The crystal structure of tear lipocalin determined in space group P21 revealed large structural deviations from the previously solved X-ray structure in space group C2, especially in the loop region and adjoining parts of the β-barrel which give rise to the ligand-binding site. These findings illustrate a novel mechanism for promiscuity in ligand recognition by the lipocalin protein family.
Keywords: apolipoprotein D; β-barrel; 1,4-butanediol; induced fit; tear lipocalin; von Ebner’s gland protein
Abstract
Tear lipocalin (TLC) with the bound artificial ligand 1,4-butanediol has been crystallized in space group P21 with four protein molecules in the asymmetric unit and its X-ray structure has been solved at 2.6 Å resolution. TLC is a member of the lipocalin family that binds ligands with diverse chemical structures, such as fatty acids, phospholipids and cholesterol as well as microbial siderophores and the antibiotic rifampin. Previous X-ray structural analysis of apo TLC crystallized in space group C2 revealed a rather large bifurcated ligand pocket and a partially disordered loop region at the entrace to the cavity. Analysis of the P21 crystal form uncovered major conformational changes (i) in β-strands B, C and D, (ii) in loops 1, 2 and 4 at the open end of the β-barrel and (iii) in the extended C-terminal segment, which is attached to the β-barrel via a disulfide bridge. The structural comparison indicates high conformational plasticity of the loop region as well as of deeper parts of the ligand pocket, thus allowing adaptation to ligands that differ vastly in size and shape. This illustrates a mechanism for promiscuity in ligand recognition which may also be relevant for some other physiologically important members of the lipocalin protein family.
1. Introduction
The lipocalins are a functionally diverse family of secretory ligand-binding proteins that were initially identified in animals and plants (Flower, 1996 ▶) and that have more recently also been found in bacteria (Bishop, 2000 ▶). Members of this protein family are characterized by a highly conserved three-dimensional fold comprising an eight-stranded antiparallel β-barrel with a ligand pocket at one end and a C-terminal α-helix attached to its side (Flower, 2000 ▶; Skerra, 2000 ▶). At the open end four structurally variable loops connect each pair of neighbouring β-strands and form the entrance to the binding site. Despite the common fold, the amino-acid sequences of lipocalins show only weak similarity, with mutual sequence identities that are often below 20%. Lipocalins play a role in the transport and storage of poorly soluble or chemically sensitive compounds such as vitamins, steroids and secondary metabolites. Some members of this family also have specific physiological functions, for example in olfaction, prostaglandin synthesis, invertebrate colouration and in regulation of the immune response (Åkerström et al., 2006 ▶; Breustedt et al., 2006 ▶).
Human tear lipocalin (TLC; also known as lipocalin 1, Lcn1 or von Ebner’s gland protein) is the major protein in tear fluid and is expressed by several secretory glands and tissues (Redl, 2000 ▶). In biological samples TLC is found in complex with diverse endogenous ligands, including fatty acids, phospholipids and cholesterol (Glasgow et al., 1995 ▶). Furthermore, TLC has been shown to scavenge microbial siderophores (Fluckinger et al., 2004 ▶) and also to bind the antibiotic drug rifampin (Gasymov, Abduragimov, Gasimov et al., 2004 ▶) as well as several synthetic fluorescent probes in vitro (Gasymov et al., 1999 ▶). Its ability to complex a large number of compounds belonging to different chemical classes is unusual among lipocalins, but has also been observed for the closely related odorant-binding proteins (Briand et al., 2002 ▶; Vincent et al., 2000 ▶) and for α1-acid glycoprotein (Israili & Dayton, 2001 ▶; Schönfeld et al., 2008 ▶).
The physiological role of TLC seems to lie in preserving the integrity of the tear film, with its peculiar aqueous–lipid interface, and in scavenging potentially harmful lipophilic substances, thereby protecting epithelial surfaces (Redl, 2000 ▶). In addition, its siderophore-binding activity may suppress microbial spreading (Fluckinger et al., 2004 ▶). Recently, a cell-surface receptor was identified that promotes internalization and subsequent degradation of TLC, possibly including bound ligands, and thus may be involved in detoxification processes (Wojnar et al., 2001 ▶, 2003 ▶).
A previously elucidated crystal structure of TLC in its apo state (space group C2) revealed that the ligand pocket of this lipocalin is unusually large and possesses a bifurcated shape (Breustedt et al., 2005 ▶), hence explaining its observed promiscuity for a series of different ligands. However, three loops at the entry to the ligand pocket and the single intramolecular disulfide bond joining Cys61 and Cys153 did not show continuous electron density, indicating elevated flexibility. Here, we report the 2.6 Å resolution X-ray structural analysis of TLC in a new crystal form (space group P21) which contains four well ordered molecules in the asymmetric unit and reveals important details of the loop region around the ligand pocket as well as the precise conformation of the polypeptide C-terminus.
2. Materials and methods
2.1. Protein production, crystallization and data collection
Recombinant human TLC, comprising residues 5–157 of the mature protein with the free Cys101 substituted by Ser, was produced in Escherichia coli and purified by means of streptavidin-affinity chromatography via a C-terminal Strep-tag II and gel filtration as described previously (Breustedt et al., 2005 ▶). Crystallization trials were carried out according to the hanging-drop vapour-diffusion technique by mixing 3 µl protein solution (10 mg ml−1, dialyzed against 10 mM Tris–HCl pH 8.0) with 1 µl precipitant solution on a siliconized glass cover slip, followed by equilibration against 0.5 ml precipitant solution. Single crystals were obtained in the presence of 27%(w/v) PEG 3350, 200 mM NaCl, 0.8%(v/v) 1,4-butanediol, 100 mM Tris–HCl pH 8.0 after about two weeks at 293 K and were harvested directly from the drop using nylon loops (Hampton Research, Laguna Niguel, California, USA), followed by freezing in a 100 K nitrogen stream (Oxford Cryosystems, Oxford, England). The crystals were generally highly mosaic and only one single crystal gave a diffraction pattern that could be processed. A native data set was collected on a MAR Research imaging-plate detector (Hamburg, Germany) using monochromated Cu Kα radiation from an RU-300 rotating-anode generator (Rigaku, Tokyo, Japan) equipped with Confocal Max-Flux Optics (Osmic, Troy, Michigan, USA). Data were processed with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶) and the space group was determined to be P21 (Table 1 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell (2.69–2.60 Å).
| Data collection | |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 63.91, b = 34.17, c = 143.48, β = 90.35 |
| Resolution (Å) | 20.0–2.60 |
| Total reflections | 238974 |
| Unique reflections | 19767 |
| Completeness (%) | 99.6 (97.0) |
| 〈I〉/〈σ(I)〉 | 27.3 (5.8) |
| Redundancy | 5.7 (4.8) |
| Mosaicity (°) | 0.466 |
| Rmerge | 0.061 |
| Refinement | |
| R/Rfree | 22.9/27.4 |
| Protein residues visible | |
| Chain A | 143 |
| Chain B | 140 |
| Chain C | 137 |
| Chain D | 137 |
| Ligand molecules | 4 |
| Solvent molecules | 185 |
| R.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.299 |
| Average B values (Å2) | |
| Protein | 43.6 |
| Ligand | 46.5 |
| Solvent | 46.1 |
| Ramachandran plot (%) | |
| Most favoured region | 91.0 |
| Additionally allowed region | 8.3 |
| Generously allowed region | 0.6 |
| Disallowed region | 0.0 |
2.2. Structure determination, model building and refinement
The crystal structure of TLC was solved by molecular replacement with Phaser (McCoy et al., 2005 ▶) using the atomic coordinates of its previously described crystal structure in a different space group (PDB entry 1xki; Breustedt et al., 2005 ▶). A truncated version of this model (residues 14–24, 38–55, 64–104 and 110–150), lacking three of the four loops at the open end of the β-barrel, was used for the successful search, revealing four different polypeptide chains in the asymmetric unit. A composite OMIT map, calculated from the molecular-replacement solution using CNS (Brünger et al., 1998 ▶), indicated considerable reorientation of the β-strands B, C and D in all four monomers. Thus, the corresponding residues were deleted from the starting model and individually rebuilt de novo. The composite OMIT map also showed well defined additional electron density in the ligand pocket for each of the four protein molecules, indicating the presence of a bound ligand, which was subsequently identifed as 1,4-butanediol. Simulated annealing using CNS proved to be helpful during early stages of model building. The missing residues were added in several sessions of manual rebuilding with QUANTA (Oldfield, 2001 ▶), alternating with crystallographic refinement cycles using CNS, by applying loose noncrystallographic symmetry restraints between monomers A, B, C and D. Finally, 1,4-butanediol was built into the electron density within each ligand pocket. Water molecules were added where the 2F o − F c and F o − F c difference Fourier maps revealed densities higher than 1.0σ and 3.5σ, respectively, if stereochemically plausible. Graphical representations, secondary-structure assignments and superpositions were made with PyMOL (DeLano, 2002 ▶), DSSP (Kabsch & Sander, 1983 ▶) and LSQMAN (Kleywegt et al., 2001 ▶), respectively. Pairwise Cα-atom distances were calculated with RMSPDB (Kleywegt et al., 2001 ▶). The coordinates of the TLC structure have been deposited in the Protein Data Bank (Berman et al., 2000 ▶) under accession code 3eyc.
3. Results
3.1. Crystallization, structure solution and quality of the final model
Crystals of bacterially produced human TLC (Breustedt et al., 2006 ▶) belonging to space group P21 were obtained using PEG 3350 as the main precipitant in the presence of NaCl and 1,4-butanediol at pH 8.0. The X-ray structure was solved by molecular replacement using the published crystal structure of recombinant TLC in space group C2 (Breustedt et al., 2005 ▶) followed by refinement to a resolution of 2.6 Å (Table 1 ▶). The final model consists of four monomers in the asymmetric unit (Fig. 1 ▶). The quality of the electron-density map allowed us to unambiguously build residues Val13–Pro155 for polypeptide chain A, in contrast to the previously described crystal structure (1xki), where density was missing for the segments Asp25–Met31, Leu56–Ile57 and Leu105–His106. In chains C and D, however, the tips of loops 1 and 4 at the open end of the β-barrel showed only weak electron density, indicating partial occupancy, and were thus omitted from the model. Also, a short stretch of electron density was missing in chain B for residues Glu45–Gly47 at the closed end of the β-barrel. Furthermore, the N-terminal residues Ala5–Gln11 (or Asp12) and several residues at the C-terminus (mostly comprising the Strep-tag II, which had been introduced for affinity purification; Schmidt & Skerra, 2007 ▶) were disordered in all four polypeptide chains. Pairwise superposition of the 58 conserved Cα positions of the β-barrel (Breustedt et al., 2005 ▶; Skerra, 2000 ▶) for the individual polypeptide chains led to r.m.s.d. values of 0.13 Å or lower, while an r.m.s.d. value of up to 0.25 Å was calculated when all visible Cα atoms were superimposed (Fig. 1 ▶ b).
Figure 1.
Three-dimensional structure of human TLC with bound 1,4-butanediol crystallized in space group P21. (a) Ribbon diagram of the four TLC monomers in the asymmetric unit (chain A, magenta; chain B, blue; chain C, green; chain D, orange). The four ligand molecules are displayed in sphere style in grey and the unit-cell boundary is indicated. (b) Stereo representation of the four superimposed TLC monomers A–D (same colours as in a). Molecules were superimposed using the Cα positions of the 58 conserved β-barrel residues (Breustedt et al., 2005 ▶). The ligand 1,4-butanediol and the disulfide bond joining Cys61 and Cys153 are depicted in stick representation.
3.2. Comparison with TLC crystallized in space group C2
As expected, the fold of TLC crystallized in space group P21 is generally similar to that of the previously described X-ray structure of apo TLC in space group C2 (Figs. 2 ▶ and 3 ▶). Both reveal a central β-barrel motif with a C-terminal α-helix attached to its side (Figs. 2 ▶ a and 3 ▶ a). However, in contrast to the C2 structure, all four loops at the open end of the β-barrel were clearly defined in the new crystal form, at least for protein monomers A and B. Furthermore, the disulfide bond linking Cys61 and Cys153, which was missing from the old structure, could be unambiguously modelled for all four chains. Since the loop segments were slightly better defined in the electron density for molecule A than for the other polypeptides in the P21 structure, this chain will be used in the following for comparison with the C2 structure.
Figure 2.
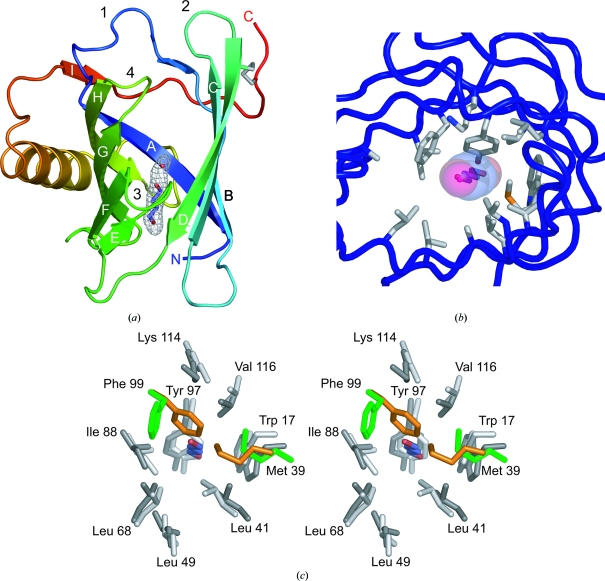
Details of TLC in the P21 crystal structure. (a) Ribbon diagram of TLC (chain A) with bound 1,4-butanediol and a hydrogen-bonded water molecule (stick representation; 2F o − Fc electron-density map contoured at 1.0σ). TLC exhibits the typical lipocalin fold, with an eight-stranded antiparallel β-barrel forming a central cavity, which is open to the upper end, and an α-helix attached to its side. The N- and C-termini of the polypeptide chain, the eight β-strands A–H, the extraneous β-strand I and the four loops at the open end of the β-barrel are labelled. The disulfide bond that attaches the C-terminal peptide segment to β-strand D is shown in stick representation (grey). (b) Residues involved in the interaction with the ligand (view from the top). 1,4-Butanediol and a hydrogen-bonded water molecule are represented as solid sticks with a translucent surface and the side chains of residues that are in direct contact with the ligand are depicted in stick representation. (c) Stereoview illustrating the rearrangement of the side chains that line the ligand pocket between apo TLC and the ligand complex (view similar to that in b). Molecules were superimposed using the Cα positions of the 58 conserved β-barrel residues (as in Fig. 1 ▶ b). While residues Trp17, Leu41, Leu49, Leu68, Ile88, Tyr97, Lys114 and Val116 assume virtually identical side-chain conformations in the P21 structure (dark grey) as in the C2 structure (light grey), residues Met39 and Phe99, which protrude into the cavity in the C2 structure (orange), have undergone structural reorientation in the P21 structure (green) and create space for the ligand (C, blue; O, red).
Figure 3.
Comparison of the TLC structures obtained from two different crystal forms. Molecules were superimposed using the Cα positions of the 58 conserved β-barrel residues (as in Fig. 1 ▶ b). (a) Superposition of TLC crystallized in space group P21 (chain A, blue) and in space group C2 (PDB code 1xki; gold, with modelled residues in grey; Breustedt et al., 2005 ▶). The bound 1,4-butanediol and the hydrogen-bonded water molecule are represented as spheres, while the conserved disulfide bond is shown in stick representation (white). Most of the β-strands are elongated towards the open end of the β-barrel in the P21 structure. (b) Pairwise Cα-atom distances between the two different TLC crystal structures after superposition of the 58 conserved Cα positions of the β-barrel, resulting in an overall Cα r.m.s.d. value of 1.86 Å (β-strands A–H are labelled as bars; loops 1, 2, 3 and 4 connect strands A and B, C and D, E and F, and G and H, respectively). The largest deviations between the two crystal structures occur in β-strands B, C and D and in the neighbouring loops 1 and 2 at the open end of the β-barrel. (c) The electrostatic interactions that may trigger the two alternative conformations of loop 1. In the P21 structure (dark grey) residues Glu27 and Lys108 (green) at the tips of the adjacent loops 1 and 4 make an electrostatic contact that fixes the ‘open’ conformation of loop 1 (blue). In contrast, the ‘closed’ conformation of this loop in the C2 structure (light grey; loop 1 coloured gold, modelled residues coloured yellow) is stabilized by a similar interaction between residues Glu34 and Lys114 (orange). (d) Flexibility of the region around the disulfide bridge. While the disulfide bond between Cys61 and Cys153 is well ordered in the P21 structure (dark grey; loop 2 and the C-terminal peptide segment coloured blue, side chains coloured violet), only residue Cys61 was resolved in the C2 structure (light grey; loop 2 and the C-terminal peptide segment coloured gold, modelled residues coloured yellow, Cys side chain coloured orange) and appears shifted outward by 5.6 Å compared with the P21 structure (red dotted line between the corresponding Cα positions). Nevertheless, the Cα distance between Cys61 in the C2 structure and Cys153 in the superimposed P21 structure is only 0.3 Å larger than the distance between Cys61 and Cys153 within the P21 structure (green dotted lines). By assuming a more extended backbone conformation at residue Glu151 (side chain displayed in blue), the (unresolved) C-terminal peptide segment of the C2 structure could easily span the 0.3 Å distance and move Cys153 sufficiently close to Cys61, indicating that the disulfide bond is probably also formed in the C2 crystal (despite the differing conformation of loop 2) but is not visible in the electron density owing to local structural disorder.
Upon superposition of the 58 conserved Cα positions of the β-barrel (see above), the P21 and C2 structures showed an r.m.s.d. value of 1.86 Å (Fig. 3 ▶ b). This value is much higher than the r.m.s.d. values calculated above for the individual chains in the P21 crystal form and approaches the values for superposition between different members of the lipocalin family (Skerra, 2000 ▶). This deviation is the consequence of major conformational changes of β-strands B, C and D and consequently also of loops 1 and 2 at the open end of the β-barrel. A lower r.m.s.d. value of 1.27 Å was calculated when all 127 visible Cα atoms were superimposed, which arises from the fact that (outside the 58 conserved Cα positions) the orientation of the α-helix relative to the β-barrel as well as the conformations of loops 3 and 4 match closely between the two crystal structures (Figs. 3 ▶ a and 3 ▶ b).
In the new crystal structure, loop 1, which is disordered to a large extent in the C2 structure, runs almost diametrally across the opening of the calyx (Fig. 3 ▶ c). Notably, residues Leu33–Val36, which point towards the central axis of the β-barrel in the C2 structure, form a short β-sheet with the upper part of β-strand C in the P21 structure (Fig. 3 ▶ a). As a result, the Cα position of Glu34 is displaced outwards by 10.6 Å compared with the C2 structure, which thus leads to an opening of the cavity for the ligand (Figs. 3 ▶ a and 3 ▶ c). Interestingly, in the C2 structure this residue is involved in an electrostatic interaction with the side chain of Lys114, which seems to fix the ‘closed’ conformation of loop 1. Conversely, the ‘open’ conformation of this loop in the P21 structure appears to be stabilized by an alternative electrostatic interaction between residue Glu27 at the N-terminal side of loop 1 and Lys108, which is located at the tip of loop 4 (Fig. 3 ▶ c).
The Cα position of Gly59 at the tip of loop 2 is shifted about 9.9 Å towards the central axis of the β-barrel in the P21 structure compared with the C2 structure (Fig. 3 ▶ a). Owing to the concomitant reorientation of β-strands C and D, the Cα position of Cys61 also moves by ∼5.6 Å. In the P21 structure, the disulfide bond between Cys61 and Cys153 at the C-terminus of the polypeptide chain is clearly defined (Fig. 2 ▶ a), whereas this feature as well as all C-terminal residues starting from Glu151 did not give rise to electron density in the C2 structure. Notably, after superposition of the two structures, Cys61 of the C2 structure is located only 0.3 Å further away from Cys153 of the P21 structure than the actual disulfide-bonding partner of the latter (with Cα—Cα distances of 6.1 and 6.4 Å, respectively; Fig. 3 ▶ d). While in the P21 structure the C-terminal peptide segment containing Cys153, which packs against the side of the β-barrel, is bent upwards by about 90° at residue Glu151, a more extended conformation would be possible in the C2 crystal structure, thus also allowing formation of this disulfide bridge. Probably, this part was invisible in the C2 data set because of local structural flexibility as a consequence of different crystal packing.
Further to the altered orientations of β-strands B, C and D (Fig. 3 ▶ b), most of the secondary-structure elements appear to be extended in the P21 structure compared with the C2 structure. β-Strands A–I consist of a total of 74 amino acids in the P21 structure, contrasting with only 61 residues in the C2 structure. Interestingly, the additional segments with β-sheet conformation arise at the open end of the β-barrel at the entry to the ligand pocket.
3.3. The cavity and complexation of 1,4-butanediol
TLC is known to bind a variety of different ligands in the low-micromolar affinity range, with chemical structures ranging from lipids and cholesterol to the macrocyclic antibiotic rifampin (Gasymov, Abduragimov, Gasimov et al., 2004 ▶). Unfortunately, corresponding cocrystallization trials remained unsuccessful (Breustedt et al., 2005 ▶). However, in the present crystal structure additional electron density clearly appeared in the ligand pocket for each of the four monomers and was identified as 1,4-butanediol, which was present in the precipitant solution, with a hydrogen-bonded water molecule.
The mode of interaction between TLC and 1,4-butanediol is comparable with that between retinol-binding protein (RBP), a prototypic human lipocalin, and its ligand retinol (Cowan et al., 1990 ▶). 1,4-Butanediol is almost perfectly located at the symmetry axis of the β-barrel, at the centre of the cavity (Fig. 2 ▶ a). However, it is buried even deeper than retinol in RBP. In fact, this artificial ligand is in close contact with the hydrophobic core region at the bottom of the β-barrel, which is formed by several bulky apolar residues, including the side chains of Trp17 and Tyr97 (Figs. 2 ▶ b and 2 ▶ c). Notably, residues Met39 and Phe99, which protrude into the cavity and separate its two lobes in the C2 crystal structure, have undergone reorientation to create the space occupied by the new ligand. The Cα position of residue Met39 is shifted by 2.2 Å, which is accompanied by the outward movement of β-strand B (see above), while the side chain of Phe99 is rotated by Δχ1 ≃ 120° and Δχ2 ≃ 90°. In fact, a similar mode of binding as for 1,4-butanediol can be envisaged for the hydrophobic tail groups of fatty acids or their derivatives, i.e. the physiological ligands of TLC.
4. Discussion
In contrast to many other lipocalins, such as RBP (Raghu & Sivakumar, 2004 ▶) and apolipoprotein D (ApoD; Eichinger et al., 2007 ▶), TLC exhibits promiscuous binding behaviour and can complex a broad range of compounds belonging to different chemical classes (Redl, 2000 ▶). The first hints regarding the structural basis underlying this phenomenon became apparent from the crystallographic analysis of apo TLC crystallized in space group C2, which indicated that this lipocalin possesses a surprisingly large and bifurcated ligand pocket (Breustedt et al., 2005 ▶). The P21 crystal form analysed here furthermore reveals high conformational flexibility in the loop region at the entry to the ligand pocket, in several strands of the β-barrel surrounding the cavity and also for internal side chains, thus allowing structural adaptation to ligands that can differ vastly in size and shape. This mechanism, which enables ligand promiscuity in the case of TLC, has not been described for other members of the lipocalin family to date.
The number of lipocalins which have been crystallized and analyzed in different space groups, thus yielding independent views of the protein structure, is still growing (Flower, 2000 ▶). Some of these lipocalins appear to be rather rigid, resulting in essentially identical structures being obtained from different crystal forms. Examples are human lipocalin 2 (Lcn2, also known as siderocalin or neutrophil gelatinase-associated lipocalin; NGAL), which was crystallized in monomeric and dimeric states (Goetz et al., 2000 ▶) and with several different siderophore ligands (Clifton et al., 2009 ▶), and RBP, which yielded very similar structures when crystallized in trigonal and orthorhombic forms, irrespective of the presence of retinoid ligands (Zanotti et al., 1993 ▶).
On the other hand, bovine β-lactoglobulin (Blg), a lipocalin that is structurally related to TLC (Breustedt et al., 2005 ▶) and crystallizes in at least six different crystal forms (Aschaffenburg et al., 1965 ▶), exhibits a high degree of conformational flexibility in its loop region (Jameson et al., 2002 ▶; Kontopidis et al., 2004 ▶). Similar to the situation observed here for the two independently solved TLC crystal structures, the C-terminal peptide segment of Blg, as well as the disulfide bond that links this segment to β-strand D, were poorly ordered in some crystal forms while showing well defined electron density in others (Qin et al., 1998 ▶). Even within the same crystal form, Blg appeared to be conformationally flexible in a pH-dependent manner: loop 3, which acts like a lid on top of the ligand pocket at pH 6.2, undergoes a major conformational change as the pH is raised to 7.1 or 8.2, resulting in better accessibility of the central cavity. This mechanism, the so-called Tanford transition (Sakurai & Goto, 2006 ▶), was postulated to have functional implications for the reversible binding and release of ligands (Qin et al., 1998 ▶).
Even though a similar pH-dependent movement of loop 3 (and also loop 4) has been described on the basis of circular-dichroism and tryptophan-fluorescence measurements for TLC (Gasymov, Abduragimov, Yusifov et al., 2004 ▶), the differences between the two TLC crystal structures discussed here are probably not pH-induced as these crystals were obtained at virtually identical pH values [pH 7.9 for the C2 structure (Breustedt et al., 2005 ▶) and pH 8.0 for the P21 structure]. Nevertheless, loop flexibility might be of relevance for the physiological function of this lipocalin. Circular-dichroism experiments suggested that lipid binding is associated with the reversible formation of additional β-sheet structure and a more rigid state of the protein (Gasymov et al., 1998 ▶). Accordingly, we observed an ∼20% higher β-sheet content, especially at the open end of the β-barrel, for TLC in complex with 1,4-butanediol compared with apo TLC. Furthermore, the entire loop region, which was seen to be partially disordered in the crystal structure of the apoprotein, appears to be more rigid in the complex structure, although the elevated B factors of some loop residues still indicate increased flexibility. Also, an influence of the molecular packing in the P21 crystal form on the conformation of the loop region seems unlikely because the four independent proteins in the asymmetric unit exhibit almost identical structures.
The conformational change may be triggered to some extent by complex formation with the new ligand. While the bound 1,4-butanediol itself is located far away from the loop region, its accommodation is accompanied by two significant side-chain rearrangements in the deeper part of the β-barrel (Fig. 2 ▶ c). In particular, Met39 is pushed outward and with it the entire β-strand B. Its Cα position is shifted by 2.2 Å which, in an indirect manner, could cause conformational changes both in loop 1, which connects strands A and B, and in the neighbouring strands C and D as well as their connecting loop 2. Nevertheless, the extent of these rearrangements is astonishing. Thus, the specific molecular environment in the previously described C2 crystal form has most likely additionally favoured the elevated and probably inherent structural flexibility of TLC observed there.
In the present crystal structure 1,4-butanediol is deeply buried within the β-barrel, similar to the fatty-acid tail of laurate bound to the lipocalin C8γ (Chiswell et al., 2007 ▶), yet laterally shifted, and forms direct contacts with Trp17 and Tyr97 at the bottom of the cavity. This ligand-binding mode could not be predicted based on the C2 crystal structure, in which the side chains of residues Met39 and Phe99 protrude into the cavity and lead to a bifurcation at its bottom, thereby shielding the aromatic side chain of Trp17. Obviously, the TLC ligand pocket is not only larger compared with other lipocalins (Breustedt et al., 2005 ▶), but can also considerably adapt to ligands via side-chain rearrangements, mainly involving residue Phe99. Notably, Tyr/Trp-fluorescence titration of recombinant TLC (Breustedt et al., 2006 ▶) with 1,4-butanediol in a concentration range up to 15 µM did not indicate binding (data not shown), suggesting that this artificial ligand is recognized with poor affinity and mainly owes its presence to its high concentration in the crystallization solution.
Nevertheless, it is not clear from structural comparison alone whether the conformational rearrangement of Phe99 is caused by the presence of the ligand. Spectroscopic measurements in solution indicated that Trp17 is equally buried in apo TLC and its ligand complexes (Gasymov et al., 1998 ▶), which is consistent with the structures discussed here. While Trp17 is buried by the neighbouring side chains of Met39 and Phe99 in the apoprotein, it is shielded from solvent by the bound 1,4-butanediol in the holo structure. Consequently, it would be conceivable that the observed structural rearrangements in the cavity are indeed ligand-induced, as generally postulated previously (Gasymov et al., 1998 ▶). This finding would be in contrast to the situation observed for odorant-binding proteins, a subset of lipocalins that exhibit high sequence similarity to TLC (45.2% identity between human OBPIIaα and TLC) and also bind a broad spectrum of ligands (Briand et al., 2002 ▶). In fact, crystallographic analysis of porcine OBP in complex with a variety of odorant molecules revealed that none of these ligands induced considerable side-chain rearrangements in the cavity compared with the apoprotein (Vincent et al., 2000 ▶).
Taken together, we have shown that the loop region and adjoining parts of the β-barrel exhibit considerable conformational flexibility, thus allowing structural adaptation of TLC to various natural and artificial ligands, including 1,4-butanediol. This illustrates a new mechanism for promiscuity in ligand recognition that may also be relevant for other physiologically important members of the lipocalin protein family.
Supplementary Material
PDB reference: tear lipocalin, 3eyc, r3eycsf
Acknowledgments
The authors wish to thank Ina Theobald and Irmgard Neumaier for the preparation of mutant streptavidin Sepharose and Andreas Eichinger for advice in using RMSPDB. This work was financially supported by Pieris AG, Freising, Germany.
References
- Åkerström, B., Borregaard, N., Flower, D. A. & Salier, J.-S. (2006). Lipocalins. Georgetown: Landes Bioscience.
- Aschaffenburg, R., Green, D. W. & Simmons, R. M. (1965). J. Mol. Biol.13, 194–201. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Bishop, R. E. (2000). Biochim. Biophys. Acta, 1482, 73–83. [DOI] [PubMed]
- Breustedt, D. A., Korndörfer, I. P., Redl, B. & Skerra, A. (2005). J. Biol. Chem.280, 484–493. [DOI] [PubMed]
- Breustedt, D. A., Schönfeld, D. L. & Skerra, A. (2006). Biochim. Biophys. Acta, 1764, 161–173. [DOI] [PubMed]
- Briand, L., Eloit, C., Nespoulous, C., Bezirard, V., Huet, J. C., Henry, C., Blon, F., Trotier, D. & Pernollet, J. C. (2002). Biochemistry, 41, 7241–7252. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chiswell, B., Lovelace, L. L., Brannen, C., Ortlund, E. A., Lebioda, L. & Sodetz, J. M. (2007). Biochim. Biophys. Acta, 1774, 637–644. [DOI] [PubMed]
- Clifton, M. C., Corrent, C. & Strong, R. K. (2009). Biometals, 22, 557–564. [DOI] [PubMed]
- Cowan, S. W., Newcomer, M. E. & Jones, T. A. (1990). Proteins, 8, 44–61. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, California, USA.
- Eichinger, A., Nasreen, A., Kim, H. J. & Skerra, A. (2007). J. Biol. Chem.282, 31068–31075. [DOI] [PubMed]
- Flower, D. R. (1996). Biochem. J.318, 1–14. [DOI] [PMC free article] [PubMed]
- Flower, D. R. (2000). Biochim. Biophys. Acta, 1482, 46–56. [DOI] [PubMed]
- Fluckinger, M., Haas, H., Merschak, P., Glasgow, B. J. & Redl, B. (2004). Antimicrob. Agents Chemother.48, 3367–3372. [DOI] [PMC free article] [PubMed]
- Gasymov, O. K., Abduragimov, A. R., Gasimov, E. O., Yusifov, T. N., Dooley, A. N. & Glasgow, B. J. (2004). Biochim. Biophys. Acta, 1688, 102–111. [DOI] [PubMed]
- Gasymov, O. K., Abduragimov, A. R., Yusifov, T. N. & Glasgow, B. J. (1998). Biochim. Biophys. Acta, 1386, 145–156. [DOI] [PubMed]
- Gasymov, O. K., Abduragimov, A. R., Yusifov, T. N. & Glasgow, B. J. (1999). Biochim. Biophys. Acta, 1433, 307–320. [DOI] [PubMed]
- Gasymov, O. K., Abduragimov, A. R., Yusifov, T. N. & Glasgow, B. J. (2004). Biochemistry, 43, 12894–12904. [DOI] [PubMed]
- Glasgow, B. J., Abduragimov, A. R., Farahbakhsh, Z. T., Faull, K. F. & Hubbell, W. L. (1995). Curr. Eye Res.14, 363–372. [DOI] [PubMed]
- Goetz, D. H., Willie, S. T., Armen, R. S., Bratt, T., Borregaard, N. & Strong, R. K. (2000). Biochemistry, 39, 1935–1941. [DOI] [PubMed]
- Israili, Z. H. & Dayton, P. G. (2001). Drug Metab. Rev.33, 161–235. [DOI] [PubMed]
- Jameson, G. B., Adams, J. J. & Creamer, L. K. (2002). Int. Dairy J.12, 319–329.
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Kleywegt, G. J., Zou, J.-Y., Kjeldgaard, M. & Jones, T. A. (2001). International Tables for Crystallography, Vol. F, edited by M. G. Rossmann & E. Arnold, pp. 353–356. Dordrecht: Kluwer Academic Publishers.
- Kontopidis, G., Holt, C. & Sawyer, L. (2004). J. Dairy Sci.87, 785–796. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Oldfield, T. J. (2001). Acta Cryst. D57, 82–94. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Qin, B. Y., Bewley, M. C., Creamer, L. K., Baker, H. M., Baker, E. N. & Jameson, G. B. (1998). Biochemistry, 37, 14014–14023. [DOI] [PubMed]
- Raghu, P. & Sivakumar, B. (2004). Biochim. Biophys. Acta, 1703, 1–9. [DOI] [PubMed]
- Redl, B. (2000). Biochim. Biophys. Acta, 1482, 241–248. [DOI] [PubMed]
- Sakurai, K. & Goto, Y. (2006). J. Mol. Biol.356, 483–496. [DOI] [PubMed]
- Schmidt, T. G. M. & Skerra, A. (2007). Nature Protoc.2, 1528–1535. [DOI] [PubMed]
- Schönfeld, D. L., Ravelli, R. B., Mueller, U. & Skerra, A. (2008). J. Mol. Biol.384, 393–405. [DOI] [PubMed]
- Skerra, A. (2000). Biochim. Biophys. Acta, 1482, 337–350. [DOI] [PubMed]
- Vincent, F., Spinelli, S., Ramoni, R., Grolli, S., Pelosi, P., Cambillau, C. & Tegoni, M. (2000). J. Mol. Biol.300, 127–139. [DOI] [PubMed]
- Wojnar, P., Lechner, M., Merschak, P. & Redl, B. (2001). J. Biol. Chem.276, 20206–20212. [DOI] [PubMed]
- Wojnar, P., Lechner, M. & Redl, B. (2003). J. Biol. Chem.278, 16209–16215. [DOI] [PubMed]
- Zanotti, G., Ottonello, S., Berni, R. & Monaco, H. L. (1993). J. Mol. Biol.230, 613–624. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: tear lipocalin, 3eyc, r3eycsf
PDB reference: tear lipocalin, 3eyc, r3eycsf