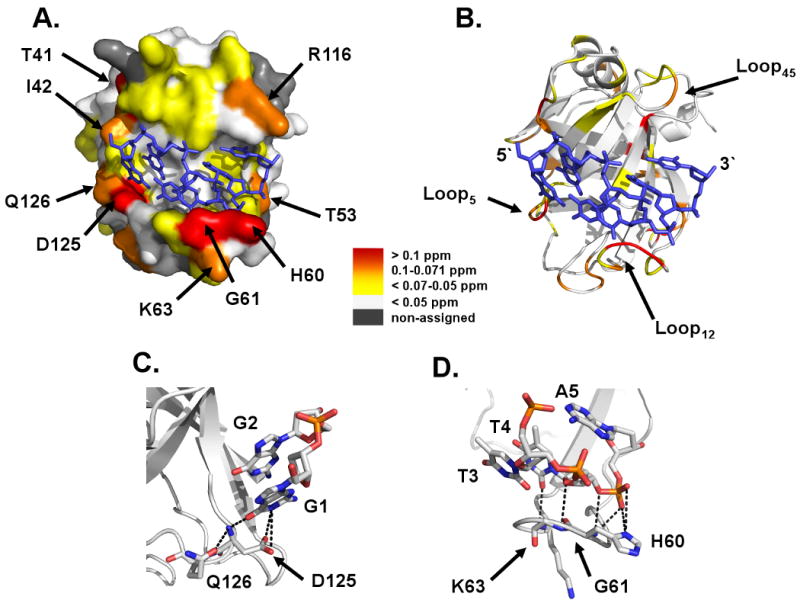
A) Surface and B) ribbon representations of the Pot1pN/d(GGTTAC) crystal complex highlighting regions of no (white), weak (yellow), moderate (orange) and strong (red) MCSP differences between isolated Pot1pN and in the context of Pot1-DBD. Amino acids of interest are labeled and the bound oligonucleotide, d(GGTTAC), is represented by a blue stick model. Ribbon and stick model representation of residues within the C) G1/G2 and D) T3/T4 binding pockets that display MCSP differences between Pot1pN in isolation and in the context of Pot1-DBD. Hydrogen bonding contacts from these residues (< 4 Å) are represented by black dashed lines and all atoms represented by stick model have been colored as follows: white = carbon, orange = phosphorous, red = oxygen, and blue = hydrogen. All figures were made using Pymol version 1.0 {DeLano, 2002 #649}.
MCSP analysis indicates a rearrangement of the ssDNA-binding surface of Pot1pN in Pot1-DBD
The lack of pronounced widespread chemical shift perturbations in the Pot1pN domain of Pot1-DBD indicates that the observed changes in biochemical activity arise from limited alterations in structural character. The backbone assignments of isolated Pot1pN have been previously reported (28) and the structure of the Pot1pN/d(GGTTAC) complex is known (17). This information allows for a more detailed per residue analysis of the perturbations that occur within Pot1pN to specify the regions of structural alteration. Mapping of the MCSPs onto the structure of the Pot1pN/d(GGTTAC) complex (17) indicates that the majority of strong and moderate perturbations occur within residues found in the ssDNA-binding cleft (Figure 4AB). The first group of clustered perturbations localizes to two residues found within the G1/G2 binding pocket: Asp 125 and Gln 126. Asp 125 hydrogen bonds directly with the base of guanosine at position 1 (Figure 4C) (17), while Gln 126, which is not directly involved in making contact with the bound oligonucleotide, hydrogen bonds with the amide backbone of Asp 125 and helps to stabilize the protein structure in that region (Figure 4C) (17). The second group of strong to moderate perturbations is found within the structured loop connecting β1 and β2 (Loop12, Figure 4BD), which forms the centerpiece of the T3/T4 binding pocket (17). Specifically, we observed perturbations in the backbone amides of select residues that directly hydrogen bond to the base and phosphate backbone of T4 (Gly 61 and Lys 63), as well as the phosphate backbone of A5 (His 60) (Figure 4D). Finally, we also observed a few strong to moderate perturbations in non-ssDNA contacting residues: Thr 41, Ile 42, Thr 53 and Arg 116 (Figure 4A). Though these residues do not directly contact the ssDNA, they are located proximal to the perturbed binding pockets present in the ssDNA-binding interface. Taken together, these studies indicate that localized structural changes occur in the G1/G2 and T3/T4 binding pockets of the Pot1pN domain of Pot1-DBD relative to isolated Pot1pN.
Alanine scanning mutagenesis indicates that the overall thermodynamic profile of the ssDNA-binding cleft in Pot1-DBD is similar to that of isolated Pot1pN
To further explore the impact of the structural perturbations that occur within the G1/G2 and T3/T4 binding pockets, we performed alanine scanning mutagenesis of select ssDNA-binding residues in the context of both isolated Pot1pN and Pot1-DBD (Figure 5A). Specifically, we targeted residues in isolated Pot1pN that participate in stacking/van der Waals interactions (Phe 88, Leu 122 and Tyr 115), as well as those residues that hydrogen bond with the ssDNA (Thr 62, Asp 64 and Ser 123) within each respective binding pocket (G1/G2 (Figure 5B), T3/T4 (Figure 5C) and A5/C6 (Figure 5D)). As expected based on the interactions observed in the Pot1pN/d(GGTTAC) crystal complex, all of the residues mutated contribute to the binding of Pot1pN to d(GGTTAC) (Figure 5A and Table 3). Our results indicate that both Leu 122, Phe 88 and Tyr 115, which provide foundations for stacking interactions within the G1/G2, T3/T4 and A5/C6 binding pockets, respectively, are essential for ssDNA binding (Figure 5A-D). Residues involved in hydrogen bonding in the T3/T4 binding pocket were found to display almost identical contributions to the binding free energy (Thr 62 and Asp 64). Interestingly, the Ser 123/G2 hydrogen bond interaction within the G1/G2 pocket appears to contribute less to affinity than other residues studied (Figure 5B).
Figure 5.
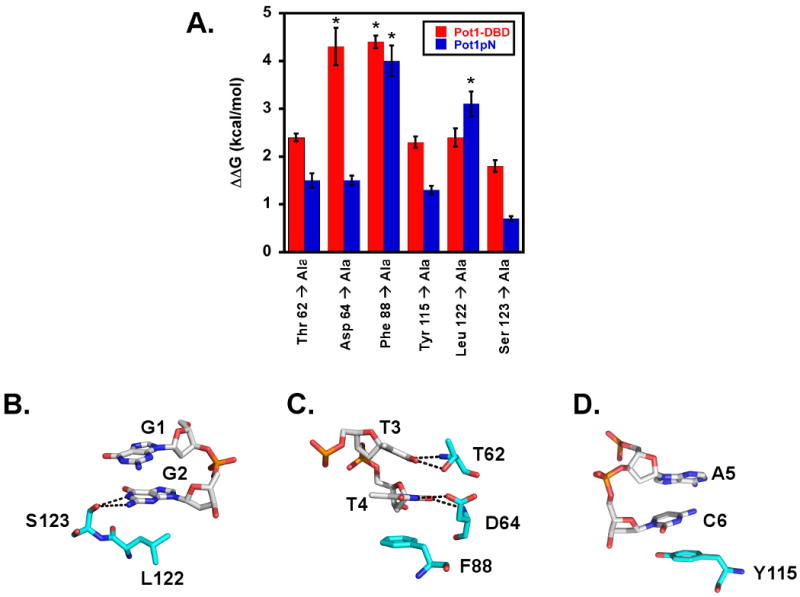
Alanine scanning mutagenesis studies targeting residues within the ssDNA-binding interface of isolated Pot1pN and in the context of Pot1-DBD reveal a similar thermodynamic profile. A) Plot of ΔΔG values as a function of Pot1pN (blue) and Pot1-DBD (red) ssDNA-binding interface alanine mutants. ΔΔG values for alanine mutants were calculated as follows:
ΔΔG=RTln(KDsub//KDd(GGTTAC) (for Pot1pN) or ΔΔG=RTln(KDsub//KDd(GGTTACGGTTAC) (for Pot1-DBD), where R = 1.9872 cal/mol*K, T = 277.15 K and KDd(GGTTAC) = 30 nM KDd(GGTTACGGTTAC) = 1.2 nM. Asterisks represent binding reactions for which binding was observed, but saturation was not reached. Stick model representation of targeted residues within the B) G1/G2, C) T3/T4 and D) A5/C6 binding pockets of Pot1pN. Hydrogen bonding contacts arising from these residues (< 4 Å) are represented by black dashed lines and all atoms represented by stick model have been colored as follows: White (DNA) or cyan (protein) = carbon, orange = phosphorous, red = oxygen and blue = hydrogen. Individual amino acids and nucleotides have been labeled according to their respective one letter amino acid or nucleotide code and numeric position. All figures were made using Pymol version 1.0 {DeLano, 2002 #649}.
Table 3.
KD and free energy changes for various ssDNA-binding interface alanine mutants of Pot1pN bound to cognate d(GGTTAC).
| Pot1pN Construct | KD (nM)a | Fold Changeb |
|---|---|---|
| Pot1pN_WT | 30±20 | --- |
| Pot1pN_T62A | 500±50 | 20 |
| Pot1pN_D64A | 400±30 | 10 |
| Pot1pN_F88Ac | ≥38000 | >1300 |
| Pot1pN_Y115A | 300±20 | 10 |
| Pot1pN_L122Ac | >9000 | >300 |
| Pot1pN_S123A | 100±10 | 3 |
Filter-binding experiments were conducted at 15 mM sodium chloride, 20 mM potassium phosphate, 3 mM BME, pH 8.0 at 4° C. Errors represent the standard deviation of equilibrium binding constants obtained from three independent binding experiments.
Fold difference is relative to Pot1pN_WT.
Saturation was not observed for these weak Pot1pN/ssDNA interactions. KD values were calculated using a synthetic data point as described in the materials and methods and represent a lower limit for binding.
These same mutations were also important to the ssDNA-binding affinity of Pot1-DBD (Table 4 and Figure 5A). Overall, most of the residues had a similar effect on binding in Pot1pN and Pot1-DBD (ΔΔGPot1-DBD-ΔΔGPot1pN < 1 kcal/mol), suggesting that the binding cleft is largely preserved. However, we found that the contributions of a few of these residues to the free energy of ssDNA binding were differentially altered with respect to isolated Pot1pN (Table 3). The most striking change occurred when the Asp 64 to Ala mutation was examined in the context of Pot1-DBD, which exhibited a ∼ 3 kcal/mol change relative to Pot1pN (ΔΔGPot1-DBD-ΔΔGPot1pN)(Figure 5A). The dramatic shift in the thermodynamic importance of this residue may be related to structural perturbations within the T3/T4 binding pocket of Pot1-DBD as detected in our MCSP analysis and is likely a function of the altered specificity profile within Pot1-DBD. However, since the majority of residues showed little alteration and we observe modest structural changes via MCSP analysis, these results support a model in which the overall binding surface observed in the Pot1pN/d(GGTTAC) crystal structure is largely preserved in the context of Pot1-DBD, with some subtle rearrangements in localized regions.
Table 4.
KD and free energy changes for various ssDNA-binding interface alanine mutants of Pot1-DBD bound to cognate d(GGTTACGGTTAC).
| Pot1-DBD Construct | KD (nM)a | Fold Changeb |
|---|---|---|
| Pot1-DBD_WT | 1 ±0.1 | --- |
| Pot1-DBD_T62A | 90±3 | 90 |
| Pot1-DBD_D64Ac | >3000 | >3000 |
| Pot1-DBD_F88Ac | >3000 | >3000 |
| Pot1-DBD_Y115A | 80±4 | 80 |
| Pot1-DBD_L122A | 90±7 | 90 |
| Pot1-DBD_S123A | 30±2 | 30 |
Filter binding experiments were conducted in 400 mM sodium chloride, 20 mM potassium phosphate, 3 mM BME, pH 8.0 at 4° C. Errors represent the standard error of equilibrium binding constants obtained from three independent binding experiments.
Fold difference is relative to wild type Pot1-DBD (Pot1-DBD_WT).
Saturation was not observed for these weak Pot1-DBD/ssDNA interactions. KD values were calculated using a synthetic data point as described in the materials and methods and represent a lower limit for binding.
MCSP analysis suggests the presence of a domain/domain interaction interface in the Pot1pN domain of Pot1-DBD
In addition to the localized changes within the ssDNA-binding cleft of the Pot1pN domain in Pot1-DBD, we also detected a second group of perturbed residues distant from the ssDNA-binding residues discussed in the previous section (Figure 6AB). These residues are located on a single face of Pot1pN, with the majority located in the structured loop connecting β3 and β4 (Loop34) (Gln 91, Asn 93, Asp 94, Val 97, Lys 99), with the remaining two residues (Ser 139 and Thr 142) located in the non-regular structured loop connecting β5 with the C-terminal helix (LoopPostβ5; Figure 6AB). The crystal structure of the Pot1pN/d(GGTTAC) complex indicates that these residues are distinct from the ssDNA-binding interface (17). Therefore, we hypothesized that these perturbations arise from a domain/domain interface between the Pot1pN and Pot1pC domains within Pot1-DBD. The presence of such an interaction surface provides a means to alter the binding properties of both domains relative to their isolated forms through an allosteric type of mechanism.
Figure 6.
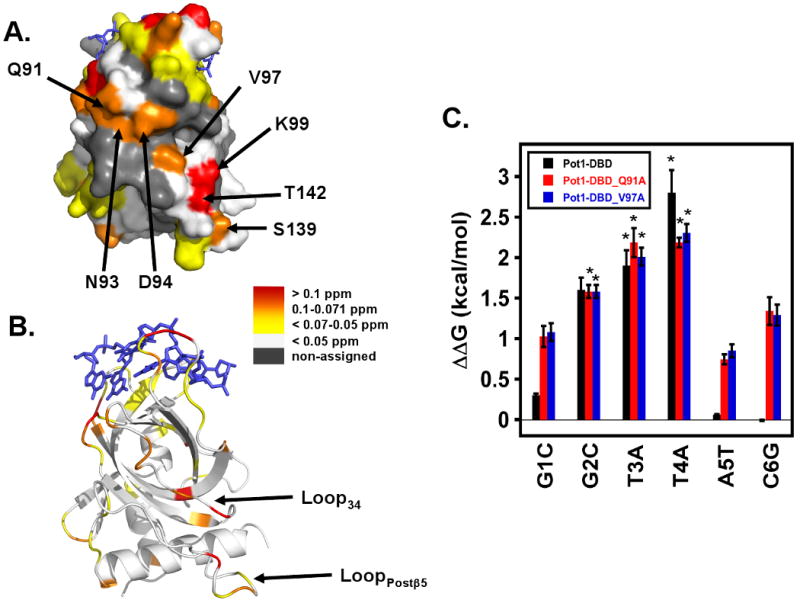
MCSP analysis reveals a putative domain/domain interface on the Pot1pN domain of Pot1-DBD. A) Surface and B) ribbon representations of the Pot1pN/d(GGTTAC) crystal complex highlighting regions of no (white), weak (yellow), moderate (orange) and strong (red) MCSP differences between Pot1pN in isolation in the context of Pot1-DBD. Amino acids of interest are labeled and the bound oligonucleotide, d(GGTTAC), is represented by a blue stick model. Loops involved in forming the putative interdomain interface have been labeled. C) Plot of ΔΔG values as a function of individual nucleotide substitution for Pot1-DBD (black), Pot1-DBD_Q91A (red) and Pot1-DBD_V97A (red). ΔΔG values for each nucleotide substitution were calculated as follows:
ΔΔG=RTln(KDsub//KDd(GGTTACGGTTAC)), where R = 1.9872 cal/mol*K, T = 277.15 K and KDd(GGTTACGGTTAC) = 670 pM for Pot1-DBD, 7.0 nM for Pot1-DBD_Q91A and 9.6 nM for Pot1-DBD_V97A. Asterisks represent binding reactions for which binding was observed, but saturation was not reached. All figures were made using Pymol version 1.0 {DeLano, 2002 #649}.
We were not able to detect a Pot1pN/Pot1pC interaction directly in trans using solution based NMR based chemical shift perturbation methods either in the presence of absence of ssDNA (data not shown) suggesting that the interactions of the Pot1pC and Pot1pN domains within Pot1-DBD are weak. To probe this potential interface, we instead performed alanine mutagenesis of residues present in Loop34 to see if disruption of this putative Pot1pN/Pot1pC interaction leads to changes in the ssDNA-binding activity of Pot1-DBD. Of the residues targeted, only Pot1-DBD_Q91A and Pot1-DBD_V97A were soluble and monomeric in solution (data not shown). Pot1-DBD_Q91A and Pot1-DBD_V97A displayed a 10- and 15-fold reduction in binding affinity for d(GGTTACGGTTAC), respectively, when compared with wild type Pot1-DBD, suggesting that they contribute to the overall ssDNA-binding activity (24) (Table 5). We also tested whether disruption of this interface altered the specific nucleotide recognition profile for Pot1-DBD by measuring the impact on binding affinity for individual complementary nucleotide substitutions within the first hexameric core repeat of d(GGTTACGGTTAC). Interestingly, we found that the thermodynamic profile describing the specific recognition of individual nucleotides by Pot1-DBD was significantly altered in the context of both Pot1-DBD_Q91A and Pot1-DBD_V97A. Specifically, in addition to the trinucleotide GTT recognized by wildtype Pot1-DBD, these mutant proteins displayed an added preference for guanosine at position 1, adenosine at position 5 and cytosine at position 6 (Table 5 and Figure 6C). These results point to a reversion to isolated Pot1pN binding characteristics for the Pot1pN domain upon disruption of this putative domain/domain interface. These data lead us to test whether the decoupling of Pot1pN and Pot1pC within Pot1-DBD also causes a change in sequence length preference to accommodate non-overlapping binding sequences for Pot1pN and Pot1pC. We examined binding to both a 15 nucleotide sequence d(GGTTACGGTTACGGT) and a 24 nucleotide sequence d(GGTTACGGTTAC)2 (Table 5). Surprisingly, we found > 100-fold tighter binding affinity for both oligonucleotides in the in the context of Pot1-DBD_Q91A and Pot1-DBD_V97A but not for wild-type Pot1-DBD (Table 5 and Supplemental Table 1). These results suggest that interaction between the two domains within Pot1-DBD negatively impacts the free energy of binding, as well as alters the specific recognition profile exhibited by Pot1-DBD.
Table 5.
KD and free energy changes of Pot1-DBD_Q91A and Pot1-DBD_V97A binding to various oligonucleotides.
| Pot1-DBD_Q91A Oligonucleotide Name | Oligonucleotide Sequence | KD (nM)a | ΔΔG (kcal/mol)b | Fold Changec |
|---|---|---|---|---|
| 12mer | GGTTACGGTTAC | 7±0.6 | --- | --- |
| 12mer_G1C | CGTTACGGTTAC | 50±6 | 1±0.1 | 7 |
| 12mer_G2Cd | GCTTACGGTTAC | >100 | >2 | 10 |
| 12mer_T3Ad | GGATACGGTTAC | >400 | >2 | 60 |
| 12mer_T4Ad | GGTAACGGTTAC | >400 | >2 | 60 |
| 12mer_A5T | GGTTTCGGTTAC | 30±2 | 1±0.1 | 4 |
| 12mer_C6G | GGTTAGGGTTAC | 80±10 | 1±0.1 | 10 |
| 15mer | GGTTACGGTTACGGT | 0.04±0.003 | -3±0.1 | 200 |
| 24mer | (GGTTACGGTTAC)2 | 0.06±0.003 | -3±0.1 | 100 |
| Pot1-DBD_V97A Oligonucleotide Name | Oligonucleotide Sequence | KD (nM)a | ΔΔG (kcal/mol)b | Fold Changec |
| 12mer | GGTTACGGTTAC | 10±1 | --- | --- |
| 12mer_G1C | CGTTACGGTTAC | 70±7 | 1±0.1 | 7 |
| 12mer_G2Cd | GCTTACGGTTAC | >200 | >2 | 20 |
| 12mer_T3Ad | GGATACGGTTAC | >400 | >2 | 40 |
| 12mer_T4Ad | GGTAACGGTTAC | >600 | >2 | 60 |
| 12mer_A5T | GGTTTCGGTTAC | 50±4 | 1±0.1 | 5 |
| 12mer_C6G | GGTTAGGGTTAC | 100±10 | 1±0.1 | 10 |
| 15mer | GGTTACGGTTACGGT | 0.04±0.004 | -3±0.1 | 300 |
| 24mer | (GGTTACGGTTAC)2 | 0.06±0.004 | -3±0.1 | 200 |
Due to the extremely tight binding affinity of Pot1-DBD for ssDNA, filter-binding experiments were conducted at 400 mM sodium chloride, 20 mM potassium phosphate, 3 mM BME, pH 8.0 at 4° C. Errors represent the standard error of equilibrium binding constants obtained from three independent binding experiments.
ΔΔG = RTln(KD/KD12mer), R = 1.9872 cal mol-1 K-1, T = 277.15 K
Fold difference is relative to Pot1-DBD mutants binding to d(GGTTACGGTTAC).
Saturation was not observed for these weak Pot1-DBD/ssDNA interactions. KD values were calculated using a synthetic data point as described in the materials and methods and represent a lower limit for binding.
Discussion
The ssDNA-binding activity of Pot1-DBD results from the non-additive functions of its constitutive Pot1pN and Pot1pC domains
Like Pot1pN, the C-terminal domain of Pot1-DBD is capable of independently binding telomeric ssDNA and exhibits ssDNA-binding activity that is distinct from both Pot1pN and Pot1-DBD. We find that Pot1pC binds to a minimal nonameric oligonucleotide, d(GGTTACGGT), with a binding affinity of 400 nM at 50 mM NaCl (Table 1). This result was surprising; as Pot1-DBD recognizes a minimal dodecamer (24) and Pot1pN in isolation recognizes a minimal hexamer (23), we expected Pot1pC to also bind a hexamer. (24). Surprisingly, we found that the overall binding affinity of Pot1-DBD for d(GGTTACGGTTAC) (46 pM) was substantially weaker than the sum of the energetic interactions of Pot1pN and Pot1pC in isolation with their respective minimal cognate oligonucleotide substrates (-8.1 kcal/mol + -8.5 kcal/mol = -16.6 kcal/mol or 80 fM). If the mode of binding were conserved, we would expect the Pot1-DBD/d(GGTTACGGTTAC) interaction to potentially be tighter than 80 fM due to additional energetic contributions arising from the chelate effect (31). This unexpected behavior does not appear to result from an incomplete ssDNA-binding site for both Pot1pN and Pot1pC in the 12mer oligonucleotide since increasing the oligonucleotide length lead to only modest changes in overall binding affinity (Supplemental Table 1). We also observed non-additivity of Pot1pN and Pot1pC in the context of Pot1-DBD with regard to their respective individual nucleotide recognition patterns when compared with Pot1-DBD. Pot1pN and Pot1pC recognize, d(GGTTAC) (23) and d(GGTTACGGT), respectively which are markedly altered in the context of Pot1-DBD, d(GGTTACGGTTAC) (24) (specifically recognized nucleotides in bold). Taken together, these data suggest that the ssDNA-binding activity of Pot1-DBD is not a simple sum of its constitutive parts. Rather, our results indicate that the ssDNA-binding activity of Pot1-DBD is altered relative to the individual activities of isolated Pot1pN and Pot1pC, suggesting that their arrangement and interactions within Pot1-DBD influences the binding characteristics of each domain.
The observed changes in ssDNA-binding activity are not due to the global refolding of the isolated domains
Our MCSP analysis indicates that the altered ssDNA-binding activities of isolated Pot1pC and Pot1pN relative to Pot1-DBD, originates from subtle, rather than dramatic, changes in their respective structures (Figure 3 and Supplemental Figure 2). Our MCSP studies suggest that the structural character of the majority of amide backbones in isolated Pot1pN and Pot1pC are conserved in the context of Pot1-DBD. Of the residues that are perturbed within the Pot1pN domain of Pot1-DBD, the majority are localized to the ssDNA-binding interface defined by the Pot1pN/d(GGTTAC) crystal complex (17). Alanine scanning mutagenesis of the DNA-binding surface reveals relatively few changes in the contributions of these residues to the overall binding affinity when independent Pot1pN and Pot1-DBD are compared (Tables 3 and 4 and Figure 5A). These NMR and mutagenesis studies indicate that the ssDNA-binding cleft of the Pot1pN domain of Pot1-DBD is quite similar to that of the isolated protein. These results suggest that slight adjustments in the molecular details governing the interactions of the protein and oligonucleotide lead to the altered ssDNA-binding features of Pot1-DBD relative to isolated Pot1pN and Pot1pC.
Subtle rearrangements of the G1/G2 and T3/T4 binding pockets within ssDNA-binding interface in Pot1pN contribute to the altered ssDNA-binding activity of Pot1-DBD
Pot1-DBD exhibits a weakened preference for G1 and A5 relative to Pot1pN in isolation (24). Specific recognition of G1 by Pot1pN alone is thought to result from hydrogen bonding interactions formed between Asp 125 and G1 within the G1/G2 binding pocket (23). Our MCSP analysis indicates that Asp 125 is strongly perturbed in the context of Pot1-DBD (Figure 4A). These data suggest that structural alteration of this binding pocket in Pot1-DBD disrupts hydrogen-bonding interactions resulting in the decreased preference for G1. In contrast to G1 recognition, the specific recognition of A5 arises from the internucleotide interactions of the adenosine base with the phosophodiester backbones of T3 and T4 (23). We find that residues present in Loop12, which forms the core of the T3/T4 binding pocket, are strongly perturbed in Pot1-DBD, indicating that the T3/T4 binding pocket is altered in the context of Pot1-DBD (Figure 4A). Dynamics studies of isolated Pot1pN indicate that Loop12 is unstructured in the absence of bound oligonucleotide and formation of the T3/T4 binding pocket involves the co-folding of Loop12 and d(GGTTAC) (32). These results support a model in which the T3/T4 binding pocket in Pot1-DBD is present in an alternate conformation that no longer provides for the specificity-dictating interactions between the phosphodiester backbones of T3/T4 with A5 (Supplemental Figure 1A). Further evidence for an altered T3/T4 binding pocket comes from our alanine scanning studies, which show an increase in the contributions of Asp 64 (which hydrogen bonds to the base of T4 (Supplemental Figure 1C)) to the overall binding affinity in Pot1-DBD (Figure 5A). Additionally, these alterations clarify why the importance of the non-canonical hydrogen bonding interaction formed between the exocyclic amine at position 2 of G2 and the carboxyl at position 4 in T4 in Pot1pN is no longer required for high-affinity binding in Pot1-DBD (24). Alteration of the G1/G2 and T3/T4 binding pockets is not unexpected as the ssDNA-binding cleft in Pot1pN is a highly malleable surface that can accommodate several alternative oligonucleotide conformations with varying degrees of thermodynamic change (28).
Evidence for a putative domain/domain interface that mediates interactions between Pot1pN and Pot1pC within Pot1-DBD
In addition to the perturbation of residues within the ssDNA-binding interface of Pot1pN, a second region of perturbation was observed in residues that are distant (> 10Å) from the ssDNA-binding interface within Loop34 of Pot1pN (Figure 6AB). These residues define a putative surface that facilitates interaction between the Pot1pN and Pot1pC domains in Pot1-DBD. Support for this hypothesis can be obtained from comparisons with the DBD from human Pot1, HsPot1_v2. HsPot1_v2 contains two OB-fold domains (OB1 and OB2) that associate with each other via an extensive domain/domain interface in which hydrophobic residues in Loop34 (residues 69-75) provide a surface that facilitates the interaction of OB1 and OB2 (18). Clustal (33) sequence alignments reveal that the hydrophobic character of the Loop34 in HsPot1_v2 is conserved in the Pot1pN domain of Pot1-DBD (Supplemental Figure 5). Consistent with this hypothesis, targeted disruption of this putative domain/domain interface by alanine substitution leads to changes in both the binding affinity, specific nucleotide recognition exhibited by Pot1-DBD (Table 5). Mutation of these interface residues also alters the oligonucleotide length preference of Pot1-DBD to instead include the complete binding sequences for both Pot1pN and Pot1pC, as both mutants display a > 100-fold enhancement for these extended sequences relative to d(GGTTACGGTTAC) (Table 5),. In contrast, wild-type Pot1-DBD, which maintains this putative domain/domain interface, binds both the 12mer and 15mer oligonucleotides with relatively similar affinities (Supplemental Table 1). Finally, the disruption of the interface results in a > 10-fold weaker binding affinity relative to wild-type Pot1-DBD binding to its cognate 12mer sequence (40-60 pM for Pot1-DBD_Q91A/ or Pot1-DBD_V97A/d(GGTTACGGTTACGGT) compared to the reported 670 pM (24) for Pot1-DBD/d(GGTTACGGTTAC)). These results suggest that the domain/domain interactions of Pot1pN and Pot1pC within Pot1-DBD lead to their altered binding characteristics relative to its constitutive domains.
In addition to residues in Loop34, the domain/domain interface in HsPot1_v2 also contains residues found in Loop12 (Supplemental Figure 5). Unlike the conservation observed in Loop34, Clustal alignments indicate that Loop12 is only modestly conserved in the context of Pot1-DBD. This lack of conservation is consistent with our MCSP studies that indicate minimal perturbation of these residues. These results imply that only certain aspects of the interface presented in HsPot1_v2 are retained in the context of Pot1-DBD. The lack of complete conservation between the domain/domain interfaces in HsPot1_v2 and Pot1-DBD and the decreased points of contact between Pot1pN and Pot1pC domains in Pot1-DBD suggest a more flexible interdomain interface relative to HsPot1. Such an interface may allow for the reorientation of the Pot1pN and Pot1pC domains with respect to one another, while still maintaining the important domain/domain interactions, to accommodate the heterogeneous spacer elements in S. pombe telomeres.
Pot1DBD ssDNA-binding activity may be regulated by interdomain allostery
Our results suggest that the interactions formed between the Pot1pN and Pot1pC domains within Pot1-DBD leads to the alteration of their individual ssDNA-binding activities, ultimately resulting in the ssDNA-binding activity exhibited by Pot1-DBD. Such domain/domain interactions in the context of the TEP family have not been reported. However, the regulation of biochemical function through bimolecular protein/protein interactions is not unprecedented in the TEP family. The Oxytrichia nova TEP family member, TEBP, is a multimeric protein formed from two distinct polypeptide subunits (α and β), which exhibit ssDNA-binding activity independently of each other or together as a heterodimeric complex. In isolation the TEBPα subunit (a tandem repeat of OB-folds) recognizes d(TTTTGGGG) (specifically recognized residues in bold) with an apparent KD of 125 nM (39). Addition of TEBPβ (a single OB-fold) leads to an alteration of the ssDNA-binding activity of TEBPα. Formation of the TEBPαβ heterodimer leads to a dramatic shift in the binding affinity (from 125 nM to 2.8 nM), oligonucleotide sequence recognized, minimal oligonucleotide length required, and alteration of the specific recognition profile exhibited by TEBPα to d(GGGGTTTTGGGG) in TEBPαβ (specifically recognized residues in bold) (40, 41). Protein/protein interactions have also been found to be important in altering the ssDNA-binding activity of the human Pot1 protein (42). In the absence of the telomere-associated protein, TPP1, HsPot1 binds to telomere ends and blocks the nucleotide addition activity of telomerase. Interaction of TPP1 with HsPot1 leads to a change in the ssDNA-binding activity of HsPot1 (42). Association with TPP1 also appears to alter HsPot1 interaction with telomerase resulting in an increase in the processive nature of telomerase nucleotide repeat addition. Though protein/protein mediated allostery is commonly observed in the TEP family, our studies indicate that TEP proteins may allosterically regulate their biological functions by manipulating interdomain interactions. Further insight into how these interactions might be harnessed to regulate biological function and telomere end structure is a developing area of investigation.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Geoff Armstrong and the Rocky Mountain Regional 900 MHz NMR Facility at the University of Colorado, Denver Health Sciences Center (NIH-GM68928) for aiding our NMR data collection at 900 MHz. We also wish to thank Cristina Sandoval and Andrew Garst for their work on developing protocols for the expression and purification of Pot1pC, Michelle Turco for her work on developing, expressing and purifying Pot1pN and Pot1-DBD ssDNA-binding interface alanine mutants and Kelsey Chow for her work on developing, expressing and purifying the Pot1-DBD domain/domain interface mutants. We would like to thank Dr. Olke Uhlenbeck (Northwestern University) for thoughtful discussions on our data and Lisa Warner for critical review of this manuscript.
Abbreviations
- BME
β-mercaptoethanol
- BSA
bovine serum albumin
- DBD
ssDNA-binding domain
- β
β-strand
- DTT
dithiothreitol
- EMSA
electrophoretic mobility shift assay
- HSQC
heteronuclear single quantum coherence
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- KD
apparent equilibrium binding constant
- MCSP
minimal chemical shift perturbation
- OB-fold
oligonucleotide/oligosaccharide-binding fold
- Pot1-DBD
residues 1-389 of full length S. pombe Pot1
- Pot1pC
residues 178-389 of full length S. pombe Pot1
- Pot1pN
residues 1-185 of full length S. pombe Pot1
- ssDNA
single-stranded deoxyribonucleic acid
- SpPot1
Schizosaccharyomyces pombe protection of telomeres 1
- ssDNA
single-stranded deoxyribonucleic acid
- TEP
telomere-end protection
Footnotes
We acknowledge the NRSA Postdoctoral Fellowship GM-071257 (to J.E.C.), National Institutes of Health Training Appointment (NIH) GM065103-06 (to S.E.A.), National Institutes of Health Training Appointment (NIH) (to N.E.G.), NIH GM-059414 (to D.S.W.), University of Colorado Cancer Center (to D.S.W.), and National Science Foundation (NSF) MCB-0617956 (to D.S.W.) for funding this research.
Supporting Information: Purification of Pot1pC, 15N-1H HSQC analysis of Pot1pN/Pot1pC/Pot1-DBD and sequence comparisons of Pot1-DBD and HsPot1_v2 is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 3.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 4.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 5.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 6.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 8.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaug AJ, Podell E, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang G, Cech TR. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 12.Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 14.Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat Struct Mol Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- 15.Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J Mol Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 16.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 17.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the crystal structure of the Pot1 bound to telomeric single-stranded DNA (Protection of Telomeres)-ssDNA complex. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 18.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 19.Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Mol Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 22.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 23.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 24.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: The telomeric single-stranded DNA-binding activity of Pot11-389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of hetergeneous fission yeast telomeres. J Biol Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 26.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 27.Eldridge AM, Wuttke DS. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucl Acids Res. 2008;36:1624–1633. doi: 10.1093/nar/gkn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croy JE, Fast JL, Grimm NE, Wuttke DS. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 30.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Mol Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 31.Wyman J, Gill SJ. Binding and linkage: Functional chemistry of biological macromolecules. University Science Books; 1990. [Google Scholar]
- 32.Croy JE, Wuttke DS. Insights into the dynamics of specific telomeric single-stranded DNA recognition by Pot1pN. J Mol Biol. 2009;387(4):935–948. doi: 10.1016/j.jmb.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace LM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasan J, Hu S, Hrabal R, Zhu Y, Komvies EA, Ni F. Thrombin-bound structure of an EGF subdomain from human thrombomodulin determined by transferred nuclear Overhauser effects. Biochemistry. 1994;33:13553–13560. doi: 10.1021/bi00250a007. [DOI] [PubMed] [Google Scholar]
- 36.White CE, Hunter MJ, Meininger DP, White LR, Komives EA. Large-scale expression, purification and characterization of small fragments of thrombomodulin: the roles of the sixth domain and of methionine 388. Protein Eng. 1995;8:1177–1187. doi: 10.1093/protein/8.11.1177. [DOI] [PubMed] [Google Scholar]
- 37.Wood MJ, Sampoli Benitez BA, Komives EA. Solution structure of the smallest cofactor-active fragment of thrombomodulin. Nat Struct Mol Biol. 2000;7:200–204. doi: 10.1038/73302. [DOI] [PubMed] [Google Scholar]
- 38.Buck M, Xu W, Rosen MK. A two-state allosteric model for autoinhibition rationalizes WASP signal integration and targeting. J Mol Biol. 2004;338:271–285. doi: 10.1016/j.jmb.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Classen S, Lyons D, Cech TR, Schultz SC. Sequence-specific and 3′-end selective single-strand DNA binding by the Oxytricha nova telomere end binding protein alpha subunit. Biochemistry. 2003;42:9269–9277. doi: 10.1021/bi0273718. [DOI] [PubMed] [Google Scholar]
- 40.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang G, Cech TR. Oxytricha telomere-binding protein: DNA-dependent dimerization of the alpha and beta subunits. Proc Natl Acad Sci USA. 1993;90:6056–6060. doi: 10.1073/pnas.90.13.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL molecular graphics system. Scientific, D., Ed.; San Carlos, CA U.S.A: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


