New classes of helix–helix interactions in protein structures are reported in which interactions only occur at the terminal regions or between the terminal region of one helix and the middle region of another helix.
Keywords: helix-packing interactions, helix–helix interfaces, residue surface areas, side-chain lengths, interhelical hydrophobic contacts, interhelical distances
Abstract
Helix–helix interactions are important for the structure, stability and function of α-helical proteins. Helices that either cross in the middle or show extensive contacts between each other, such as coiled coils, have been investigated in previous studies. Interactions between two helices can also occur only at the terminal regions or between the terminal region of one helix and the middle region of another helix. Examples of such helix pairs are found in aquaporin, H+/Cl− transporter and Bcl-2 proteins. The frequency of the occurrence of such ‘end-to-end’ (EE) and ‘end-to-middle’ (EM) helix pairs in protein structures is not known. Questions regarding the residue preferences in the interface and the mode of interhelical interactions in such helix pairs also remain unanswered. In this study, high-resolution structures of all-α proteins from the PDB have been systematically analyzed and the helix pairs that interact only in EE or EM fashion have been extracted. EE and EM helix pairs have been categorized into five classes (N–N, N–C, C–C, N–MID and C–MID) depending on the region of interaction. Nearly 13% of 5725 helix pairs belonged to one of the five classes. Analysis of single-residue propensities indicated that hydrophobic and polar residues prefer to occur in the C-terminal and N-terminal regions, respectively. Hydrophobic C-terminal interacting residues and polar N-terminal interacting residues are also highly conserved. A strong correlation exists between some of the residue properties (surface area/volume and length of side chains) and their preferences for occurring in the interface of EE and EM helix pairs. In contrast to interacting non-EE/EM helix pairs, helices in EE and EM pairs are farther apart. In these helix pairs, residues with large surface area/volume and longer side chains are preferred in the interfacial region.
1. Introduction
The major secondary-structural element found in protein structures is the α-helix. In the taxonomy of protein structures the α-class is one of the most important groups and in this class of protein structures the core of the protein consists exclusively of α-helices (Brändén & Tooze, 1999 ▶). Experimental and computational studies have investigated the residue preferences in different positions within an α-helix (Aurora & Rose, 1998 ▶; Aurora et al., 1994 ▶; Ballesteros et al., 2000 ▶; Chakrabarti & Pal, 2001 ▶; Chakrabartty & Baldwin, 1995 ▶; Cochran & Doig, 2001 ▶; Creamer & Rose, 1992 ▶; Engel & DeGrado, 2004 ▶; Iqbalsyah & Doig, 2004 ▶; Kumar & Bansal, 1998 ▶; Lacroix et al., 1998 ▶; Penel et al., 1999 ▶; Presta & Rose, 1988 ▶; Richardson & Richardson, 1988 ▶; Rohl et al., 1996 ▶; Sankararamakrishnan & Vishveshwara, 1992 ▶). The interaction between a pair of α-helices has long been a subject of study (Crick, 1953 ▶) and these interactions have been characterized using interhelical angles and interhelical distances (Chothia et al., 1981 ▶). Some of the well known models that explain helix–helix packing include the ‘knobs-in-holes’ (Crick, 1953 ▶), ‘ridges-in-grooves’ (Chothia et al., 1981 ▶), ‘close-packed sphere’ (Richmond & Richards, 1978 ▶) and ‘helix lattice superposition’ (Walther et al., 1996 ▶) models. The packing of and interactions between helices have also been studied in membrane proteins and compared with those of globular proteins (Adamian & Liang, 2001 ▶; Bowie, 1997 ▶; Gimpelev et al., 2004 ▶). It was shown that the distribution of helix-packing angles in membrane proteins is very different from that of soluble proteins (Bowie, 1997 ▶). The nature and distribution of the amino acids that mediate helix–helix interactions have also been investigated in soluble and membrane α-bundle proteins (Adamian et al., 2003 ▶; Adamian & Liang, 2001 ▶, 2002 ▶; Eilers et al., 2002 ▶; Javadpour et al., 1999 ▶; Walters & DeGrado, 2006 ▶). Hydrophobic residues seem to dominate the helix–helix interfaces of soluble proteins and transmembrane helices were shown to pack more tightly than those in soluble proteins. Several sequence motifs, such as the GXXG motif, ‘serine zipper’ and ‘polar clamp’, have been identified to mediate helix–helix interactions in membrane proteins (Adamian & Liang, 2002 ▶; Senes et al., 2004 ▶). Recently, small and weakly polar residues have been shown to be conserved as a group in the helix–helix interface of two major families of helical membrane proteins, namely GPCRs (Liu et al., 2004 ▶) and major intrinsic proteins (Bansal & Sankararamakrishnan, 2007 ▶). Tools have been developed to analyze and characterize α-helices and helix–helix packing in proteins (Burba et al., 2006 ▶; Mezei & Filizola, 2006 ▶). The role of charged residues in the stability of helical coiled coils has been investigated experimentally (Litowski & Hodges, 2002 ▶; Straussman et al., 2007 ▶; Zhou et al., 1994 ▶). Alanine-scanning mutagenesis and sedimentation-equilibrium ultracentrifugation techniques have been used to understand the energetic principles of helix–helix interactions in glycophorin A transmembrane-helix dimerization (Doura & Fleming, 2004 ▶; Fleming & Engelman, 2001 ▶).
Thus far, both computational analysis and experimental studies have focused on understanding helix pairs that interact in a coiled-coil fashion or that cross in the middle, in which the faces of both helices interact extensively with each other. However, there are cases in protein structures in which helix pairs interact exclusively in the terminal regions and only in the terminal regions. For example, in channel proteins such as aquaporin (Agre & Kozono, 2003 ▶) and H+/Cl− exchanger (Dutzler et al., 2002 ▶) the ends of two helices interact within the transmembrane region and such an arrangement has been shown to be functionally significant. We also have examples in which the end of one helix interacts with the face of another helix, as observed in the structures of Bcl-2 proteins (Petros et al., 2004 ▶) which mediate apoptosis. Nearly two decades ago, Murzin and Finkelstein recognized such helix packings in their classification using a quasi-spherical polyhedra model (Murzin & Finkelstein, 1988 ▶). In their analysis of interacting helix pairs, Reddy and Blundell eliminated those helix pairs which interacted only in the N-terminal or C-terminal regions (Reddy & Blundell, 1993 ▶). To our knowledge, there is no systematic study of helix pairs in which interaction occurs only at the ends of the two helices or between the end of one helix and the face of another helix. Several questions remain unanswered for such types of helix pairs. We do not know whether these end-to-end (EE) and end-to-middle (EM) helix pairs are observed as frequently as the coiled-coil helix pairs or helix pairs interacting in the ridges-in-grooves model. How does the interaction pattern of these helix pairs differ from those already characterized? Are there any residues that show a preference for occurring in the interface of EE and EM helix pairs and if so how do they differ from those helix pairs analyzed in earlier studies? Without answering the above questions, understanding of helix–helix interactions would be incomplete. In this paper, we have extracted high-resolution α-class protein structures from the Protein Data Bank (Berman et al., 2000 ▶) and have systematically identified and analyzed such helix pairs. Five different categories were classified and characterized. Three of them interact only through their terminal regions and in the remaining two interaction occurs between the N-terminus or C-terminus of one helix and the middle region of another helix. We have analyzed the residue preferences in the interacting region and also examined the residue–residue interactions that occur in the interfaces of such helix pairs. Our results show that residues with long side chains are involved in interactions with the terminal regions. The modes of interaction in EE and EM helix pairs are distinct from those in helix pairs that have been characterized previously in soluble and membrane proteins.
2. Methods
2.1. Classification of end-to-end and end-to-middle helix pairs
Protein structures belonging to the ‘all-α’ category as per the SCOP classification (Murzin et al., 1995 ▶) were extracted from the Protein Data Bank (PDB; Berman et al., 2000 ▶). Only those structures with a resolution of 2.5 Å or better and an R factor of 0.3 or better were considered for analysis. We used CD-HIT (Li & Godzik, 2006 ▶; Li et al., 2001 ▶, 2002 ▶) to remove redundant proteins. The nonredundant set contained 558 protein structures with 673 chains and the sequence identity of any two polypeptide chains was less than 25% (extracted in September 2006). Since ‘membrane and cell-surface proteins and peptides’ form a separate category in SCOP, the chosen data set did not contain any membrane proteins. The helical regions for each protein structure were obtained from ‘HELIX’ records of the PDB files and only those helices with a minimum length of eight residues were considered. We used the following steps to identify the ‘end-to-end’ and ‘end-to-middle’ helix pairs in each PDB structure.
Step 1. The helix axis of each helix was determined using the parametric least-squares fit algorithm (Christopher et al., 1996 ▶). All possible helix pairs were considered for a given structure if they were separated by at least 15 residues. This is to ensure that the interactions observed in a helix pair are not a consequence of their proximity in the primary sequence. Interhelical distances (d) for each helix pair were calculated using the helix axes. We also determined whether the helix pairs have at least one atomic contact. Interhelical atomic contact is defined if the distance between two atoms is less than the sum of their van der Waals radii plus 0.6 Å. This is the same criteria as used by Chothia (1975 ▶) in analysis of helix pairs. Those helix pairs with d ≤ 15 Å and with at least one interhelical atomic contact were filtered out and we found 5725 helix pairs.
Step 2. End-to-end and end-to-middle helix pairs were selected by analyzing the interhelical atomic contacts and the angle between the two helix axes. Each helix was divided into three regions. The first four and the last four residues were considered as the N-terminal and C-terminal regions, respectively. The residues between the N-terminal and C-terminal regions were considered to lie in the middle region.
Depending upon the nature of the contacts, end-to-end helix pairs have been classified into three different types: N–N, N–C and C–C helix pairs. An N–N helix pair is defined if the following criteria are satisfied: (i) there must be at least one interhelical atomic contact between the N-terminal regions of the two helices and (ii) the interhelical atomic contacts should only occur in the N-terminal regions of the two helices under consideration. A similar criterion is applied to define C–C helix pairs. If there is at least one interhelical atomic contact between the N-terminal region of one helix and the C-terminal region of another helix and if all the interhelical atomic contacts occur only between these two regions, the helix pair is defined as an N–C pair.
Two classes are defined for end-to-middle helix pairs: N–MID and C–MID helix pairs. Here, at least one interhelical atomic contact must occur between the N-terminal region or C-terminal region of one helix and the middle region of another helix and interhelical contacts must only be observed between these two regions. Such helix pairs are classified as N–MID or C–MID helix pairs, respectively.
The end-to-end and end-to-middle helix pairs thus obtained were further refined using a criterion based on the crossing angle (ω) between the helix pairs. The crossing angle is the angle between the two helix axes: ω = 0° corresponds to parallel helices and ω = ±180° indicates that the helices are antiparallel to each other. For coiled-coil helix pairs, the favoured angle for ω is 20° (Crick, 1953 ▶). For helix pairs that interact in the ridges-in-grooves model, a high peak in the distribution of crossing angles is observed in the range −40 to −50° (Chothia et al., 1981 ▶). We wanted to ensure that no helix pair is chosen whose interaction could arise from a coiled-coil arrangement or from interactions that could be described as the ridges-in-grooves model. Therefore, we discarded N–N and C–C helix pairs whose crossing angles fall above −45° (0° ≥ ω > −45°) or below +45° (0 ≤ ω < 45°). Similarly, N–C helix pairs were excluded if −180° ≥ ω ≥ −135° or 180° ≤ ω ≤ 135°. N–MID and C–MID helix pairs were only considered for further analysis if −45° ≤ ω ≤ −135° or 45° ≥ ω ≥ 135°. These criteria helped to avoid parallel orientation of N–N and C–C helix pairs and antiparallel orientation of N–C helix pairs. In the case of N–MID and C–MID pairs both parallel and antiparallel orientations are avoided.
2.2. Calculation of single-residue propensities
When all of the five different classes of helix pairs are considered, residues from one helix interact with one of the three regions (N-terminal, C-terminal or middle region) of another helix. For example, in N–N, N–C and N–MID helix pairs residues from one helix interact with the N-terminal region of another helix. Similarly, interactions with the C-terminal region are observed in C–C, N–C and C–MID helix pairs. In both N–MID and C–MID helix pairs the N-terminal or C-terminal regions of one helix interact with the middle region of another helix. It would be interesting to determine whether there is any preference for residues to interact with any of the three regions. Single-residue propensity values for each amino acid interacting with the N-terminal region were calculated from the above helix pairs using
where P i (R) is the single-residue propensity of an amino acid i that takes part in interhelical contacts with the particular region R (in this case, R represents the N-terminal region of the helix) and f i (R) is the fraction of amino acid i interacting with the same region R (N-terminal region) in N–N, N–C or N–MID helix pairs. In the case of N–N helix pairs both helices are considered individually since the N-terminal region of one helix interacts with the N-terminal region of the second helix. f i G is the fraction of the same amino acid involved in interaction with another residue irrespective of the secondary structures in which the interacting residues are present and this calculation was carried out for the entire data set of 558 protein structures. In some examples, the two helices of EE and EM helix pairs could come from two different polypeptide chains. In such cases, if the polypeptides are identical we considered only one of them when computing the global frequency f i G. The single-residue propensities of those residues interacting with the C-terminal and middle regions were calculated using the same approach.
2.3. Conservation of interacting residues in EE and EM helix pairs
The sequence of each protein with one or more EE or EM helix pairs was considered as a query sequence for a PSI-BLAST (Altschul et al., 1997 ▶) search. At least four iterations were carried out in PSI-BLAST and homologous sequences were extracted. CD-HIT (Li & Godzik, 2006 ▶; Li et al., 2001 ▶, 2002 ▶) was used to remove redundant sequences at an 80% cutoff level. The multiple sequence alignment obtained using ClustalW (v.1.83; Higgins et al., 1991 ▶) was then processed using JalView (Clamp et al., 2004 ▶). The conservation of each residue in the query sequence was calculated from the multiple sequence alignment. For this purpose, amino acids were grouped together based on their chemical properties as follows: acidic, Asp and Glu; basic, Arg, Lys and His; large polar, Asn and Gln; small polar, Ser, Thr and Cys; aromatic, Phe, Trp, Tyr and His; hydrophobic, Ala, Leu, Val, Ile and Met; others, Gly and Pro. In this analysis, there were protein sequences in which the conservation of every position exceeded 50%. We only considered those protein sequences in which the overall conservation was less than 50% but the conservation of interacting residues in EE and EM helix pairs exceeded 80%. We also excluded those cases where the interacting residues are close to cofactors or ligands (within 4.0 Å) bound to the protein molecule.
3. Results
3.1. End-to-end and end-to-middle helix pairs in protein structures
End-to-end (EE) helix pairs were classified into three groups: N–N, N–C and C–C helix pairs. The nomenclature was chosen to indicate the region of interhelical atomic contact. For end-to-middle (EM) helix pairs, two classes (N–MID and C–MID) were categorized, similar to the EE helix pairs. For all five classes, helix pairs were discarded if there were any interhelical atomic contacts outside the specific regions. The number of helix pairs observed in each class after applying the contact and angle criteria is given in Table 1 ▶. In total, 736 examples belonging to five different classes of EE and EM helix pairs were observed and each class contained at least ∼100 helix pairs (Table 1 ▶). The maximum number of examples (239) was found for the N–C category.
Table 1. End-to-end and end-to-middle helix pairs in protein structures.
| Percentage of interacting residues† | |||||
|---|---|---|---|---|---|
| Class | No. of helix pairs | No. of proteins | Polar | Nonpolar | Proline |
| N–N | 150 | 98 | 59.0 | 29.8 | 7.2 |
| C–C | 122 | 96 | 32.9 | 61.5 | 0.0 |
| N–C | 239 | 145 | 49.6 | 41.1 | 4.2 |
| N–MID | 99 | 81 | 45.6 | 49.0 | 3.1 |
| C–MID | 126 | 101 | 35.1 | 63.5 | 0.0 |
Polar, Asp, Glu, Arg, Lys, His, Asn, Gln, Ser, Thr and Cys; nonpolar: Ala, Leu, Ile, Val, Met, Phe, Tyr and Trp.
The number of helix pairs that can be described to have EE or EM interhelical interactions is about 13% of the total number of helix pairs and this proportion is significant (736 of 5725). Examples from each class of EE helix pairs and EM helix pairs are shown in Figs. 1 ▶ and 2 ▶, respectively. It is interesting to note that in 222 cases the helices forming such pairs come from two different polypeptide chains of the same protein.
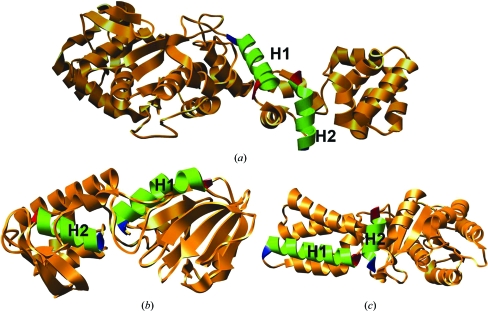
Figure 1.
Representative examples for each class of EE helix pairs. (a) C–C helix pair in the structure of a glutamyl tRNA synthetase (PDB code 1g59; 2.4 Å resolution). The C-terminal region of helix H1 (residues 306–321 of chain A) interacts with the C-terminal region of helix H2 (residues 344–356 of chain A). (b) N–N helix pair in the structure of a bacterial DNA glycosylase enzyme (PDB code 1r2y; 2.34 Å resolution). The N-terminal regions of helix H1 (residues 3–19 of chain A) and helix H2 (residues 173–184 of chain A) interact with each other. (c) N–C helix pair observed in the crystal structure of a conserved GTPase (PDB code 1j8m; 2.0 Å resolution). The N-terminal region of helix H1 (residues 20–39 in chain F) and the C-terminal region of helix H2 (residues 254–264 in chain F) make interhelical contacts. In this figure and in the subsequent figures the first and the last residues of a helix are displayed in blue and red, respectively, to indicate the N- and C-terminal regions and some helices are omitted for clarity. All molecular images were created using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (Pettersen et al., 2004 ▶).
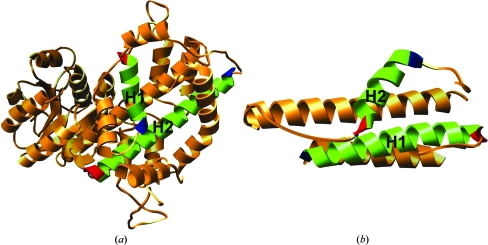
Figure 2.
Representative examples for the two classes of EM helix pairs. (a) N–MID helix pair observed in the structure of 6-phosphogluconate dehydrogenase (PDB code 2pgd; 2.0 Å resolution). The N-terminal region of helix H1 (residues 278–291) interacts with the middle region of the second helix H2 (residues 315–348). (b) C–MID pair in the crystal structure of HEPN domain protein (PDB code 1o3u; 1.75 Å resolution). The C-terminal and the middle regions of helix H1 (residues 19–43 of chain A) and helix H2 (residues 65–77 of chain A) are involved in interhelical contacts.
We have analyzed the nature of the interacting residues in the five different classes of helix pairs. Depending upon the region of interaction, differences are observed in the nature of the interacting residues. Residues interacting with the N-terminus tend to be more polar. This is evident in the N–N class of helix pairs, in which nearly 60% of the interacting residues are polar (Table 1 ▶). On the other hand, residues interacting with the C-terminus are found to be more hydrophobic. In C–C helix pairs more than 60% of the residues can be described as nonpolar. In the case of N–C, N–MID and C–MID helix pairs the interacting residues imply that the two sets of residues interact with two different regions. Polar interacting residues are relatively common in N–C and N–MID helix pairs (∼50% and ∼46%, respectively), in which the residues from the C-terminus or the middle region of an α-helix interact with the N-terminus. Similarly, when the C-terminus is involved a higher number of interacting residues are found to be hydrophobic, as in the case of C–MID and N–C helix pairs. Proline is only found to be involved in interaction when at least one of the interacting regions of the helix is N-terminal, i.e. in N–N, N–C and N–MID helix pairs. Residues interacting with the middle region tend to be nonpolar in C–MID pairs (∼63% nonpolar versus ∼35% polar) and are relatively more polar in N–MID helix pairs (∼50% nonpolar versus ∼46% polar).
When an interacting residue is polar it does not necessarily imply that it only participates in polar interactions. For example, we have observed at least three different types of interactions for arginine when it occurs as an interacting residue in EE/EM helix pairs. The side chain of Arg from one helix can participate in a hydrogen-bond interaction with the backbone carbonyl O atom of another helix. The acyl portion of the long Arg side chain can take part in hydrophobic interactions with an aliphatic residue of the interacting helix. Arg is also observed to form salt-bridge interactions with acidic residues in EE/EM helix pairs. Table 1 only ▶ presents data about the nature of the interacting residues for each class of EE and EM helix pairs and not about the nature of the interactions in which the interacting residues participate.
3.2. Single-residue propensities
We have calculated single-residue propensities to determine whether any residue or groups of residues show a preference for interaction with any particular region of the helix. The propensities were calculated as described in §2.2.
3.2.1. Residues interacting with the C-terminal region
We have calculated the single-residue propensities of residues involved in interaction with the C-terminal region of a helix separately for the C–C, N–C and C–MID classes of helix pairs (Table 2 ▶). For C–C helix pairs interacting residues from both C-terminal regions were considered. For N–C and C–MID helix pairs residues from the N-terminal and middle regions, respectively, that interact with the C-terminal region of the partner helix were considered for this purpose. When a residue from the N-terminus interacts with the C-terminus of another helix, the preferred residue mostly seems to be polar. Ser, His, Arg, Lys, Asp and Asn residues from the N-terminus show a higher propensity for interaction with the C-terminal region of the second helix. This is the only class in which Pro has a preference for interaction with the C-terminus. This is obviously a consequence of its high preference for ocurring at the beginning of an α-helix (Aurora & Rose, 1998 ▶). In the case of C–C helix pairs, a number of hydrophobic residues (Leu, Ile, Met and Trp) show higher propensity values for interaction with the C-terminal region of another helix. A similar pattern is also observed for C–MID helix pairs. Hydrophobic residues from the middle region of one helix (Leu, Ile, Met, Phe and Trp) prefer to interact with the C-terminal region of another helix. Only the residues Trp and Arg are commonly observed to have a higher preference for interaction with the C-terminal region across all three classes of helix pairs. Basic residues generally show a high preference for interaction with the C-terminus in all three classes.
Table 2. Single-residue propensities of residues interacting with the C-terminus of EE and EM helix pairs.
Single-residue propensity values for each residue interacting with the C-terminal region of another helix were calculated using (1) (see §2.2). Propensity values greater than 1.1 in all three classes are shown in italics. Propensity values greater than 1.1 in two classes are shown in bold italics. Propensity values greater than 1.1 in only one class are shown in bold.
| Residue | C–C† | C–MID‡ | N–C§ |
|---|---|---|---|
| Gly | 18 (1.01) | 3 (0.20) | 16 (0.78) |
| Ala | 23 (0.86) | 17 (0.78) | 15 (0.44) |
| Val | 25 (1.00) | 18 (0.88) | 19 (0.71) |
| Leu | 68 (1.73) | 40 (1.24) | 27 (0.62) |
| Ile | 27 (1.22) | 23 (1.27) | 19 (0.76) |
| Met | 16 (1.88) | 19 (2.69) | 6 (0.69) |
| Thr | 11 (0.67) | 9 (0.67) | 21 (1.04) |
| Ser | 13 (0.83) | 8 (0.62) | 22 (1.27) |
| Cys | 3 (0.60) | 4 (0.93) | 5 (1.00) |
| Tyr | 15 (1.09) | 12 (1.05) | 18 (1.28) |
| Trp | 9 (1.59) | 8 (1.70) | 11 (1.76) |
| Phe | 17 (1.02) | 22 (1.61) | 19 (1.05) |
| His | 10 (1.20) | 6 (0.88) | 16 (1.92) |
| Arg | 25 (1.38) | 25 (1.69) | 25 (1.14) |
| Lys | 17 (1.04) | 21 (1.56) | 22 (1.14) |
| Glu | 4 (0.21) | 8 (0.51) | 21 (1.05) |
| Asp | 7 (0.46) | 5 (0.39) | 23 (1.43) |
| Gln | 11 (0.94) | 13 (1.37) | 9 (0.77) |
| Asn | 6 (0.53) | 7 (0.76) | 13 (1.15) |
| Pro | 0 (0.00) | 0 (0.00) | 30 (2.19) |
| Total | 325 | 268 | 357 |
Number of residues from the C-terminal region of one helix interacting with the C-terminal region of the second helix; in this class of helix pairs interacting residues from both helices were considered.
Number of residues from the middle region of one helix interacting with the C-terminal region of another helix.
Number of residues from the N-terminal region of one helix interacting with the C-terminal region of another helix.
3.2.2. Residues interacting with the N-terminal region
The single-residue propensities of residues participating in interhelical atomic contacts with the N-terminal region of a helix were calculated separately for N–N, N–C and N–MID helix pairs (Table 3 ▶). For N–N helix pairs interacting residues from both N-terminal regions were considered. In the case of N–C and N–MID helix pairs residues from the C-terminal and middle regions, respectively, that interact with the N-terminal region of the second helix were considered. In all three classes we observed a preference for polar residues to interact with the N-terminal region. Surprisingly, both acidic and basic residues showed a higher preference for interaction with the N-terminus. The residues Lys, Glu and Gln showed higher propensity values for interaction with the N-terminal region in all three classes of helix pairs. In the N–N helix pairs several more polar residues (Ser, His, Asp and Asn) have a higher preference for interaction with the N-terminus. The only additional polar residue that showed a higher preference for interaction with the N-terminal region of another helix in N–MID and N–C helix pairs was Arg. In addition to the polar residues, several hydrophobic residues (Met, Leu and Phe) also prefer to interact with the N-terminus if they come from the middle or C-terminal region of another helix. Proline was observed to interact with the N-terminal region in N–N pairs. As mentioned previously, proline has a preference for ocurring at the beginning of an α-helix (Aurora & Rose, 1998 ▶). When the N-terminus of one helix interacts with the same region of another helix, prolines in both N-terminal regions could interact with each other or with other residues.
Table 3. Single-residue propensities of residues interacting with the N-terminus of EE and EM helix pairs.
Single-residue propensity values for each residue interacting with the N-terminal region of another helix were calculated using (1) (see §2.2). For other explanations, see the headnote of Table 2 ▶.
| Residue | N–N† | N–MID‡ | N–C§ |
|---|---|---|---|
| Gly | 15 (0.74) | 3 (0.28) | 20 (1.04) |
| Ala | 14 (0.46) | 10 (0.63) | 18 (0.62) |
| Val | 9 (0.31) | 7 (0.47) | 11 (0.41) |
| Leu | 20 (0.44) | 17 (0.73) | 65 (1.52) |
| Ile | 13 (0.51) | 13 (1.00) | 18 (0.75) |
| Met | 9 (0.92) | 15 (2.96) | 14 (1.50) |
| Thr | 18 (0.98) | 7 (0.73) | 15 (0.86) |
| Ser | 24 (1.36) | 10 (1.10) | 8 (0.47) |
| Cys | 3 (0.53) | 0 (0.00) | 7 (1.27) |
| Tyr | 22 (1.38) | 8 (0.98) | 11 (0.74) |
| Trp | 4 (0.59) | 6 (1.82) | 4 (0.65) |
| Phe | 20 (1.04) | 20 (2.02) | 17 (0.94) |
| His | 21 (2.24) | 4 (0.80) | 8 (0.88) |
| Arg | 19 (0.91) | 20 (1.87) | 31 (1.58) |
| Lys | 24 (1.28) | 12 (1.24) | 29 (1.62) |
| Glu | 38 (1.77) | 22 (1.98) | 25 (1.23) |
| Asp | 29 (1.67) | 5 (0.54) | 17 (1.04) |
| Gln | 17 (1.28) | 9 (1.31) | 22 (1.77) |
| Asn | 27 (2.12) | 3 (0.44) | 13 (1.05) |
| Pro | 27 (1.76) | 0 (0.00) | 0 (0.00) |
| Total | 373 | 191 | 353 |
Number of residues from the N-terminal region of one helix interacting with the N-terminal region of the second helix; in this class of helix pairs interacting residues from both helices were considered.
Number of residues from the middle region of one helix interacting with the N-terminal region of another helix.
Number of residues from the C-terminal region of one helix interacting with the N-terminal region of another helix.
3.2.3. Residues interacting with the middle region
The preferences for amino acids to interact with the middle region of an α-helix were determined by calculating the single-residue propensities of the residues interacting in the helical middle region in N–MID and C–MID helix pairs (Table 4 ▶). This analysis was carried out separately for N–MID and C–MID helix pairs, in which residues from the N-terminal and C-terminal region, respectively, of one helix interact with the middle region of another helix. As in the previous cases, when the interacting residues belong to the N-terminal region they are most likely to be polar (His, Glu, Asp and Asn). Residues from the C-terminal region are mostly hydrophobic (Ala, Leu, Ile and Met) when they interact with the middle region of the second helix. The only common residues that show a higher preference in both N–MID and C–MID helix pairs are Trp and Phe. Again, proline has higher propensity value for occurring in the N-terminal region of N–MID helix pairs and participating in interhelical contacts with the middle region of the second helix. As noted previously, this preference could be a consequence of the general preference of this residue for occurring at the beginning of an α-helix (Aurora & Rose, 1998 ▶).
Table 4. Single-residue propensities of residues interacting with the middle region of EE and EM helix pairs.
Single-residue propensity values for each residue interacting with the middle region of another helix were calculated using (1) (see §2.2). Propensity values greater than 1.1 in both classes are shown in italics. Propensity values greater than 1.1 in only one class are shown in bold.
| Residue | N–MID† | C–MID‡ |
|---|---|---|
| Gly | 5 (0.55) | 3 (0.26) |
| Ala | 8 (0.60) | 19 (1.16) |
| Val | 14 (1.13) | 12 (0.78) |
| Leu | 20 (1.02) | 44 (1.81) |
| Ile | 7 (0.64) | 16 (1.18) |
| Met | 1 (0.23) | 15 (2.85) |
| Thr | 8 (1.00) | 5 (0.49) |
| Ser | 5 (0.64) | 7 (0.72) |
| Cys | 1 (0.40) | 3 (0.93) |
| Tyr | 9 (1.31) | 7 (0.81) |
| Trp | 5 (1.76) | 7 (2.00) |
| Phe | 13 (1.57) | 19 (1.84) |
| His | 11 (2.68) | 4 (0.76) |
| Arg | 8 (0.89) | 7 (0.62) |
| Lys | 3 (0.36) | 4 (0.38) |
| Glu | 12 (1.30) | 11 (0.95) |
| Asp | 9 (1.19) | 8 (0.85) |
| Gln | 3 (0.51) | 8 (1.11) |
| Asn | 9 (1.61) | 2 (0.26) |
| Pro | 11 (1.63) | 0 (0.00) |
| Total | 162 | 201 |
Number of residues from the N-terminal region of one helix interacting with the middle region of the second helix.
Number of residues from the C-terminal region of one helix interacting with the middle region of another helix.
In summary, the interacting residues from C–C and C–MID helix pairs are found to be mostly hydrophobic and prefer to interact with the C-terminal or middle region of the second helix. On the other hand, the residues that interact with the N-terminus are often found to be polar in nature and this is true for N–N, N–MID and N–C helix pairs, in which the interacting residue comes from another N-terminus or the middle region or C-terminus of another helix, respectively. In the case of N–MID and N–C helix pairs the interacting residues from the N-terminus are found to be predominantly polar whether they interact with the middle or C-terminal region of the partner helix.
3.3. Conservation of interacting residues
We have analyzed the conservation of interacting residues in EE and EM helix pairs as described in §2.3. For the majority of proteins that contain at least one EE/EM helix pair, we have obtained at least 25 nonredundant homologous protein sequences that were subsequently used for multiple sequence alignment (MSA). The conservation of each amino acid in the query protein was determined from MSA. 565 of the interacting residues from all five classes of EE and EM helix pairs show a conservation of 80% or more. Within this set, 337 residues are more than 80% conserved, while the overall conservation of each individual protein is less than 50%. This seems to unambiguously indicate that these interacting residues may be structurally and/or functionally important. We refer to such residues as highly conserved interacting residues (HCIs). We have specifically focused on these residues in order to determine the nature of conserved residues in different classes of EE/EM helix pairs (Table 5 ▶). However, it should be kept in mind that this analysis does not provide information on whether the actual interaction itself is conserved in the EE/EM helix pairs. For such an analysis, knowledge of structures of all the protein sequences used in the conservation analysis would be essential.
Table 5. Highly conserved interacting residues in EE and EM helix pairs.
An interacting residue is highly conserved only if its conservation is >80% and the overall conservation of the protein to which it belongs is less than 50%. For groupings of amino acids, see §2 and Table 1 ▶.
| Helix pairs | Conserved interacting residues | Polar (%) | Nonpolar (%) |
|---|---|---|---|
| N–N | 51 (acidic, 10; basic, 9; large polar, 2; small polar, 5; aromatic, 9; hydrophobic, 13; proline, 3) | 26 (51) | 22 (43) |
| N–MID (MID)† | 52 (acidic, 3; basic, 15; large polar, 2; small polar, 3; aromatic, 8; hydrophobic, 21) | 23 (44) | 29 (58) |
| N–C (C)† | 33 (acidic, 3; basic, 5; large polar, 1; small polar, 3; aromatic, 5; hydrophobic, 16) | 12 (36.3) | 21 (63.7) |
| Total | 136 (acidic, 16; basic, 29; large polar, 5; small polar, 11; aromatic, 22; hydrophobic, 50; proline, 3) | 61 (45) | 72 (53) |
| C–C | 48 (acidic, 1; basic, 3; large polar, 1; small polar, 1; aromatic, 4; hydrophobic, 38) | 6 (12.5) | 42 (87.5) |
| C–MID (MID)† | 45 (acidic, 1; basic, 6; large polar, 1; small polar, 1; aromatic, 6; hydrophobic, 30) | 9 (20) | 36 (80) |
| N–C (N)† | 45 (acidic, 4; basic, 6; large polar, 1; small polar, 4; aromatic, 9; hydrophobic, 18; proline, 3) | 15 (33.3) | 27 (60) |
| Total | 138 (acidic, 6; basic, 15; large polar, 3; small polar, 6; aromatic, 19; hydrophobic, 86; proline, 3) | 30 (22) | 105 (76) |
| N–MID (N)‡ | 32 (acidic, 5; basic, 3; small polar, 1; aromatic, 6; hydrophobic, 13; proline, 4) | 9 (28) | 19 (59) |
| C–MID (C)‡ | 31 (acidic, 2; large polar, 1; aromatic, 2; hydrophobic, 26) | 3 (9.7) | 28 (90.3) |
| Total | 63 (acidic, 7; basic, 3; large polar, 1; small polar, 1; aromatic, 8; hydrophobic, 39; proline, 4) | 12 (19) | 47 (75) |
Residues from N–MID and N–C helix pairs interacting with the N-terminus are considered. These residues interact with the N-terminus of the first helix through the middle [N–MID (MID)] or C-terminal [N–C (C)] region of the second helix.
Residues from C–MID and N–C helix pairs interacting with the C-terminus are considered. These residues interact with the C-terminus of the first helix through the middle [C–MID (MID)] or N-terminal [N–C (N)] region of the second helix.
Residues from N–MID and C–MID helix pairs interacting with the middle region of an α-helix are considered. These residues interact with the middle region of the first helix through the N-terminal [N–MID (N)] or C-terminal [C–MID (C)] regions of the second helix.
It is interesting to note that the HCIs interacting with the C-terminal regions are predominantly hydrophobic. This is especially true for both C–C and C–MID helix pairs, in which more than 80% of the HCIs interacting with the C-terminal regions are nonpolar. Comparison of residues that interact with the N- and C-terminal regions indicates that HCIs that interact with N-terminal regions have a higher percentage of polar residues. In the N–N, N–MID and N–C classes of helix pairs between ∼35 and ∼50% of HCIs are observed to be polar and to interact with the N-terminal region (Table 5 ▶). Only ∼12 to ∼33% of HCIs interacting with the C-terminal regions are polar. A similar trend is observed for HCIs that interact with the middle region of a helix (∼10 to ∼28% are polar). In summary, a larger number of hydrophobic residues that interact with the C-terminal or middle regions of a helix are conserved in EE and EM helix pairs. As far as residues that interact with the N-terminal region are concerned, more polar residues are conserved. The difference could be a consequence of the fact that polar residues are generally observed to be the interacting residues in the N-terminal region, while they are hydrophobic in the C-terminal region.
4. Discussion
In this paper, a systematic analysis has been carried out on helix pairs that interact exclusively in the terminal regions or between a terminal region and a middle region. By choosing a minimum of a 15-residue separation, we have ensured that the observation of interactions between the helix pairs does not arise from their proximity in the primary sequence. The crossing-angle criteria helped us to eliminate those helix pairs that are suspected to interact either in a coiled-coil fashion or in the ridges-in-grooves model. We also discarded the helix pairs if one helix makes interactions with more than one region of the other helix. We have observed that in 736 of 5725 helix pairs interhelical contacts are only observed between the terminal regions or between the terminal region of one helix and the middle region of another helix. Nearly 13% of the total helix pairs interact in an end-to-end or an end-to-middle fashion. To the best of our knowledge, this is the first study that has systematically identified, characterized and analyzed the interhelical interactions in such EE and EM helix pairs. Without understanding such helix pairs, knowledge of how helices interact amongst themselves will be incomplete. The present study has attempted to fill this gap.
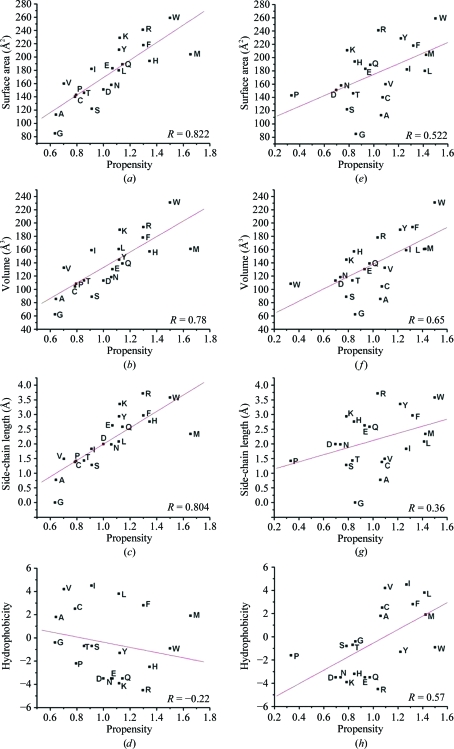
We then investigated whether there was any correlation between single-residue propensity values and various properties of individual residues. Single-residue propensities were calculated separately for the 736 EE/EM helix pairs and the 4989 interacting non-EE/EM helix pairs and the method used was similar to that described in §2.2 (1). The residue surface area (Miller et al., 1987 ▶), residue volume (Chothia, 1975 ▶), side-chain length (Levitt, 1976 ▶) and hydrophobicity (Kyte & Doolittle, 1982 ▶) of individual residues were considered in this analysis (Fig. 3 ▶). For EE and EM helix pairs, strong positive correlation was observed between the propensity values of different residues and the surface area (Fig. 3 ▶ a), volume (Fig. 3 ▶ b) and side-chain length (Fig. 3 ▶ c) of residues. The correlation coefficients were 0.82, 0.78 and 0.80 for surface area, volume and side-chain length. This indicates that residues with larger surface area and volume and longer side chains have an overwhelming preference to occur in the interface of EE and EM helix pairs. On the other hand, there is a negative correlation (correlation coefficient −0.22) between the hydrophobicity and residue-propensity values (Fig. 3 ▶ d). However, if only residues interacting with the C-terminal region are considered, the relationship may not hold good since the residues that interact with the C-terminal region are found to be largely hydrophobic. A similar analysis for all interacting non-EE/EM helix pairs was also carried out for comparison purposes. In the set of interacting non-EE/EM helix pairs the correlation between the propensity values and the surface area/volume was found to be weaker (Figs. 3 ▶ e and 3 ▶ f) and a poor correlation was observed with the length of the side chains (Fig. 3 ▶ g). The correlation coefficients for surface area, volume and length of the side chain were 0.55, 0.65 and 0.36, respectively. This shows that the relationship between the residues that prefer to occur in the interface of interacting non-EE/EM helix pairs and the residue properties (surface area, volume and length of the side chain) is weak. However, the correlation between hydrophobicity and the preference of the residue to occur in the interface of non-EE/EM helix pairs is better than that observed for EE and EM helix pairs (Fig. 3 ▶ h). The results of these studies reiterate the point that the preferences for residues to occur in the interface of EE and EM helix pairs are distinct and that residues with larger surface area/volumes and longer side chains and that are not necessarily hydrophobic seem to be required for interfacial residues in such helix pairs. The residues Arg, Lys, Glu, Gln, Met, Phe, Trp and Tyr readily fulfil these criteria.
Figure 3.
Plots showing the relationships between various properties of residues and their propensities to occur in the interface of EE and EM helix pairs (a–d) and interacting non-EE/EM helix pairs (e–h) of soluble proteins. Residue propensities are plotted against surface area (a, e), residue volume (b, f), length of the side chain (c, g) and hydrophobicity (d, h). The correlation coefficient between the residue propensity and the property under consideration is displayed in each graph. The function ‘linear fit’ available in Origin v.6.1 Pro (http://www.originlab.com) was used to fit the data and calculate the correlation coefficients.
What causes the necessity for longer and larger residues at the interfaces of these helix pairs? To answer this question, we calculated the average minimum Cα—Cα distance in the EE and EM helix pairs. This distance (7.53 Å) is ∼1 Å larger than that observed in helix pairs (6.49 Å) that do not interact in EE or EM fashion. A two-sample t-test shows that this difference is statistically significant (P < 0.001). This implies that the helix–helix separation is higher in EE and EM helix pairs. Moreover, there are only 6.37 atomic contacts per helix pair on average when the helix pairs belong to one of the five classes of EE and EM pairs. A similar analysis reveals that the average number of contacts per helix pair for helix pairs that do not belong to one of the EE/EM classes is 26.12. The greater average Cα—Cα distance and the average number of atomic contacts per helix pairs implies that in helix pairs belonging to the EE or EM categories the interacting helices are farther apart and the interface is smaller. This explains the preference for larger residues with longer side chains; such residues could act as ‘long arms’ between the interacting helix pairs that are farther away. Residues such as arginine and lysine have the advantage of participating in both hydrophobic and polar interactions. The methylene groups in the aliphatic portion of their long side chains can take part in hydrophobic contacts with hydrophobic residues or with the methylene groups of other polar residues. Their polar ends can make polar contacts that can either be hydrogen bonds or salt bridges.
5. Conclusions
In this paper, we have identified helix pairs that interact exclusively between their terminal regions or between the terminal and middle regions. They occur in more than 10% of all helix pairs observed. The interface of EE and EM helix pairs is more polar if they interact with the N-terminal region and more hydrophobic if the interaction is with the C-terminal region. In N–C and N–MID helix pairs an intermediate character is observed in the interface. This is also reflected in the conservation analysis. More polar residues are conserved if they interact with the N-terminal region and the majority of conserved interacting residues are hydrophobic when the interaction is with the C-terminal region. In EE and EM helix pairs the helices are separated by longer distances. The residue preferences for occurring in the interface differ distinctly compared with those found in soluble and membrane proteins. The observed EE and EM helix pairs are likely to be structurally and functionally important in their respective proteins. With a substantial number of examples, the present study confirms the existence of this new category of helix pairs separate from those already recognized and investigating the structural and functional significance of such pairs will be the focus of structural biologists in the future.
Acknowledgments
We thank Tuhin Kumar Pal, Dilraj Lama and Alok Jain for their help during the course of this work. This work was supported by a research grant to RS from the Ministry of Human Resources and Development (MHRD), Government of India. RS is a Joy Gill Chair Professor at IIT-Kanpur. TSG and SKC thank the institute for a fellowship. We thank all members of our group for discussions.
References
- Adamian, L., Jackups, R. Jr, Binkowski, T. A. & Liang, J. (2003). J. Mol. Biol.327, 251–272. [DOI] [PubMed]
- Adamian, L. & Liang, J. (2001). J. Mol. Biol.311, 891–907. [DOI] [PubMed]
- Adamian, L. & Liang, J. (2002). Proteins, 47, 209–218. [DOI] [PubMed]
- Agre, P. & Kozono, D. (2003). FEBS Lett.555, 72–78. [DOI] [PubMed]
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Aurora, R. & Rose, G. D. (1998). Protein Sci.7, 21–38. [DOI] [PMC free article] [PubMed]
- Aurora, R., Srinivasan, R. & Rose, G. D. (1994). Science, 264, 1126–1130. [DOI] [PubMed]
- Ballesteros, J. A., Deupi, X., Olivella, M., Haaksma, E. E. J. & Pardo, L. (2000). Biophys. J.79, 2754–2760. [DOI] [PMC free article] [PubMed]
- Bansal, A. & Sankararamakrishnan, R. (2007). BMC Struct. Biol.7, 27. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Bowie, J. U. (1997). J. Mol. Biol.272, 780–789. [DOI] [PubMed]
- Brändén, C. & Tooze, J. (1999). Introduction to Protein Structure. New York: Garland.
- Burba, A. E. C., Lehnert, U., Yu, E. Z. & Gerstein, M. (2006). Bioinformatics, 22, 2735–2738. [DOI] [PubMed]
- Chakrabarti, P. & Pal, D. (2001). Prog. Biophys. Mol. Biol.76, 1–102. [DOI] [PubMed]
- Chakrabartty, A. & Baldwin, R. L. (1995). Adv. Protein Chem.46, 141–176. [PubMed]
- Chothia, C. (1975). Nature (London), 254, 304–308. [DOI] [PubMed]
- Chothia, C., Levitt, M. & Richardson, D. (1981). J. Mol. Biol.145, 215–250. [DOI] [PubMed]
- Christopher, J. A., Swanson, R. & Baldwin, T. O. (1996). Comput. Chem.20, 339–345. [DOI] [PubMed]
- Clamp, M., Cuff, J., Searle, S. M. & Barton, G. J. (2004). Bioinformatics, 20, 426–427. [DOI] [PubMed]
- Cochran, D. A. E. & Doig, A. J. (2001). Protein Sci.10, 1305–1311. [DOI] [PMC free article] [PubMed]
- Creamer, T. P. & Rose, G. D. (1992). Proc. Natl Acad. Sci. USA, 89, 5937–5941. [DOI] [PMC free article] [PubMed]
- Crick, F. H. C. (1953). Acta Cryst.6, 689–697.
- Doura, A. K. & Fleming, K. G. (2004). J. Mol. Biol.343, 1487–1497. [DOI] [PubMed]
- Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. (2002). Nature (London), 415, 287–294. [DOI] [PubMed]
- Eilers, M., Patel, A. B., Liu, W. & Smith, S. O. (2002). Biophys. J.82, 2720–2736. [DOI] [PMC free article] [PubMed]
- Engel, D. E. & DeGrado, W. F. (2004). J. Mol. Biol.337, 1195–1205. [DOI] [PubMed]
- Fleming, K. G. & Engelman, D. M. (2001). Proc. Natl Acad. Sci. USA, 98, 14340–14344. [DOI] [PMC free article] [PubMed]
- Gimpelev, M., Forrest, L. R., Murray, D. & Honig, B. (2004). Biophys. J.87, 4075–4086. [DOI] [PMC free article] [PubMed]
- Higgins, D. G., Bleasby, A. J. & Fuchs, R. (1991). CABIOS, 8, 189–191. [DOI] [PubMed]
- Iqbalsyah, T. M. & Doig, A. J. (2004). Protein Sci.13, 32–39. [DOI] [PMC free article] [PubMed]
- Javadpour, M. M., Eilers, M., Groesbeek, M. & Smith, S. O. (1999). Biophys. J.77, 1609–1618. [DOI] [PMC free article] [PubMed]
- Kumar, S. & Bansal, M. (1998). Proteins, 31, 460–476. [DOI] [PubMed]
- Kyte, J. & Doolittle, R. F. (1982). J. Mol. Biol.157, 105–132. [DOI] [PubMed]
- Lacroix, E., Viguera, A. R. & Serrano, L. (1998). J. Mol. Biol.284, 173–191. [DOI] [PubMed]
- Levitt, M. (1976). J. Mol. Biol.104, 59–107. [DOI] [PubMed]
- Li, W. Z. & Godzik, A. (2006). Bioinformatics, 22, 1658–1659. [DOI] [PubMed]
- Li, W. Z., Jaroszewski, L. & Godzik, A. (2001). Bioinformatics, 17, 282–283. [DOI] [PubMed]
- Li, W. Z., Jaroszewski, L. & Godzik, A. (2002). Bioinformatics, 18, 77–82. [DOI] [PubMed]
- Litowski, J. R. & Hodges, R. S. (2002). J. Biol. Chem.277, 37272–37279. [DOI] [PubMed]
- Liu, W., Eilers, M., Patel, A. B. & Smith, S. O. (2004). J. Mol. Biol.337, 713–729. [DOI] [PubMed]
- Mezei, M. & Filizola, M. (2006). J. Comput. Aided Mol. Des.20, 97–107. [DOI] [PubMed]
- Miller, S., Janin, J., Lesk, A. M. & Chothia, C. (1987). J. Mol. Biol.196, 641–656. [DOI] [PubMed]
- Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995). J. Mol. Biol.247, 536–540. [DOI] [PubMed]
- Murzin, A. G. & Finkelstein, A. V. (1988). J. Mol. Biol.204, 749–769. [DOI] [PubMed]
- Penel, S., Hughes, E. & Doig, A. J. (1999). J. Mol. Biol.287, 127–143. [DOI] [PubMed]
- Petros, A. M., Olejniczak, E. T. & Fesik, S. W. (2004). Biochim. Biophys. Acta, 1644, 83–94. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem.25, 1605–1612. [DOI] [PubMed]
- Presta, L. G. & Rose, G. D. (1988). Science, 240, 1632–1641. [DOI] [PubMed]
- Reddy, B. V. B. & Blundell, T. L. (1993). J. Mol. Biol.233, 464–479. [DOI] [PubMed]
- Richardson, J. S. & Richardson, D. C. (1988). Science, 240, 1648–1652. [DOI] [PubMed]
- Richmond, T. J. & Richards, F. M. (1978). J. Mol. Biol.119, 537–555. [DOI] [PubMed]
- Rohl, C. A., Chakrabartty, A. & Baldwin, R. L. (1996). Protein Sci.5, 2623–2637. [DOI] [PMC free article] [PubMed]
- Sankararamakrishnan, R. & Vishveshwara, S. (1992). Int. J. Pept. Protein Res.39, 356–363. [DOI] [PubMed]
- Senes, A., Engel, D. E. & DeGrado, W. F. (2004). Curr. Opin. Struct. Biol.14, 465–479. [DOI] [PubMed]
- Straussman, R., Ben-Ya’acov, A., Woolfson, D. N. & Ravid, S. (2007). J. Mol. Biol.366, 1232–1242. [DOI] [PubMed]
- Walters, R. F. S. & DeGrado, W. F. (2006). Proc. Natl Acad. Sci. USA, 103, 13658–13663. [DOI] [PMC free article] [PubMed]
- Walther, D., Eisenhaber, F. & Argos, P. (1996). J. Mol. Biol.255, 536–553. [DOI] [PubMed]
- Zhou, N. E., Kay, C. M. & Hodges, R. S. (1994). J. Mol. Biol.237, 500–512. [DOI] [PubMed]