Abstract
Class D β-lactamases hydrolyze β-lactam antibiotics by using an active site serine nucleophile to form a covalent acyl-enzyme intermediate, and subsequently employ water to deacylate the β-lactam and release product. Class D β-lactamases are carboxylated on the ε-amino group of an active site lysine, with the resulting carbamate functional group serving as a general base. We discovered that substitutions of the active site serine and lysine in OXA-1 β-lactamase, a monomeric class D enzyme, significantly disrupt catalytic turnover. Substitution of glycine for the nucleophilic serine (S67G) results in an enzyme that can still bind substrate but is unable to form a covalent acyl-enzyme intermediate. Substitution of the carboxylated lysine (K70), on the other hand, results in enzyme that can be acylated by substrate, but is impaired for deacylation. We employed the fluorescent penicillin BOCILLIN FL™ to show that three different substitutions for K70 (alanine, aspartate and glutamate) accumulate significant acyl-enzyme intermediate. Interestingly, BOCILLIN FL™ deacylation rates vary depending on the identity of the substituting residue, from t1/2 ≈ 60 min for K70A to undetectable deacylation for K70D. Tryptophan fluorescence spectroscopy was used to confirm that these results are applicable to natural (i.e. non-fluorescent) substrates. Deacylation by K70A, but not K70D or K70E, can be partially restored by the addition of short-chain carboxylic acid mimetics of the lysine carbamate. In conclusion, we establish the functional role of the carboxylated lysine in OXA-1 and highlight its specific role in acylation and deacylation.
Members of the β-lactamase (E.C. 3.2.5.6) family of enzymes—comprising more than 700 known enzymes—hydrolyze the lactam ring of penicillin, cephalosporin and carbapenem antibiotics, rendering them incompetent for antibacterial activity. Four classes (A–D) of β-lactamases have been described based on sequence homology and general substrate specificity (1). Three of the classes (A, C and D) employ covalent catalysis and form an acyl-enzyme intermediate between the β-lactam and a conserved active site serine (2). Class B enzymes, on the other hand, use a Zn+2 ion to stabilize both the attacking water and the hydrolysis transition state (2).
The general outline of events that accompany β-lactamase-mediated covalent catalysis are known. After the association of enzyme and substrate, an active site base deprotonates a serine, activating it for nucleophilic attack on the β-lactam carbonyl (3). The anionic tetrahedral transition state that subsequently forms is stabilized by two main-chain amide hydrogens in a manner reminiscent of the serine proteases (e.g. chymotrypsin). The collapse of this transition state results in an intermediate in which the β-lactam forms an ester (acyl) linkage with the attacking serine. General base mediated activation of an active site water molecule then leads to hydrolysis of the ester bond, releasing the product and restoring the serine to its original state. In the kinetic scheme shown, E represents free enzyme, S and P represent the β-lactam substrate
| (Eq. 1) |
and product, E:S is the non-covalent Michaelis complex, and E-S is the acyl enzyme intermediate. Binding, acylation and deacylation are described by the rate constants k1, k2, and k3 respectively.
For all three of the β-lactamase classes that follow this scheme (A, C and D), the identification of the general base has been very challenging. In class A, multiple approaches implicated a ubiquitously conserved glutamate (position 166 in TEM-1) as the ultimate base in both the acylation and deacylation reactions (4). This residue does not activate the serine nucleophile directly, but rather acts through a water molecule that bridges the two residues (5). Computational analysis suggests that this pathway exists in competition with another mechanism, in which E166 acts through K73 (consensus numbering) rather than the water (6). In class C, the identity of the general base has not been fully elucidated, with arguments for (7, 8) and against (9–11) a tyrosine (Y150), a highly conserved lysine (K315) (12), and even the lactam carboxylate (13, 14).
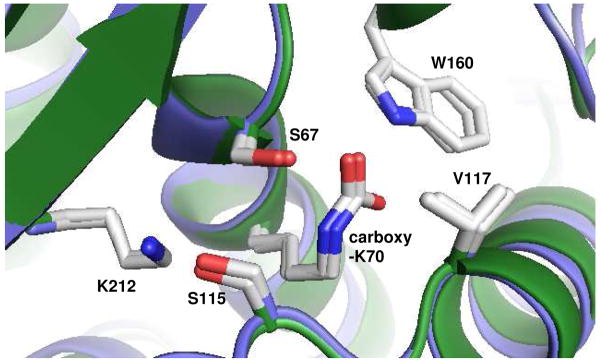
The mechanism underlying hydrolysis by class D enzymes was also unresolved until the discovery of an unusual N-carboxylation modification of the conserved active site lysine (K70) in two class members, OXA-10 (15, 16) and OXA-1 (17). Structural analysis shows that in both enzymes, the resulting carbamate anion forms a hydrogen bond with the serine nucleophile (Figure 1). This observation strongly suggested that this carboxy-lysine may be the agent that activates the attacking groups in both the acylation and deacylation reactions (15–18). This hypothesis has been supported by mutagenesis studies in both enzymes (16, 19). The importance of the carboxy-lysine modification also helps explain another unusual feature of the class D β-lactamases. The OXA enzymes, as they are sometimes known, have a much more hydrophobic active site compared to the class A and C enzymes (15, 20). It has been suggested that non-polar active site residues are necessary to lower the pKa of the lysine ε–amino group, allowing it to be deprotonated at physiological pH values and thereby able to form the carbamate through attack on carbon dioxide (16). One of the important hydrophobic active site residues in OXA-1 and OXA-10 is V117, the side-chain of which lies proximal to the ε–amino group of K70. Among the more than 100 members of the class D β-lactamase family, the position homologous to V117 is always occupied by valine, isoleucine or rarely, leucine.
Figure 1.
Three-dimensional alignment of key active site residues of OXA-1 and OXA-10. The six residues shown (S67, K70, S115, V117, W160 and K212; OXA-1 numbering) were used to align OXA-1 (1M6K) (17) and OXA-10 (1K55) (16) using Pymol (56).
The carbamate moiety of K70 is stabilized by a number of hydrogen bonds, most notably to the serine nucleophile and the side-chain nitrogen of a highly conserved tryptophan (W160 in OXA-1). While carboxylated amines are normally labile, these cooperative interactions favor carbamate formation enough to lower the CO2 Kd to ~ 1 μM (16, 19). For both OXA-1 and OXA-10, CO2 can be removed only by dialysis in low pH buffer and under vacuum conditions. Such treatment eliminates enzymatic activity towards β-lactams, but activity can be fully restored by the addition of bicarbonate (16, 18, 19).
The importance of the carbamate in class D oxacillinases can also be inferred from the role of an essentially identical carboxylated lysine in the β-lactam sensor protein family. β-lactam sensors such as BlaR1 from Staphylococcus aureus share a high degree of sequence and tertiary structure homology with class D lactamases (21, 22). The sensor/signaling function of these proteins depends on their ability to acylate β-lactam substrates, but unlike β-lactamases they are unable to deacylate them (22–25). The sensor proteins accumulate acyl-lactam substrates in their active site, and therefore serve as natural deacylation-deficient analogs of the class D β-lactamases. It is thought that the carboxylated lysine of BlaR1 plays the role of general base in the acylation step as it does in the class D enzymes. Interestingly, it has been proposed that after acylation, the carboxylated lysine decarboxylates, and is therefore unable to activate a water for the deacylation reaction (22, 26, 27).
The removal of a β-lactamase’s serine nucleophile or its postulated general base by mutagenesis has been used extensively to create enzyme-substrate complexes at various stages in the catalytic cycle. In the case of substitution for the nucleophile, it is possible to trap the substrate in a state that mimics the pre-acylation Michaelis complex (13, 28). On the other hand, when the general base is eliminated, the enzyme becomes deacylation deficient, allowing one to trap the β-lactam in the active site as an acyl-intermediate. The latter approach was applied to numerous class A (4, 29–37) and class C (13) enzymes. Structural studies of enzymes with substrates trapped in either the pre-acylation stage or the acyl-intermediate stage have led to a much deeper understanding of how various enzymes distinguish substrates specifically, how natural mutations lead to enhanced spectrum hydrolytic activities and to the details of catalytic turnover mechanisms. In this study, we demonstrate that these approaches are applicable to the class D β-lactamase OXA-1, with implications for future structural studies.
Materials and Methods
Mutagenesis
Amino acid substitutions at position S67 and K70 were generated using PCR overlap extension (38). Two rounds of PCR were conducted using mutant oligonucleotides and Phusion High Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) to produce the desired point mutation in the blaOXA-1 gene. PCR products were directionally subcloned into the BamHI and NdeI sites of pET24a(+) (Novagen, Madison, WI) and mutant plasmids were transformed into NEB 5-α competent Escherichia coli cells (New England Biolabs). After verification of the plasmid constructs, each was transformed into E. coli strain BL21 DE3 (K70E and S67G) or strain BL21 DE3 pLysS (K70D and K70A) for expression.
Expression and Purification
E. coli cells containing the wild-type or mutant blaOXA-1 genes were grown in LB media containing 25 μg/mL kanamycin to an optical density (OD600) of ~ 0.8. Protein expression was induced with 100 μM isopropyl β-D-1-thiogalactopyranoside (IPTG; Calbiochem) for 2 hours. Cells were harvested by centrifugation (9500 × g for 20 min, 25°C, Sorvall SLA-3000 rotor) and frozen at −20°C to improve cell lysis. Frozen cells from 1–2 liters of culture were thawed in ~ 20 mL 50 mM NaH2PO4, 1 mM EDTA, pH 7.0 supplemented with 100 μL HALT Protease Inhibitor Cocktail (Thermo Fischer Scientific, Rockford, IL). In the case of K70E and S67G, a final concentration of 1.0 mg/mL lysozyme (Sigma, St. Louis, MO) was also added to achieve lysis. Chromosomal DNA was eliminated using a final concentration of 2.5 μg/mL DNAse I and 2 mM MgCl2. The resulting lysate was clarified by centrifugation (26900 × g for 30 min, 4°C, Sorvall SS-34 rotor) and dialyzed against 4 L of 5 mM NaH2PO4, pH 5.8. The dialysis retentate was purified according to a previous method (19). The lysate was applied to a CM-32 carboxymethyl-cellulose (Whatman, Kent, UK) column (1.5 cm × 15 cm) equilibrated with 5 mM NaH2PO4, pH 5.8. A linear gradient of 5 mM NaH2PO4, pH 5.8 to 50 mM NaH2PO4, pH 7.0 was used to elute the protein. Pure fractions (>95% purity by SDS-PAGE) of eluted OXA-1 β-lactamase were combined and concentrated to ~5 mg/mL (17) using Amicon Ultra 10 kDa MWCO centrifugal filtration devices (Millipore, Billerica, MA). Aliquots of protein were snap frozen with liquid nitrogen and stored at −80°C until use.
Cibacron Blue 3GA Titrations
Tryptophan fluorescence quench of OXA-1 by Cibacron Blue 3GA was carried out in a Photon Technology International QuantaMaster™ 7 Fluorimeter. Proteins (0.4 μM) were titrated with increasing concentrations of Cibacron Blue from 0–8 μM. Tryptophan fluorescence (FI) was fit to the following equation:
| (Eq. 2) |
where Fmax is the initial tryptophan fluorescence, Fchange is the total fluorescence quench, and Kd is the dissociation constant.
Fluorescent SDS-PAGE Assay for Acyl-Enzyme Intermediates
Acyl-enzyme intermediates were detected using the fluorescent substrate BOCILLIN FL™ (Invitrogen, Carlsbad, CA). Aliquots of 2.2 μg of each protein were incubated with 50 μM BOCILLIN FL™ in 50 mM NaH2PO4, pH 7.0 for 5 min, followed by a 250-fold excess of ampicillin (12.5 mM). A second set of the OXA-1 variants were treated the same way, except the order of addition of ampicillin and BOCILLIN FL™ was reversed. Following the incubations, SDS sample buffer was added to each tube and all samples were separated by 10% SDS-PAGE. Gels were illuminated at 365 nm and imaged using UVP Bioimaging Systems Epichemi 3 Darkroom with Biochemi Camera Kit (Ultraviolet Products, Upland, CA). Gels were subsequently stained with Coomassie Brilliant Blue G.
Electrospray Ionization Mass Spectrometry (ESI-MS)
ESI-MS was used to determine the mass of wild-type and mutant forms of OXA-1 in the absence and presence of ampicillin. Each protein (40 μM) was incubated with 40 mM ampicillin for 15 min in 10 mM phosphate buffered saline, pH 7.4. This mixture was equilibrated with 0.1% trifluoroacetic acid and desalted using a C18 ZipTip (Millipore, Bedford, MA). Samples were placed on ice and analyzed within 10 min. Spectra of enzyme with and without ampicillin were generated on an Applied Biosystems (Framington, MA) Q-STAR XL quadrupole-time-of-flight (TOF) mass spectrometer equipped with a nanospray source. Experiments were performed by diluting the sample with 50% acetonitrile/0.1% formic acid to a concentration of 10 μM. This solution was then infused at a rate of 0.5 μL/min and data were collected for 2 min. Spectra were deconvoluted using the Applied Biosystems Analyst program.
Active Site Titration
The acylation stoichiometry of K70D was assessed by treating the substrate ampicillin as an irreversibly binding inhibitor following previous methods (22, 39). Aliquots of K70D protein at 13 μM were incubated with varying levels of ampicillin ranging from 1.3–65 μM for 2 hours to assure complete acylation. BOCILLIN FL™ was added to samples at 160 μM and incubated for 1 hour, at which point the reactions were quenched with SDS sample buffer. Samples were separated by 10% SDS-PAGE and the resulting gel was illuminated at 365 nm and imaged. Fluorescence intensity was quantified with Vision Works LS imaging software (Ultraviolet Products) and plotted versus the [ampicillin] to [enzyme] ratio.
Michaelis-Menten Steady-State Kinetics
Kinetic analysis was carried out in 50 mM NaH2PO4, pH 7.0, at room temperature in a Beckman DU-800 spectrophotometer. Measurements were carried out for nitrocefin and ampicillin using the Δε [molar absorption coefficient (M−1 · cm−1)] values of 17400 (λ = 472 nm) and −900 (λ = 235 nm), respectively (19). The average of three measurements of initial velocity was plotted as a function of substrate concentration, and the Km and kcat values were determined by non-linear least squares regression to the Michaelis-Menten-Henri equation.
Deacylation Rate Determination
Deacylation rate constants (k3) were determined by tryptophan fluorescence [excitation = 280 nm (0.4 nm slit width), emission = 330 nm (26 nm slit width)]. Variant proteins (0.4 μM) were incubated in 50 mM NaH2PO4, pH 7.0 with 300-fold excess substrate in a 1 cm pathlength quartz cell and allowed to fully acylate (2–10 min). A small amount of wild-type enzyme was added (20 nM) to hydrolyze excess substrate. The reporter dye Cibacron Blue 3GA was added prior to wild-type addition at 3.7 μM. Samples were incubated at 25°C and fluorescence intensity was measured at varying time intervals. Fluorescence intensity (FI) was fit to a first-order exponential rate equation:
| (Eq. 3) |
where k is the deacylation rate constant (k3), Fmax is fluorescence intensity at time 0, and Fbase is the fluorescence intensity at the end of the experiment. To measure the effects of buffer or small molecule carboxylates, the above assay was repeated in four different buffers: 50 mM Tris-HCl, pH 7.4; 50 mM NaH2PO4, pH 7.4; 50 mM NaH2PO4, 9 mM sodium acetate, pH 7.4; or 50 mM NaH2PO4, 9 mM sodium propanoate, pH 7.4.
Results
We used PCR mutagenesis to prepare substitutions for the serine at position 67 (S67G) and the carboxy-lysine at position 70 (K70A, K70D and K70E) of OXA-1 β-lactamase. Each of these variants was expressed in E. coli and purified to homogeneity using carboxymethyl cation exchange chromatography. All of the protein preparations gave similar yields. All four substitutions resulted in a dramatic drop in hydrolysis efficacy for both ampicillin and the chromogenic cephalosporin nitrocefin in phosphate buffer, pH 7.0 (Table 1). For the K70 variants, turnover of ampicillin was not observed (e.g. kcat < 0.002 s−1) even after a 30 minute incubation with very high concentrations of the enzyme (8 μM). Interestingly, for all of the variants except K70D, low levels of nitrocefinase activity were still observed, indicating that neither residue is absolutely required for catalytic turnover of that cephalosporin. The kcat value for this residual activity, however, was greatly reduced for all mutants compared to wild-type (>2000-fold lower). These results are consistent with the roles for S67 (nucleophile) and carboxy-K70 (general base) predicted by Sun et al (17), and suggested by homology with other β-lactamases.
Table 1.
Kinetic constants for OXA-1 wild-type and mutants
| Nitrocefin |
Ampicillin |
|||||
|---|---|---|---|---|---|---|
| Protein | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
| wild-type | 110 ± 10 | 1.9 ± 0.3 | 61 ± 11 | 500 ± 48 | 13 ± 4 | 40 ± 12 |
| S67G | 0.052 ± 0.003 | 61 ± 8 | 0.00086 ± 0.00010 | < 0.002 | ND | ND |
| K70A | 0.024 ± 0.003 | 0.72 ± 0.33 | 0.033 ± 0.016 | < 0.002 | ND | ND |
| K70E | 0.020 ± 0.001 | 1.2 ± 0.3 | 0.017 ± 0.004 | < 0.002 | ND | ND |
| K70D | < 0.002 | ND | ND | < 0.002 | ND | ND |
ND – not determined
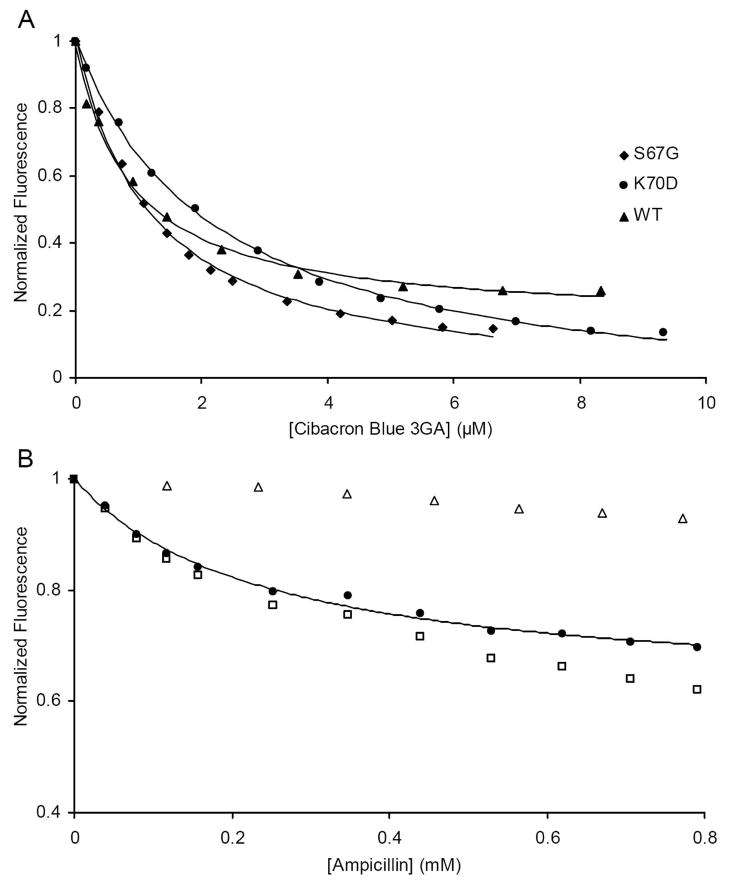
In order to ensure that the significant loss of hydrolytic activity was not caused by the misfolding of the mutants, we used fluorescence spectroscopy to observe the binding of known class D active site probes. Two of the five tryptophan residues of OXA-1 are located in the first layer of active site residues, and one of those (W160) forms a critical stabilizing hydrogen bond with the carbamate group of carboxy-K70. The presence of these residues suggested we may be able to observe the binding of ligands or substrates to OXA-1 β-lactamase by monitoring tryptophan fluorescence. Therefore, the effect of the anthraquinone dye Cibracron Blue 3GA on OXA-1 fluorescence was observed. This hydrophobic dye was shown previously to bind specifically to the active site of another class D β-lactamase (OXA-2) with low micromolar affinity (40). We found that this dye quenched wild-type OXA-1 tryptophan emission. The magnitude of this quench is exceptionally large (over 90% at saturation), and fast (complete in less than the one second response time of the instrument). This observed lowering of signal was almost entirely the result of a loss of intensity rather than any shift of the wavelength of maximal emission (data not shown). The quench was fit to an inverted hyperbola single-site binding model and yielded a Kd of 0.86 ± 0.09 μM (Figure 2A). This indicates that the affinity of Cibacron Blue with OXA-1 was similar to that seen with OXA-2 (1.2 μM) (40). The dye gave nearly identical quenching results with all four OXA-1 mutants, and yielded Kd values that remained relatively unchanged (S67G, 0.90 ± 0.11 μM; K70A, 0.88 ± 0.15 μM; K70E, 2.64 ± 0.16 μM; K70D 2.17 ± 0.12 μM; data not shown). These results were taken as evidence that the mutants form an active site that is structurally similar to that of the wild-type enzyme.
Figure 2.
(A) Intrinsic tryptophan fluorescence quench of OXA-1 by Cibacron Blue 3GA. Tryptophan quench was fit to an inverted hyperbola to determine the Kd values. (B) Intrinsic tryptophan quench of S67G by ampicillin. Non-specific tryptophan quench and inner filter effects were observed by direct addition of ampicillin to tryptophan (△) and used to correct the S67G quench (□). The corrected data set (●) was fit to an inverted hyperbola to determine the Kd.
The successful use of Cibacron Blue to probe the active sites of the β-lactamases compelled us to try the same assay with β-lactam substrates. While evidence suggested that ampicillin was not converted to product by the four variant enzymes, it was not yet known if this β-lactam could still bind in the active site. The addition of ampicillin to S67G did indeed cause a quench of fluorescence intensity, albeit a much smaller one (~ 30%; Figure 2B). Because of the large concentration of ampicillin that was required to saturate the enzyme, it was necessary to control for inner filter effect and dilution by also titrating ampicillin against tryptophan as a free amino acid. The effect of Cibacron Blue on free tryptophan fluorescence was subtracted from the S67G signal change, resulting in a saturable quench and a Kd of 235 ± 29 μM. While this indicates that the ampicillin interaction with S67G is much weaker than that seen for Cibacron Blue, it is only 10-fold higher than the known Km of ampicillin for wild-type OXA-1. This suggests that the S67G mutant, which is not expected to form an acyl-enzyme intermediate, is able to interact with substrate in a manner that may resemble a pre-catalytic Michaelis complex. It is therefore possible that the S67G mutant may be useful for X-ray crystallographic studies of such pre-catalytic complexes.
The addition of ampicillin to any of the K70 mutants did not result in a saturable quench of their tryptophan fluorescence (data not shown). This could be interpreted in two ways: i) either the β-lactam antibiotic cannot bind in the active site, or ii) it can bind but does not change the tryptophan fluorescence. In the latter case, the loss of the carbamate hydrogen bond to W160 in the K70 mutants might alter the microenvironment around the tryptophan residue and thereby render its fluorescence emission insensitive to substrate binding. While ampicillin did not appreciably quench the fluorescence of any of the K70 mutants, it did almost fully block the fluorescence quench that normally accompanies the addition of Cibacron Blue (data not shown). This competitive inhibition of dye binding shows that the K70 mutants are still able to bind β-lactam antibiotics, but that the environment of the W160 residue (and thus the protein fluorescence) in the ligand-bound K70 mutants is not altered by this binding event.
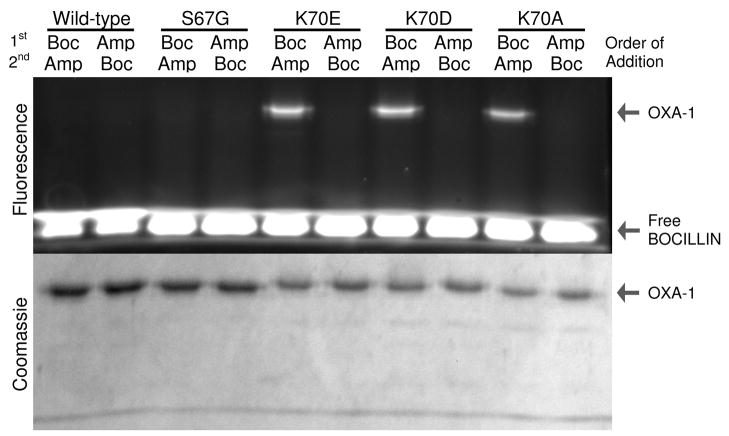
The competitive block of dye binding to the K70 mutants could be caused by ampicillin forming a non-covalent complex or a covalent acyl-enzyme intermediate. In class A and C enzymes, it has been shown that substitution for the suspected general base results in acylation-competent/deacylation-deficient enzymes that accumulate acylated intermediates. Our expectation of such a result in OXA-1 was tempered by the case of another class D enzyme, OXA-10, in which an alanine substitution for the carboxy-lysine led to an enzyme unable to acylate any substrate, including the highly reactive nitrocefin (16). In order to determine if any of the K70 mutants were able to form acyl-intermediates, we employed the highly-fluorescent penicillin β-lactam BOCILLIN FL™ (41). After a five minute incubation of BOCILLIN FL™ with wild-type OXA-1 and all four mutants, the proteins were separated from excess free BOCILLIN FL™ by SDS-PAGE. As seen in Figure 3, all three of the K70 mutants displayed fluorescence labeling, while the wild-type and S67G did not. This indicates that loss of the carboxy-lysine in OXA-1 does not prevent acylation of penicillin substrates, but likely slows down deacylation enough to allow accumulation of the acyl-intermediate. This contrasts with the complete lack of acylation activity seen with OXA-10 (16), suggesting active site conformational/stability differences between the two enzymes.
Figure 3.
Detection of BOCILLIN/OXA-1 acyl-enzyme intermediates. Proteins were incubated with excess BOCILLIN FL™ (5 min) and separated by SDS-PAGE. Excess ampicillin was incubated for 5 minutes either after (lanes 1, 3, 5, 7, 9) or before (lanes 2, 4, 6, 8, 10) the BOCILLIN FL™ incubation. BOCILLIN FL™ fluorescence was imaged under ultraviolet light (365 nm) prior to Coomassie staining.
In order to confirm these findings, we carried out the labeling experiment twice. In one case, a large excess of ampicillin was preincubated for five minutes with each protein before the addition of BOCILLIN FL™ (Figure 3, lanes 2, 4, 6, 8, 10), while in the other case, the order of addition was reversed (lanes 1, 3, 5, 7, 9). The complete competition of BOCILLIN FL™ labeling in the former case confirmed active site labeling, while the strong signal observed in the latter (addition of excess ampicillin after BOCILLIN FL™) suggests that the acyl-intermediate is relatively long-lived. The lack of any labeling of wild-type protein was interpreted as rapid turnover of BOCILLIN FL™ with no accumulation of a covalent intermediate. The lack of labeling with S67G was interpreted as loss of acylation activity upon removal of the nucleophilic alcohol. As a final control, we repeated the experiment with BOCILLIN FL™ that had been pre-treated with a trace of wild-type OXA-1. The complete lack of labeling of any of the proteins in this case (data not shown) demonstrated that the formation of the covalent intermediate requires an intact β-lactam ring, and therefore is not due to a side-reaction with some portion of the BOCILLIN’s bodipy fluorophore.
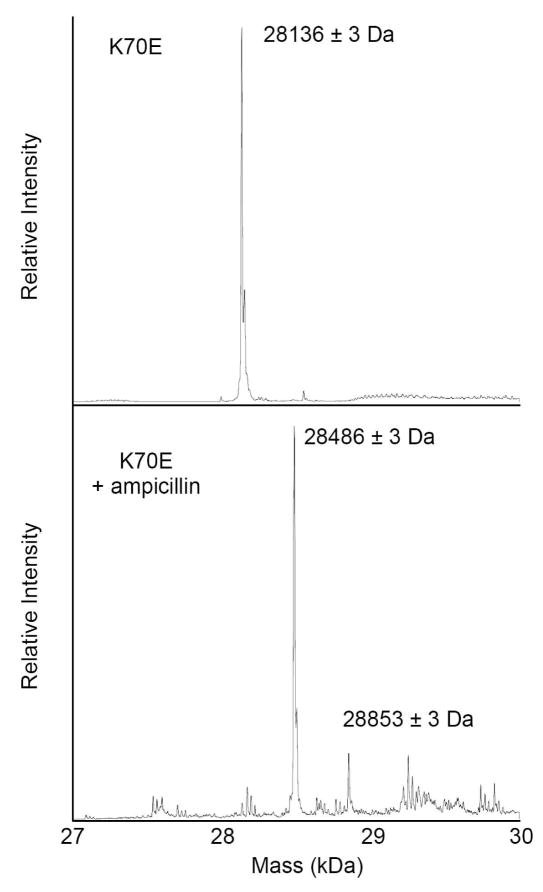
At this point, it was important for us to determine whether the accumulation of covalent intermediates with the K70 mutants was simply an artifact of the unique structure of BOCILLIN FL™ or was also observed with natural substrates. We used electrospray ionization mass-spectrometry (ESI-MS) to determine if such intermediates could be observed with ampicillin. We observed that all four proteins (wild-type, K70A, K70D, K70E) generated single peaks when no substrate was added, though each was ~127 Da smaller than expected (K70E results shown in Figure 4). This discrepancy is within 4 Da of the value expected if the N-terminal methionine was cleaved off during expression or purification. This methionine was engineered onto the protein in place of the secretion signal peptide to allow cytoplasmic expression of the mature OXA-1 protein. Its loss means that our wild-type preparation is likely identical to the native protein. A ten minute incubation of excess ampicillin had no effect on the mass of the wild-type protein. The same treatment of the K70 mutants, however, resulted in a near complete elimination of the apo-protein peak, and the concomitant appearance of a peak 349–350 Da larger (results for K70E shown in Figure 4). This shift matches the size of an ampicillin acyl moiety (349.4 Da), confirming our hypothesis, and also suggesting that there is no fragmentation of the substrate (beyond lactam cleavage) as is sometimes seen with mechanism-based inhibitors. The disappearance of the apo-peak suggests that all molecules in the protein preparation are competent to form the intermediate, a result confirmed by titration experiments (see below).
Figure 4.
Deconvoluted ESI-MS spectra of OXA-1 K70E alone and after incubation with ampicillin.
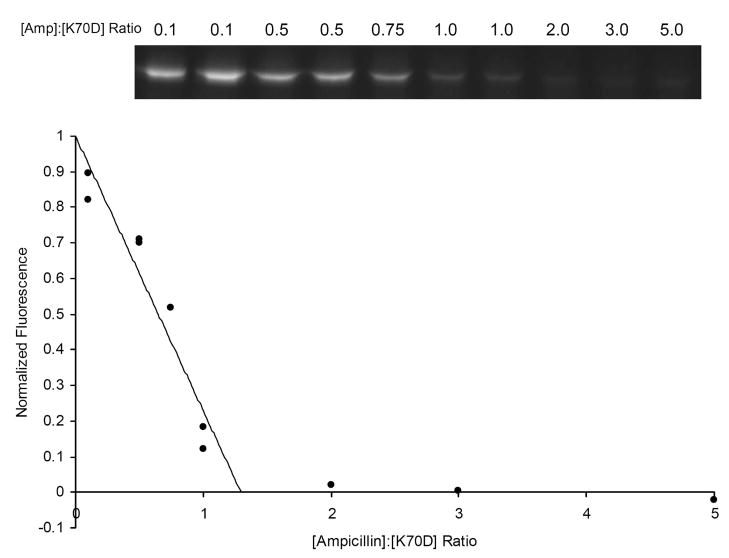
Active site titrations have been used to show that acylating inhibitors bind stoichiometrically to penicillin-binding proteins, β-lactam sensors and β-lactamases (22, 39). We incubated OXA-1 K70D with a range of ampicillin concentrations ranging from sub-stoichiometric to 5-fold excess. After a time period sufficient for full acylation, excess BOCILLIN FL™ was added to acylate and thereby quantitate the empty active sites. After separating the labeled proteins by SDS-PAGE, the fluorescence intensity of the protein bands was determined and plotted as a function of the ampicillin: protein ratio. As Figure 5 shows, the ampicillin: protein ratio that fully blocks BOCILLIN FL™ binding is approximately 1.3: 1.0. This low value is consistent with a negligible rate of turnover, as seen with the inhibitor aztreonam and the β-lactamase ACT-1 (39). In comparison, the BlaR1 protein—not considered a catalytic enzyme—turns over approximately six molecules of substrate before full inactivation is reached (22).
Figure 5.
OXA-1 K70D titrated with ampicillin. After acylation with increasing concentrations of ampicillin, open active sites were monitored by the addition of excess BOCILLIN FL™ and visualized by SDS-PAGE. The intensity of each fluorescent protein band was quantitated and plotted as a function of ampicillin: OXA-1 ratio.
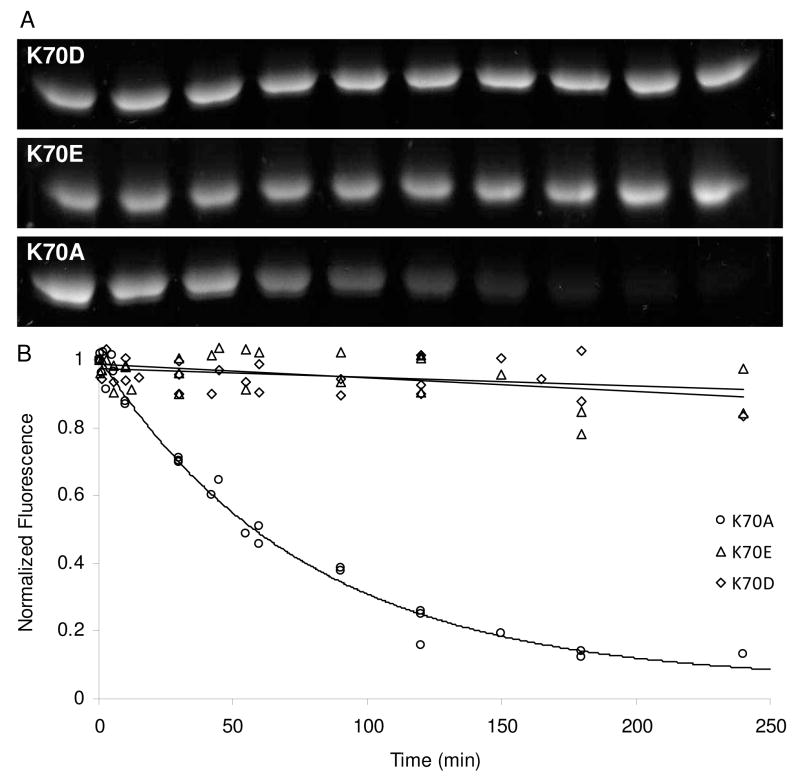
To determine the extent of the deacylation deficiency in each of the K70 variants, we measured the stability of the complexes over time using a modified form of the BOCILLIN/SDS-PAGE method described above. After incubation of a particular mutant with BOCILLIN FL™ to allow formation of the acyl-enzyme intermediate, a trace of wild-type enzyme is added to degrade excess intact BOCILLIN FL™. From this point on, any protein that deacylates will remain non-fluorescent, and quantitation of the fluorescent protein bands over time allows us to follow first-order deacylation kinetics. We found variability among the mutants; K70E and K70D deacylated very slowly, if at all, in three hours, while the K70A mutant displayed exponential decay that was complete within 2–3 hours (Figure 6).
Figure 6.
BOCILLIN FL™ deacylation rates of three different K70 mutants assessed by SDS-PAGE. (A) BOCILLIN FL™ complexes with K70 mutants were separated by SDS-PAGE and the fluorescence was visualized with ultraviolet light (365 nm). (B) Fluorescence intensity was quantitated and plotted as a function of time (three separate trials combined). K70D and K70E were fit by linear regression and K70A was fit to a single exponential decay.
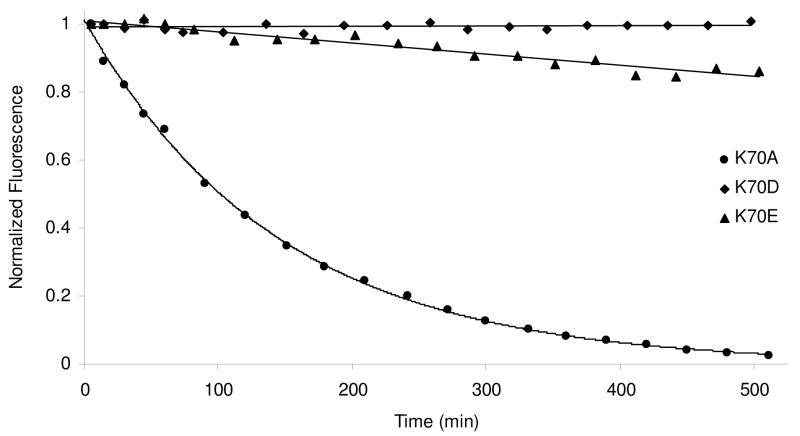
Again we felt that it was necessary to determine if the relative deacylation rates of these three mutants were influenced by the bodipy modification of BOCILLIN FL™. Moreover, we found that the gel assays were not quite sensitive enough to allow us to confidently assess the rates of the very slowly deacylating variants (K70D and K70E). We therefore developed a real-time assay for the deacylation of natural substrates. Because most β-lactam drugs are not fluorescent, and we know that the presence of ampicillin in the active site has no effect on the tryptophan fluorescence of the K70 mutants, we exploited the large change in fluorescence observed between the ampicillin-bound and Cibacron Blue-bound states of the mutants. Each K70 mutant was incubated with a large excess of ampicillin, and after sufficient time for acylation had passed, a trace of wild-type OXA-1 and a saturating concentration of Cibacron Blue were added simultaneously. As with the BOCILLIN/gel assay above, the wild-type enzyme eliminates free intact β-lactam, thus preventing any further reaction with the fluorescent substrate. Upon deacylation and product release, the empty active site becomes occupied by Cibacron Blue and displays a large drop in tryptophan fluorescence. Figure 7 illustrates that this approach does give higher resolution data with less “noise” and thereby allows better estimation of rate constants. Interestingly, it appears that K70D has not deacylated at all after three hours, while the K70E protein shows approximately 10% breakdown over that time period. As with the BOCILLIN FL™ assay, K70A is the fastest to deacylate (t1/2 = 100 min).
Figure 7.
Ampicillin deacylation rates of three different K70 mutants measured by tryptophan fluorescence. Deacylation was detected indirectly by observing the Cibacron Blue 3GA quench that occurs after hydrolyzed ampicillin has left the active site. K70D and K70E deacylation were fit by linear regression and K70A deacylation was fit to a single exponential decay.
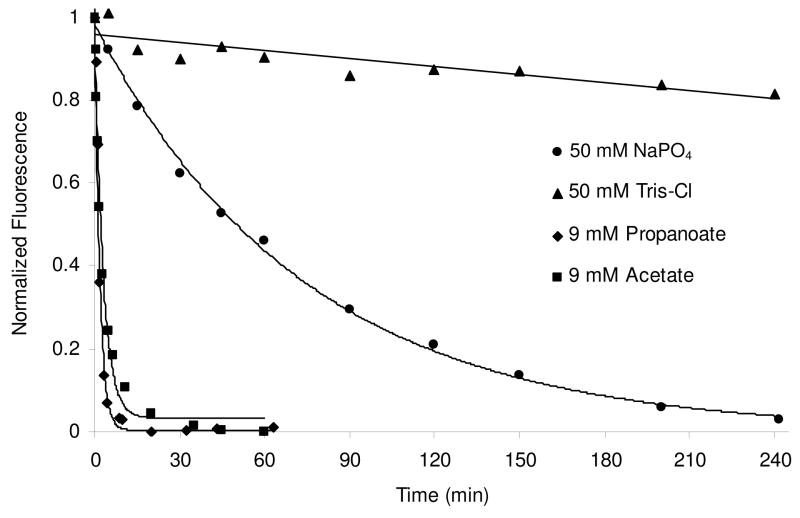
Interestingly, the rate of deacylation from K70A, but not K70D or K70E, was somewhat dependent on the choice and concentration of buffer used. Higher concentrations of phosphate or the addition of 9 mM sodium bicarbonate moderately increased the rate (Supporting information Figure S1 and Table 2), while Tris-Cl buffer significantly slowed it (Figure 8 and Table 2). We hypothesized that the alanine side chain might leave enough room for small anionic molecules to enter the active site and act as a general base for deacylation. Conversely, small cationic species such as Tris might be able to enter the active site, but would not likely be able to abstract a proton from the deacylating water. In further support of this, we found that short-chain carboxylates, which might enter the active site and fill the position normally occupied by the missing carboxy-lysine, were even more effective at accelerating deacylation (Figure 8). The addition of 9 mM acetate for instance, accelerated the K70A deacylation rate to 0.0056 ± 0.0004 s−1 (compared to 0.00023 ± 0.00001 s−1 for phosphate buffer alone), while the same concentration of propanoate gave an even higher rate of 0.0096 ± 0.0006 s−1. Propanoate had no effect on the negligible deacylation rate of OXA-1 K70D (data not shown). Longer acyl carboxylates such as butanoate and pentanoate gave no rate enhancement over phosphate buffer alone. The deacylation rate constants for various buffers, OXA-1 variants and assays are summarized in Table 2.
Table 2.
Deacylation rates of OXA-1 K70 mutants
| Substrate | Buffer | Deacylation rate k3 (s−1) | ||
|---|---|---|---|---|
| K70A | K70E | K70D | ||
| Bocillina | 50 mM NaH2PO4, pH 7.0 | 0.00022±0.00002 | < 4.0 × 10−5 | < 4.0 × 10−5 |
| Ampicillinb | 50 mM NaH2PO4, pH 7.0 | 0.00012±0.00001 | < 4.0 × 10−5 | < 4.0 × 10−5 |
| 50 mM NaH2PO4, 9 mM Na2CO3, pH 7.0 | 0.00090±0.00004 | < 4.0 × 10−5 | < 4.0 × 10−5 | |
| Ampicillinb | 50 mM NaH2PO4, pH 7.4 | 0.00023±0.00001 | ND | ND |
| 100 mM NaH2PO4, pH 7.4 | 0.00034±0.00001 | ND | ND | |
| 50 mM Tris-Cl, pH 7.4 | < 4.0 × 10−5 | ND | ND | |
| 50 mM NaH2PO4, 9 mM Na acetate, pH 7.4 | 0.0056 ± 0.0004 | < 4.0 × 10−5 | < 4.0 × 10−5 | |
| 50 mM NaH2PO4, 9 mM Na propanoate, pH 7.4 | 0.0096 ± 0.0006 | < 4.0 × 10−5 | < 4.0 × 10−5 | |
Measured by SDS-PAGE assay.
Measured by competition with Cibacron Blue 3GA. ND – not determined
Figure 8.
The ampicillin deacylation rate for K70A is dependent on the identity of the buffer and the presence of short alkyl carboxylates. Ampicillin deacylation was monitored indirectly using Cibacron Blue 3GA quench of tryptophan fluorescence in 50 mM Tris-Cl; 50 mM NaH2PO4; 50 mM NaH2PO4, 9 mM acetate; and 50 mM NaH2PO4, 9 mM propanoate (all buffers were pH 7.4).
Discussion
Through site-directed mutagenesis of two separate active site residues, we generated OXA-1 variants that arrest the catalytic mechanism at the pre-acylation stage (S67G) or the acyl intermediate stage (K70D). In both cases, the results we find are similar to those seen when homologous substitutions are introduced in class A and C β-lactamases. It is expected that these variants will be very useful for the study of OXA-1/substrate complexes, as they have been in class A and class C.
While the loss of the serine nucleophile in the S67G mutant resulted in a large diminution of the catalytic turnover rate, residual activity against nitrocefin remained. This result is reminiscent of studies involving class A (42, 43) and class C (44, 45) β-lactamases in which similar variants maintain measurable (albeit highly diminished) activity. The loss of the β carbon and its alcohol at position 67 may leave enough room for a water (or hydroxide ion), which could potentially attack the β-lactam carbonyl carbon directly. This scenario has been supported by structural analysis of the S70G mutant of TEM-1 (46), and has been observed in the mechanistically-related serine proteases (47). In several cases, it was shown that residual activity is the result of a small sub-population of wild-type enzyme produced through the mistranslation of the mutant codon (43, 48). This is not likely the case for OXA-1 S67G, however, as the nitrocefinase activity we observe displays a different Km compared to that seen with wild-type (49).
The substitution of alanine, glutamate or aspartate for K70 in OXA-1 has an enormous effect on the deacylation rate constant of OXA-1. While it is not known whether acylation or deacylation is rate-limiting for wild-type OXA-1, the deacylation rate can be no slower than the kcat, which is close to 500 s−1 for ampicillin. With an ampicillin deacylation rate constant of 0.00012 ± 0.00001 s−1 for the K70A mutant (the fastest of the three variants made), it is therefore possible to conclude that the substitution results in a rate decrease of at least 4 × 106 fold. Such changes are consistent with the effects of substitutions for the general base in other β-lactamase classes (4, 37), and further support the idea that the carbamate of carboxy-K70 is responsible for activation of the deacylating water.
Elimination of a β-lactamase’s general base can affect both acylation and deacylation rates (4). The accumulation of acyl-intermediates in the K70 variants, however, strongly suggests that such substitutions lower the deacylation rate much more than the acylation rate. Similar results for Glu166 mutants in class A have been used to argue for an asymmetric mechanism in which Glu166 is the general base for deacylation, while another residue plays that role for activation of the serine nucleophile (37). It has been noted, however, that a lack of change in a rate constant after substitution of so-called “essential” catalytic residues must be interpreted cautiously (2, 49, 50). Enzyme active sites are often highly plastic, and the maintenance of catalysis (or in this case, acylation) upon removal of an important functional residue may simply reflect adaptations within the active site that make up for the missing functionality (49). Alternatively, the variant enzyme may adopt an entirely different mechanistic strategy, such as when mutation of a β-lactamase’s nucleophile leads to non-covalent direct hydrolysis [see above, and (46)]. In light of these arguments, the fact that acylation activity is not eliminated in the K70 variants does not preclude a general base role for carboxy-K70 during the activation of S67. How does that nucleophile still carry out acylation when the carbamate is not present? First, it should be noted that the action of a general base is only part of the complex array of strategies used by an enzyme to induce catalysis. The anionic tetrahedral transition state is likely still stabilized by the backbone amide hydrogens of residues S67 and A215, and the side-chain of K212. The deprotonation of S67 is thought to be enhanced by the positive charge of K212 and the positive helical dipole of a helix A3 (17, 51); these effects may still be present despite the absence of carboxy-K70. Moreover, in the spatial arrangement of the active site, K212 is closer to S67 than it is to the expected position of a deacylating water, and this may account for the preferential enhancement of acylation over deacylation in the absence of K70. While there are no other residues that appear to be close enough to act as a general base, it is possible that the loss of the K70 side-chain, the binding of substrate, or both, induces a new active site architecture that allows such a residue to fill in for the carbamate (52).
The fact that the K70A substitution allows acylation of substrates in OXA-1, while the same mutant in OXA-10 does not (16), is puzzling. An overlay of the two enzyme structures shows that the key catalytic residues S67, K70, S115, V117, W160 (154 in OXA10) and K212 (205) are located in the same positions (Figure 1; rmsd = 0.28 Å for the six residues listed above). There are several residues in the active site that are not conserved, including A215 (F208 in OXA-10), S120 (F120) and L73 (N73). S120 hydrogen bonds to carboxy-K70 in OXA-1, and N73 interacts with an active site water in OXA-10. It is therefore likely that subtle differences in the positions of these residues and water molecules after substrate binding, but before acylation, are responsible for this difference in activity between OXA-1 and OXA-10. It should also be noted that members of the OXA-1 subclass of oxacillinases lack an arginine corresponding to R250 in OXA-10 (17). In many β-lactamase/substrate complexes, this arginine forms a critical salt bridge to the carboxylate common to all β-lactam substrates. It is therefore possible that the mode of substrate binding is significantly different in OXA-1 compared to those enzymes that contain an arginine at this position.
Our result showing that the lack of the carboxy-lysine affects deacylation is reminiscent of the activity of the naturally deacylation deficient β-lactam sensor, BlaR1. Acylation of the sensor protein has been shown to lead to decarboxylation of the lysine carbamate, with the concomitant loss of its ability to activate the water for the deacylation reaction (22). Cha et al. used chemical modification to replace the BlaR1 carboxy-lysine with a carboxylate analog, which, unlike the natural carbamate, cannot undergo decarboxylation. This modification was shown to impart deacylation activity upon BlaR1, in essence creating a novel catalyst (26). These findings, taken together with the results reported here, further support the notion that the presence of the carbamate modification is particularly important for promoting the deprotonation of the deacylating water. We therefore predict that other substitutions in class D β-lactamases that lead to destabilization of the carboxy-lysine may also lead to deacylation deficiency due to the loss of the carbamate.
Our observation that an alanine substitution for K70 maintained a significant deacylation rate, while substitution with glutamate or aspartate did not, was initially surprising. We originally hypothesized that the K70A variant, with only the relatively unreactive methyl group, would be the least likely to be able to activate a deacylating water. The K70E and K70D mutants, we reasoned, were more likely to show some deacylation activity, as the carboxylate group might be able to act as a general base. Substitution of aspartate for E166 in the class A enzyme from Bacillus cereus, for instance, results in higher residual activity than seen with other non-acidic substitutions (53). The fact that we find the opposite result suggests that the small alanine side chain leaves enough space to accommodate a phosphate group (from the buffer) which can then serve as the general base. The full negative charges of the aspartate and glutamate mutants would not tolerate such an ion either sterically or electronically. Our observation that short-chain fatty acid mimetics of the carboxy-lysine (acetate and propanoate) were even more effective at enhancing deacylation in K70A, but not K70D or K70E, strongly argues in favor of this hypothesis. The deacylation rate enhancement was not observed with butanoate or pentanoate, both of which would likely be too large to fit in the cavity created by the mutation of lysine to alanine.
Lastly, we have extended the utility of two previously known β-lactamase active site probes, BOCILLIN FL™ and Cibacron Blue 3GA. While BOCILLIN FL™ has been used as an active site probe of the naturally deacylation deficient penicillin-binding proteins (PBPs) (41, 54) and BlaR1 (22, 23) sensors, we have shown that this probe can be useful for quantitation of deacylation rates of β-lactamases that have been catalytically compromised by mutagenesis. In particular, our data showing that the deacylation rates for three different K70 mutants are very similar for ampicillin and BOCILLIN FL™ lends support to the use of the latter as a faithful mimic of natural penicillin substrates. It is likely that the bodipy modification does not clash with any structural features that are found in class D active sites. The use of Cibacron Blue 3GA as a specific ligand for class D β-lactamases has been limited to affinity chromatography for purification, and protein-induced shifts in dye absorbance (40). The discovery that Cibacron Blue strongly quenches tryptophan fluorescence of OXA-1 makes possible a sensitive and facile test for proper active site formation in mutant enzymes. We have also demonstrated its utility for the measurement of deacylation rates of natural substrates, allowing such measurements when the use of a fluorescently-modified probe such as BOCILLIN FL™ is problematic. The extraordinary extent of the tryptophan fluorescence quench—over 90% at saturation—is no doubt due in large part to the proximity of W160 to K70 and S67. It should be noted additionally, that another highly conserved tryptophan, W102, is also in the first layer of active site residues. Moreover, this residue undergoes a > 9 Å shift upon the binding of meropenem in OXA-13 (55), which may also explain such a large change in fluorescence. The ubiquity of tryptophan at position 160 in all class D β-lactamases suggests that Cibacron Blue may be a useful probe for all class members. Indeed, we have observed that the affinity of the ligand and its extent of tryptophan quench are essentially identical when used with the class D carbapenemase, OXA-24 (data not shown).
In conclusion, our analysis highlights the key role of carboxy-K70 in OXA-1 β-lactamase. The observation that substitutions at this position result in deacylation-deficient variants illuminates the role of the unique carbamate in the mechanism, and also provides a powerful tool for future structural studies of the acyl-enzyme intermediate state. Future studies will help elucidate whether this mutational effect can be applied broadly throughout the class D β-lactamase family, or is unique to the OXA-1 subfamily.
Supplementary Material
Abbreviations
- LB
Luria Bertani
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ESI-MS
Electrospray Ionization Mass Spectrometry
Footnotes
This research was supported by National Institutes of Health grant 1R15AI082416-01 (D.A.L.). The Veterans Affairs Merit Review Program, Geriatric Research Education and Clinical Care VISN 10, and the National Institutes of Health (RO1 AI072219) supported R.A.B.
Supporting Information Available
Plots of ampicillin deacylation time-courses for K70A, K70E and K70D in the absence and presence of bicarbonate are provided as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 3.Ledent P, Raquet X, Joris B, Van Beeumen J, Frere JM. A comparative study of class-D beta-lactamases. Biochem J. 1993;292(Pt 2):555–562. doi: 10.1042/bj2920555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillaume G, Vanhove M, Lamotte-Brasseur J, Ledent P, Jamin M, Joris B, Frere JM. Site-directed mutagenesis of glutamate 166 in two beta-lactamases. Kinetic and molecular modeling studies. J Biol Chem. 1997;272:5438–5444. doi: 10.1074/jbc.272.9.5438. [DOI] [PubMed] [Google Scholar]
- 5.Minasov G, Wang X, Shoichet BK. An ultrahigh resolution structure of TEM-1 β-lactamase suggests a role for Glu166 as the general base in acylation. J Am Chem Soc. 2002;124:5333–5340. doi: 10.1021/ja0259640. [DOI] [PubMed] [Google Scholar]
- 6.Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. Ab initio QM/MM study of class A β-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J Am Chem Soc. 2005;127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 7.Dubus A, Ledent P, Lamotte-Brasseur J, Frere JM. The roles of residues Tyr150, Glu272, and His314 in class C beta-lactamases. Proteins. 1996;25:473–485. doi: 10.1002/prot.7. [DOI] [PubMed] [Google Scholar]
- 8.Oefner C, D’Arcy A, Daly JJ, Gubernator K, Charnas RL, Heinze I, Hubschwerlen C, Winkler FK. Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis. Nature. 1990;343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Minasov G, Roth TA, Prati F, Shoichet BK. The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J Am Chem Soc. 2006;128:2970–2976. doi: 10.1021/ja056806m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato-Toma Y, Iwashita T, Masuda K, Oyama Y, Ishiguro M. pKa measurements from nuclear magnetic resonance of tyrosine-150 in class C β-lactamase. Biochem J. 2003;371:175–181. doi: 10.1042/BJ20021447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubus A, Normark S, Kania M, Page MG. The role of tyrosine 150 in catalysis of beta-lactam hydrolysis by AmpC beta-lactamase from Escherichia coli investigated by site-directed mutagenesis. Biochemistry. 1994;33:8577–8586. doi: 10.1021/bi00194a024. [DOI] [PubMed] [Google Scholar]
- 12.Gherman BF, Goldberg SD, Cornish VW, Friesner RA. Mixed quantum mechanical/molecular mechanical (QM/MM) study of the deacylation reaction in a penicillin binding protein (PBP) versus in a class C β-lactamase. J Am Chem Soc. 2004;126:7652–7664. doi: 10.1021/ja036879a. [DOI] [PubMed] [Google Scholar]
- 13.Beadle BM, Trehan I, Focia PJ, Shoichet BK. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC β-lactamase. Structure. 2002;10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 14.Bulychev A, Massova I, Miyashita K, Mobashery S. Nuances of Mechanisms and Their Implications for Evolution of the Versatile β-Lactamase Activity: From Biosynthetic Enzymes to Drug Resistance Factors. J Am Chem Soc. 1997;119:7619–7625. [Google Scholar]
- 15.Maveyraud L, Golemi D, Kotra LP, Tranier S, Vakulenko S, Mobashery S, Samama JP. Insights into class D beta-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8:1289–1298. doi: 10.1016/s0969-2126(00)00534-7. [DOI] [PubMed] [Google Scholar]
- 16.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc Natl Acad Sci U S A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of beta-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maveyraud L, Golemi-Kotra D, Ishiwata A, Meroueh O, Mobashery S, Samama JP. High-resolution X-ray structure of an acyl-enzyme species for the class D OXA-10 beta-lactamase. J Am Chem Soc. 2002;124:2461–2465. doi: 10.1021/ja016736t. [DOI] [PubMed] [Google Scholar]
- 19.Leonard DA, Hujer AM, Smith BA, Schneider KD, Bethel CR, Hujer KM, Bonomo RA. The role of OXA-1 beta-lactamase Asp(66) in the stabilization of the active-site carbamate group and in substrate turnover. Biochem J. 2008;410:455–462. doi: 10.1042/BJ20070573. [DOI] [PubMed] [Google Scholar]
- 20.Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D beta-lactamase OXA-10. Nat Struct Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 21.Zhu YF, Curran IH, Joris B, Ghuysen JM, Lampen JO. Identification of BlaR, the signal transducer for beta-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 beta-lactamase (class D) of Salmonella typhimurium. J Bacteriol. 1990;172:1137–1141. doi: 10.1128/jb.172.2.1137-1141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated beta-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J Am Chem Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 23.Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. Resistance to beta-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J Biol Chem. 2003;278:18419–18425. doi: 10.1074/jbc.M300611200. [DOI] [PubMed] [Google Scholar]
- 24.Kerff F, Charlier P, Colombo ML, Sauvage E, Brans A, Frere JM, Joris B, Fonze E. Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis. Biochemistry. 2003;42:12835–12843. doi: 10.1021/bi034976a. [DOI] [PubMed] [Google Scholar]
- 25.Wilke MS, Hills TL, Zhang HZ, Chambers HF, Strynadka NC. Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus. J Biol Chem. 2004;279:47278–47287. doi: 10.1074/jbc.M407054200. [DOI] [PubMed] [Google Scholar]
- 26.Cha J, Mobashery S. Lysine N(zeta)-decarboxylation in the BlaR1 protein from Staphylococcus aureus at the root of its function as an antibiotic sensor. J Am Chem Soc. 2007;129:3834–3835. doi: 10.1021/ja070472e. [DOI] [PubMed] [Google Scholar]
- 27.Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Discrete steps in sensing of beta-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc Natl Acad Sci U S A. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalp M, Sheri A, Buynak JD, Bethel CR, Bonomo RA, Carey PR. Efficient inhibition of class A and class D beta-lactamases by Michaelis complexes. J Biol Chem. 2007;282:21588–21591. doi: 10.1074/jbc.C700080200. [DOI] [PubMed] [Google Scholar]
- 29.Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 30.Padayatti PS, Helfand MS, Totir MA, Carey MP, Hujer AM, Carey PR, Bonomo RA, van den Akker F. Tazobactam forms a stoichiometric trans-enamine intermediate in the E166A variant of SHV-1 beta-lactamase: 1.63 Å crystal structure. Biochemistry. 2004;43:843–848. doi: 10.1021/bi035985m. [DOI] [PubMed] [Google Scholar]
- 31.Shimamura T, Ibuka A, Fushinobu S, Wakagi T, Ishiguro M, Ishii Y, Matsuzawa H. Acyl-intermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J Biol Chem. 2002;277:46601–46608. doi: 10.1074/jbc.M207884200. [DOI] [PubMed] [Google Scholar]
- 32.Knox JR, Moews PC, Escobar WA, Fink AL. A catalytically-impaired class A beta-lactamase: 2 Å crystal structure and kinetics of the Bacillus licheniformis E166A mutant. Protein Eng. 1993;6:11–18. doi: 10.1093/protein/6.1.11. [DOI] [PubMed] [Google Scholar]
- 33.Padayatti PS, Helfand MS, Totir MA, Carey MP, Carey PR, Bonomo RA, van den Akker F. High resolution crystal structures of the trans-enamine intermediates formed by sulbactam and clavulanic acid and E166A SHV-1 beta-lactamase. J Biol Chem. 2005;280:34900–34907. doi: 10.1074/jbc.M505333200. [DOI] [PubMed] [Google Scholar]
- 34.Jelsch C, Mourey L, Masson JM, Samama JP. Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 Å resolution. Proteins. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 35.Helfand MS, Totir MA, Carey MP, Hujer AM, Bonomo RA, Carey PR. Following the reactions of mechanism-based inhibitors with beta-lactamase by Raman crystallography. Biochemistry. 2003;42:13386–13392. doi: 10.1021/bi035716w. [DOI] [PubMed] [Google Scholar]
- 36.Adachi H, Ohta T, Matsuzawa H. Site-directed mutants, at position 166, of RTEM-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J Biol Chem. 1991;266:3186–3191. [PubMed] [Google Scholar]
- 37.Escobar WA, Tan AK, Fink AL. Site-directed mutagenesis of beta-lactamase leading to accumulation of a catalytic intermediate. Biochemistry. 1991;30:10783–10787. doi: 10.1021/bi00108a025. [DOI] [PubMed] [Google Scholar]
- 38.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauvois C, Ibuka AS, Celso A, Alba J, Ishii Y, Frere JM, Galleni M. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob Agents Chemother. 2005;49:4240–4246. doi: 10.1128/AAC.49.10.4240-4246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monaghan C, Holland S, Dale JW. The interaction of anthraquinone dyes with the plasmid-mediated OXA-2 beta-lactamase. Biochem J. 1982;205:413–417. doi: 10.1042/bj2050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CC, Smith TJ, Kapadia G, Wasch S, Zawadzke LE, Coulson A, Herzberg O. Structure and kinetics of the β-lactamase mutants S70A and K73H from Staphylococcus aureus PC1. Biochemistry. 1996;35:12251–12258. doi: 10.1021/bi961153v. [DOI] [PubMed] [Google Scholar]
- 43.Jacob F, Joris B, Frere JM. Active-site serine mutants of the Streptomyces albus G beta-lactamase. Biochem J. 1991;277(Pt 3):647–652. doi: 10.1042/bj2770647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beadle BM, Shoichet BK. Structural bases of stability-function tradeoffs in enzymes. J Mol Biol. 2002;321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 45.Goldberg SD, Iannuccilli W, Nguyen T, Ju J, Cornish VW. Identification of residues critical for catalysis in a class C β-lactamase by combinatorial scanning mutagenesis. Protein Sci. 2003;12:1633–1645. doi: 10.1110/ps.0302903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stec B, Holtz KM, Wojciechowski CL, Kantrowitz ER. Structure of the wild-type TEM-1 beta-lactamase at 1.55 Å and the mutant enzyme Ser70Ala at 2.1 Å suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Crystallogr D Biol Crystallogr. 2005;61:1072–1079. doi: 10.1107/S0907444905014356. [DOI] [PubMed] [Google Scholar]
- 47.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 48.Toth MJ, Murgola EJ, Schimmel P. Evidence for a unique first position codon-anticodon mismatch in vivo. J Mol Biol. 1988;201:451–454. doi: 10.1016/0022-2836(88)90152-0. [DOI] [PubMed] [Google Scholar]
- 49.Peracchi A. Enzyme catalysis: removing chemically ‘essential’ residues by site-directed mutagenesis. Trends Biochem Sci. 2001;26:497–503. doi: 10.1016/s0968-0004(01)01911-9. [DOI] [PubMed] [Google Scholar]
- 50.Knowles JR. Tinkering with enzymes: what are we learning? Science. 1987;236:1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- 51.Moews PC, Knox JR, Dideberg O, Charlier P, Frere JM. Beta-lactamase of Bacillus licheniformis 749/C at 2 Å resolution. Proteins. 1990;7:156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- 52.Steyaert J, Hallenga K, Wyns L, Stanssens P. Histidine-40 of ribonuclease T1 acts as base catalyst when the true catalytic base, glutamic acid-58, is replaced by alanine. Biochemistry. 1990;29:9064–9072. doi: 10.1021/bi00490a025. [DOI] [PubMed] [Google Scholar]
- 53.Gibson RM, Christensen H, Waley SG. Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73. Biochem J. 1990;272:613–619. doi: 10.1042/bj2720613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hujer AM, Kania M, Gerken T, Anderson VE, Buynak JD, Ge X, Caspers P, Page MG, Rice LB, Bonomo RA. Structure-activity relationships of different beta-lactam antibiotics against a soluble form of Enterococcus faecium PBP5, a type II bacterial transpeptidase. Antimicrob Agents Chemother. 2005;49:612–618. doi: 10.1128/AAC.49.2.612-618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pernot L, Frenois F, Rybkine T, L’Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. Crystal structures of the class D beta-lactamase OXA-13 in the native form and in complex with meropenem. J Mol Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- 56.DeLano WL. DeLano Scientific LLC. Palo Alto; CA, USA: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.