Abstract
This article presents the latest developments in neuroscience information dissemination through the SenseLab suite of databases: NeuronDB, CellPropDB, ORDB, OdorDB, OdorMapDB, ModelDB and BrainPharm. These databases include information related to: (i) neuronal membrane properties and neuronal models, and (ii) genetics, genomics, proteomics and imaging studies of the olfactory system. We describe here: the new features for each database, the evolution of SenseLab’s unifying database architecture and instances of SenseLab database interoperation with other neuroscience online resources.
Keywords: neuroscience, databases, SenseLab, neuroinformatics, Human Brain Project
INTRODUCTION
The SenseLab suite of databases (www.senselab.med.yale.edu/senselab), supported in part by the Human Brain Project [1, 2], is an online resource for neuroscience information. SenseLab was developed to disseminate information related to neuronal membrane properties with special emphasis on the olfactory system. SenseLab currently contains seven databases: NeuronDB, CellPropDB, ModelDB, Olfactory Receptor Database (ORDB), OdorDB, OdorMapDB and, most recently, the pilot BrainPharm database. These databases combine to provide neuronal, genomics/genetics, proteomics and imaging information to the neuroscience research community and the public.
Over the years, SenseLab’s databases and information pages have garnered several million hits and unique visits. A logging system surveys and analyzes the usage of SenseLab resources (http://senselab.med.yale.edu/senselab/site/log/reports/) [3]. Analyses of visitor IP (Internet Protocol) addresses indicate that SenseLab’s databases have been accessed extensively by people across the world. These resources have been used in neuroscience research, teaching, thesis-writing and database design. The number of web page requests has significantly increased in the past two years.
One of the unique features of SenseLab is that all the databases are implemented in a unifying database architecture—EAV/CR (Entity Attribute Values with Classes and Relationships) [4]. This architecture helps assure that new types of information can be added to each database with few, if any, programming modifications. In the EAV/CR schema, an ‘entity’ or object is described by ‘attributes’ for which ‘values’ are stored. Complex values can be stored as entities with their own attributes. For example, if a certain laboratory is responsible for work related to olfactory receptors, then Laboratory, which is an ‘attribute’ for the ‘entity’ Olfactory Receptor, is itself stored as an independent entity. This ensures that the Laboratory ‘entity’ can have its own set of ‘attributes’, such as name, address, phone number, etc. The EAV/CR architecture has also facilitated the addition of new databases in SenseLab.
Several publications and book chapters have described the evolution of SenseLab over the years [5-18]. This article will describe recent developments in SenseLab’s information dissemination, automatic deposition of information into its databases, and interoperation between SenseLab databases and other neuroscience databases.
EAV/CR: NEW DEVELOPMENTS IN SenseLab ARCHITECTURE
The EAV/CR [4] data storage approach was devised to address the issues of data integration and software maintenance in SenseLab [19], with a view to potential applicability to other bioscience databases. EAV/CR applications have evolved to facilitate data navigation and flexible, ad-hoc, metadata searching [11]. In NeuronDB (see next section), for example, one can navigate from a link to a neuron to access that neuron’s properties. If a computational neuronal model exists for this neuron (including its properties), then the property page for this neuron provides a link to computational models of the neuron in ModelDB (described later). Each class can also have sub-classes that are stored independently as classes. Adding more classes or categories (data specialization) to any of the classes in the database does not require additional programming. The metadata-driven approach to creating web interfaces (on the fly) in the EAV/CR framework automatically adapts to new classes and to the relationships between these classes. Thus, EAV/CR facilitates database adaptability for the continually evolving domain of neuroscience.
The EAV/CR framework has also been extended to databases outside the SenseLab project. It has been used in the creation of the Neuroscience Database Gateway (NDG) http://ndg.sfn.org, a registry of web resources related to neuroscience. SenseLab has been actively participating in database integration experiments with NDG through the development of a knowledge-integration mechanism known as the ndg.disco interoperability framework. Use of ndg.disco involves identifying and creating workflows between databases. These workflows may include integrator systems such as database mediators, ontological servers, text search engines or gateways. Other resources, BIRN-Mediator (http://www.nbirn.net/research/data_integration/data_mediation_FAQ.shtm), BIRN Bonfire (http://www.nbirn.net/research/ontology/index.shtm) and the BrainML/Metal (http://brainml.org), have contributed to this integration project.
One specific use of the ndg.disco framework is seen in the Entrez LinkOut Broker. Entrez is the NCBI information-dissemination web-portal. NCBI houses comprehensive listings of published and deposited information related to genes, proteins, journal articles, etc. The LinkOut Broker allows different databases that provide links to Entrez to be, in return, linked to by Entrez wherever relevant. For example, in SenseLab, ORDB entries for chemosensory receptors are linked to accession numbers in Entrez (PubMed and GenBank entries). In return, when a user accesses a journal article in Entrez, the web page for that journal article in PubMed also provides a link back to the page containing the relevant chemosensory receptor in ORDB. The NDG web resource is used as a mediator for all neuroscience-based databases and Entrez: every database registered at NDG can take advantage of this facility and use the LinkOut Broker utility to define bi-directional links to Entrez.
NEURONDB AND CELLPROPDB
Both NeuronDB (http://senselab.med.yale.edu/senselab/NeuronDB) and CellPropDB (http://senselab.med.yale.edu/senselab/CellPropDB) are repositories of published research information related to three neuronal membrane properties: voltage gated conductances, neurotransmitter receptors and neurotransmitter substances. Whereas CellPropDB provides this information with reference to a neuron as a whole, the information in NeuronDB is localized to specific neuronal compartments. The EAV/CR schema methodology allows users of CellPropDB and NeuronDB access to similar information, the differences are only in the granularity of information sought (neuron as a whole versus at the compartmental level). Both databases present information related to 20 neurons and 11, recently added interneurons, which represent every major region on the brain (Figure 1). Information stored in the two databases can be presented to the end user in the brain region-neuron-property hierarchy.
Figure 1.
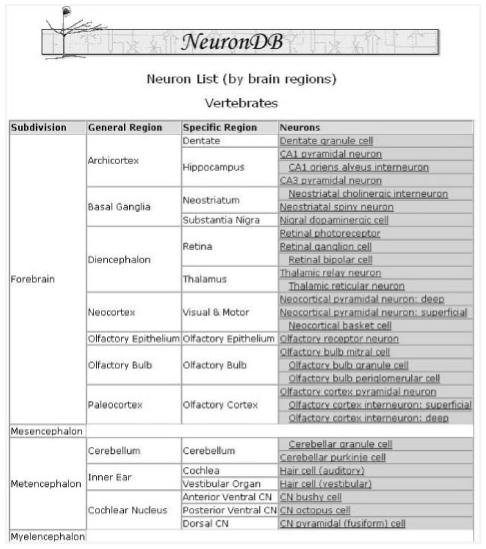
List of neurons and interneurons included in NeuronDB. A similar figure can be found in CellPropDB. The columns in the figure show the region-sub-region-neuron-property hierarchy.
Figure 2 shows the information in NeuronDB presented for each neuronal compartment with annotations for the relevant publication entered by the domain experts. Both CellPropDB and NeuronDB databases are not necessarily repositories of all the published information. They seek to present first-principles perspectives of neuronal membrane properties for a fixed set of neurons.
Figure 2.

Information presented in NeuronDB. The columns indicate the receptor, current, transmitter properties for the distal apical dendrite (Dad) compartment of the Ca1 Pyramidal Neuron.
The continuing maintenance of NeuronDB will be facilitated by information-extraction tools such as NeuroText [13]. NeuroText mines the abstracts of neuroscience journal articles and is designed to help the database curator make a decision as to whether an article should be included in NeuronDB. NeuroText uses indexing, context matching, concept based scoring, hierarchical matching, lexical parsing and supervised and unsupervised learning. Each processed abstract is marked-up to inform the domain expert as to how NeuroText arrived at its suggestion as to whether to populate NeuronDB and CellPropDB. NeuroText also dynamically creates a form, which the domain expert can use to evaluate the text-mining results, make changes dynamically and directly deposit the information into the databases if desired.
One of the first instances of interoperability in neuroinformatics involved NeuronDB. NeuronDB currently interoperates with CCDB (Cell Centered Data Base—http://ccdb.ucsd.edu) housed at the University of California at San Diego. While NeuronDB provides information related to neuronal membrane properties for a fixed set of neurons in specific regions of the brain, CCDB provides microscopy imaging data related to these neurons [8].
A pilot tool NeuroExtract has been developed [20] in conjunction with NeuronDB to perform text-based integrated queries from generalized sources of data, which contain neuroscience concepts only as sub-categories. NeuroExtract uses concepts stored in NeuronDB and CellPropDB to query and extract information from other neuroscience sources and sources that contain neuroscience information as sub-categories.
ORDB AND ODORDB
ORDB and OdorDB along with OdorMapDB (described in following section) are dedicated to disseminating genetic, genomic, proteomic and imaging information related to the olfactory system. Olfactory Receptors (ORs) are a class of GTP-Binding Protein Coupled Receptors (GPCR). GPCRs are membrane receptors that are responsible for myriad functions that involve biological cell membranes, and which find direct applications in the pharmaceutical industry [21]. The study of ORs has gained momentum in the decade and a half since the first OR was identified in a seminal paper by Linda Buck and Richard Axel [22]. Buck and Axel received the Nobel Prize in medicine in 2004 for this work and subsequent contributions to the field of chemosensory research. ORDB plays a key role in the olfactory receptor research field. It also serves as a warehouse of information related to fungal receptors, pheromone receptors, insect olfactory receptors and mammalian olfactory and taste receptors. ORDB represents the work of more than 100 laboratories worldwide. It contains information for more than 40 different species. OR-like genes (judging by sequence similarities) can be found in different tissues in different organs.
Functional analyses of olfactory receptors involve experimentally identifying specific odors excite or inhibit specific ORs. Research on OR function has shown [23] that there exists a many-to-many relationship between ORs and odorous molecules (more than one OR is likely to be excited by an odor and more than one odor compound is likely to excite an OR). OdorDB is a repository of odors that have been shown to excite olfactory receptors. ORDB interoperates with OdorDB: OdorDB contains a list of odors with links to the ORs (listed in ORDB) they are known to excite. Similarly, for each OR, the odors that excite these ORs are listed under the attribute ‘Ligand’.
The information stored in ORDB and OdorDB is constantly being updated. ORDB has grown from a list of a thousand ORs and other chemosensory receptors that were reported in GenBank to more than thirteen thousand such receptors in early 2007. The large increase in the list of receptors in the last few years has been largely due to the publications of drafts of mammalian genomes: humans, mouse, rat and chimpanzee [24-33]. The continuous and rapid update of ORDB was possible due to AutoPop [15], an automatic information deposition tool that interoperated between sources of olfactory receptors, such as GenBank, SwissProt and EMBL and ORDB. AutoPop is a web-based tool that extracts information relevant to ORDB from web pages using HTML or XML file parsers by mapping the information from the source databases to corresponding attributes (descriptors for each OR or chemosensory receptor) in ORDB and then directly depositing this extracted information into the database.
Since ORDB is the centralized repository of chemosensory receptor information, ORDB curators and scientific coordinators were charged with the task of performing a sequence analysis of OR genes identified from different groups from the mouse and human genomes. Two web pages (http://senselab/senselab/ORDB/files/humanorseqanal.html) and (http://senselab/senselab/ORDB/files/mouseorseqanal.html) illustrate the results of this endeavor. The web pages contain different color mark-ups to identify sequences that were full-length, partial and pseudogenic. Links to GenBank and EMBL web pages (wherever available) are provided for each OR gene.
Functional analysis of olfactory receptors is often a challenging task. Fewer than 50 odor compounds have been identified as having an excitatory or inhibitory effect on ORs. The specific details of the experimental challenges in performing functional studies [34] are beyond the scope of this article. A related difficulty is testing odors out of several thousand known to identify which odor might excite a particular OR. Experimentalists often have to rely on modeling and theoretical binding studies to identify a select number of odor-ligands, which might successfully bind and excite or inhibit OR function. One attribute added to ORDB is ‘OR Model’ which provides a link to a picture or a movie file of a computationally determined OR model and an odor-molecule computationally docked in the OR binding region. A PubMed link to functional studies if any for an OR allows an end user to access all the information for that particular OR or chemosensory receptor. Figure 3 shows the web page for a single OR gene entry.
Figure 3.
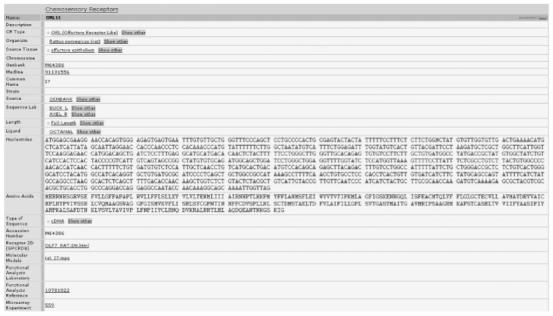
Entry in ORDB. Rat ORI7 was one of the first cloned and identified. The figure shows the attributes and values for this OR. Values that appear as links are also stored as attributes. The last attribute in the figure provides a link to an entry in ORMicroarrayDB with links to the results of microarray experiments for this OR.
Recently, a multi-attribute search has been instituted in ORDB, using search capabilities already developed for other SenseLab databases. The EAV/CR schema on which SenseLab is based ensures that any new feature instituted for one database is easily applicable to other databases. This search feature is critical in a database that has seen an almost exponential increase in deposition of gene information for chemosensory receptors. Figure 4 shows sample search pages for ORDB. The search results are available in HTML (with links to entries), text and XML formats. The search page in ORDB can be accessed at http://senselab/senselab/ORDB/eavXSearch.asp and a tutorial describing the use of the program is available at the following web page: http://senselab/senselab/ORDB/search_tutorial.asp.
Figure 4.
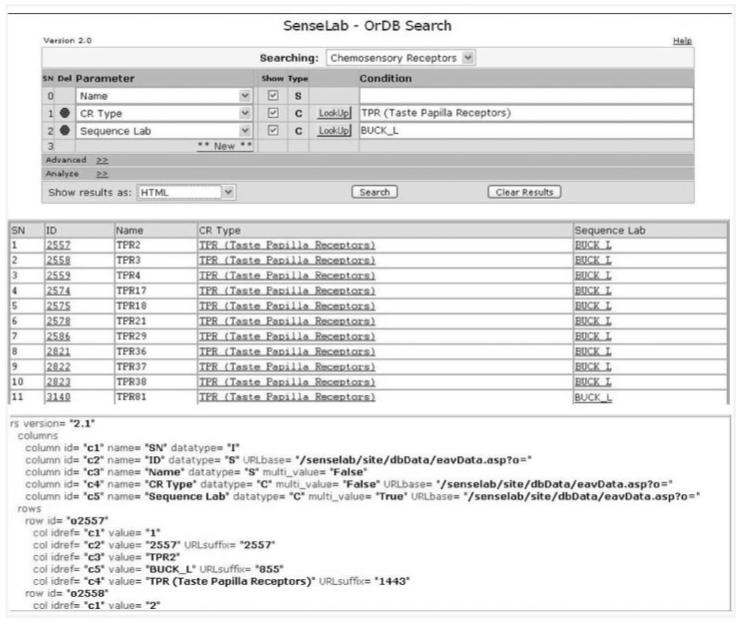
Results of the multi-attribute search in ORDB. This search feature appears in all SenseLab databases. The end user here wishes to find all taste receptors that have been explored by the laboratory of Dr Linda Buck. The figure also shows partial results in both HTML and XML formats.
The most recent attribute added to ORDB is a link to a microarray experiment for every OR gene-entry, if such a microarray experiment has been performed. The link in the ‘Microarray Experiment’ attribute leads the user to the Olfactory Receptor Microarray Database (ORMD- http://neurolab.med.yale.edu/ormd), which contains descriptions of results from microarrays containing this specific gene.
OdorDB contains a list of 45 odors. As mentioned previously, OdorDB is not a repository of all known odor compounds but tracks the research related to the functional analysis of olfactory receptors. The list of odors in OdorDB (saved as entities) is also stored in ORDB under the attribute heading ‘Ligand’. Clicking on the ‘OR by Ligand’ link takes the user to a page containing this list of odors with numbers besides them. The numbers indicate how many ORs this odor-ligand is known to excite. By clicking the number-link, the user can access a page with a list of the ORs; clicking each of the OR links takes the user to the OR entry in ORDB. Though these entries are accessed through ORDB, the mark-up and colors appear as if the information is stored separately in OdorDB. Much like the entries that are dynamically presented in CellPropDB and NeuronDB discussed in the previous section, each data source, therefore, appears to operate independently even though the actual data are shared.
Within OdorDB, users can access the odors based on the ORs they are known to excite. Odors are also stored based on specific features of these compounds: hydrocarbon features, cyclic, straight-chain or branched, and by their functional groups. Figure 3 shows the web-page for entry related to octanal. The entity ‘octanal’ is described in terms of its Chemical Abstracts Service (CAS) number, its chemical formula, its functional group (useful for searches), its hydrocarbon feature designation, a diagram of its structural formula and a link to a map (if available) of glomerular activity in the olfactory bulb. The last attribute allows OdorDB to interoperate with OdorMapDB described in the next section.
ODORMAPDB
The sense of smell in animals originates in the olfactory sensory neurons (OSNs) in the neuroepithelium, which is at the roof of the nasal cavity. Axons from OSNs cross the cribriform bone and project onto the olfactory bulb (OB), the first relay station of olfactory signals in the brain. Projections of the sensory neurons have been illustrated in the OB as well as the olfactory cortex [35, 36]. More than a decade before olfactory receptor genes were first identified [22], odor-induced spatial patterns of neural activity in the OB had been discovered by researchers using odor mapping techniques where a series of sequential coronal OB sections were examined and the activity in the roughly spherical glomerular layer was plotted in a 2-dimensional flat map [37]. This technique has since been extended to a broad range of OB activities, including neuronal uptake of 2-deoxyglucose (2DG), expression of immediate early gene c-fos and fMRI. [12, 38, 39] Odor maps are a medium by which functional imaging data can be presented. The maps describe the unique spatial activity patterns in the glomerular layer of the main OB in animals exposed to odor stimuli. OdorMapDB (http://senselab.med.yale.edu/senselab/OdorMapDB) is a data repository for the odor maps generated by researchers at Yale University and at other laboratories [9].
The goal of archiving odor maps is to create a resource of functional properties of the OB that is available to the olfactory research community as well as the public. OdorMapDB is a web-accessible database. Images are stored as binary data-type in an Oracle database management system. Each odor map ‘entity’ is described using the ‘attributes’: species, age, odor type, concentration, mapping method, etc. It is integrated with two other SenseLab olfactory databases, i.e. ORDB and OdorDB (described in the previous section). These three databases taken as a whole provide a multi-faceted knowledge-based informatics tool to help address the mechanisms underlying the olfactory perception.
OdorMapDB continues to evolve in terms of both its content and database schema. The database now contains fMRI odor maps for isoamyl acetate, aliphatic aldehyde homologues, ketones and odor mixtures. In addition to fMRI and 2DG maps, OdorMapDB now archives c-fos odor maps [39]. The database schema of OdorMapDB has been expanded to include the original 3D data from which the maps are derived. This allows users to examine the odor-induced neural activity that occurred in the entire bulb. OdorMapDB also archives fMRI data obtained from the accessory-OB. This is a unique bulbar region responsible for pheromone perception. The accessory-OB plays an important role in an organism’s social and reproductive behaviors [6].
OdorMapDB interoperates with a large 2DG odor map database developed at the University of California at Irvine [40]. 2DG, c-fos and fMRI approaches examine different neuronal properties. Odor maps from these studies are therefore not redundant. They offer different perspectives in an attempt to answer the same questions—the response in the olfactory bulb to olfactory stimuli. Odor map databases face a challenge to archive the different types of odor maps in a cooperative fashion. OdorMapDB serves as a useful informatics tool to help explain the signal encoding mechanism of smell perception.
MODELDB
Neuroscience investigators computationally recreate published computer models for several reasons: to validate models a models reproducibility, to understand models by comparing effects of parameter changes with intuitive predictions, to explore neuronal processes by using models beyond their original application, to modify models to test alternative hypotheses, or to reuse models or components of models as building blocks in other models. Efforts to facilitate the creation and storage of computational neuronal models on-line in web-based databases1 [10, 14, 41-45] and in distributed sharing through web resources [46] have been ongoing for at least a decade. ModelDB is the result of one such approach.
One of the strengths of the EAVCR system (described earlier in this work) is the ability for new categories of classes and attributes to be created as the database population grows, with low maintenance requirements for maintaining the existing web pages as the database evolves. ModelDB has expanded from four models in 2000 to 270 models of neurons, neuronal components and neuronal networks as of early 2007. A partial list of model entries appears in Figure 5. Recently, ModelDB has see increased usage [47-51]2 due to the collaborations within the neuroscience community along with a willingness to contribute models to this resource. An editorial in the Journal of Computational Neuroscience recommended using ModelDB to store models for all modeling articles published in that journal. Spurred by a data sharing policy adopted by molecular biologists for the last two decades, ModelDB developers recently suggested that computational neuroscientists adopt a policy on model sharing. When a modeling paper is accepted for publication, a request is made to the authors to deposit the model’s source code in ModelDB’s publicly accessible model repositories.
Figure 5.
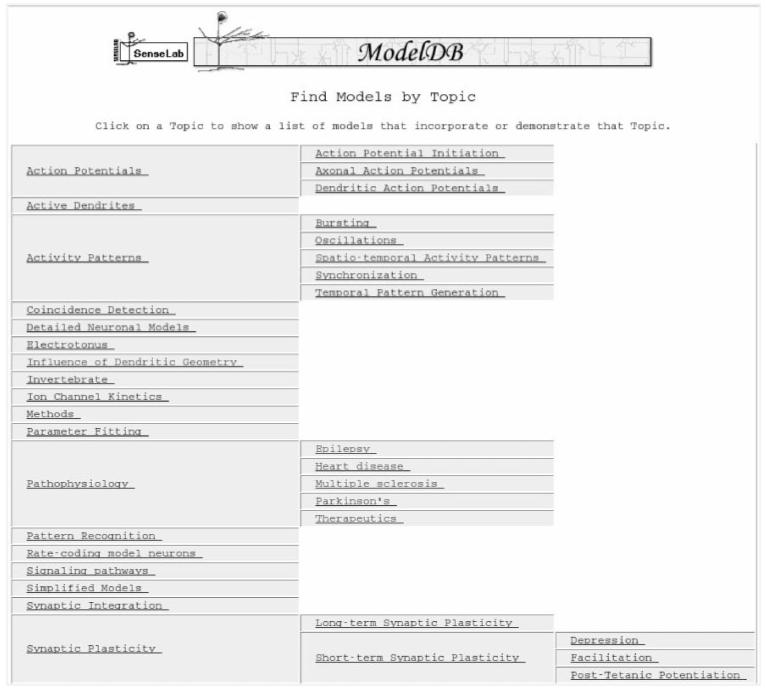
A partial list of model Topics in ModelDB.
ModelDB is now also a test database for the Entrez-LinkOut broker. This is illustrated in Figure 6. The original Hodgkin-Huxley computational model can be found in PubMed by entering ‘hodgkin a,huxley a, 500’ in PubMed. This PubMed web page also contains ‘Links’ → ‘Link out’ (Figure 6 upper left) link. When this link is clicked, the Link-out resources for that journal article are displayed (Figure 6 lower left). Clicking on ‘Neuroscience Related Data’ shows a link to an NDG (among links to other sources that reference this article, Figure 6 lower right) web page. The NDG page contains links to the Hodgkin-Huxley model in ModelDB. Clicking on this link then shows the model description page (Figure 6 upper right) where the user can download or autolaunch the model using the NEURON neuronal model simulator.
Figure 6.
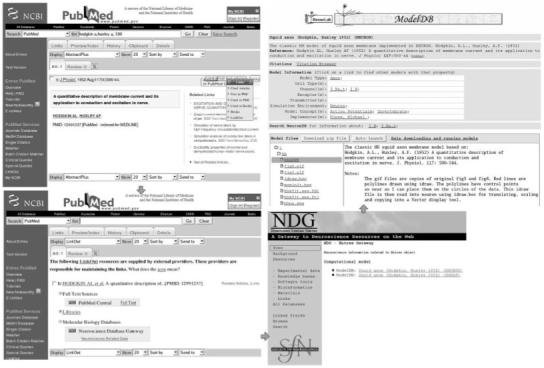
Entrez-LinkOut broker used for ModelDB. When the link to the PUBMED entry in the upper right hand panel is clicked, it opens the page for the original Hodgkin-Huxley computational model in PUBMED shown in the upper left hand panel. The lower left hand panel displays the LinkOut resources in PUBMED for NDG as a link. When that link is clicked, it results in the Hodgkin-Huxley model link in NDG. The lower right panel shows that the Hodgkin-Huxley model is stored in both ModelDB and the SNNAP database housed at University of Texas Medical School in Houston, Texas. Clicking on the link for ModelDB in NDG then results in the original page illustrated in the upper right hand panel.
A new area that ModelDB hopes to leverage the EAV/CR framework is by interoperating with databases of journals articles. The ‘Publication’ class in ModelDB, whose instances contains citations to 31,000 scientific articles [52], allows the browsing of citations associated with model and the citation’s bibliographies. One new attribute for ModelDB papers, the digital object identifier (doi), stores a unique code supplied by journals that identifies each paper. We plan on adding new tables that will map the name or abbreviated name of the journal as supplied in the citation to the journals URL. The doi, if included in paper attributes, is sufficient to link to at least the abstract of the paper, which journals uniformly supply free of charge. This will allow modelers to view abstracts or the full text of the paper, if available, while browsing citations in ModelDB. With the shift in the field toward open access of articles (usually after a delay of a year) many journals will make papers freely available.
BRAINPHARM
BrainPharm, a pilot database under development, was recently added to SenseLab. It is primarily concerned with the management of knowledge associated with neurological disorders with emphasis on Alzheimer’s Disease, Parkinson’s Disease and epilepsy. BrainPharm was created to store research-information on drugs for the treatment of different neurological disorders, identify agents that act on neuronal receptors and differentiate between signal transduction pathways in the normal and diseased brain. The flexibility of the EAV/CR schema that allows users access to all of SenseLab functionality enables searches for drug actions at the level of key molecular constituents, neuronal compartments and individual neurons (in NeuronDB). Also provided are links to ModelDB that allow users to run neuronal model simulation packages that differentiate processes between normal and diseased brains. Much of BrainPharm is still under development. The section on Parkinson’s Disease currently contains information related to drugs that act during several specific stages of the Dopamine neurotransmitter mechanism; the section on Epilepsy section contains links to a table containing of 23 drugs used in Epilepsy treatment and the different types of symptoms of epilepsy that each drug is known to affect. These symptoms range from: Epilepsy without seizures, infantile spasms, Lennox-Gastaut, generalized tonic-clonic seizure, myoclonic seizures and partial seizures.
The most development in BrainPharm is in the area of Alzheimer’s Disease. The next section describes in detail how by using technologies of the Semantic Web, BrainPharm seeks to interoperate with Alzforum (www.alzforum.org), a web resource for Alzheimer’s Disease information, through a pilot project AlzPharm. BrainPharm interoperates with NeuronDB, which has to this point disseminated information related to properties of healthy neurons, and extends it to include diseased cells in that it describes the extracellular elements such as β-amyloid precursor proteins—the primary constituent of plaques, and intracellular elements such as tau proteins. BrainPharm contains lists of neurons affected by Alzheimer’s Disease elements with the information being presented as annotated links to the literature related to the neuronal compartments where these elements are expressed. The EAV/CR schema mentioned earlier in this article allows for the easy updating of information in BrainPharm.
SEMANTIC WEB DEVELOPMENT IN SenseLab
This section describes the application of the Semantic Web, an evolving extension of the World Wide Web (http://www.w3.org/2001/sw/) for knowledge representation in SenseLab. This is being explored as a research project and will aid in extending SenseLab’s knowledge environment to become more semantically interoperable with other neuroscience knowledge sources available over the Internet. The EAV/CR schema on which SenseLab’s architecture is based bears some resemblance to the RDF (Resource Description Framework) schema. RDF is an ontological language for knowledge representation. Semantic Web supports RDF. When the SenseLab project was initiated (more than 10 years ago), RDF was a relatively new standard and there were not many software tools available for manipulating data stored in RDF format. Both EAV/CR and RDFS model concepts and their instances are in a format called a ‘triple’. The following example illustrates an RDF triple:
<http://en.wikipedia.org/wiki/Dopamine, http://en.wikipedia.org/wiki/Function, http://en.wikipedia.org/wiki/Neurotransmitter>
The above triple expresses the notion that Dopamine has the Function of being a Neurotransmitter. In the Semantic Web, each component of the triple is identified using a Uniform Resource Identifier (URI) (details can be found at the following web site: http://www.w3.org/Addressing/). When resources have the same URI, they are assumed to be the same entity, and any data about that entity can be merged. In RDF, as triple statements become connected they form a directed labeled graph. The EAV/CR triple, on the other hand, consists of an ‘entity’ which is described by ‘attributes’ for which ‘values’ exist as described earlier in this article (See the first section). RDF may also be extended by another ontology language called Ontology Web Language or OWL (http://www.w3.org/TR/owl-features/), which is also supported by the Semantic Web. It extends RDF by adding vocabulary that describes the relations of cardinality and equality among classes and properties. The application of the Semantic Web in the neuroscience domain is actively being pursued by the Semantic Web for Health Care and Life Sciences Interest Group (HCLSIG) (http://www.w3.org/2001/sw/hcls/). The HCLSIG is established by the World Wide Web Consortium (W3C) (http://www.w3.org/).
SenseLab in OWL/RDF
We have begun the process of converting some of the SenseLab databases into OWL or RDF. This allows us to explore the representation of knowledge at different levels of expressiveness and use a range of tools to query and integrate knowledge in different representation formats. We converted a subset of NeuronDB into the target OWL ontology that was created manually using the ontology editor Protégé (http://protege.stanford.edu/). We mapped NeuronDB from its relational (EAV/CR) form to OWL using a tool called ‘D2RQ’ (http://sites.wiwiss.fu-berlin.de/suhl/bizer/D2RQ/). D2RQ provides a high-level mapping specification language for converting the relational database structure to RDF or OWL structure. To demonstrate the operation of this OWL data set with other related OWL data sets, we also converted a portion of CoCoDat [53] (a relational database based at the University of Dusseldorff, Germany) into OWL using D2RQ. Then we demonstrated the use of OWL’s constructs such as ‘sameAs’ to create a bridge between the NeuronDB and CoCoDat OWL ontologies. We also demonstrated how to use nRQL, which is an OWL query language supported by the OWL reasoner ‘Racer’ (http://www.cs.concordia.ca/~haarslev/racer/racer-queries.pdf), to query bridged ontologies. In [54], we describe this OWL-based integration approach.
Recently, we have converted the new prototype version of BrainPharm (discussed in the previous section) into RDF. The conversion was implemented using the XML Stylesheet Transformation or XSLT (http://www.w3.org/TR/xslt), which was used to transform a portion of BrainPharm data exported in EDSP/XML format into the target RDF/XML structure. In addition, we have manually created an ontology using RDF Schema (RDFS) which is available through Protégé. The ontology represents information about the effect of drugs on pathological (or molecular) mechanisms (which involve neuronal properties such as receptors, currents and neurotransmitters) mediating the pathological changes in various neurological disorders such as Alzheimer’s Disease (AD). Figure 7 shows the ontology diagram of BrainPharm. The diagram depicts a class-relationship graph representing the various classes as nodes and their relationships as edges. It also shows examples of class instances. In terms of understanding a drug’s effect on the molecular mechanism of Alzheimer’s Disease, these example instances tell us the following:
‘the drug Nifedipine reduces the pathological effect of beta Amlyloid on the I Calcium current of CA1 pyramidal neuron’.
Figure 7.
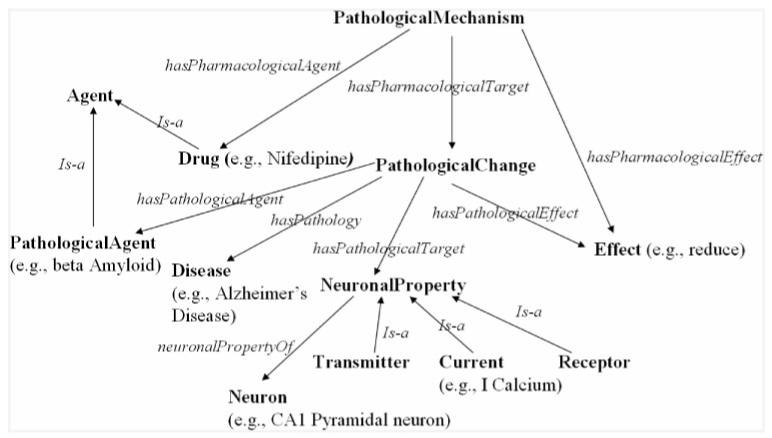
BrainPharm ontology diagram.
To explore the capabilities provided by RDFS to make inferences, we used Oracle Database 10g’s RDF Data Model (http://www.oracle.com/technology/tech/semantic_technologies). We loaded the converted RDF data set of BrainPharm (and the corresponding RDF Schema) into the Oracle RDF database. In addition, we loaded into this Oracle RDF database another existing RDF data set (with RDF Schema) provided by the pilot version of SWAN (Semantic Web Applications in Neuromedicine) [55]. The pilot data set contains a small subset of data extracted from AlzForum [56], which has to do with research hypotheses and the associated publications relating to both basic and clinical research studies of Alzheimer’s Disease. We have built a Web-based application called ‘AlzPharm’ [56], which allows the user to access and query the BrainPharm and SWAN RDF data in an integrated fashion. In [56], we described in detail the Semantic Web development of BrainPharm and demonstrated how to use the inference capability provided by Oracle’s RDF query language to retrieve from SWAN annotated publications (which is defined as a subclass of publications in the SWAN RDF schema) that are associated with a particular AD drug target.
We plan to continue to expand the Semantic Web development in SenseLab. We will also make our effort visible to HCLSIG, as we have been actively involved in many HCLSIG activities such as teleconferences and face-to-face meetings. In addition, we have made our RDF/OWL data sets accessible through the HCLSIG Wiki site.
CONCLUSION
SenseLab as a neuroscience information dissemination resource has through its seven database continued to evolve, expand and diversify. SenseLab’s involvement in projects such as NDG and NIF, interoperability and integrated querying through the web medium will only serve to make neuroscience information to the research community.
Acknowledgements
The authors wish to thank the following sources for the continued development of SenseLab. This research is supported in part by grants P01 DC04732, P20 LM07253, T15 LM07056, NS11613, K25 HG02378, R01 DA021253, G08 LM05583, T15 LM07056, K22 LM008422 and P20 LM07253 from the National Institute of Health, NIH contract N01 DA-BAA-5-7753, by NIH Grants and by grants DBI-0135442 from the National Science Foundation.
Footnotes
ModelDB:http://senselab.med.yale.edu/senselab/modeldb
Visiome:http://platform.visiome.org/index.html
Biomodels:http://www.ebi.ac.uk/biomodels/
Papers listed here are restricted to those with authors outside the ModelDB development group.
Contributor Information
Chiquito J. Crasto, Chiquito Joaqium Crasto, Ph.D. was an Associate Research Scientist at the Yale Center for Medical Informatics and Department of Neurobiology at the Yale University School of Medicine. Dr. Crasto’s research involves database development, computational studies of chemosensory receptors, data and text mining, and neuroinformatics. He is the Scientific Coordinator of the Olfactory Receptor Database. Dr. Crasto is currently an Assistant Professor at the Howell and Elizabeth Heflin Center for Human Genetics, University of Alabama at Birmingham School of Medicine
Luis N. Marenco, Luis Marenco, M.D. is an Assistant Professor at the Yale Center for Medical Informatics at the Yale University School of Medicine. Dr Marenco’s research interests lie in developing evolvable Semantic data models, and their application in the medical and biological sciences. Dr. Marenco is the chief developer for SenseLab
Nian Liu, Nian Liu, Ph.D. is an Associate Research Scientist at the Center for Medical Informatics at the Yale University School of Medicine. His research interests include functional brain mapping of the olfactory system, content-based imaging data retrieval, and usage pattern analysis of the Web-based neuroscience resources.
Thomas M. Morse, Thomas Marston Morse, Ph.D. whose main project involves ModelDB, is an Associate Research Scientist in the Department of Neurobiology at Yale University School of Medicine. His research interests include neuroinformatics and a broad-range study of computational neuroscience models with a special interest in single cell and neuronal network models of electrical activity
Kei-Hoi Cheung, Kei-Hoi Cheung, Ph.D. is an Associate Professor at the Yale Center for Medical Informatics at the Yale University School of Medicine. His research interests lie in data and tool interoperation involving the use of a variety of technologies including the Semantic Web technology.
Peter C. Lai, Peter C. Lai is currently the Systems Administrator for Information Technology Services at Simon’s Rock College of Bard. His research interests include computational molecular dynamics and developing laboratory information management systems
Gautam Bahl, Gautam Bahl, M.D. is a diagnostic radiology resident at the Wayne State University/Detroit Medical Center. His research interests include medical image processing and magnetic resonance imaging. Dr. Bahl has been the curator of Olfactory Receptor Database.
Peter Masiar, Peter Masiar, M.S. is a Computer Programmer and Applications Developer at the Yale Center for Medical Informatics, Yale University School of Medicine, with more than 20 years of experience developing database and web-based applications.
Hugo Y.K. Lam, Hugo Y.K. Lam is a Ph.D. student of the Interdepartmental Ph.D. Program in Computational Biology and Bioinformatics at Yale University. His research interests include the use of Semantic Web to facilitate data integration in the life sciences domain
Ernest Lim, Ernest Lim is a programmer at the Yale Center for Medical Informatics. His efforts have been geared toward the development and implementation of ontologies in the neuroscience domain.
Prakash Nadkarni, Prakash Nadkarni, M.D. is Associate Professor at the Yale Center for Medical Informatics. His interests are biomedical databases, text processing and metadata-driven software architectures. Dr. Nadkarni is the creator of the EAV/CR database schema.
Michele Migliore, Michele Migliore, Ph.D. is a senior research scientist at the Institute of Biophysics of the National Research Council (Palermo, Italy), and a visiting Scientist in the Department of Neurobiology at the Yale University School of Medicine (New Haven, CT, USA). His research interests are focused on the realistic modeling of neurons and neuronal networks to investigate the microscopic processes underlying higher brain functions.
Perry L. Miller, Perry L. Miller, MD, Ph.D. is Professor of Anesthesiology and Molecular, Cellular, and Developmental Biology, as well as the Director of the Center for Medical Informatics at Yale University. His research interests include clinical, genome, and neuroinformatics, and he is particularly interested in informatics projects at the intersections of these areas
Gordon M. Shepherd, Gordon M. Shepherd, M.D., D. Phil. is a Professor of Neurobiology at the Yale Center for Medical Informatics. He is the originator of the SenseLab project. His interests include exploration of the olfactory system, neuroscience and neuroinformatics. Dr. Shepherd is the author of several textbooks on Neurobiology and is the editor of the seminal work “Synaptic Organization of the Brain”
References
- 1.Koslow SH, Subramaniam S. Databasing the Brain: From Data to Knowledge (Neuroinformatics) Wiley-Liss; Hoboken, NJ: 2005. [Google Scholar]
- 2.Kèotter R. Neuroscience Databases: A Practical Guide. Kluwer Academic; Boston: 2003. [Google Scholar]
- 3.Liu N, Marenco L, Miller PL. ResourceLog: an embeddable tool for dynamically monitoring the usage of web-based bioscience resources. J Am Med Inform Assoc. 2006;13:432–7. doi: 10.1197/jamia.M2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadkarni PM, Marenco L, Chen R, et al. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999;6:478–93. doi: 10.1136/jamia.1999.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migliore M, Shepherd GM. Opinion: an integrated approach to classifying neuronal phenotypes. Nat Rev Neurosci. 2005;6:810–8. doi: 10.1038/nrn1769. [DOI] [PubMed] [Google Scholar]
- 6.Xu F, Schaefer M, Kida I, et al. Simultaneous activation of mouse main and accessory olfactory bulbs by odors or pheromones. J Comp Neurol. 2005;489:491–500. doi: 10.1002/cne.20652. [DOI] [PubMed] [Google Scholar]
- 7.Migliore M, Hines ML, Shepherd GM. The role of distal dendritic gap junctions in synchronization of mitral cell axonal output. J Comput Neurosci. 2005;18:151–61. doi: 10.1007/s10827-005-6556-1. [DOI] [PubMed] [Google Scholar]
- 8.Marenco L, Wang TY, Shepherd G, et al. QIS: a framework for biomedical database federation. J Am Med Inform Assoc. 2004;11:523–34. doi: 10.1197/jamia.M1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu N, Xu F, Marenco L, et al. Informatics approaches to functional MRI odor mapping of the rodent olfactory bulb: OdorMapBuilder and OdorMapDB. Neuroinformatics. 2004;2:3–18. doi: 10.1385/NI:2:1:003. [DOI] [PubMed] [Google Scholar]
- 10.Hines ML, Morse T, Migliore M, et al. ModelDB: a database to support computational neuroscience. J Comput Neurosci. 2004;17:7–11. doi: 10.1023/B:JCNS.0000023869.22017.2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marenco L, Tosches N, Crasto C, et al. Achieving evolvable Web-database bioscience applications using the EAV/CR framework: recent advances. J Am Med Inform Assoc. 2003;10:444–53. doi: 10.1197/jamia.M1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, Liu N, Kida I, et al. Odor maps of aldehydes and esters revealed by functional MRI in the glomerular layer of the mouse olfactory bulb. Proc Natl Acad Sci USA. 2003;100:11029–34. doi: 10.1073/pnas.1832864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crasto CJ, Marenco LN, Migliore M, et al. Text mining neuroscience journal articles to populate neuroscience databases. Neuroinformatics. 2003;1:215–37. doi: 10.1385/NI:1:3:215. [DOI] [PubMed] [Google Scholar]
- 14.Migliore M, Morse TM, Davison AP, et al. ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics. 2003;1:135–9. doi: 10.1385/NI:1:1:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crasto C, Marenco L, Miller P, et al. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;30:354–60. doi: 10.1093/nar/30.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepherd GM. Supporting databases for neuroscience research. J Neurosci. 2002;22:1497. doi: 10.1523/JNEUROSCI.22-05-01497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller PL, Nadkarni P, Singer M, et al. Integration of multidisciplinary sensory data: a pilot model of the human brain project approach. J Am Med Inform Assoc. 2001;8:34–48. doi: 10.1136/jamia.2001.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skoufos E, Marenco L, Nadkarni PM, et al. Olfactory receptor database: a sensory chemoreceptor resource. Nucleic Acids Res. 2000;28:341–3. doi: 10.1093/nar/28.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd GM, Healy MD, Singer MS, et al. SenseLab: a project in multidisciplinary, multilevel sensory integration. In: Koslow EH, Huerta MF, editors. Neuroinformatics: An Overview of the Human Brain Project. Lawrence Erlbaum; New York: 1997. pp. 21–56. [Google Scholar]
- 20.Crasto C, Masier P, Shepherd G, et al. NeuroExtract: facilitating neuroscience-oriented retrieval from broadly-focused bioscience databases using text-based query mediation. JAMIA. 2006;14(3):355–60. doi: 10.1197/jamia.M2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr Med Chem. 2000;7:861–88. doi: 10.2174/0929867003374534. [DOI] [PubMed] [Google Scholar]
- 22.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 23.Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–11. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 24.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. ProcNatlAcadSciUSA. 2004;101:2584–9. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glusman G, Yanai I, Rubin I, et al. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs T, Glusman G, Horn-Saban S, et al. The human olfactory subgenome: from sequence to structure and evolution. Hum Genet. 2001;108:1–13. doi: 10.1007/s004390000436. [DOI] [PubMed] [Google Scholar]
- 27.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101:2156–61. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–33. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 29.Young JM, Friedman C, Williams EM, et al. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–46. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- 30.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0018. RESEARCH0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quignon P, Kirkness E, Cadieu E, et al. Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol. 2003;4:R80. doi: 10.1186/gb-2003-4-12-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quignon P, Giraud M, Rimbault M, et al. The dog and rat olfactory receptor repertoires. Genome Biol. 2005;6:R83. doi: 10.1186/gb-2005-6-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100:12235–40. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malnic B, Hirono J, Sato T, et al. Combinatorial receptor codes for odors. Cell. 1999;96:713–23. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 35.Mombaerts P, Wang F, Dulac C, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–86. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 36.Zou Z, Horowitz LF, Montmayeur JP, et al. Genetic tracing reveals a stereotyped sensory map in the olfactory cortex. Nature. 2001;414:173–9. doi: 10.1038/35102506. [DOI] [PubMed] [Google Scholar]
- 37.Stewart WB, Kauer JS, Shepherd GM. Functional organization of rat olfactory bulb analysed by the 2-deoxyglucose method. J Comp Neurol. 1979;185:715–34. doi: 10.1002/cne.901850407. [DOI] [PubMed] [Google Scholar]
- 38.Jourdan F. Spatial dimension in olfactory coding: a representation of the 2-deoxyglucose patterns of glomerular labeling in the olfactory bulb. Brain Res. 1982;240:341–4. doi: 10.1016/0006-8993(82)90232-3. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer ML, Young DA, Restrepo D. Olfactory finger-prints for major histocompatibility complex-determined body odors. J Neurosci. 2001;21:2481–7. doi: 10.1523/JNEUROSCI.21-07-02481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson BA, Farahbod H, Saber S, et al. Effects of functional group position on spatial representations of aliphatic odorants in the rat olfactory bulb. J Comp Neurol. 2005;483:192–204. doi: 10.1002/cne.20415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson BE, Healy MD, Nadkarni PM, et al. ModelDB: an environment for running and storing computational models and their results applied to neuroscience. J Am Med Inform Assoc. 1996;3:389–98. doi: 10.1136/jamia.1996.97084512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirsky JS, Nadkarni PM, Healy MD, et al. Database tools for integrating and searching membrane property data correlated with neuronal morphology. J Neurosci Methods. 1998;82:105–21. doi: 10.1016/s0165-0270(98)00049-1. [DOI] [PubMed] [Google Scholar]
- 43.Usui S. Visiome: neuroinformatics research in vision project. Neural Netw. 2003;16:1293–300. doi: 10.1016/j.neunet.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Sivakumaran S, Hariharaputran S, Mishra J, et al. The database of quantitative cellular signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics. 2003;19:408–15. doi: 10.1093/bioinformatics/btf860. [DOI] [PubMed] [Google Scholar]
- 45.Le Novere N, Bornstein B, Broicher A, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–91. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cannon RC, Howell FW, Goddard NH, et al. Non-curated distributed databases for experimental data and models in neuroscience. Network. 2002;13:415–28. [PubMed] [Google Scholar]
- 47.Akemann W, Knopfel T. Interaction of Kv3 potassium channels and resurgent sodium current influences the rate of spontaneous firing of Purkinje neurons. J Neurosci. 2006;26:4602–12. doi: 10.1523/JNEUROSCI.5204-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter DA, Byrne JH. Short-term plasticity in a computational model of the tail-withdrawal circuit in Aplysia. Neurocomputing. 2006;70:1993–9. doi: 10.1016/j.neucom.2006.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyhrfjeld-Johnsen J, Santhakumar V, Morgan RJ, et al. Topological determinants of epileptogenesis in large-scale structural and functional models of the dentate gyrus derived from experimental data. J Neurophysiol. 2006 doi: 10.1152/jn.00950.2006. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Ascoli GA. Computational simulation of the input-output relationship in hippocampal pyramidal cells. J Comput Neurosci. 2006;21:191–209. doi: 10.1007/s10827-006-8797-z. [DOI] [PubMed] [Google Scholar]
- 51.van Welie I, Remme MW, van Hooft JA, et al. Different levels of Ih determine distinct temporal integration in bursting and regular-spiking neurons in rat subiculum. JPhysiol. 2006;576:203–14. doi: 10.1113/jphysiol.2006.113944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davison AP, Morse TM, Migliore M, et al. Semi-automated population of an online database of neuronal models (ModelDB) with citation information, using PubMed for validation. Neuroinformatics. 2004;2:327–32. doi: 10.1385/NI:2:3:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dyhrfjeld-Johnsen J, Maier J, Schubert D, et al. CoCoDat: a database system for organizing and selecting quantitative data on single neurons and neuronal microcircuitry. J Neurosci Methods. 2005;141:291–308. doi: 10.1016/j.jneumeth.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Lam YKH, Marenco L, Clark T, et al. BMC Bioinformatics. Suppl 3. Vol. 8. 2007. AlzPharm: integration of neurogeneration data using RDF; p. S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Y, Kinoshita J, Wu E, et al. SWAN: a distributed knowledge infrastructure for alzheimer disease research. Journal of Web Semantics. 2006;4 [Google Scholar]
- 56.Kinoshita J, Fagan A, Ewbank D, et al. Alzheimer research forum live discussion: insulin resistance: a common axis linking Alzheimer’s, depression, and metabolism? J Alzheimers Dis. 2006;9:89–93. doi: 10.3233/jad-2006-9110. [DOI] [PubMed] [Google Scholar]


