The structures of five forms of d-alanine-d-alanine ligase from T. thermophilus HB8 showed a cumulative conformational change of the molecular structure through the induced rotation of the central domain in concert with a local conformational change of three loops. The active-site structures shed light on the catalytic mechanism and the roles of the conformational change.
Keywords: d-alanine-d-alanine ligase, Thermus thermophilus HB8, catalytic mechanism, conformational change
Abstract
d-Alanine-d-alanine ligase (Ddl) is one of the key enzymes in peptidoglycan biosynthesis and is an important target for drug discovery. The enzyme catalyzes the condensation of two d-Ala molecules using ATP to produce d-Ala-d-Ala, which is the terminal peptide of a peptidoglycan monomer. The structures of five forms of the enzyme from Thermus thermophilus HB8 (TtDdl) were determined: unliganded TtDdl (2.3 Å resolution), TtDdl–adenylyl imidodiphosphate (2.6 Å), TtDdl–ADP (2.2 Å), TtDdl–ADP–d-Ala (1.9 Å) and TtDdl–ATP–d-Ala-d-Ala (2.3 Å). The central domain rotates as a rigid body towards the active site in a cumulative manner in concert with the local conformational change of three flexible loops depending upon substrate or product binding, resulting in an overall structural change from the open to the closed form through semi-open and semi-closed forms. Reaction-intermediate models were simulated using TtDdl-complex structures and other Ddl structures previously determined by X-ray methods. The catalytic process accompanied by the cumulative conformational change has been elucidated based on the intermediate models in order to provide new insights regarding the details of the catalytic mechanism.
1. Introduction
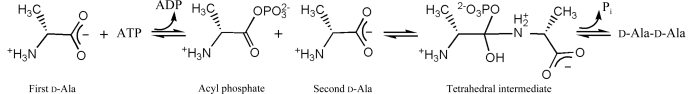
d-Ala-d-Ala ligase (Ddl), one of the key enzymes in the peptidoglycan-biosynthetic pathway, catalyzes the condensation of two d-Ala molecules using ATP. The mechanism proposed for d-Ala-d-Ala synthesis is shown in Fig. 1 ▶ (Mullins et al., 1990 ▶). The carboxylate O atom of the first d-Ala performs a nucleophilic attack on the γ-phosphate of ATP to produce an acylphosphate and ADP. Next, the acyl C atom of the acylphosphate is attacked by the amino group (nucleophile) of the second d-Ala to yield a tetrahedral intermediate. Finally, the intermediate releases phosphate to yield d-Ala-d-Ala.
Figure 1.
Proposed mechanism for d-Ala-d-Ala synthesis.
The cell-wall peptidoglycan polymer is produced through cross-linking of peptidoglycan monomer units (van Heijenoort, 2001 ▶; Welzel, 2005 ▶): the glycan parts of the monomer units are polymerized to yield linear glycan chains through a transglycosylation reaction and the peptide parts of the glycan chains are covalently linked to the neighbouring peptide parts through a transpeptidation reaction. These reactions, which are both crucial for the biosynthesis of the peptidoglycan polymers, are catalyzed by a single enzyme: bifunctional transglycosylase/transpeptidase. d-Ala-d-Ala constitutes the terminus of the peptide part of the peptidoglycan monomer unit and is involved in the transpeptidation reaction as the substrate. The penultimate d-Ala is linked to the neighbouring peptide and the terminal d-Ala is released. The terminal d-Ala-d-Ala moiety is thus directly involved in the cross-linking of the peptidoglycan monomer units.
Many efforts have been made to develop antibiotics which inhibit peptidoglycan biosynthesis, as peptidoglycan polymers play a critical role in maintenance of the cell-wall structure. β-Lactam antibiotics are covalently bound to the active site of the transpeptidase, resulting in inactivation of the enzyme, and have long been used for the treatment of infectious diseases (Knox et al., 1996 ▶). However, methicillin-resistant Staphylococcus aureus (MRSA), which shows multi-drug resistance to β-lactam antibiotics, has emerged (Spratt, 1994 ▶). A glycopeptide antibiotic, vancomycin, was isolated and has been used as a drug of last resort. Vancomycin becomes hydrogen bonded to the terminal d-Ala-d-Ala of the growing peptidoglycan polymers, inhibiting the transpeptidation reaction and further cross-linking of the peptidoglycan polymers. However, vancomycin-resistant strains have also appeared. In these strains, d-Ala-d-lactate replaces the terminal d-Ala-d-Ala of the peptidoglycan polymers and prevents vancomycin from binding to the terminal peptide (Davies, 1994 ▶).
Ddl and its homologue d-Ala-d-lactate ligase have been studied by X-ray crystallographic methods as these enzymes play key roles in the biosynthesis of the bacterial cell and are interesting targets for drug discovery. To date, the crystal structures of Ddls from Escherichia coli (Fan et al., 1994 ▶, 1997 ▶), S. aureus (Liu et al., 2006 ▶), Thermus caldophilus (Lee et al., 2006 ▶) and Helicobacter pylori (Wu et al., 2008 ▶) and those of d-Ala-d-lactate ligases from Leuconostoc mesenteroides (Kuzin et al., 2000 ▶) and Enterococci faecium (Roper et al., 2000 ▶) have been reported in order to elucidate the structural and chemical bases of enzymatic catalysis and vancomycin resistance. These enzymes, which belong to the ATP-dependent carboxylate-amine/thiol ligase superfamily together with glutathione synthetase, biotin carboxylase and carbamoyl phosphate synthetase (Galperin & Koonin, 1997 ▶; Denessiouk et al., 1998 ▶), consist of N-terminal, central and C-terminal domains. ATP is bound to the interface between the central and C-terminal domains and the substrate d-Ala molecules are located at the intersection between the three domains. The central domain and the flexible loop have been suggested to move towards the C-terminal domain to close the active site based on structural comparison of Ddls (Liu et al., 2006 ▶; Lee et al., 2006 ▶). The structure of Ddl complexed with the transition-state analogue (phosphorylated phosphinate) and with ADP allowed elucidation of the enzyme–substrate interactions and the proposal of a mechanism for the catalysis (Fan et al., 1994 ▶, 1997 ▶).
Ddl from T. thermophilus HB8 (TtDdl), which was overexpressed in E. coli, contains 319 residues and has a molecular mass of 34 670 Da. In order to elucidate the conformational change, the substrate recognition, the enzyme–substrate interactions and the mutual positioning between substrates with reference to the reaction mechanism, we have determined the crystal structures of unliganded TtDdl, TtDdl–adenylyl imidodiphosphate (AMP-PNP), TtDdl–ADP, TtDdl–ADP–d-Ala and TtDdl–ATP–d-Ala-d-Ala. The structures first revealed that the enzyme changes its structure in a cumulative manner upon binding of a substrate or product. Four conformational states (open, semi-open, semi-closed and closed forms) were identified and the reaction catalyzed by TtDdl is considered to proceed through these states. The reaction-intermediate models shed light on the detailed catalytic process accompanied by the cumulative conformational change. Here, we report an X-ray structural study of TtDdl, which belongs to the ATP-dependent carboxylate-amine/thiol ligase superfamily, and its complexes.
2. Experimental methods
2.1. Expression and purification
The structural gene for TtDdl was inserted between the NdeI and the BamHI restriction sites of plasmid pET-11a (Novagen). The resulting expression plasmid (pTtDdl) was used to transform E. coli strain BL21 (DE3) (Novagen). The transformant was cultured at 310 K in 6 l Luria broth supplemented with ampicillin (50 µg ml−1; Miller, 1972 ▶). When the optical density at 600 nm (OD600) of the medium reached 0.6, protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM. Cells were harvested by centrifugation after 16 h and stored at 193 K until use. Selenomethionine (SeMet) substituted TtDdl was prepared by overexpressing pTtDdl in E. coli B834 (DE3) cells grown in the presence of selenomethionine.
The cells were suspended in 20 mM Tris–HCl pH 8.0 containing 5 mM 2-mercaptoethanol and 50 mM NaCl and then disrupted by sonication. The cell lysate was incubated at 343 K for 10 min and then ultracentrifuged (200 000g) for 60 min at 277 K. Solid ammonium sulfate was added to the resulting supernatant to a final concentration of 1.5 M. The precipitate was dissolved in 20 mM Tris–HCl pH 8.0 and then applied onto an ion-exchange column (Resource Q, GE Healthcare) equilibrated with the same buffer. The fractions containing TtDdl were desalted and applied onto a hydroxyapatite CHT10-I column (Bio-Rad) equilibrated with 10 mM sodium phosphate pH 7.0. The flowthrough fractions containing TtDdl were pooled and concentrated by ultrafiltration. The concentrated solution was applied onto a gel-filtration column (HiLoad 16/60 Superdex 75pg, GE Healthcare) equilibrated with 20 mM Tris–HCl pH 8.0 containing 150 mM NaCl. The peak fractions were concentrated and stored at 277 K.
2.2. Crystallization and data collection
Unless stated otherwise, crystallizations were performed at 293 K using the hanging-drop vapour-diffusion method by equilibration of a drop containing 1 µl 11.6 mg ml−1 protein solution and 1 µl reservoir solution against 500 µl reservoir solution. Complex crystals were prepared by a cocrystallization method. The unliganded (SeMet) TtDdl was crystallized using protein solution and a reservoir solution containing 14%(w/v) polyethylene glycol 4000 and 100 mM NaCl in 100 mM MES buffer pH 5.8. Crystals of TtDdl–AMP-PNP were obtained using the sitting-drop method by mixing protein solution containing 10 mM AMP-PNP with reservoir solution composed of 16% PEG 3350 and 100 mM ammonium sulfate in 100 mM bis-tris buffer pH 5.8. TtDdl–ADP was crystallized by the sitting-drop method by mixing protein solution containing 10 mM ATP with reservoir solution composed of 16% PEG 3350 and 100 mM magnesium formate in 100 mM bis-tris buffer pH 5.6. This implies that the enzyme has ATPase activity that hydrolyzes the bound ATP to ADP and phosphate, as observed in S. faecalis Ddl (Neuhaus, 1962a ▶,b ▶), as the cocrystallization of TtDdl with an excess of ATP and magnesium formate gave crystals of a 1:1 complex of TtDdl and ADP. Crystals of TtDdl–ADP–d-Ala were obtained using protein solution containing 100 mM d-Ala and 10 mM ADP and the same reservoir solution as for the ADP complex. TtDdl–ATP–d-Ala-d-Ala was crystallized using protein solution containing 50 mM d-Ala-d-Ala and 10 mM ATP and a reservoir solution composed of 0.91 M K2HPO4, 0.49 M NaH2PO4 and 20% glycerol.
For the cryogenic experiment involving SPACE (Ueno et al., 2004 ▶), the crystals were flash-cooled to 100 K in a nitrogen stream. TtDdl–ADP and TtDdl–ATP–d-Ala-d-Ala crystals were flash-cooled without a cryoprotectant, whereas the other crystals were briefly soaked in a cryoprotectant solution containing 20–30%(v/v) ethylene glycol and flash-cooled. All diffraction data were collected on the BL26B2 beamline (Ueno et al., 2006 ▶) at SPring-8, Hyogo, Japan and were processed with the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). MAD data for unliganded (SeMet) TtDdl were collected at three different wavelengths. The crystals of unliganded TtDdl and TtDdl–AMP-PNP belonged to space group C2, with one dimer and one molecule per asymmetric unit. The crystals of TtDdl–ADP and TtDdl–ADP–d-Ala were isomorphous with space group P212121, with two dimers per asymmetric unit. The crystals of TtDdl–ATP–d-Ala-d-Ala belonged to space group P41212, with one dimer per asymmetric unit.
2.3. Structure determination and refinement
The scaling of the data and map calculations were performed with the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The structure of the unliganded (SeMet) TtDdl was determined at 2.3 Å resolution by the multiple-wavelength anomalous diffraction (MAD) phasing method with SOLVE/RESOLVE (Terwilliger & Berendzen, 1999 ▶), followed by automatic model tracing with the program ARP/wARP (Morris et al., 2002 ▶). The structure was then manually completed with the program XTALVIEW (McRee, 1999 ▶) and refined with CNS (Brünger et al., 1998 ▶). Solvent molecules were picked up from the σA-weighted F o − F c map. The structures of the complexes with the ligands were determined at 1.9–2.6 Å resolution by means of the molecular-replacement program MOLREP using the structure of the unliganded TtDdl monomer as a search model. Models were built with the program XTALVIEW and refinements were carried out with CNS. When the R-factor value decreased below 30%, simulated annealing 2F o − F c maps were calculated to assign the bound ligands to the residual electron densities. Solvent molecules were picked up from the σA-weighted F o − F c map. The data-collection and refinement statistics are summarized in Table 1 ▶. Structure diagrams were drawn using PyMOL (DeLano, 2002 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Unliganded (SeMet) TtDdl | |||||||
|---|---|---|---|---|---|---|---|
| TtDdl–AMP-PNP | TtDdl–ADP | TtDdl–ADP–D-Ala | TtDdl–ATP–D-Ala-D-Ala | Peak | Edge | Remote | |
| Diffraction data | |||||||
| Space group | C2 | P212121 | P212121 | P41212 | C2 | ||
| Unit-cell parameters | |||||||
| a (Å) | 167.7 | 69.1 | 69.0 | 95.0 | 165.1 | 165.3 | 165.2 |
| b (Å) | 56.7 | 101.1 | 105.4 | 95.0 | 56.4 | 56.5 | 56.4 |
| c (Å) | 141.8 | 197.5 | 199.8 | 192.0 | 142.1 | 142.3 | 142.2 |
| α (°) | 90 | 90 | 90 | 90 | 90 | 90 | 90 |
| β (°) | 108.6 | 90 | 90 | 90 | 109.1 | 109.1 | 109.1 |
| γ (°) | 90 | 90 | 90 | 90 | 90 | 90 | 90 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.9790 | 0.9792 | 0.90000 |
| Resolution (Å) | 50.0–2.6 | 50.0–2.2 (2.28–2.20) | 50.0–1.9 (1.97–1.90) | 50.0–2.3 (2.38–2.30) | 50.0–2.3 (2.38–2.30) | 50.0–2.4 (2.49–2.40) | 50.0–2.3 (2.38–2.30) |
| Total No. of reflections | 197257 | 642627 | 792120 | 689431 | 278449 | 246471 | 277518 |
| No. of unique reflections | 38994 | 68003 | 115070 | 39856 | 55134 | 48674 | 54945 |
| Redundancy | 5.0 (5.1) | 9.5 (6.0) | 6.9 (5.4) | 17.3 (8.9) | 5.1 (4.9) | 5.1 (5.4) | 5.1 (4.9) |
| Completeness (%) | 99.6 (99.0) | 95.4 (69.8) | 99.8 (97.9) | 99.6 (96.2) | 99.9 (100.0) | 99.8 (100.0) | 99.9 (99.6) |
| I/σ(I) | 25.7 (3.2) | 30.1 (4.7) | 23.2 (4.1) | 34.8 (3.7) | 26.2 (5.5) | 29.0 (6.0) | 25.6 (4.6) |
| Rmerge† (%) | 3.9 (29.6) | 4.9 (21.9) | 7.8 (31.7) | 7.0 (32.6) | 5.1 (28.0) | 5.3 (28.2) | 5.5 (35.6) |
| Refinement | |||||||
| Resolution limits (Å) | 47.9–2.6 | 49.40–2.2 | 47.9–1.9 | 47.9–2.3 | 44.1–2.3 | ||
| R factor (%) | 23.0 | 21.5 | 20.7 | 23.1 | 24.3 | ||
| Rfree (%) | 28.0 | 26.2 | 23.9 | 26.6 | 29.0 | ||
| No. of solvent atoms | 81 | 352 | 643 | 194 | 204 | ||
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.008 | 0.006 | 0.008 | 0.008 | 0.007 | ||
| Bond angles (°) | 1.4 | 1.3 | 1.4 | 1.4 | 1.4 | ||
| Mean B factors (Å2) | |||||||
| Main-chain atoms | 46.8 | 35.8 | 22.2 | 40.6 | 43.6 | ||
| Side-chain atoms | 48.9 | 38.5 | 25.6 | 42.8 | 46.4 | ||
| Heteroatoms | 100.5 | 37.4 | 19.2 | 37.4 | |||
| Water atoms | 34.0 | 36.2 | 28.4 | 37.1 | 39.8 | ||
| Ramachandran plot¶ (%) | |||||||
| Most favoured | 89.2 | 90.3 | 92.0 | 91.9 | 89.3 | ||
| Additionally allowed | 10.0 | 9.0 | 7.3 | 7.7 | 9.5 | ||
| Generously allowed | 0.4 | 0.3 | 0.0 | 0.0 | 0.8 | ||
| Disallowed | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | ||
R
merge = 
 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity for multiple measurements.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity for multiple measurements.
Stereochemistry was assessed by Procheck (Laskowski et al., 1993 ▶)
2.4. Enzyme assay
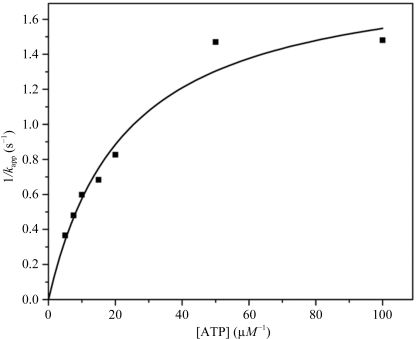
Enzyme activity was measured on the basis of ADP formation using a pyruvate kinase–lactate dehydrogenase coupled assay (Seelig & Meister, 1985 ▶). The reaction mixture comprised 20 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 2.0 mM phosphoenolpyruvate, 0.2 mM NADH, 25 units ml−1 lactate dehydrogenase and 25 units ml−1 pyruvate kinase. The assay was initiated by adding 20 µl TtDdl solution (20 mM HEPES pH 7.5, 17.33 µg ml−1 TtDdl) to 200 µl reaction buffer at 297 K and was monitored at 340 nm. The kinetic mechanism for Ddl was determined to be an ordered Ter-Ter mechanism (Mullins et al., 1990 ▶). At a saturating concentration of d-Ala, this mechanism is represented by the Michaelis–Menten-type equation
where K m,ATP is the Michaelis constant for ATP. It has been reported that the reaction is inhibited by a high concentration of d-Ala through binding of the product d-Ala-d-Ala to the Ddl–ATP complex (Mullins et al., 1990 ▶). Therefore, the rates of ADP formation were first measured by varying d-Ala (0.1–1.0 mM) with a fixed concentration of ATP (5.0–100.0 µM). The double-reciprocal plots constituted a family of nonlinear lines. The velocities of ADP formation under d-Ala saturation conditions were then obtained by extrapolating the curves to 1/[d-Ala] = 0 and plotted against [ATP] (Fig. 2 ▶). From the saturation curve in Fig. 2 ▶, K m,ATP, V max and k cat were calculated to be 23.0 ± 3.7 µM, 95.2 ± 7.7 nM s−1 and 1.90 ± 0.1 s−1, respectively.
Figure 2.
Plot of 1/k app versus ATP concentration for TtDdl.
3. Results and discussion
3.1. Overall structure
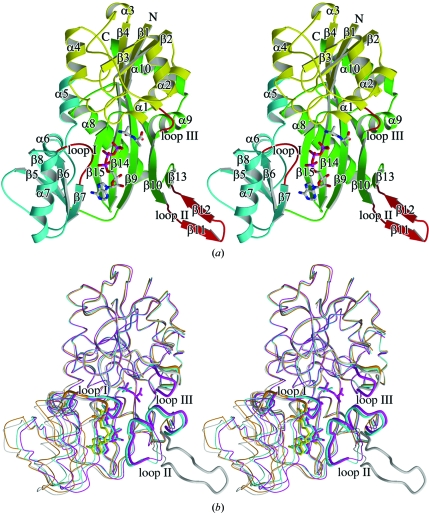
The molecular structure of TtDdl is shown in Fig. 3 ▶(a) together with the secondary-structure assignments made using the program DSSP (Kabsch & Sander, 1983 ▶). The polypeptide chain of TtDdl is folded into a dimeric form with twofold symmetry. The molecule is divided into three domains characterized by an α/β structure: an N-terminal domain (Met1–Gly104), a central domain (Ala105–Leu192) and a C-terminal domain (Ser193–Thr319). The long and wide crevice that accommodates ATP is located between the central and C-terminal domains and is encircled by the three domains, with the bottom of the crevice made up of three antiparallel β-strands (β9, β14 and β15) of the C-terminal domain (Fig. 3 ▶ a). The wall of the crevice is studded with loop I (Ala155–Val161), loop II (Tyr220–Ala234; ω-loop) and loop III (Phe289–Ser293), which are involved in the recognition of ATP and/or d-Ala.
Figure 3.
Overall structure of TtDdl. ATP and d-Ala-d-Ala are shown as ball-and-stick models. (a) Ribbon structural drawing of unliganded TtDdl in the open form. ATP and d-Ala-d-Ala are modelled into the active site by superposing TtDdl–ATP–d-Ala-d-Ala onto unliganded TtDdl. The N-terminal, central and C-terminal domains and loops (loops I, II and III) are shown as yellow, cyan, green and red ribbons, respectively. β-Strands are denoted by β1–β15 and α-helices by α1–α10. The N- and C-termini are labelled. (b) Superposed backbone structures. The molecular structures of unliganded TtDdl in the open form (grey), TtDdl–ADP in the semi-open form (orange), TtDdl–ADP–d-Ala in the semi-closed form (cyan) and TtDdl–ATP–d-Ala-d-Ala in the closed form (magenta) forms were superimposed by fitting the Cα atoms of the N- and C-terminal domains. The loops are represented by bold lines.
3.2. Cumulative conformational change
The crystal structures of unliganded TtDdl, TtDdl–AMP-PNP, TtDdl–ADP, TtDdl–ADP–d-Ala and TtDdl–ATP–d-Ala-d-Ala revealed that the enzyme changes its molecular conformation from the open form in unliganded TtDdl to the closed form in TtDdl–ATP–d-Ala-d-Ala in a cumulative manner (Fig. 3 ▶ b). The molecular structure of TtDdl is considered to be composed of two rigid bodies: the central domain and the N-terminal plus C-terminal domains. The overall conformational change results from a cumulative rotation of the central domain towards the C-terminal domain in accord with the local conformational change in loops I–III as shown in Table 2 ▶. The unliganded TtDdl, which has the same primary and tertiary structures as those of Ddl from T. caldophilus (Lee et al., 2006 ▶), is in an open form: a long and wide crevice formed at the interface between the central and C-terminal domains is exposed to the solvent region. TtDdl–AMP-PNP is in an open conformation with an overall structure that is quite similar to that of the unliganded form.
Table 2. Conformational change in TtDdl.
| Loop conformational change† | ||||||
|---|---|---|---|---|---|---|
| Ligands | Molecule | Overall conformation | Rotation angle of the central domain (°) | Loop I | Loop II | Loop III |
| None | A, B, C | Open | ||||
| AMP-PNP | A, B, C | Open | 0.2 | − | − | − |
| ADP | A, B, D | Semi-open | 4.0 | + | − | − |
| C | Semi-closed | 10.1 | + | + | − | |
| ADP, D-Ala | A, D‡ | Semi-closed | 10.3 | + | + | − |
| ATP, D-Ala-D-Ala | A, B | Closed | 13.6 | + | + | + |
A minus sign indicates that the loop conformation is the same as that of the unliganded enzyme in the open form. A plus sign indicates that the loop changes its conformation and approaches the ligands bound to the active site.
The conformation of molecules B and C is the semi-open form because D-Ala is not bound to these molecules.
The crystal structure of TtDdl–ADP showed that there are four independent molecules in the asymmetric unit, of which three are in the same conformation, with the remaining one being in a different conformation. The central domain in the former conformation (the semi-open form) rotates by about 4.0° as a rigid body towards the C-terminal domain, with local conformational change of loop I compared with that in the unliganded form (Fig. 3 ▶ b and Table 2 ▶). The accessible surface area of ADP is 102 Å2. In contrast, the latter conformation (the semi-closed form) exhibits a 10.1° rotation of the central domain towards the C-terminal domain, with local conformational changes of loops I and II (the ω-loop; Fig. 3 ▶ b). Loop II, which protrudes as a long two-stranded β-sheet towards the solvent region in unliganded TtDdl (Fig. 3 ▶ a), is folded back towards the crevice and covers it. The bound ADP is almost completely shielded from the solvent region; the accessible surface area of ADP is reduced to 16.0 Å2. It is possible that the semi-open form is in equilibrium with the semi-closed form in solution, although the two conformers trapped in the same asymmetric unit are affected by crystal-packing interactions. ADP behaves like part of the central domain; the arrangement of ADP with respect to the central domain is the same in the two conformers.
There are four independent molecules in the asymmetric unit of TtDdl–ADP–d-Ala. Two of them are in the semi-closed form; the Cα atoms of the molecules superimpose onto those of TtDdl–ADP in the semi-closed form with an average r.m.s. deviation of 0.40 Å. The remaining molecules do not bind d-Ala and are in the semi-open form. The addition of d-Ala to TtDdl–ADP stabilizes the semi-closed form. TtDdl–ATP–d-Ala-d-Ala is in a closed form; the central domain rotates by a further 3.5° towards the C-terminal domain compared with that of TtDdl–ADP–d-Ala in the semi-closed form (Fig. 3 ▶ b). Additionally, loop III changes its main-chain conformation to shield the C-terminal d-Ala (second d-Ala) of d-Ala-d-Ala from the solvent region.
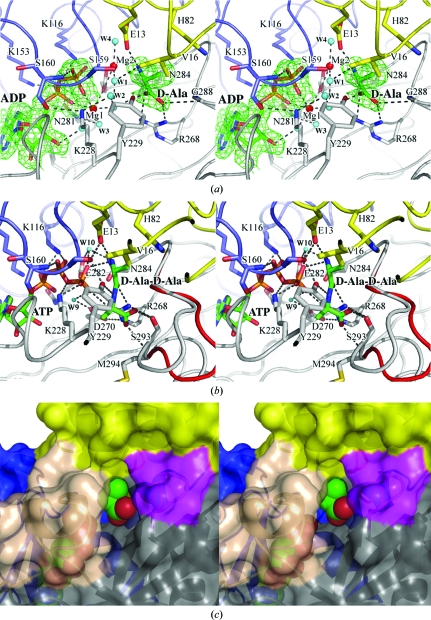
3.3. Active-site structure of TtDdl–ADP–d-Ala
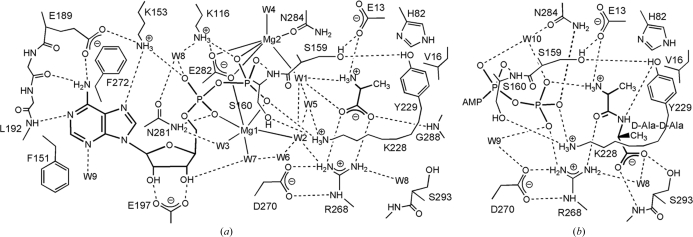
The structure and the hydrogen-bonding scheme of the active site are shown in Figs. 4 ▶(a) and 5 ▶(a), respectively. With the exception of the loop residues, the active-site residues maintain the same locations regardless of whether the enzyme is unliganded or liganded. The loop I residues Ser159 and Ser160 and the loop II residues Lys228 and Tyr229 approach the ADP β-phosphate and d-Ala to interact with them. The diphosphate of ADP interacts with Lys116 and Ser160 (loop I) of the central domain, Lys228 (loop II) and Asn281 of the C-terminal domain and two Mg ions (Mg1 and Mg2; Figs. 4 ▶ a and 5 ▶ a). The substrate d-Ala is bound to the first d-Ala-binding site (Kuzin et al., 2000 ▶; Roper et al., 2000 ▶; Liu et al., 2006 ▶). The protonated amino group of d-Ala is hydrogen bonded to Glu13. The carboxylate group interacts with the guanidino group of Arg268 and the main-chain NH group of Gly288. Arg268 and Gly288 form an oxyanion hole to stabilize the tetrahedral intermediate (Fig. 1 ▶; Kuzin et al., 2000 ▶; Fan et al., 1997 ▶).
Figure 4.
Stereo diagram of the active site in TtDdl. Each figure is viewed from the solvent side (the entrance to the active site). The active-site residues are represented by stick models. Water molecules are drawn as cyan circles. The N-terminal, central and C-terminal domains are drawn in yellow, blue and grey, respectively. Dotted lines represent coordination or hydrogen bonds. (a) Active site of TtDdl–ADP–d-Ala. ADP and d-Ala are shown as stick models. Mg ions are represented by red circles. The 2F o − F c electron-density map contoured at 1.0σ was calculated using data to 1.9 Å resolution. (b) Active site of TtDdl–ATP–d-Ala-d-Ala. ATP and d-Ala-d-Ala are shown as stick models. Loop III in TtDdl–ADP–d-Ala (red) is superimposed. (c) Surface representation of the d-Ala channel in TtDdl–ADP–d-Ala. Loops II and III are shown in wheat and magenta, respectively. d-Ala and ADP are drawn as CPK models. Loops II and III are predominantly involved in the formation of the d-Ala channel.
Figure 5.
Schematic diagram showing hydrogen bonds and coordinate interactions. Putative hydrogen-bond interactions are shown by dotted lines if the acceptor and donor are less than 3.5 Å apart. (a) TtDdl–ADP–d-Ala complex. Coordinate bonds between Mg ions and the ligands are less than 2.6 Å apart. (b) TtDdl–ATP–d-Ala-d-Ala. The AMP moiety of ATP and the residues interacting with it are omitted because the interactions are essentially the same for TtDdl–ADP–d-Ala and TtDdl–ATP–d-Ala-d-Ala.
The complex is in the semi-closed form and the active-site cavity is not completely shielded from the solvent, with an ASA of 14.4 Å2 when d-Ala is bound. There is a d-Ala channel that directly leads to the second d-Ala-binding site adjacent to the first d-Ala-binding site (Fig. 4 ▶ c). Loop III is a constituent of the channel and the tip of the loop may act as the door to the channel through which the second d-Ala enters the channel and reaches the second d-Ala-binding site. When the active-site structure of TtDdl–ADP–d-Ala is compared with that of TtDdl–ATP–d-Ala-d-Ala or E. coli Ddl–ADP–phosphorylated phosphinate, the substrate d-Ala in TtDdl–ADP–d-Ala fits well with the corresponding d-Ala bound to the first d-Ala-binding site. The coordinates of the two Mg ions interacting with ADP diphosphate in TtDdl–ADP–d-Ala are quite similar to those of the corresponding ions in E. coli Ddl–ADP–phosphorylated phosphinate (Fan et al., 1994 ▶).
3.4. Active-site structure of TtDdl–ATP–d-Ala-d-Ala
The structure and hydrogen-bonding scheme of the active site are shown in Figs. 4 ▶(b) and 5 ▶(b), respectively. In addition to loops I and II, loop III changes its conformation, approaches the active site and closes the entrance of the d-Ala channel (Fig. 4 ▶ c), indicating that the induced fitting of loop III to encapsulate ligands in the crevice plays an important role in the enzyme action. d-Ala-d-Ala bound to the first and second d-Ala-binding sites is almost completely shielded from the solvent region; the ASA of d-Ala-d-Ala is 0.5 Å2. The protonated amino group of d-Ala-d-Ala forms a salt bridge with the γ-phosphate and the carboxylate O atom of Glu13. The amide C=O makes a hydrogen bond to the guanidino group of Arg268. The amide NH group interacts with the hydroxy group of Tyr229, which forms a hydrogen-bonded triad together with Ser159 and Glu13 (Fan et al., 1994 ▶). The methyl group of the second d-Ala makes a contact with the aliphatic chain of Lys228. The carboxylate is directed towards the N-terminal side of α-helix α9 and interacts with the hydroxy group of Ser293 (loop III) and the main-chain NH group of Met294. The first d-Ala of d-Ala-d-Ala faces the γ-phosphate of ATP, with a distance of 3.77 Å between the carbonyl O atom and the P atom.
The Cα atoms of TtDdl–ATP–d-Ala-d-Ala superimposed on those of TtDdl–ADP–d-Ala show an excellent overlap between the ADP moiety of ATP and ADP and between the N-terminal d-Ala (first d-Ala) of d-Ala-d-Ala and d-Ala. With the exceptions of Lys190 and Asn281, the residues interacting with ADP or ATP are completely or strongly conserved in Ddls or d-Ala-d-lactate ligases for which structures have been determined. Superimposition of the active-site structures of TtDdl–ATP–d-Ala-d-Ala and E. coli Ddl–ADP–phosphorylated phosphinate (tetrahedral intermediate analogue; Fan et al., 1994 ▶) shows that the ADP moiety of ATP and d-Ala-d-Ala occupy similar positions to those of ADP and the phosphinate moiety of phosphorylated phosphinate in E. coli Ddl, respectively.
3.5. Mechanistic implications
The kinetic mechanism of d-Ala-d-Ala ligase has been determined to be an ordered Ter-Ter mechanism: ATP is the first substrate to bind, d-Ala then binds to the first d-Ala site as the second substrate and finally d-Ala binds to the second d-Ala site as the third substrate (Mullins et al., 1990 ▶). The reaction mechanism, including the stereochemistry, has been proposed on the basis of the X-ray structure of E. coli Ddl–ADP–phosphorylated phosphinate (Fan et al., 1994 ▶). However, the X-ray structures of various complexes of TtDdl have provided new insights regarding the involvement of the cumulative conformational change in the catalytic process.
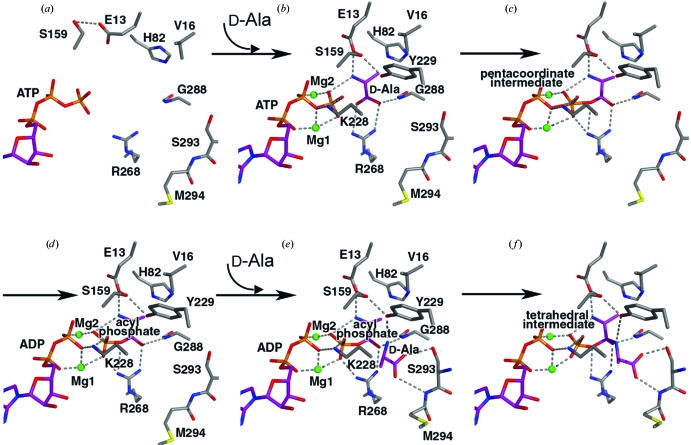
Firstly, ATP is bound to the large crevice between the central and N-terminal domains in the open form. Simultaneously, the central domain rotates towards the C-terminal domain. The active-site structure of TtDdl–ATP was modelled by replacing ADP of TtDdl–ADP in the semi-open form with ATP (Fig. 6 ▶ a). TtDdl–ATP might be in equilibrium between the semi-open and semi-closed forms of the enzyme, as inferred from the crystal structure of TtDdl–ADP (Fig. 3 ▶ a). Next, it is possible that the first d-Ala binds to the semi-open form rather than the semi-closed form of the ATP complex, as d-Ala can easily access the first d-Ala site exposed to the solvent region. The complex will be then converted to the semi-closed form observed in TtDdl–ADP–d-Ala. The TtDdl–ATP–d-Ala complex in the semi-closed form was modelled by replacing ADP in TtDdl–ADP–d-Ala by ATP in TtDdl–ATP–d-Ala-d-Ala (Fig. 6 ▶ b). In this model, ATP is relocated adjacent to the bound d-Ala to provide a favourable arrangement for phosphate transfer from ATP to d-Ala. The electrophilicity of the P atom in ATP γ-phosphate is enhanced by the interactions of the γ-phosphate O atoms with the two Mg ions, Lys228, Arg268 and the protonated amino group of the first d-Ala. One of the carboxylate O atoms of d-Ala, the γ-P atom and the ether O atom between the β-phosphate and γ-phosphate are in line. The carboxylate O atom then makes an inline attack on the γ-P atom to form the pentacoordinate transition state with trigonal bipyramidal geometry, which would be stabilized through maintenance of the phosphate interactions as much as possible (Fig. 6 ▶ c). The geometrical constraint on the transition state makes the P atom move slightly towards the carboxylate O atom to loosen the opposite P—O bond, which is polarized by a Mg ion (Harutyunyan et al., 1996 ▶). ADP is then released to produce the phosphorylated d-alanyl intermediate (acylphosphate) in the semi-closed form (Fig. 6 ▶ d). The two Mg ions and the protonated amino group of Lys228 bridge the ADP β-phosphate and the released phosphate to weaken the repulsive interaction between them.
Figure 6.
Catalytic process based on the X-ray structures of TtDdl complexes. Putative hydrogen bonds and coordinate bonds are shown by dotted lines. The C atoms of the substrates and active-site residues are shown in purple and grey, respectively. (a) TtDdl complex with ATP in the semi-open form. (b) TtDdl complex with ATP and d-Ala in the semi-closed form. (c) TtDdl complex with a trigonal bipyramid pentacoordinate intermediate in the semi-closed form. (d) TtDdl complex with ADP and acyl phosphate in the semi-closed form. (e) TtDdl complex with ADP, acyl phosphate and d-Ala in the closed form. (f) TtDdl complex with ADP and a tetrahedral intermediate.
The second d-Ala can access the second d-Ala-binding site adjacent to the acylphosphate bound to the first d-Ala-binding site through the d-Ala channel because loop III, which acts as the door to the channel, is open in the semi-closed form. ADP and acylphosphate in Fig. 6 ▶(d) were modelled into TtDdl–ATP–d-Ala-d-Ala with ATP and the first d-Ala of d-Ala-d-Ala removed, producing the model structure of TtDdl–ADP–acylphosphate–d-Ala in the closed form (Fig. 6 ▶ e). Ser293 belonging to loop III approaches the second d-Ala to interact with it. The protonated amino group of the second d-Ala resides on the carbonyl C atom of the acylphosphate from the solvent side and forms a hydrogen bond to Tyr229. It is likely that the protonated amino group of the second d-Ala is deprotonated by Tyr229, which forms a triad with Ser159 and Glu13. The deprotonated amino group then makes a nucleophilic attack on the carbonyl C atom of the acylphosphate to form the tetrahedral intermediate (Fig. 6 ▶ f). The intermediate is stabilized by the interactions with the oxyanion hole (main-chain NH of Gly288 and side chain of Arg268; Fan et al., 1997 ▶; Kuzin et al., 2000 ▶). Finally, the phosphate is released from the acylphosphate to yield d-Ala-d-Ala.
In summary, we have determined the crystal structure of TtDdl as unliganded TtDdl, TtDdl–AMP-PNP, TtDdl–ADP, TtDdl–ADP–d-Ala and TtDdl–ATP–d-Ala-d-Ala at 2.3, 2.6, 2.2, 2.2, 1.9 and 2.3 Å resolution, respectively. The enzyme shows a cumulative conformational change of the molecular structure through the induced rotation of the central domain in concert with a local conformational change of three loops. The X-ray structures of various conformational states shed light on the catalytic mechanism and the roles of the cumulative conformational change.
Supplementary Material
PDB reference: unliganded TtDdl, 2yzg, r2yzgsf
PDB reference: TtDdl–AMP-PNP, 2yzn, r2yznsf
PDB reference: TtDdl–ADP, 2zdg, r2zdgsf
PDB reference: TtDdl–ADP–d-Ala, 2zdh, r2zdhsf
PDB reference: TtDdl–ATP–d-Ala-d-Ala, 2zdq, r2zdqsf
Acknowledgments
The authors wish to thank Shinya Sato, Masami Nishida, Yumiko Inoue and Kayoko Matsumoto for protein expression and purification, and Yuka Nonaka for data collection at SPring-8.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davies, J. (1994). Science, 264, 375–382. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Denessiouk, K. A., Lehtonen, J. V. & Johnson, M. S. (1998). Protein Sci.7, 1768–1771. [DOI] [PMC free article] [PubMed]
- Fan, C., Moews, P. C., Walsh, C. T. & Knox, J. R. (1994). Science, 266, 439–443. [DOI] [PubMed]
- Fan, C., Park, I. S., Walsh, C. T. & Knox, J. R. (1997). Biochemistry, 36, 2531–2538. [DOI] [PubMed]
- Galperin, M. Y. & Koonin, E. V. (1997). Protein Sci.6, 2639–2643. [DOI] [PMC free article] [PubMed]
- Harutyunyan, E. H., Kuranova, I. P., Vainshtein, B. K., Hohne, W. E., Lamzin, V. S., Dauter, Z., Teplyakov, A. V. & Wilson, K. S. (1996). Eur. J. Biochem.239, 220–228. [DOI] [PubMed]
- Heijenoort, J. van (2001). Nat. Prod. Rep.18, 503–519. [DOI] [PubMed]
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Knox, J. R., Moews, P. C. & Frere, J. M. (1996). Chem. Biol.3, 937–947. [DOI] [PubMed]
- Kuzin, A. P., Sun, T., Jorczak-Baillass, J., Healy, V. L., Walsh, C. T. & Knox, J. R. (2000). Structure, 8, 463–470. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lee, J. H., Na, Y., Song, H. E., Kim, D., Park, B. H., Rho, S. H., Im, Y. J., Kim, M. K., Kang, G. B., Lee, D. S. & Eom, S. H. (2006). Proteins, 64, 1078–1082. [DOI] [PubMed]
- Liu, S., Chang, J. S., Herberg, J. T., Horng, M. M., Tomich, P. K., Lin, A. H. & Marotti, K. R. (2006). Proc. Natl Acad. Sci. USA, 103, 15178–15183. [DOI] [PMC free article] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed]
- Miller, J. (1972). Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press.
- Morris, R. J., Perrakis, A. & Lamzin, V. S. (2002). Acta Cryst. D58, 968–975. [DOI] [PubMed]
- Mullins, L. S., Zawadzke, L. E., Walsh, C. T. & Raushel, F. M. (1990). J. Biol. Chem.265, 8993–8998. [PubMed]
- Neuhaus, F. C. (1962a). J. Biol. Chem.237, 778–786. [PubMed]
- Neuhaus, F. C. (1962b). J. Biol. Chem.237, 3128–3135. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Roper, D. I., Huyton, T., Vagin, A. & Dodson, G. (2000). Proc. Natl Acad. Sci. USA, 97, 8921–8925. [DOI] [PMC free article] [PubMed]
- Seelig, G. F. & Meister, A. (1985). Methods Enzymol.113, 379–390. [DOI] [PubMed]
- Spratt, B. G. (1994). Science, 264, 388–393. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Ueno, G., Hirose, R., Ida, K., Kumasaka, T. & Yamamoto, M. (2004). J. Appl. Cryst.37, 867–873.
- Ueno, G., Kanda, H., Hirose, R., Ida, K., Kumasaka, T. & Yamamoto, M. (2006). J. Struct. Funct. Genomics, 7, 15–22. [DOI] [PubMed]
- Welzel, P. (2005). Chem. Rev.105, 4610–4660. [DOI] [PubMed]
- Wu, D., Zhang, L., Kong, Y., Du, J., Chen, S., Chen, J., Ding, J., Jiang, H. & Shen, X. (2008). Proteins, 72, 1148–1160. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: unliganded TtDdl, 2yzg, r2yzgsf
PDB reference: TtDdl–AMP-PNP, 2yzn, r2yznsf
PDB reference: TtDdl–ADP, 2zdg, r2zdgsf
PDB reference: TtDdl–ADP–d-Ala, 2zdh, r2zdhsf
PDB reference: TtDdl–ATP–d-Ala-d-Ala, 2zdq, r2zdqsf
PDB reference: unliganded TtDdl, 2yzg, r2yzgsf
PDB reference: TtDdl–AMP-PNP, 2yzn, r2yznsf
PDB reference: TtDdl–ADP, 2zdg, r2zdgsf
PDB reference: TtDdl–ADP–d-Ala, 2zdh, r2zdhsf
PDB reference: TtDdl–ATP–d-Ala-d-Ala, 2zdq, r2zdqsf