Summary
Mycobacterium tuberculosis is arguably the world’s most successful infectious agent due to its ability to control its own cell growth within the host. Bacterial growth rate is closely coupled to rRNA transcription, which in E. coli is regulated through DksA and (p)ppGpp. The mechanisms of rRNA transcriptional control in mycobacteria, which lack DksA, are undefined. Here we identify CarD as an essential mycobacterial protein that controls rRNA transcription. Loss of CarD is lethal for mycobacteria in culture and during infection of mice. CarD depletion leads to sensitivity to killing by oxidative stress, starvation, and DNA damage, accompanied by failure to reduce rRNA transcription. CarD can functionally replace DksA for stringent control of rRNA transcription, even though CarD associates with a distinct site on RNA polymerase. These findings highlight a new molecular mechanism for regulating rRNA transcription in mycobacteria that is critical for M. tuberculosis pathogenesis.
Introduction
At least 30% of the world’s population is infected with latent Mycobacterium tuberculosis, which in some individuals will reactivate and cause an estimated 1.3 million deaths a year (WHO, 2009). This health crisis is exacerbated by the alarming emergence of multi-drug and extensively drug resistant strains. The development of new chemotherapeutic strategies is imperative, which requires insight into the pathways involved in M. tuberculosis infection, persistence, and drug resistance.
During infection, mycobacteria and other intracellular bacterial pathogens withstand an arsenal of host-derived mutagens that can result in DNA strand breaks, point mutations, and chromosomal deletions (Boshoff et al., 2003; Buchmeier et al., 1995; Darwin and Nathan, 2005; Hassett and Cohen, 1989). The bacteria are also starved for nutrients and oxygen such that endogenous alkylating agents damage the bacterial genome (Taverna and Sedgwick, 1996). Regardless, M. tuberculosis is able to persist for the lifetime of the host, indicating that this pathogen has substantial mechanisms to resist host inflicted starvation and damage. M. tuberculosis mutants deficient in DNA repair, amino acid metabolism, or response to hypoxia are attenuated in vivo, thus highlighting the importance of active resistance to host attacks (Darwin and Nathan, 2005; Boshoff et al., 2003; McAdam et al., 1995; Shiloh et al., 2008;).
To persist in this hostile environment, M. tuberculosis enters a state of dormancy and rapidly downregulates ribosome biogenesis to match declining translational need, a response that requires coordinate transcriptional regulation of all ribosome components (Betts et al., 2002). Bacteria accomplish this via the stringent response, a global regulatory mechanism in which transcription of stable RNAs is inhibited, in part by the production of the hyperphosphorylated guanine nucleotides ppGpp and pppGpp, ((p)ppGpp) (Avarbock et al., 2000; Chatterji and Ojha, 2001; Magnusson et al., 2005). (p)ppGpp is synthesized during nutrient, phosphate, and nucleotide deprivation, stationary phase, and alkaline shock. (p)ppGpp destabilizes the RNAP open complex, a kinetic intermediate that exists during transcriptional initiation, as well as inhibits DNA replication and other cellular processes (Magnusson et al., 2005; Nanamiya et al., 2008; Ojha et al., 2000; Primm et al., 2000; Wang et al., 2007; Haugen et al., 2008).
It has become clear that (p)ppGpp is not the only factor that participates in stringent control. In E. coli, the DksA protein potentiates the effect of (p)ppGpp by directly binding the RNAP (Paul et al., 2004). In a ΔdksA strain, rRNA transcription is insensitive to (p)ppGpp accumulation, thus leaving rRNA promoters unresponsive to changes in amino acid availability and growth rate (Paul et al., 2004). Deletion of DksA has pleiotropic effects including DNA damage sensitivity (Branny et al., 2001; Magnusson et al., 2007; Meddows et al., 2005; Trautinger et al., 2005; Webb et al., 1999). Despite the intricate functional relationship between DksA and (p)ppGpp in E. coli, obvious DksA homologs are absent from most non-proteobacteria, whereas (p)ppGpp synthetases are broadly distributed, possibly indicating that other factors exist to control ribosomal biogenesis in these organisms.
In this report, we identify carD as a widely distributed DNA damage and starvation inducible gene whose protein product binds the RNAP to control rRNA transcription in mycobacteria. CarD can complement an E. coli ΔdksA strain, but in contrast to E. coli DksA, CarD is required for mycobacterial viability under all conditions, both in culture and during all stages of mouse infection. Unlike DksA, CarD proteins interact with the N-terminus of the RNAP β subunit, a binding site shared by Transcription Repair Coupling Factor (TRCF). These data establish CarD proteins as novel, highly conserved, and essential components of a previously unrecognized mechanism of stringent control in mycobacteria.
Results
Mycobacterial carD is upregulated in response to genotoxic stress and nutrient deprivation
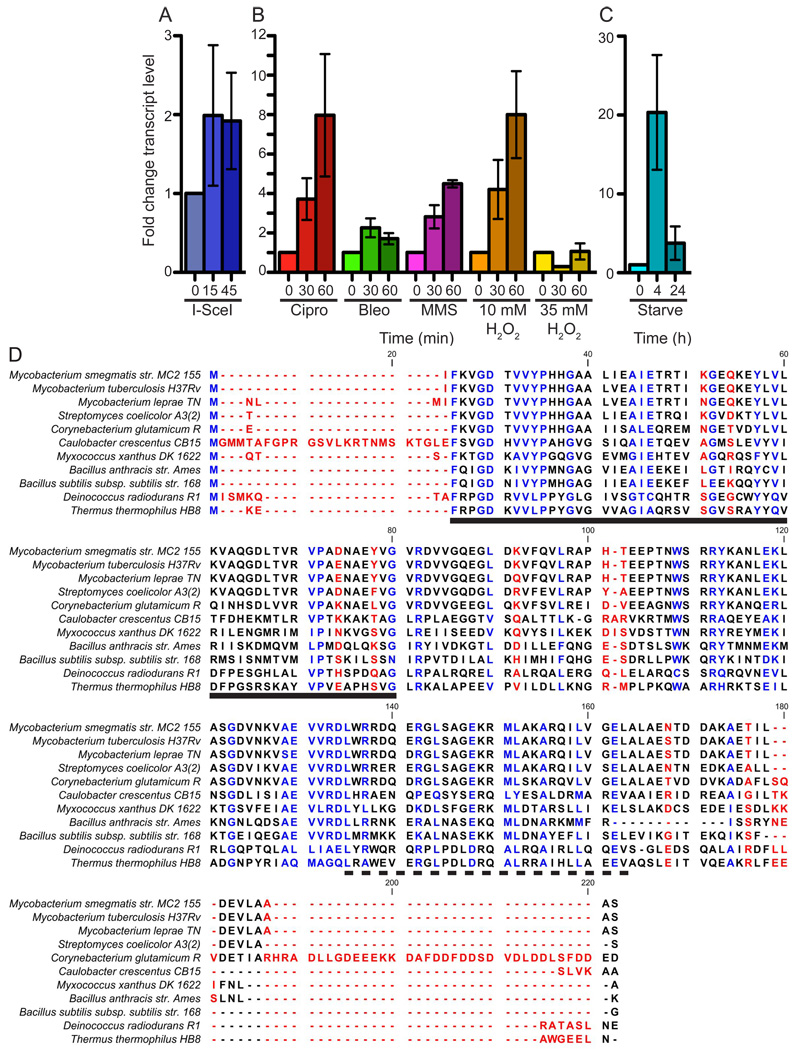
To further understand the mycobacterial response to DNA double strand breaks (DSB), we undertook a DNA microarray analysis of the transcriptional response of Mycobacterium smegmatis to DSB generated by the homing endonuclease I-SceI. The analysis compared two M. smegmatis strains: mgm181 and mgm182. Both strains are induced by anhydrotetracycline (ATc) treatment to express a hemagluttinin (HA) epitope-tagged I-SceI (Ehrt et al., 2005; Stephanou et al., 2007), but only the mgm182 genome is cleaved by I-SceI while mgm181 produces the endonuclease without a chromosomal break. One of the most highly upregulated genes in this comparison was previously uncharacterized carD (MSMEG_6077). On average, carD mRNA was upregulated 2.4 fold in bacteria undergoing chromosomal breakage (data not shown). The transcriptional induction of carD following I-SceI generated DSB was verified using quantitative real time PCR (qRT-PCR) (Figure 1A).
Figure 1. carD transcription is upregulated in response to oxidative stress, DNA damage, and starvation.
carD transcript levels in treated log phase wild-type M. smegmatis cultures were measured by qRT-PCR, normalized to sigA transcript levels, and expressed as a fold change from untreated cultures. Each experiment was done in triplicate. Graphical data in this and subsequent figures are represented as mean ± SEM.
(A) carD transcript levels during DSBs. I-SceI expression is induced at time 0.
(B) carD transcript levels during genotoxic stress. M. smegmatis Mc2155 was treated with 10 µg/ml Ciprofloxacin (Cipro), 10 µg/ml bleomycin (bleo), 0.1% MMS, and 10 or 35 mM H2O2.
(C) carD transcript levels during nutrient deprivation. M. smegmatis was washed once and starved in PBS+0.05% Tween 80.
(D) Amino acid sequence alignment of selected CarD proteins. Letter coloring indicates the sequence conservation. Blue is highly conserved (>66%) and red is not conserved (<33%). The solid black line designates the region of CarD that is homologous to the TRCF RID. The dashed black line designates the leucine zipper motif.
To determine if carD upregulation is a generalized response to genotoxins, we treated wild-type M. smegmatis cultures with double strand DNA damaging agents bleomycin and Ciprofloxacin, alkylating agent methyl methanesulphonate (MMS), and the oxidizing agent hydrogen peroxide (H2O2). We observed that carD transcript increased with all genotoxins tested except high doses of H202, with the most dramatic induction observed with Ciprofloxacin and 10mM H2O2, which induced carD transcription 8 fold compared to untreated cells (Figure 1B). M. tuberculosis carD (Rv3583c) has also been identified as a transcriptionally upregulated gene in microarray experiments following exposure to the DNA damaging agents mitomycin C, UV radiation, H2O2, and quinolone DNA gyrase inhibitors (Boshoff et al., 2004; Boshoff et al., 2003).
CarD proteins are highly conserved in all mycobacteria sequenced thus far. M. smegmatis CarD shares 98.1% and 95.7% identity with the proteins encoded by M. tuberculosis Rv3583c and M. leprae ML0320, respectively. CarD proteins are highly conserved in many eubacteria, but are absent from archaebacteria and eukaryotes (Cayuela et al., 2003) (Figure 1D and Table S1), but are of unknown function . The only available information comes from work in Myxococcus xanthus and Stigmatella aurantiaca, which each express two related CarD proteins of differing length (Cayuela et al., 2003; Nicolas et al., 1994). The shorter myxobacterial CarD protein is the same length as mycobacterial CarD and contains a conserved leucine zipper motif and N-terminal region of homology to the TRCF RNAP interaction domain (RID), but is otherwise uncharacterized (Figure 1D). The longer M. xanthus CarD protein, is necessary for the activation of numerous pathways including starvation triggered fruiting body formation (Cayuela et al., 2003; Nicolas et al., 1994). Similarily, we observed 20 fold induction of M. smegmatis carD transcription during early starvation in PBS (Figure 1C). Together, the microarray and qRT-PCR data demonstrate the upregulation of carD mRNA in response to a broad range of stresses.
CarD is essential for M. smegmatis and M. tuberculosis viability
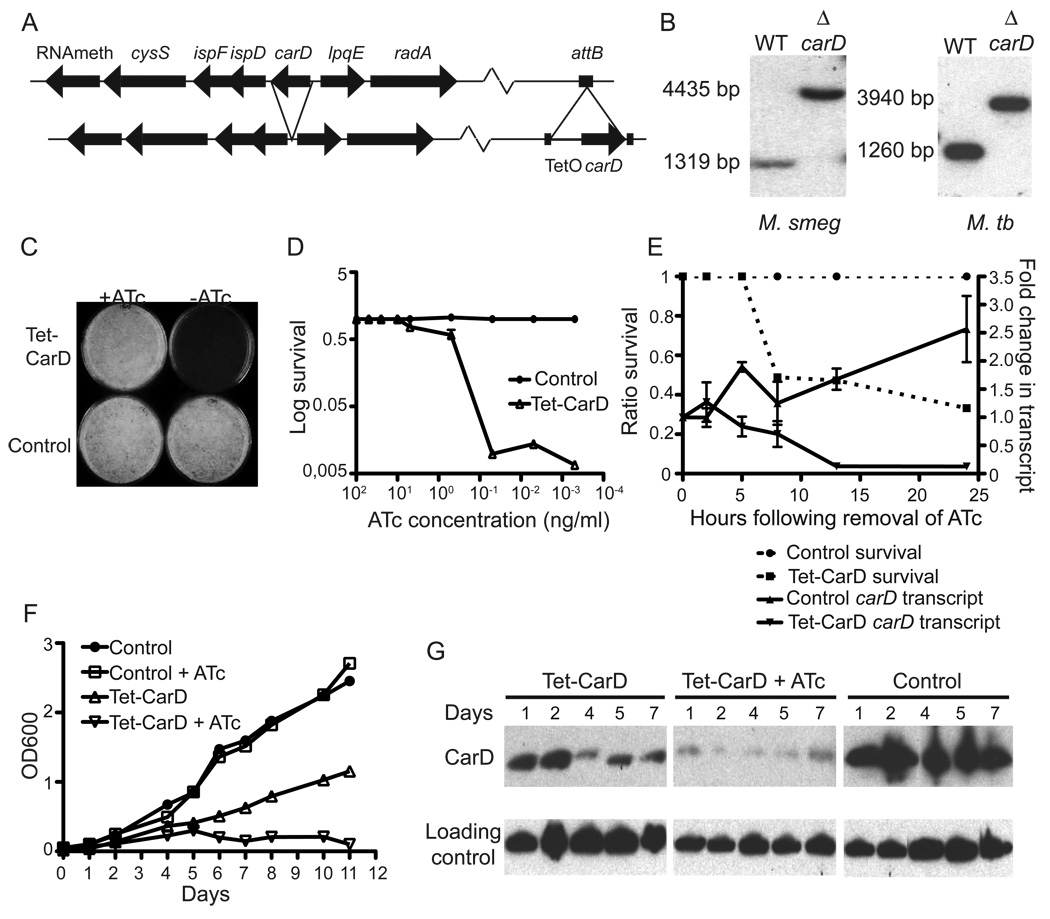
To further understand the cellular function of CarD, we undertook a genetic approach. Initial attempts to delete carD from the M. smegmatis and M. tuberculosis chromosomes were unsuccessful, strongly suggesting that CarD is essential for viability. To confirm essentiality and study CarD further, we constructed strains that allow for conditional depletion of CarD under tetracycline control. CarD expression from the attB site allowed deletion of the endogenous carD gene, thus generating the M. smegmatis and M. tuberculosis ΔcarD attb∷tetcarD strains (Figures 2A–B). To obtain conditional knockdown of M. smegmatis CarD, ΔcarD attB∷tetcarD was transformed with an episomal plasmid that directs the synthesis of TetR (Ehrt et al., 2005), a repressor that binds the operator of the tetracycline regulated promoter in the absence of anhydrotetracycline (ATc), which generated the strain mgm1703. Mgm1703 only grows well in the presence of at least 10.0 ng/ml of ATc, thus confirming that CarD is essential for viability (Figures 2C and D). A control strain, mgm1701 survives independent of ATc (Figures 2C and D). Survival assays and quantitative RT-PCR after tetracycline withdrawal showed that declining carD transcript levels directly correlated with cell death (Figure 2E).
Figure 2. CarD is essential for growth of M. tuberculosis and M. smegmatis in culture.
(A) Diagram of the carD gene region in M. smegmatis and M. tuberculosis and the construction of M. smegmatis ΔcarD attb∷tetcarD. Arrangement of M. tuberculosis ΔcarD attb∷tetcarD is identical except a hygR cassette replaces carD.
(B) Southern blot analysis of wild-type and ΔcarD attb∷tetcarD M. smegmatis (left panel) and M. tuberculosis (right panel). Wild-type M. smegmatis yields a 1319 bp band, while ΔcarD results in a 4435 bp band. Wild-type M. tuberculosis yields a 1260 bp band, while ΔcarD results in a 3940 bp band.
(C and D) Growth of M. smegmatis Tet-CarD (mgm1703) and control (mgm1701) strains on LB plates containing varying concentrations of ATc. (C) shows the growth of Tet-CarD and the control strain on plates containing either 50 ng/ml ATc or no ATc. (D) shows the ratio of survival of each strain on different concentrations of ATc as compared to in the presence of 50 ng/ml.
(E) Survival and carD transcript levels of M. smegmatis Tet-CarD and the control strain following removal of ATc. Each strain was diluted to an OD600 of 0.05 into media either with or without ATc. At each time point survival was determined by plating dilutions onto LB + 50 ng/ml ATc and is expressed as a ratio to CFUs from the respective culture in 50 ng/ml ATc (left axis, dashed lines). qRT-PCR was performed on RNA samples collected at each time point and carD transcript levels were normalized to sigA transcript levels and expressed as a fold change from time 0 (right axis, solid lines).
(F) Growth of M. tuberculosis Tet-CarD (mgm1797) and control strain (mgm1799) in 7H9 broth with or without ATc. Each strain was diluted to an OD600 of 0.05 into the appropriate media and the OD600 was measured at the designated time points. In this system, ATc represses carD expression.
(G) Western blot analysis of total protein lysates of the same cultures in (F) using rabbit polyclonal antibodies specific for CarD and DlaT (loading control).
Transformation of M. tuberculosis ΔcarD attb∷tetcarD with a plasmid that expresses revTetR (Guo et al., 2007), a repressor that binds the operator of the tetracycline regulated promoter in the presence of ATc, generated the strain mgm1797 and abolished growth in 50 ng/ml ATc (Figure 2F). Mgm1797 also grew slowly in the absence of ATc, and western blot analysis demonstrated that low CarD protein levels in mgm1797 correlated with the slow growth of the strain (Figure 2G). These experiments demonstrate that CarD is essential for growth of M. tuberculosis and M. smegmatis in culture.
CarD is necessary for survival during oxidative stress, DNA damage, and nutrient limitation
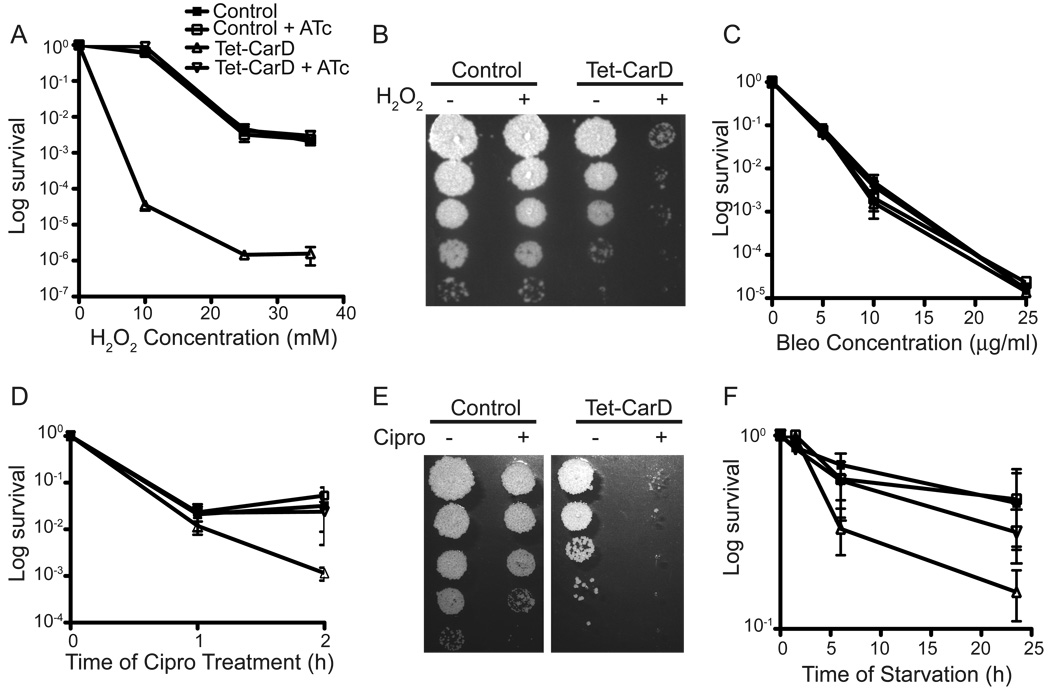
Since carD transcription is upregulated during DNA damage and nutrient deprivation, we tested whether CarD was necessary for survival under these conditions. Mgm1703 and mgm1701 cultures were diluted to an OD600 of 0.05 into media with or without 50 ng/ml ATc for 13 hours before treating with varying concentrations of H2O2 and bleomycin for 1 hour or 10 µg/ml of Ciprofloxacin for 1 and 2 hours. Thirteen hours of depletion was chosen because at this point carD RNA levels were barely detectable, but 50% of the cells were still alive (Figure 2E). We found that mycobacterial cells lacking CarD were 50,000 fold more sensitive to 10mM H2O2 and 30 times more sensitive to Ciprofloxacin treatment compared to control cells producing CarD (Figure 3A–B and D–E). CarD depletion also caused a 10 fold reduction in M. smegmatis survival during starvation (Figure 3F), but had no effect on cell survival during bleomycin treatment (Figure 3C). These data show that M. smegmatis CarD is necessary for survival during oxidative stress, double strand DNA breaks, and nutrient limitation and defines CarD as a novel mediator of mycobacterial responses to these conditions.
Figure 3. CarD is required for resistance to H2O2, Ciprofloxacin, and nutrient deprivation.
(A–E) Survival of M. smegmatis strains during genotoxic stress. The strain symbols listed in the legend in (A) are the same for all panels. Tet-CarD (mgm1703) and the control strain (mgm1701) growing in LB broth with or without ATc were treated for 1 h with varying concentrations of H2O2 (A–B), bleomycin (C), or treated for 1 and 2 hours with 10µg/ml of Ciprofloxacin (D–E). Following treatment, dilutions were plated on LB + ATc and survival is expressed as a ratio of CFUs compared to untreated cultures. (B and E) display the plating efficiency of Tet-CarD and the control strain grown in the absence of ATc after treatment with 10 mM H2O2 for 1 h (B) and 10µg/ml Ciprofloxacin for 2 h (E).
(F) Survival of M. smegmatis strains during nutrient deprivation. Tet-CarD and control strains in LB with or without ATc were washed and starved in PBS + 0.05% Tween 80 for the indicated times. Survival was determined and expressed as described above.
CarD is required for the stringent response
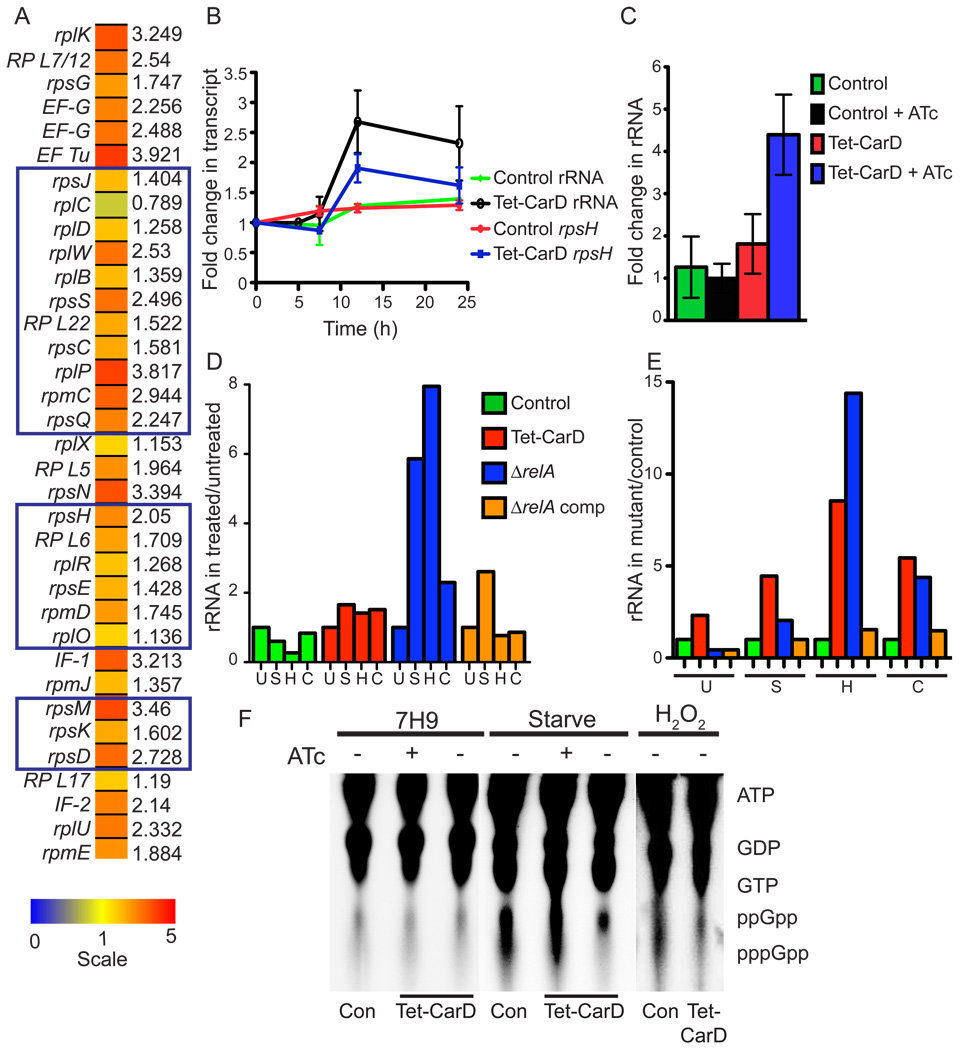
The diverse stresses that activate CarD suggest that this protein is involved in a global regulatory response. To explore the transcriptional changes caused by CarD loss, we performed whole genome transcriptional profiling on M. smegmatis cells depleted of CarD compared to CarD replete cells. The microarray analysis revealed that during CarD depletion, 193 genes were upregulated >2 fold and 176 genes were downregulated >2 fold (Table S2). The most striking result from this analysis was that 22% of the genes upregulated greater than 2 fold following CarD depletion encoded components of the translation machinery and amino acid biosynthetic pathways. This included entire operons of ribosomal proteins and translation elongation and initiation factors (Figure 4A). qRT-PCR confirmed that CarD depletion leads to an upregulation of 16S rRNA and rpsH ribosomal protein transcripts in M. smegmatis (Figure 2E and Figure 4B) and M. tuberculosis (Figure 2G and Figure 4C). Together, the microarray and qRT-PCR experiments demonstrate that CarD depletion leads to transcriptional upregulation of stable RNAs and other components of the translation machinery.
Figure 4. CarD is required for the stringent response.
(A) Heat map of upregulated ribosomal proteins and translation factors (p < 0.05) during CarD depletion in M. smegmatis. Microarray analyses compared M. smegmatis Tet-CarD (mgm1703) to the control strain (mgm1701) 13 hours after CarD depletion in LB media. The scale is shown at the bottom, gene names are listed to the left and fold changes in expression during CarD depletion compared to the control strain are to the right. Predicted operons are boxed.
(B) 16S rRNA and rpsH transcript levels in M. smegmatis strains from Figure 2E during growth in the absence of ATc, as determined by qRT-PCR and expressed as a fold change from time 0.
(C) 16S rRNA levels in M. tuberculosis strains from Figure 2F in the presence and absence of ATc, as determined by qRT-PCR. The fold change in rRNA levels at 7 days compared to day 0 is graphed.
(D and E) 16S rRNA levels in M. smegmatis control, Tet-CarD, ΔrelA, and ΔrelA complemented with a WT relA gene (ΔrelA comp) strains during starvation, oxidative, and genotoxic stress. Each strain was diluted to an OD600 of 0.05 into LB - ATc. Cultures were then left untreated (U), starved in PBS + 0.05% Tween 80 for 4 h (S), treated for 1 h with 10 mM H2O2 (H), or 2 h with 10µg/ml Ciprofloxacin (C) before collecting samples for qRT-PCR analysis of 16S rRNA. (D) shows the fold change of rRNA in each strain during treatment as compared to untreated (set to 1). Using the same data as in (D), (E) illustrates the rRNA levels in each mutant compared to the CarD+ control strain in the same conditions. In all assays, rpsH and rRNA transcript levels were normalized to sigA transcript levels.
(F) (p)ppGpp accumulation during the stringent response. M. smegmatis Tet-CarD and control strain (Con) were diluted to an OD600 of 0.05 in 7H9 broth with or without ATc for 10 h before labeling intracellular nucleotides in untreated cultures (7H9) or during nutrient deprivation (starve) and oxidative stress (H2O2) and analyzing by TLC. The migration positions of ATP, GTP, and ppGpp were determined by nonradioactive nucleotide controls.
Relaxed bacterial strains are impaired for the stringent response and fail to downregulate rRNA and ribosomal protein transcription during starvation and other stresses (Alfoldi et al., 1962; Cashel and Gallant, 1969; Nene and Glass, 1983; Tedin and Bremer, 1992). CarD depletion in nutrient rich media causes a transcriptional signature similar to the relaxed phenotype, but CarD also accumulates in response to a wide range of stresses (Figure 1), suggesting that CarD might be involved in stringent control. rRNA levels in wild-type M. smegmatis are repressed during starvation (~2 fold), H2O2 (~4 fold) and Ciprofloxacin (~1.2 fold) treatment (Figure 4D, green bars), responses that were abolished when CarD was depleted (Figure 4D, red bars). Direct comparison of rRNA levels during CarD depletion to those in control cells under each condition indicates that CarD depleted cells accumulated 4.5 fold more rRNA than wild-type cells during starvation, 8.5 fold more during oxidative stress, and 5.5 fold more during Ciprofloxacin treatment (Figure 4E).
To confirm that the regulation of rRNA during oxidative and genotoxic stress was related to the same stringent control machinery as nutrient deprivation, we analyzed rRNA levels in an M. smegmatis ΔrelA strain. Loss of RelA, an enzyme responsible for production of (p)ppGpp (Avarbock et al., 1999; Dahl et al., 2005; Dahl et al., 2003; Primm et al., 2000), abolished mycobacterial stringent control of rRNA during starvation, oxidative, and genotoxic stresses, which was reversed by expression of WT RelA (ΔrelA comp) (Figures 4D–E). Therefore, the stringent response is necessary for control of rRNA levels in mycobacteria under such conditions, and defective during CarD depletion. Unlike CarD depletion, deletion of relA did not increase rRNA levels in untreated cells and did not render M. smegmatis more sensitive to oxidative or genotoxic stress (Figure S1 and Figure 4E). These data demonstrate that CarD is necessary for the mycobacterial stringent response to starvation, oxidative, and genotoxic stresses as well as control of rRNA under normal growth conditions.
To further delineate the role of CarD in stringent control, we examined the effect of CarD depletion on the accumulation of (p)ppGpp. In untreated cultures, there was no detectable (p)ppGpp in any strain, regardless of CarD levels (Figure 4F). In wild-type cells, starvation and H2O2 both caused dramatic upregulation of (p)ppGpp. In CarD depleted cells, ppGpp still accumulated, although to a slightly lesser degree. The difference in (p)ppGpp between wild-type and CarD depleted cells is unlikely to explain the rRNA transcription deregulation observed because ppGpp accumulation during CarD depletion should lead to a partial downregulation of rRNA rather that the induction of rRNA that we observe (Figure 4). These findings indicate that ppGpp is ineffective in inducing stringent control when CarD is absent.
CarD directly interacts with the RNAP β subunit at regulated promoters
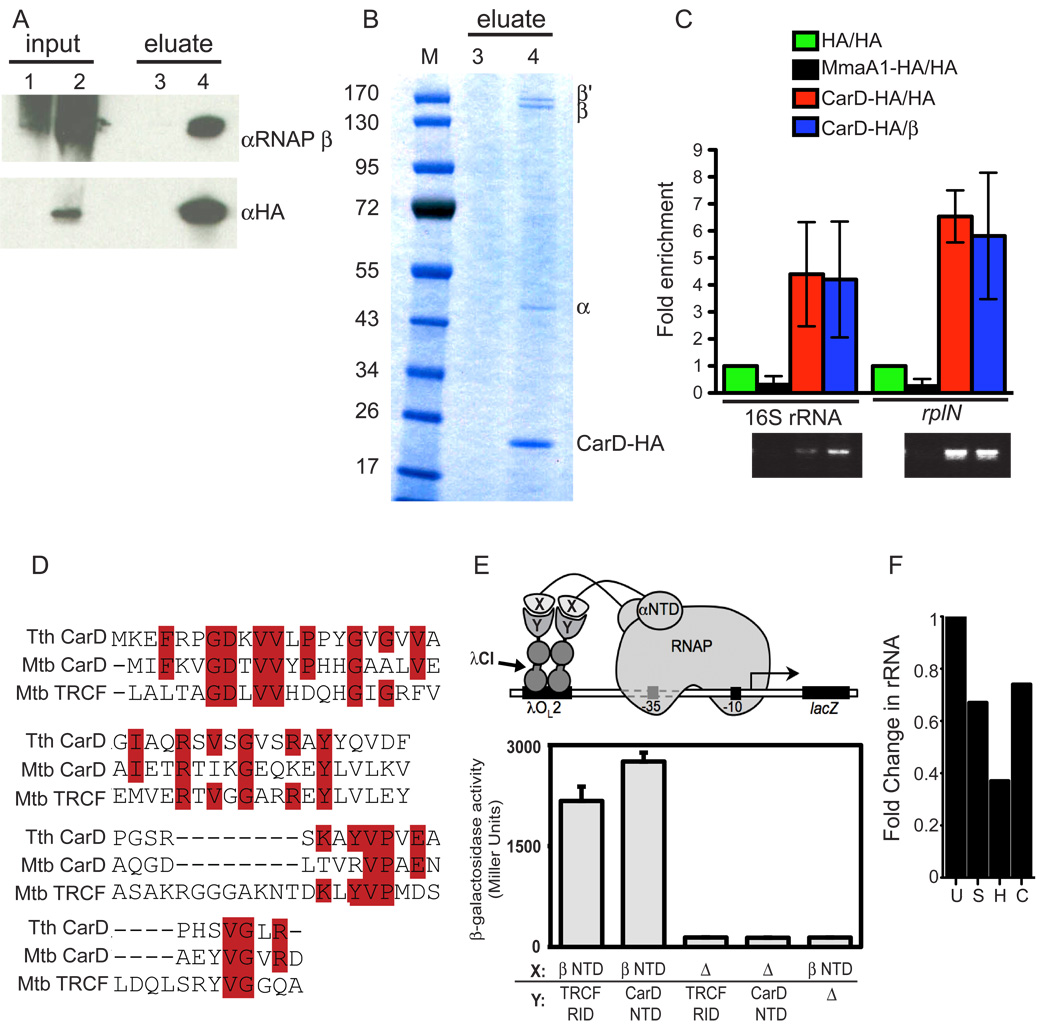
To test whether CarD regulates transcription via direct physical interactions with the RNAP, we engineered M. smegmatis ΔcarD attB∷tetcarD-HA which encodes a functional HA tagged version of CarD at the attB site. Immunoprecipitation of whole cell lysates with HA antibodies coprecipitated HA-CarD and the RNAP β subunit from the ΔcarD attB∷tetcarD-HA lysate but not from the control lysate containing untagged CarD (Figure 5A). To more comprehensively survey CarD interacting proteins in M. smegmatis, we analyzed the same eluates by SDS-PAGE and mass spectroscopy. The four major proteins detected were HA-CarD and the α, β, and β’ subunits of RNAP (Figures 5B). No other proteins on the coomassie blue stained gel were present in the same stoichiometry as the RNAP subunits, suggesting that the interaction between CarD and the RNAP is direct. These findings conclusively demonstrate that CarD forms a complex with the RNAP. Chromatin immunoprecipitation (ChIP) experiments with HA antibodies in M. smegmatis ΔcarD attB∷tetcarD-HA specifically precipitated the promoters regulating the rRNA operons or the rplN operon (encoding ribosomal proteins L14, L24, L5, and S14p/S29e) (Figure 5C), thus confirming that CarD is physically present on the RNAP complex at the rRNA and ribosomal protein loci.
Figure 5. CarD interacts directly with the N-terminus of the RNAP β subunit.
(A–B) Immunoprecipitation experiments with HA antibody in either M. smegmatis ΔcarD attb∷tetcarD (lanes 1 and 3) or M. smegmatis ΔcarD attB∷tetcarD-HA (lanes 2 and 4). (A) Inputs and eluates analyzed by western blot analysis using antibodies specific for either the RNAP β subunit or HA. (B) Eluates analyzed by coomassie blue staining. Co-precipitated RNAP β, β’, and α subunits were identified by MALDI-TOF-MS.
(C) CarD-HA and RNAP β coprecipitated fragments of DNA containing the promoters of the rRNA and rplN operons. ChIP assays were performed using antibodies specific for HA on exponentially growing M. smegmatis ΔcarD attb∷tetcarD-HA (CarD-HA/HA), M. smegmatis expressing HA alone (HA/HA), or M. smegmatis expressing MmaA1-HA (MmaA1-HA/HA) as well as using antibodies specific for RNAP β with ΔcarD attB∷tetcarD-HA cultures (CarD-HA/β). Precipitated and input DNA corresponding to the promoter regions of the rRNA and the rplN operon was determined by quantitative PCR. The graph shows the fold enrichment of DNA product compared to the amount of product precipitated from the strain expressing HA alone and agarose gel electrophoresis of PCR products. MmaA1 is a mycolic acid methyltransferase that does not interact with DNA and served as a negative control.
(D) Amino acid sequence alignment of T. thermophilus (Tth) CarD amino acids 1–60, M. tuberculosis (Mtb) CarD residues 1–60, and the M. tuberculosis TRCF RID (amino acids 516–587).
(E) T. thermophilus CarD and TRCF interact with the N-terminus of the RNAP β subunit. Cartoon depicts how contact between a protein domain (X) fused to the α subunit of E. coli RNAP and a partner domain (Y) fused to a DNA-binding protein (the CI protein of bacteriophage λ) activates transcription from test promoter placOL2–62, which bears an upstream recognition site for λCI (λOL2). In reporter strain FW102 OL2–62 (Deaconescu et al., 2006), test promoter placOL2–62 is located on an F' episome and drives the expression of a linked lacZ gene. For these experiments, the N-terminus of β (residues 10–133) was fused to α and the TRCF RID (residues 314–444) or N-terminus of CarD (residues 1–66) was fused to λCI. The bar graph shows the results of β-galactosidase assays (mean and SEM of four independent determinations) performed with FW102 OL2–62 cells that contained two compatible plasmids; one that encoded the α–β fusion protein or wild-type α (Δ), and another that encoded the λCI-TRCF RID fusion protein, the λCI-CarD NTD fusion protein or wild-type λCI (Δ).
(F) M. smegmatis Δmfd is not deficient in stringent control. M. smegmatis Δmfd was left untreated (U), starved in PBS + 0.05% Tween 80 for 4 h (S), treated for 1 h with 10 mM H2O2 (H), or 2 h with 10µg/ml Ciprofloxacin (C) before collecting samples for RNA extraction and qRT-PCR analysis of 16S rRNA. The fold change of rRNA is compared to untreated (set to 1).
The N-terminus of CarD proteins display amino acid sequence similarity to the TRCF RID (Figure 5D). TRCF, encoded by the mfd gene, revives backtracked elongating RNAPs, terminates transcription at repressor binding sites, and mediates the repair of lesions in the transcribed DNA strand by interacting with stalled transcription elongation complexes and recruiting nucleotide excision repair components to the site (Chambers et al., 2003; Deaconescu et al., 2006; Park et al., 2002; Selby and Sancar, 1995; Washburn et al., 2003; Zalieckas et al., 1998). Abundant evidence indicates that the TRCF RID interacts directly with the N-terminus of the RNAP β subunit (Deaconescu et al., 2006). We tested whether CarD interacts with the N-terminus of RNAP β using a bacterial two-hybrid assay (Dove et al., 1997; Nickels, 2009) that was previously used to detect the TRCF RID/β interaction (Deaconescu et al., 2006). Because a CarD homolog exists in Thermus thermophilus, for which a high resolution RNAP structure is available (Murakami et al., 2002), we performed the two-hybrid analysis using proteins from both M. tuberculosis and T. thermophilus. Although we were unable to detect either a CarD/β interaction or a TRCF RID/β interaction with protein sequences derived from M. tuberculosis (data not shown), we found that the N-terminal domain of T. thermophilus CarD, which shares homology with the TRCF RID and M. tuberculosis CarD (Figure 5D), interacted with the N-terminus of RNAP β (amino acids 10–133) to the same degree as did the TRCF RID (Figure 5E). Full length T. thermophilus CarD also interacted strongly with RNAP β amino acids 10–133 (data not shown). These data demonstrate that the mode of binding of CarD to RNAP is the same as TRCF.
Despite the similarities between the N-terminus of CarD and the TRCF RID, attempts to complement CarD depletion with the M. smegmatis TRCF RID were unsuccessful, indicating that RNAP binding is not sufficient for CarD activity (data not shown). Additionally, an M. smegmatis Δmfd strain does not phenocopy depletion of CarD in that TRCF is not essential in mycobacteria, loss of TRCF does not affect mycobacterial sensitivity to oxidative or genotoxic stress (Figure S1), and rRNA levels are regulated normally in Δmfd during both during normal growth and in conditions that elicit stringent control (Figures 5F). Therefore, despite the similar mechanism of interaction with the RNAP, CarD and TRCF have distinct cellular functions.
CarD can functionally replace DksA but not TRCF in E. coli
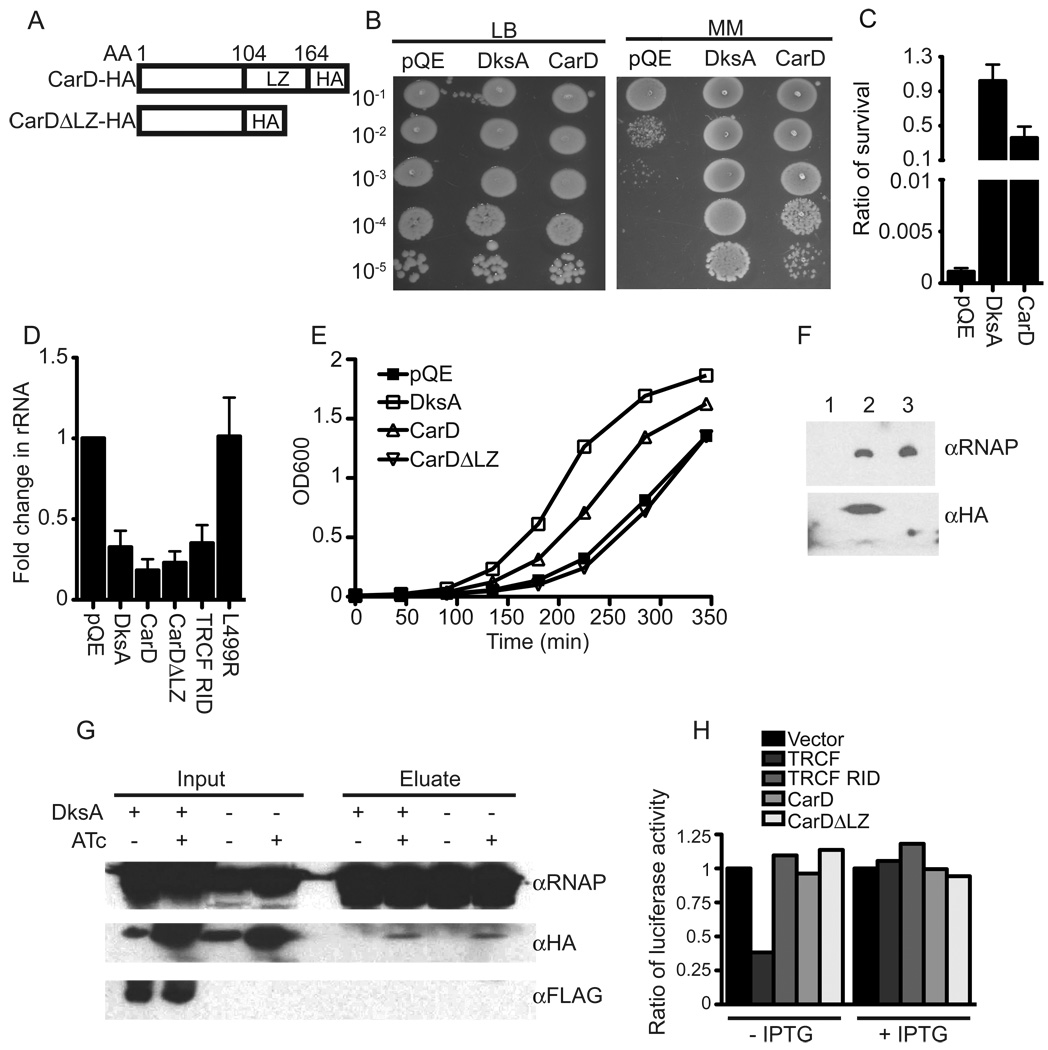
The importance of CarD in resistance to genotoxic stress and nutrient limitation, its direct association with the RNAP, and its cellular function to regulate ribosomal component transcription is reminiscent of the functions of E. coli DksA. DksA sensitizes RNAP to the action of (p)ppGpp and is essential for stringent control of transcription in E. coli (Paul et al., 2004). However, obvious DksA homologs are absent from many sequenced non-proteobacterial genomes, raising the possibility that these organisms employ distinct strategies to control transcription at ribosomal promoters. To determine if CarD is a functional homologue of DksA, we tested whether CarD could replace DksA in E. coli, assayed by rescue of the amino acid auxotrophy reported for the ΔdksA strain (Magnusson et al., 2007). Expression of M. smegmatis CarD from an E. coli promoter restored ΔdksA growth on minimal media almost to the extent as DksA itself (Figures 6B and C). CarD was also able to partially complement the small colony phenotype of the ΔdksA strain on minimal media (Figure S2). Western blot analysis with protein concentration standards showed that there were between 1000 and 3000 CarD molecules per cell in these experiments, which is comparable to the amount of endogenous E. coli DksA (data not shown) (Paul et al., 2004).
Figure 6. CarD can functionally replace DksA but not TRCF.
(A) Illustration of CarD-HA and the truncation deleting the CarD leucine zipper (CarDΔLZ-HA)
(B and C) Survival of ΔdksA complemented strains on minimal media. Dilutions of ΔdksA E. coli strains expressing DksA, CarD-HA (CarD), or vector (pQE) were plated on either LB or M9 minimal media (MM). Survival was determined by the ratio of CFUs on minimal media compared to LB.
(D) rRNA levels in ΔdksA complemented strains. E. coli ΔdksA and the indicated complemented strains were diluted into M9 minimal media for 2 h. 16S rRNA levels in each strain as determined by qRT-PCR, normalized to transcripts from the β-lactamase gene (bla) in pQE-30, and expressed as a fold change compared to levels in ΔdksA harboring the vector alone.
(E) Growth curve of ΔdksA complemented strains in LB.
(F) CarD associates with the E. coli RNAP. Immunoprecipitation experiments with HA antibody matrix of cell extracts prepared from log phase E. coli ΔdksA strains containing the vector alone (lane 1) or expressing either CarD-HA (lane 2) or CarDΔLZ-HA (lane 3). Eluates were analyzed by immunoblotting using anti-RNAP β or HA antibodies.
(G) DksA does not interact with the mycobacteria RNAP. M. smegmatis ΔcarD attB∷tetcarD-HA and M. smegmatis ΔcarD attB∷tetcarD-HA expressing FLAG-DksA were diluted to an OD600 of 0.05 in LB broth with or without ATc for 13 h before performing immunoprecipitation experiments with antibodies specific for the RNAP β subunit. Inputs and eluates were analyzed by western blot analyses using antibodies specific for RNAP β, HA, and FLAG.
(H) CarD cannot replace TRCF in the roadblock repression assay. E. coli UNCNOMFD (Δmfd) with the roadblock repression luciferase reporter construct (pRCB-KA4) and an empty pET21a vector or pET21a plasmids encoding the indicated proteins. Luciferase activities in the presence and absence of 0.5 mM IPTG shown are the average of three independent experiments and are expressed as a fraction of luciferase activity in cells harboring the empty pET21a vector (set to 1).
To test whether CarD could replace DksA in the stringent response, we measured rRNA levels during starvation and found that CarD was able to decrease the elevated rRNA levels in the ΔdksA strain during nutrient limitation (Figure 6D). The N-terminus of CarD containing the TRCF RID homology module (CarDΔLZ-HA, Figure 6A) was sufficient to compensate for dksA in terms of downregulation of rRNA, as was the E. coli TRCF RID itself (Figure 6D). In contrast, TRCF RID L499R, which does not interact with the N-terminus of the RNAP β subunit (Deaconescu et al., 2006), was inactive (Figure 6D). Overexpression of full length TRCF also did not reduce rRNA levels in ΔdksA (data not shown). These results indicate that interaction of CarD and the TRCF RID with the RNAP β subunit can reduce rRNA transcription. Despite the ability of the TRCF RID and CarDΔLZ-HA to downregulate rRNA levels in the ΔdksA strain, these proteins were not able to reproducibly complement βdksA auxotrophy (data not shown). Similarly, expression of DksA and full length CarD-HA, but not CarDΔLZ-HA, increased the growth rate of ΔdksA in LB (Figure 6E). Immunoprecipitation of CarD-HA or CarDΔLZ-HA confirmed that both proteins interact with E. coli RNAP (Figure 6F), further supporting the proposal that the N-terminus of CarD interacts with RNAP. These data clearly demonstrate that CarD can functionally replace DksA in E. coli and that interaction of either the TRCF RID or CarD at the N-terminus of the RNAP β subunit is sufficient to decrease rRNA transcription in the ΔdksA strain.
Overexpression of FLAG-DksA did not allow for deletion of carD (data not shown) in M .smegmatis, even though FLAG-DksA was able to complement a ΔdksA E. coli mutant (data not shown) and was amply expressed in M. smegmatis (Figure 6G). However, IP experiments demonstrated that E. coli DksA could not interact with the mycobacteria RNAP even when CarD was depleted, which likely explains why this protein does not replace CarD function (Figure 6G).
The data above indicate that CarD and TRCF RID share a common binding site on RNAP. To ask whether CarD could replace TRCF in displacing stalled RNAP elongation complexes, we used a roadblock repression assay (Deaconescu et al., 2006). This assay measures dissociation of a stalled RNAP at a repressor roadblock in the firefly luciferase gene. Loss of TRCF function in this assay yields higher reporter activity due to a deficiency in RNAP displacement. Only E. coli TRCF, but not TRCF RID, full length M. smegmatis CarD, or CarDΔLZ could replace TRCF in this assay (Figure 6H). This data is consistent with previous reports that interaction with the RNAP is not sufficient for RNAP displacement (Chambers et al., 2003). Thus, although CarD and TRCF RID share a common mode of interaction with RNAP and can function similarly in stringent control, CarD cannot replace the function of TRCF.
M. tuberculosis CarD is necessary for replication and persistence during infection of mice
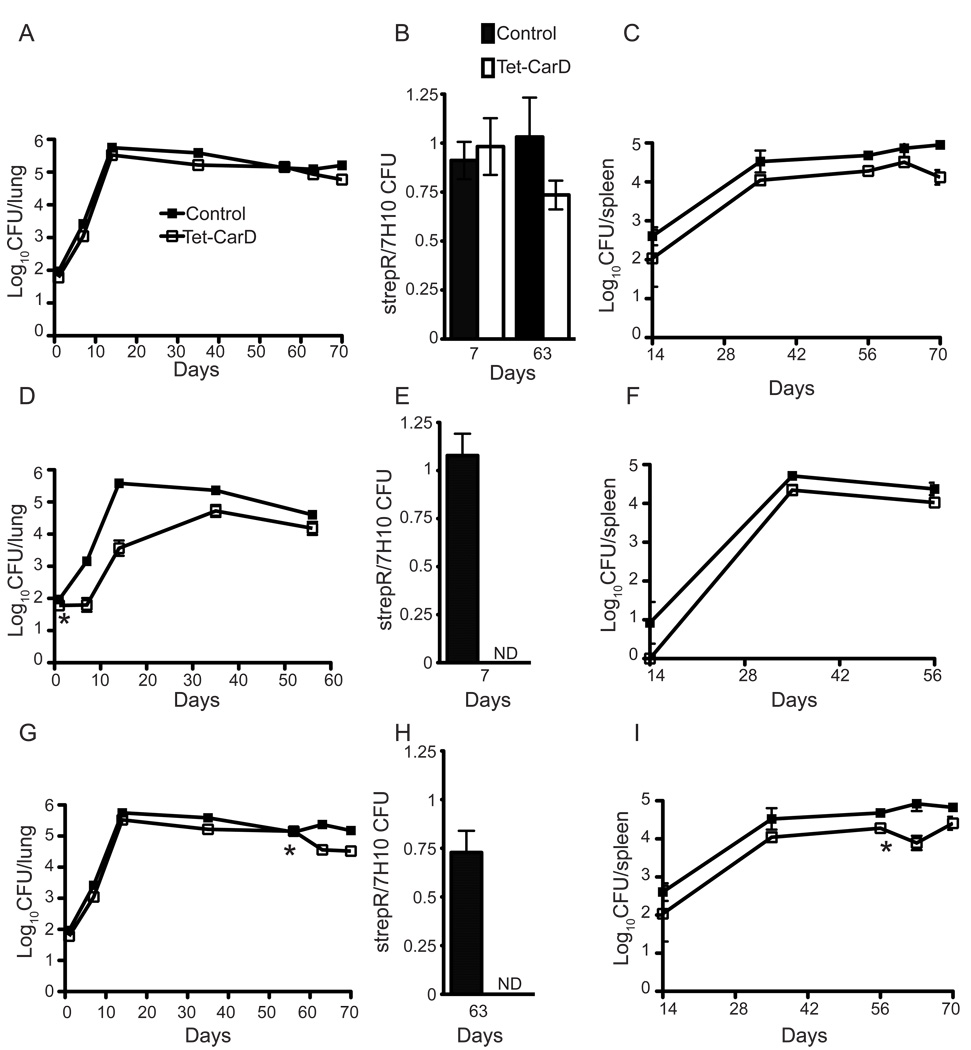
We have shown that CarD is critical for resistance to oxidative stress, DNA damage, and nutrient deprivation in culture; therefore we hypothesized that CarD would be important during M. tuberculosis pathogenesis, when host immunity imposes these stresses on the bacterium. To test this hypothesis, mice were infected with either mgm1799 or mgm1797, the latter of which allows for conditional suppression of CarD expression, and bacterial titers in the lungs and spleens were determined (Figure 7). In the absence of doxycycline, mgm1799 and mgm1797 displayed similar kinetics and virulence in mice (Figures 7A and C). Separate groups of mice received doxycycline on day 1 (acute infection, Figures 7D–F) or day 56 (chronic infection, Figures 7G–I) to determine the role of CarD in acute and persistent infection. Within a week of CarD depletion at day 1, mgm1797 cells were unable to replicate, whereas, mgm1799 cells were unaffected. Additionally, none of the surviving mgm1797 cells at day 7 retained the revTetR plasmid (Figure 7E). During persistent infection, repression of CarD by doxycycline caused a 73–75% reduction in mgm1797 titers in the lung and 50–65% decrease in the spleen (Figures 7G and I). All surviving mgm1797 cells also lost the strepR plasmid expressing revTetR, which was highly stable in both strains without doxycycline treatment (Figure 7B and H). This indicates that while some mgm1797 cells died, cells surviving doxycycline treatment had all lost the revTetR plasmid to restore CarD expression. These data demonstrate that CarD is essential not only for bacterial replication in vivo, but also during the persistent phase of infection, when bacterial titers are stable.
Figure 7. CarD is necessary for replication and persistence of M. tuberculosis in mice.
Bacterial titers in the lungs (A, D, G) and spleens (C, F, I) of C57Bl/6 mice infected with either mgm1799 (control) or mgm1797 (Tet-CarD), both carrying the revTetR on a strepR plasmid. The strain symbols listed in the legend for (A) and (B) are the same for all panels. (A–C) were given regular drinking water throughout the infection, (D–F) were administered doxycycline in their drinking water starting on day 1 (designated by the star), and (G–I) were given doxycycline in their drinking water starting on day 56 (designated by the star). (B, E, H) show the ratio of CFUs from the lung grown on 7H10 plates containing streptomycin as compared to 7H10 containing no antibiotics. ND denotes when no colonies were detected after plating 4% of the lung homogenate. Data are means ± SEM of four mice per group and time point from one of two replicate experiments.
Discussion
We have identified CarD as a novel RNAP interacting protein that controls rRNA transcription in mycobacteria both at steady state and during diverse cellular stresses including starvation, DNA damage, and oxidative stress. CarD is a new member of an expanding family of proteins and small molecule nucleotide effectors that modulate transcription without binding DNA as part of the prokaryotic global response to stress. Notably, CarD proteins are widely distributed, indicating that the mechanisms identified here are important in diverse bacterial species, including the pathogens M. tuberculosis, Bacillus anthracis, Mycobacterium leprae, Leptospira, and Clostridium.
A distinct mechanism of rRNA transcription regulation through CarD
CarD can replace some functions of DksA in E. coli and physically associate with the RNAP, but the mechanics of the CarD-RNAP interaction are distinct from DksA-RNAP. The three dimensional structure of DksA is highly similar to GreA and GreB, the transcript cleavage factors which protrude deeply into the secondary channel of RNAP (Opalka et al., 2003; Perederina et al., 2004; Stebbins et al., 1995). Structural modeling and experimental evidence support a model in which DksA exerts its effects on the RNAP through interactions within the secondary channel. In contrast, the CarD N-terminus has a TRCF RID module and interacts with the N-terminus of the RNAP β subunit, where TRCF binds. Accordingly, TRCF RID alone can mimic the effect of CarD on rRNA transcription in an E. coli ΔdksA strain. These results suggest that CarD affects rRNA transcription through a novel mechanism. Although our data identify a new molecular solution to the control of RNAP during rRNA transcription in mycobacteria, we note that a phylogenetic analysis of DksA and CarD in sequenced bacterial genomes identifies some organisms that lack both DksA and CarD, such as Staphlycoccus aureus (Table S1). This implies that these organisms have evolved yet other mechanisms to regulate rRNA levels.
Why is carD essential in mycobacteria?
Our data demonstrate that the function of CarD in mycobacteria shares many similarities to that of DksA in E. coli. However, a major difference between CarD and DksA is that CarD is essential for mycobacterial viability under all conditions, whereas, E. coli DksA is dispensable in nutrient rich media. There is emerging evidence that RNAP arrays are impediments to DNA repair and replication, and pathways that destabilize RNAP are important to maintain genomic integrity (Trautinger et al., 2005; Trautinger and Lloyd, 2002). The synergy of dksA mutations with mutations in ruvC/mfd/greA (Trautinger et al., 2005) supports the model that loss of dksA impairs viability when RNAP function and DNA repair are further compromised. Thus, one model for the essentiality of CarD in mycobacteria is that CarD loss during logarithmic growth focally increases transcription at the limited number of rDNA operons in the mycobacterial chromosome, causing a lethal impediment to replication. Although CarD of B. subtilis, which contains 10 rRNA operons, has not been examined experimentally, the gene is nonessential in saturation mutagenesis screens (Kobayashi et al., 2003).
CarD in starvation, oxidative stress, and DNA damage responses that are required for pathogenesis
Our data indicate a unique role for CarD in the stringent response to oxidative and genotoxic stresses and starvation. The stringent response in E. coli is most extensively characterized as a response to amino acid starvation. However, (p)ppGpp is produced in response to diverse cellular stresses, including hydrogen peroxide (VanBogelen et al., 1987), suggesting that downregulation of rRNA transcription is a common mechanism of adaptation to conditions that impair cell growth or translational need. Accordingly, we demonstrate that loss of a mycobacterial enzyme responsible for (p)ppGpp synthetase and hydrolase activities, designated RelA, renders mycobacteria unable to downregulate rRNA levels not only during starvation, but also during oxidative and genotoxic stress. The functions of CarD identified here are especially relevant to M. tuberculosis pathogenesis because M. tuberculosis infection is characterized by widely varying growth rates and exogenous stresses during infection. In addition, we have shown that CarD is not only essential during M. tuberculosis replication in mice, but also for persistence, a time when the bacteria are replicating slowly, if at all, but still must withstand host derived oxidative stress and starvation (Munoz-Elias et al., 2005). RelA has previously been shown to be critical for successful establishment of persistent infection in mice, also highlighting the importance of the stringent response during pathogenesis (Dahl et al., 2003). Our identification of CarD as critical to these processes suggests that targeting CarD with small molecules, possibly through disruption of the CarD/RNAP interaction, may be an effective strategy for M. tuberculosis chemotherapy. Rifampin, a critical antimycobacterial agent for treatment of Tuberculosis infection, provides a strong example of the efficacy of targeting the RNAP complex with antibiotics (Campbell et al., 2001).
In summary, we have identified CarD as a novel regulator of RNAP transcription complexes at rRNA and ribosomal protein genes both during growth and cellular stress. These studies significantly expand the known molecular mechanisms that control ribosome biogenesis in prokaryotes and identify CarD as an essential mediator of the mycobacterial stringent response to diverse cellular stresses.
Experimental Procedures
See supplemental information for more detailed methods, including media, primers, and plasmid and strain construction.
DNA Microarrays
cDNA coupled to fluorescent dyes Cy3 and Cy5 (GE Healthcare) were hybridized to gene chips spotted 4 times with oligos representing 6746 M. smegmatis ORFs (obtained from TIGR through the Pathogen Functional Genomics Resource Center). Experiments were done in triplicate and results were processed with GenePix software and analyzed in GeneSpring GX. The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numberGSE15437.
Animal infections
Before infection, exponentially replicating mgm1799 and mgm1797 growing in the absence of ATc were washed in PBS + 0.05% Tween 80, and sonicated to disperse clumps. 8–9 week old female C57Bl/6 mice (Jackson Laboratory) were exposed to 8 × 107 CFU of the appropriate strain in a Middlebrook Inhalation Exposure System (Glas-Col), which delivers 100 bacteria per animal. Groups of infected mice received 1.5 mg/ml doxycycline (Sigma) in drinking water containing 5% sucrose twice a week. Bacterial burden was determined by plating serial dilutions of lung and spleen homogenates onto either 7H10 agar plates or 7H10 plates containing streptomycin. Plates were incubated at 37°C in 5% CO2 for 3 weeks prior to counting colonies. Mouse procedures were reviewed and approved by the Institutional Animal Care and Use Committee of Sloan Kettering Institute.
Supplementary Material
Acknowledgements
The authors thank Dirk Schnappinger and Sabine Ehrt for reagents and advice about the tet system, Alex Lash for assistance with bioinformatic analyses, Seth Darst for advice and design of the thermus β fragment, Nigel Savery for materials and assistance relating to the roadblock repression assays, the Genomics Core at MSKCC for microarray hybridization and processing, and Ken Marians for critical reading of the manuscript. M. smegmatis microarrays were provided by NIAID/PFGRC. This work was supported by NIH grant GM073829 to Seth Darst, GM44025 to AH, AI064693 to MSG, and AI075805 to CLS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfoldi L, Stent GS, Clowes RC. The chromosomal site of the RNA control (RC) locus in Escherichia coli. J Mol Biol. 1962;5:348–355. doi: 10.1016/s0022-2836(62)80077-1. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39:11640–11648. doi: 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Branny P, Pearson JP, Pesci EC, Kohler T, Iglewski BH, Van Delden C. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J Bacteriol. 2001;183:1531–1539. doi: 10.1128/JB.183.5.1531-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier NA, Libby SJ, Xu Y, Loewen PC, Switala J, Guiney DG, Fang FC. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Invest. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Cayuela ML, Elias-Arnanz M, Penalver-Mellado M, Padmanabhan S, Murillo FJ. The Stigmatella aurantiaca homolog of Myxococcus xanthus high-mobility-group A-type transcription factor CarD: insights into the functional modules of CarD and their distribution in bacteria. J Bacteriol. 2003;185:3527–3537. doi: 10.1128/JB.185.12.3527-3537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription--repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol. 2005;187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE., 3rd The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Nathan CF. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect Immun. 2005;73:4581–4587. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. Silencing Mycobacterium smegmatis by using tetracycline repressors. J Bacteriol. 2007;189:4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Cohen MS. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. Faseb J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam RA, Weisbrod TR, Martin J, Scuderi JD, Brown AM, Cirillo JD, Bloom BR, Jacobs WR., Jr In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol. 2005;57:97–110. doi: 10.1111/j.1365-2958.2005.04677.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, Timm J, Botha T, Chan WT, Gomez JE, McKinney JD. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun. 2005;73:546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at a 4A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- Nene V, Glass RE. Relaxed mutants of Escherichia coli RNA polymerase. FEBS Lett. 1983;153:307–310. doi: 10.1016/0014-5793(83)80630-9. [DOI] [PubMed] [Google Scholar]
- Nickels BE. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods. 2009;47:53–62. doi: 10.1016/j.ymeth.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nicolas FJ, Ruiz-Vazquez RM, Murillo FJ. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 1994;8:2375–2387. doi: 10.1101/gad.8.19.2375. [DOI] [PubMed] [Google Scholar]
- Ojha AK, Mukherjee TK, Chatterji D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun. 2000;68:4084–4091. doi: 10.1128/iai.68.7.4084-4091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE., 3rd The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- Shiloh MU, Manzanillo P, Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–330. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst SA. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K, Bremer H. Toxic effects of high levels of ppGpp in Escherichia coli are relieved by rpoB mutations. J Biol Chem. 1992;267:2337–2344. [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen RA, Kelley PM, Neidhardt FC. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn RS, Wang Y, Gottesman ME. Role of E.coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J Mol Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R, 3rd, Foster JW. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol. 1999;34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- WHO. Global Tuberculosis Control. 2009 http://www.who.int/tb/publications/global_report/2009/en/index.html.
- Zalieckas JM, Wray LV, Jr., Ferson AE, Fisher SH. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.