Abstract
There has been a recent upsurge in the development of small-molecule inhibitors specific to DNA repair proteins or proteins peripherally involved in base excision repair and the DNA damage response. These specific, nominally toxic inhibitors are able to potentiate the effect of existing cancer cell treatments in a wide array of cancers. One of the largest obstacles to overcome in the treatment of cancer is incomplete killing with initial cancer treatments, leading to resistant cancer. The progression of our understanding of cancer and normal cell responses to DNA damage has allowed us to develop biomarkers that we can use to help us predict responses of cancers, more specifically target cancer cells and overcome resistance. Initial successes using these small-molecule DNA repair inhibitors in target-validation experiments and in the early stages of clinical trials indicate an important role for these inhibitors, and allow for the possibility of a future in which cancers are potentially treated in a highly specific, individual manner.
Keywords: Ape1 inhibitor, base excision repair, biomarkers, BRCA1, BRCA2, DNA polymerase β inhibitor, PARP inhibitor
There are several properties common across most types of cancer. They display unrestrained cell proliferation, perpetual replication, sustained angiogenesis, the ability to escape apoptosis and invasiveness. One method to fight cancer is to exploit differences between normal cells and the cancer cells so they can be selectively destroyed. Many cancers are able to avoid or escape apoptosis due to abnormal DNA damage responses [1–3]. Most types of cancer have DNA damage response deficiencies, highly proficient DNA repair mechanisms or, more often, a combination of DNA repair deficiencies and proficiencies. These innate differences have been used in the past to selectively kill cancer cells with irradiation (IR) or chemotherapies, or combinations of the two [3,4]. However, cancers are often resistant or develop resistance to these treatments due to the cancer cells’ remarkable ability to adapt their DNA damage responses to compensate for any shortcomings. Often the treatment is not selective enough towards the cancer cells, thereby causing too much toxicity to normal cells resulting in a low therapeutic index [5]. A significant number of agents used in front-line therapy include DNA-damaging agents, such that upon treatment, a wide variety of DNA damage response pathways respond to the insult. These include the base excision repair (BER), nucleotide excision repair (NER), direct repair (DR), mismatch repair (MMR), homologous recombination (HR) and non-homologous end joining (NHEJ) repair pathways. These are very specialized pathways that repair DNA damage in a highly specific manner. Although the mechanisms for some of the players will be elucidated in this review, an overview of each of the specific pathways will not be given. Readers should refer to the several comprehensive reviews recently published on these pathways [1–3,5–9].
Although we focus on the repair of certain lesions by one particular pathway, most often there is crossover, interaction and compensation within and between the various DNA repair pathways. These crossovers often allow the cancer cells to compensate and ultimately survive [10]. We are beginning to be able to overcome some of these resistances and the advantages that these cancer cells have either inherited or developed. As understanding of the DNA repair pathways has progressed, we are increasingly able to identify biomarkers that may help us better understand the response of the cancer cells to chemotherapy or DNA damage [11,12]. For example, the alkaline comet assay has been successfully used with peripheral blood lymphocytes of cancer patients to predict their response to doxorubicin and cisplatin [13]. We are also better able to use multitargeted approaches in combination with chemotherapy or chemotherapies and IR to achieve more complete responses. This can help to prevent tumors from adapting by acquiring resistance after primary treatments [8,14]. However, there are some instances of acquired resistance in cancers which, once developed, we have not been able to overcome, such as platinum resistance in ovarian and cervical cancer [15]. Additionally, some cancers, such as pancreatic cancer, still have incredibly low survival rates, so new approaches are clearly needed [16].
There has been a recent upsurge of small-molecule inhibitor candidates in DNA repair [17]. These small-molecule inhibitors are specific to players in the DNA damage response teams. Many of these inhibitors are able to inhibit the functions of the desired proteins with nominal additional toxicity, yet dramatically potentiate the effects of current cancer treatments. The following is a review of the most recent studies performed on current small-molecule inhibitors of proteins involved in BER. The review is by no means comprehensive of all the inhibitors of proteins involved in BER. Some of the inhibitors reviewed here have been used in preclinical target validation and proof-of-concept studies and were not intended to be, nor will be, developed into candidates for clinical research. However, the potential value of each inhibitor in the clinical setting will be discussed. Some of the inhibitors reviewed are currently in clinical trials (see Table 1).
Table 1.
Inhibitors in this review that are currently in clinical trials for cancer treatments according to clinicaltrials.gov [101].
| Inhibitor target |
Inhibitor | Type of cancer/condition | Drug/irradiation combination being used |
|---|---|---|---|
| PARP | ABT-888 (Abbott Laboratories) | Nonhematologic malignancies, metastatic melanoma, BRCA-deficient breast cancer, BRCA-deficient ovarian cancer | Temozolomide |
| Solid tumors or lymphoma | Cyclophosphamide | ||
| Glioblastoma multiforme | Temozolomide and irradiation | ||
| Metastatic melanoma | Temozolomide | ||
| Brain metastases | Irradiation | ||
| Leukemia | Topotecan | ||
| Metastatic or unresectable solid tumors or non-Hodgkin lymphoma | Cyclophosphamide | ||
| Advanced solid cancer | Carboplatin and paclitaxel | ||
| Lymphoma, adult solid tumor | Topotecan | ||
| Lymphoma, adult solid tumor | Irinotecan | ||
| PARP | AZD2281/KU-0059436 (AstraZeneca/KuDOS Pharmaceuticals Limited) | Triple-negative breast cancer | Cisplatin |
| Solid tumors | None | ||
| Advanced solid malignancies | None | ||
| Platinum-sensitive ovarian cancer | None | ||
| Colorectal cancer | Irinotecan | ||
| Adult solid tumor | Cisplatin, gemcitibine | ||
| Malignant solid tumors | Carboplatin, paclitaxel | ||
| BRCA1/BRCA2-associated or hereditary metastatic or unresectable breast and/or ovarian cancer | Carboplatin | ||
| Advanced solid tumors | Doxil | ||
| BRCA1-positive advanced ovarian cancer | Doxil | ||
| Known BRCA or recurrent high-grade ovarian cancer or known BRCA/triple-negative breast cancer | None | ||
| Breast neoplasms | None | ||
| Ovarian neoplasms | None | ||
| Ovarian neoplasms, BRCA1/2 | None | ||
| Malignant solid tumors | Topotecan | ||
| Pancreatic neoplasms | Gemcitabine | ||
| Melanoma neoplasms | Dacarbazine | ||
| Advanced solid tumors | Bevacizumab | ||
| PARP | AG014699 (Cancer Research UK in collaboration with Pfizer) | Locally advanced or metastatic breast cancer or advanced ovarian cancer, BRCA1/2 mutation carrier | None |
| Ape1 | TRC102/methoxyamine (Tracon Pharmaceuticals) | Neoplasm | Pemetrexed |
Targeting the base excision repair pathway
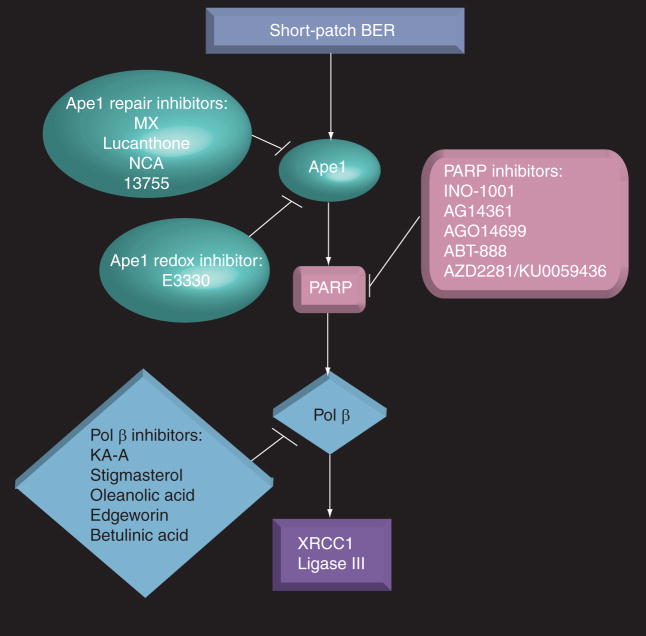
The BER pathway repairs alkylated, oxidative and IR-induced damage. There are two branches of the BER pathway, long-patch and short-patch BER, which will be discussed here. Alkylated or oxidative damage can cause single-base lesions or adducts. The BER pathway is initiated when DNA glycosylases specific to each type of lesion recognize and remove the damaged base. This generates an abasic or apurinic/apyrimidinic (AP) site. Then apurinic/apyrimidinic endo-nuclease (Ape1) processes the AP site by incising the phosphodiester backbone 5′ to the AP site. This creates a 3′-OH and a 5′-deoxyribose phosphate (dRP) end. DNA polymerase β (pol β) then inserts the correct base at the 3′-OH end and removes the 5′ dRP flap. Then DNA ligase III/XRCC1 (ligase III/x-ray complementing factor 1) seals the remaining nick (steps of BER pathway reviewed in [1,3,6,9,18]). If proteins involved in this pathway are deficient or inhibited in cancer cells and DNA damage occurs that these proteins are involved in repairing, the damage will not be processed correctly. Incomplete repair leads to cell death [1,9]. One of the mechanisms that account for this increase in cell death is the generation of double-strand breaks (DSBs) from single-strand breaks (SSBs). SSBs could occur on opposing sides of the phosphodiester backbone of DNA to create DSBs, which are lethal and often cause the induction of apoptosis in cells [4,10]. This makes inhibition of proteins involved in this pathway a desirable target in order to sensitize cells to chemotherapeutics and IR that cause DNA damage repaired by this pathway (Figure 1).
Figure 1. Inhibitors of short-patch base excision repair proteins and proteins that interact with this pathway.
Ape1: Apurinic/apyrimidinic endonuclease; BER: Base excision repair; KA-A: Kohamaic acid A; Ligase III: DNA ligase III; MX: Methoxyamine; NCA: 7-Nitroindole-2-carboxylic acid; PARP: Poly(ADP-ribose) polymerase; Pol β: DNA polymerase β; XRCC1: X-ray complementing factor 1.
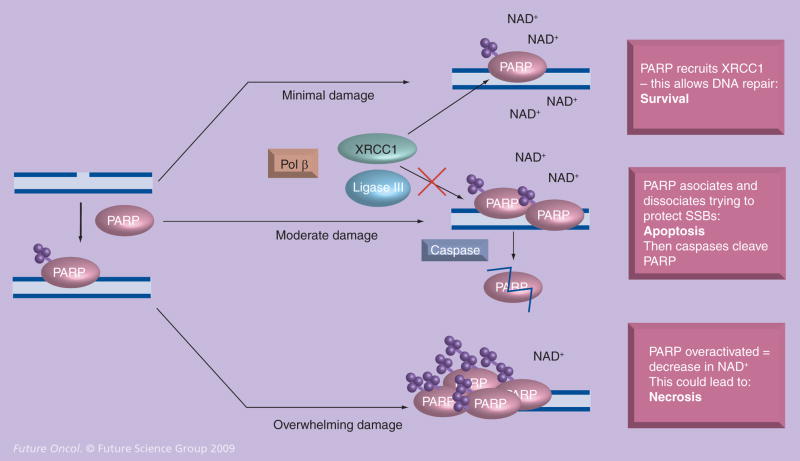
The occurrence of DSBs activates the poly(ADP-ribose) polymerase (PARP) protein, another protein involved in BER. PARP has two zinc-finger motifs in its N-terminal region that bind DNA at strand breaks. This binding activates the C-terminal end of PARP to synthesize, via the catalysis of nicotinamide adenine dinucleotide (NAD+), chains of poly(ADP-ribose)- or PAR-branched chains. The central domain of PARP, the automodification domain, is an acceptor of PAR-branched chains. Due to this automodification, PARP disassociates from the DNA [19–21]. If there is moderate damage to the DNA, PARP interacts with cell cycle checkpoint proteins and BER proteins, such as pol β, proliferating cell nuclear antigen (PCNA), ligase III and XRCC-1, to promote DNA repair, and thus cell survival. However, if the DNA damage present is acute, apoptosis can be activated, during which caspases cleave PARP. Overwhelming DNA damage can cause PARP to become overactivated and deplete NAD+, leading to cell necrosis (Figure 2) [3,22,23].
Figure 2. Proposed mechanism for PARP as a sensor to single-strand breaks.
When there are a minimal number of SSBs, PARP interacts with XRCC1, pol β and DNA ligase III to allow DNA repair. When there is moderate DNA damage, PARP associates and disassociates with the DNA, trying to protect the SSBs until they can be repaired. If the pathway is too overwhelmed, apoptosis is signaled and caspases come in to cleave PARP. If there is overwhelming damage, PARP is overactivated and depletes NAD+. This could lead to necrosis.
Ligase III: DNA ligase III; PARP: Poly(ADP-ribose) polymerase; Pol β: DNA polymerase β; SSB: Single-strand break; XRCC1: X-ray complementing factor 1.
PARP-1 knockout mice, upon first glance, appeared to be phenotypically normal. However, when the mice were challenged with DNA damage, such as that caused by IR or a standard DNA methylating agent, they were found to be extremely sensitive to these agents [19]. We begin our discussion of BER inhibitors currently being developed with PARP, as the majority of recently published data, as well as clinical trial development, focuses on PARP inhibitors.
PARP inhibitors
There has been a great hastening in recent years by pharmaceutical companies to develop highly-specific, clinically relevant PARP inhibitors. This has propelled PARP inhibitors quickly into clinical trials [17]. PARP inhibitors are one of the most promising classes of compounds for cancer therapeutics currently in development. Initial in vitro and in vivo studies indicate that adding minimally toxic levels of the new generation of very specific PARP inhibitors to existing chemotherapeutics (especially alkylating drugs, platinating drugs and topoisomerase I poisons) and IR dramatically increases sensitization of cancer cells and xenografts to the chemotherapeutic agent or IR. Perhaps most exciting, PARP inhibitors have also been able to inhibit the growth of BRCA1- and BRCA2-deficient cells and tumors selectively, while BRCA+/+ and BRCA+/− cells do not appear to be as sensitive to PARP inhibition [24,25]. BRCA1- and BRCA2-deficient cancers are some of the most difficult cancers to treat. The majority of inhibitors that are targeted at BER and have entered the clinic are designed to inhibit PARP (for a list of some of these inhibitors, see Table 1). The following five PARP inhibitors will be reviewed: INO-1001, AG14361, AG014699, ABT-888 and AZD2281 (formerly KU-0059436) (see Figure 1 ).
This is not a comprehensive review of all PARP inhibitors in development, nor will all of the PARP inhibitors reviewed here go any further in development. Rather, these inhibitors were chosen to highlight the power, promise and mechanism behind inhibition of PARP, a DNA repair protein, as a tool to fight cancer. Additionally, there are other promising PARP inhibitors, such as BiPar Science’s (CA, USA) BSI-201, which is currently in several clinical trials [101]. However, this and other inhibitors will not be reviewed as there are no peer-reviewed articles available, only abstracts from meetings. PARP inhibitors in this review that are currently in clinical trials are listed in Table 1 [101].
INO-1001
A PARP inhibitor, INO-1001, discovered by Inotek Pharmaceuticals (MA, USA), but now owned by Genentech (CA, USA), has just completed a Phase II study looking at its ability to minimize the damage caused to heart tissue and blood vessels as a result of potentially elevated levels of PARP after angioplasty. Although currently not in a clinical trial for cancer, three pre-clinical studies with INO-1001 indicate it may also have the ability to potentiate various cancer treatments [26–28].
The first study was performed on three Chinese hamster ovary (CHO) cell lines testing the ability of INO-1001 to potentiate the cytotoxicity caused by IR. A PARP-1 activity assay was performed on CHO cells and demonstrated that 95% inhibition of PARP-1 activity occurred using 10 μM INO-1001, a dose that was nontoxic to the cells as measured by colony assay. This dose was also able to enhance the sensitivity of CHO cells to IR. Brock et al. further demonstrated that doses of INO-1001 up to 100 μM did not result in a dramatic effect on cell survival [26].
The combinination of PARP inhibitors, including INO-1001, with the methylating agent temozolomide is another potential use. Temozolomide (Temodar®) is an alkylating agent currently used in combination with IR to treat patients with glioblastoma multiforme and patients with refractory anaplastic astrocytoma [29]. Temozolomide methylates DNA primarily at the N7 and O6 positions of guanine and the N3 position of adenine and BER is the primary pathway to repair these lesions [30]. The effectiveness of temozolomide is thought to depend on the O6-alkylguanine DNA methyltransferase (AGT) and the MMR status of the tumor. Cells that have high levels of AGT are able to efficiently remove the most lethal of the lesions caused by temozolomide, O6-methylguanine, allowing them to resist temozolomide cytotoxicity [31]. Unfortunately, cancer cells with normal to low levels of AGT can still develop resistance to temozolomide due to deficient MMR. Without repair of the O6-lesion by AGT, MMR exacerbates the effects of O6-methylguanine lesions caused by temozolomide. Unrepaired O6-methylguanine lesions are paired with thymine if allowed to undergo replication. MMR is recruited to fix the mismatch. However, it removes the thymine opposite the damaged guanine, then the incorrect base, thymine, is once again inserted. This futile attempt at repair can lead to an accumulation of SSBs during S-phase, leading to the signaling of programmed cell death when the lesions are too overwhelming or cannot be repaired [27]. Conversely, cells with MMR deficiency that have accumulated normally toxic levels of O6-methylguanine lesions do not undergo this futile attempt at repair and are sometimes allowed to escape death [31,32].
INO-1001 was used to partially overcome temozolomide resistance in MMR-deficient malignant glioma xenografts [27]. In this study exploring temozolomide resistance, the authors first looked at PARP-1 levels in a MMR-deficient medulloblastoma cell line after treatment with temozolomide. They found that PARP-1 activity increased after treatment, but this increase could be abrogated with the pretreatment of INO-1001. They then went on to perform an in vivo study with MMR-deficient malignant glioma tumor xenografts using temozolomide in combination with INO-1001. Some increased toxicity was observed in the mice that were treated with both temozolomide and INO-1001. This increased toxicity was most likely due to the additional lesions caused by temozolomide, N3-methyladenine and N7-methylguanine. Blocking PARP with INO-1001 would prevent the involvement of BER in the repair of these lesions, allowing accumulation of SSBs. Although the temozolomide resistance was not entirely overcome in the xenografts, there was a growth delay of 13.9–25.8 days [27].
The PARP inhibitor INO-1001 was used in a third study to potentiate the effect of doxorubicin treatment on p53-deficient tumors created using the breast cancer cell line, MDA-MB-231, and the murine mammary carcinoma, MCa-K [28]. More than 50% of tumors have defective p53. Cell cycle arrest, caused by p53, is important to DNA repair in that it allows the cells to repair damage before they re-enter the cell cycle. Defective p53 causes the cells to fail to arrest their cell cycle long enough to repair the DNA damage. This allows the damage to be perpetuated through cell cycling, often causing the initiation of apoptosis [33]. The primary mechanisms of action of doxorubicin are blocking DNA replication via intercalation of DNA and inhibition of topoisomerase II (topo II), which can lead to DSBs and apoptosis. Additionally, it has been proposed that toxic levels of reactive oxygen species (ROS) may be generated as a derivative of doxorubicin treatment, but this is observed only at very high therapeutic levels [34]. The authors of this study reported that the combination of doxorubicin and INO-1001 had a synergistic effect on p53-deficient tumor growth rate as measured by tumor growth after treatment [28]. Unfortunately, the study included p53-deficient tumors, but no wild-type tumors.
AG14361
According to Calabrese et al., the PARP inhibitor AG14361, a compound produced by Pfizer (NY, USA), is over 1000-times more potent than 3-aminobenzamide (3-AB), one of the earliest PARP inhibitors, at inhibiting PARP activity [35]. They demonstrated that AG14361 was able to inhibit 85% of PARP activity at 0.4 μM without growth rate or cytotoxic effects in two colorectal cancer cell lines, MMR-deficient LoVo and MMR-proficient SW620, and a non-small-cell lung cancer cell line, A549 [35]. AG14361 was able to potentiate the chemotherapeutic effects of temozolomide in the LoVo and A549 cell lines, but not the MMR-proficient SW620 cell line. Furthermore, AG14361 potentiated the cytotoxic effect when in combination with topotecan, a topoisomerase I inhibitor, in all three cell lines, although not as dramatically as the potentiation with temozolomide in LoVo cells. The growth of LoVo cells treated with γ-irradiation in addition to AG14361 did not recover as quickly as cells that were only irradiated. Results with γ-irradiation were not reported in the other two cell lines for this portion of the experiment. As part of the same study, in vivo experiments were performed using xenografts with LoVo and SW620 cells. The combination of temozolomide and a dose of AG14361 that itself did not affect tumor growth was able to cause significant growth delay as compared with the temozolomide alone in the MMR-deficient xenografts, and complete regression of the MMR-proficient xenografts. The authors attributed this change in outcome for the SW620 versus the in vitro experiments to the effect of AG14361 on the tumor microenvironment. Tumor growth delay was also significantly potentiated by AG14361 in combination with IR in the MMR-deficient LoVo xenografts and in both types of xenografts when combined with irinotecan, a topoisomerase I (topo I) inhibitor. The combination of IR and AG14361 was not used in the SW620 xenograft [35].
The mechanism for the potentiation of topo I poisons, such as topotecan and camptothecin, was elucidated in a study using cells from both PARP-1 wild-type mice and PARP knockout mice [36]. Cells from PARP-1 knockout mice were three times more sensitive to topotecan. Sensitization of cells from wild-type mice identical to that seen in the cells without PARP-1 was achieved by adding AG14361 to the topotecan. This confirmed that PARP-1 was an important player in protecting cells from topo I poisons and demonstrated the specificity of AG14361 for PARP-1. Smith et al. also used XRCC1-, DNA-dependent protein kinase catalytic subunit- and XRCC3-deficient CHO cell lines (EM9, V3 and irs1SF, respectively), along with their parental cell line, AA8, to test the effect of AG14361 on camptothecin-induced cytotoxicity in DNA repair-deficient cells as compared with the DNA repair-proficient parental cell line. They wanted to investigate the involvement of PARP-1 with other DNA repair proteins/pathways in response to camptothecin. All three DNA repair-deficient cell lines were significantly more sensitive to camptothecin alone as compared with the parental cell line. The HR-deficient cell line (irs1SF) was tenfold more sensitive to the camptothecin, while the BER- and NHEJ-deficient cell lines (EM9 and V3) were five- and 1.5-fold more sensitive. A significant potentiation of camptothecin cytotoxicity was observed when combined with AG14361 in both the parental and NHEJ-deficient cell lines, but not in the BER-deficient cell line. The HR-deficient cell line, irs1SF, was hypersensitive to AG14361 as a single agent, making it difficult to determine if camptothecin would be further potentiated with the PARP inhibitor [36]. A later study also found that HR-deficient cells were hypersensitive to AG14361 alone [37].
Based on the fact that AG14361 did not potentiate camptothecin-induced sensitivity in the BER-deficient cell line but did in the cell lines deficient in other repair pathways, the authors proposed the following possible mechanism. The proposed mechanism through which this PARP inhibitor potentiates camptothecin cytotoxicity is inhibition of BER. In this mechanism, topo I poisons would cause SSBs and form a cleavable complex with the 3′ phosphate end of the DNA. PARP-1, in turn, would bind to the 5′OH end of DNA. PARP-1 would then undergo automodification and recruit XRCC1. The XRCC1 would then recruit tyrosyl DNA phosphodiesterase-1 (TDP-1), which would remove the topo I and create a 3′ OH end that would be converted to a 5′ phosphate by polynucleotide kinase (PNK), also recruited by XRCC1. The final chore for the XRCC1 would be to act as a scaffolding protein allowing pol β to fill in the gap and ligase III to ligate the gap [36]. The EM9 cells utilized here are XRCC1 deficient, and would therefore not be able to perform the actions described above. In the absence of XRCC1, PARP inhibitors could not enhance camptothecin-induced cytotoxicity, underscoring the importance of PARP/BER interactions.
In response to IR, PARP-1 is involved in upregulating NF-κB (NF-κ-light-chain-enhancer of activated B cells) activity [38]. Studies were performed with mouse embryonic fibroblasts (MEFs) that were either proficient or deficient in NF-κB [39]. Veuger et al. knocked NF-κB down by transfecting the cells with small interfering RNAs (siRNA). AG14361 was able to sensitize the cells proficient in NF-κB, but not the cells deficient in NF-κB, to IR. These results indicated that PARP signaling through NF-κB activity is important following IR-induced cell death [39].
Most interestingly, AG14361 was used successfully as a single agent in BRCA2-deficient cells and tumors [24]. Patients who have inherited a BRCA1 or BRCA2 mutation on one allele have a higher risk of developing ovarian or breast cancer, along with other cancers, because if the remaining functional allele mutates to a nonfunctional form, cells with the deficient BRCA1 or BRCA2 have genomic instability that can result in tumor development [40]. BRCA1- and BRCA2-deficient cells are deficient in HR [41]. This study used the PARP inhibitor AG14361, along with other PARP inhibitors, to take advantage of the HR defect that selectively targets the BRCA2-deficient cells and BRCA-2-deficient tumors from the cells and tumors that have functioning BRCA2. First, the authors tested the hypothesis that HR-deficient cells would not be able to withstand the amount of DNA damage incurred in the absence of PARP activity. Using CHO cell lines that were deficient in HR, they treated the XRCC2-deficient (irs1) cells and XRCC3-deficient (irs1SF) cells with the PARP inhibitors 3-AB, 1,5-dihydroxyisoquinoline (ISQ) and AG14361. The HR-deficient cells were sensitive to the PARP inhibitors and the sensitivity was reduced when XRCC2 and XRCC3 were added back to the cells, thereby restoring their HR function. Small, interfering RNAs were used to knockdown the expression of BRCA2 in two breast cancer cell lines, one with wild-type p53 (MCF7) and one with mutated p53 (MDA-MB-231). The transfected cells were then treated with AG14361 and another PARP inhibitor, NU1025. Colony assays demonstrated a significant decrease in the colony formation from AG14361- and NU1025-treated cells in which the BRCA2 was knocked down as compared with the cells with normal levels of BRCA2, regardless of p53 status. Lastly, the authors inoculated mice with BRCA2-deficient V-C8 cells or BRCA2-complement cells, V-C8 + B2, to form xenografts, then treated the mice with AG14361. AG14361 did not slow the growth of the xenograft in the tumor line that expressed wild-type BRCA2. However, three out of five of the BRCA2-deficient xenografts showed a response to AG14361, with one tumor appearing to disappear completely [24]. This was one of two studies published concurrently in the journal Nature showing a great effect of PARP inhibitors alone on BRCA1- and BRCA2-deficient cells and tumors [24,25].
AG014699
AG014699 is a PARP inhibitor that was developed in a collaboration between Agouron Pharmaceuticals (CA, USA, a division of Pfizer), Cancer Research UK and Newcastle University (UK) [42]. It is the first PARP inhibitor to enter into a clinical trial. AG014699 is the phosphate salt of a derivative of AG14361 (AG14447), which was discussed above [43]. According to the clinicaltrials.gov website [101], there is one current clinical trial of this drug in advanced breast or ovarian cancer with BRCA1 or BRCA2 mutations (Table 1). In a previous clinical trial for AG014699, patients with various malignancies were given temozolomide and AG014699 to determine the best doses for the combination. They determined that the PARP inhibitor was able to be administered in doses that gave no symptomatic toxicities with the inhibitor alone and at levels that demonstrated inhibition of PARP in the tumor. Additionally, patients were able to tolerate the full dose of temozolomide in addition to AG014699. Patients with metastatic melanoma and a desmoid tumor showed responses that ranged from partial to one complete response. Other patients with melanoma, prostate cancer, pancreatic cancer and leiomyosarcoma experienced some stabilization after treatment with the combination [42].
Another study demonstrated that AG014699 was able to potentiate the effect of topotecan and temozolomide in neuroblastoma cell lines at levels that produced a greater than 97% PARP activity inhibition with no changes in cell growth or toxicity in the AG014699-only treated cells. There was an AG014699-induced enhancement of growth delay in two types of neuroblastoma xenografts treated with temozolomide. A tumor growth delay was also observed in a neuroblastoma xenograft experiment when the mice were treated with topotecan in combination with AG014699. In summary, AG014699 potentiated the growth delay of both temozolomide and topotecan in neuroblastoma xenografts using amounts of inhibitor that had little added toxicity or growth delay as a single agent [44].
ABT-888
ABT-888, a cyclic amine-containing benzimidazole carboxamide PARP inhibitor discovered by Abbott Laboratories (IL, USA), is currently in 11 clinical trials (Table 1) [101]. In one study, researchers demonstrated that this PARP inhibitor had the ability to potentiate temozolomide in a mouse melanoma model and rat glioma model. The same study also demonstrated that ABT-888 potentiated cisplatin, carboplatin and cyclophosphamide in a BRCA1-deficient breast carcinoma xenograft model [45]. ABT-888 has also been shown to potentiate IR in a human colon cancer cell line [45] and a lung cancer cell line, along with tumor growth delay in a lung cancer xenograft model [46]. ABT-888 has a long half-life that allows it to remain in the cells longer and convert SSBs to more lethal DSBs [47]. Acquisition of temozolomide resistance is an ongoing issue in the treatment of cancer [31]. Unfortunately, temozolomide-resistant tumor lines created via gradual glioblastoma xenograft exposure to temozolomide were unaffected by the addition of ABT-888, whereas the temozolomide effect in previously unexposed glioblastoma xenografts was potentiated by ABT-888 [48]. This indicates the possibility that only glioblastoma patients without previous exposure to temozolomide will be able to benefit from the combination of temozolomide and ABT-888.
AZD2281/KU-0059436
AZD2281 is a PARP inhibitor that was first developed by KuDOS Pharmaceuticals (Cambridge, UK) and given the name KU-0059436 (Figure 1). However, when AstraZeneca (London, UK) acquired KuDOS Pharmaceuticals, the name was changed to AZD2281 [17]. AZD2281 is currently in 20 clinical trials related to cancer treatments (Table 1) [101]. In preliminary studies, AZD2281 was able to significantly delay tumor growth in combination with temozolomide in a colon cancer tumor model, potentiate the effect of methyl methanesulfonate (MMS), an alkylating drug, in colon cancer cells, and as a single agent increase the cytotoxicity in two BRCA1-deficient breast cancer cell lines [49]. Further studies were carried out in BRCA1- and BRCA2-deficient cells. Interestingly, the BRCA2-deficient cell lines showed a high degree of sensitivity to temozolomide alone, indicating that these cells are highly sensitive to DSBs. In the same study, AZD2881 was able to inhibit the growth of BRCA2-deficient cell lines at doses that were minimally toxic to the cells. Additionally, AZD2881 in combination with cisplatin in BRCA2-deficient cells resulted in synergistic effects on cytotoxicity, but no synergy in the BRCA-2-proficient cells [50].
In another study, AZD2281 was able to cause growth arrest and even shrinkage of BRCA1-deficient tumors in mice without any unwanted toxic side effects [51]. However, once the AZD2281 was removed, the tumors began to grow. The tumors were allowed to grow to the size they originally were when the AZD2281 was added, then treated with another course of the PARP inhibitor. After the second exposure to AZD2281, the tumors were no longer sensitive to its effects. The investigators were able to determine the mechanism of this resistance to be P-glycoprotein (P-gp) overexpression. They hypothesized that this resistance could be overcome via the use of tariquidar, a P-gp inhibitor, and are currently testing this hypothesis. AZD2281 was also used in this study to potentiate cisplatin and carboplatin in the treatment of mammary tumors. Although there was an increase in survival when compared with the mice treated only with a platinating agent, this effect was transient. If allowed to grow, the tumors would relapse over time. Additionally, this study ascertained that AZD2281-treated mice were not able to tolerate their usual dosage of cisplatin treatment when AZD2281 was present, even though those levels were nontoxic by themselves [51]. In a third study, AZD2281 at nontoxic levels increased the sensitivity of three out of four glioma cell lines to IR. However, this sensitization with AZD2281 did not occur when cell cycle arrest was induced with aphidicolin. Lastly, the study showed that the repair of the DNA breaks caused by IR was delayed with the addition of AZD2281 [52].
Acquired resistance to PARP inhibitors
Resistances that develop in previously treated tumors is a potential obstacle in the use of PARP inhibitors. In the study by Clarke et al. the PARP inhibitor ABT-888 was not able to overcome temozolomide resistance in glioblastoma xenografts previously exposed to the alkylating agent [48]. Also, BRCA1-deficient xenografts were no longer sensitive to AZD2281 used as a single agent in xenografts created from the cells of previously exposed xenografts [51]. A paired study in Nature elucidates a discovered mechanism of acquired cisplatin and PARP inhibitor resistance. As previously described, BRCA2-deficient tumors are sensitive to PARP inhibitors, while wild-type BRCA2 tumors have limited, if any, sensitivity to PARP inhibitors. These investigators found that previous exposure of tumors to cisplatin or PARP inhibitors sometimes caused secondary mutations in BRCA2 that could create a frameshift in the open reading frame of BRCA2. This frameshift often reverted the BRCA2-deficient tumor to a wild-type or novel functional form of BRCA2 that was resistant to cisplatin and PARP inhibitors. This secondary mutation and resultant acquired resistance was able to be predicted by the restored ability of tumor cells to form RAD51 foci after DNA damage induced by IR [53]. In response to DNA damage, wild-type BRCA2 interacts with RAD51 and localizes RAD51 to the site of DSBs to allow repair via HR [55]. Edwards et al. proposed that a possible way to overcome the acquired resistance would be to prevent HR-mediated DSB repair by treating patients with proteasome inhibitors the would prevent the recruitment of RAD51 by BRCA2 [53].
In summary, the PARP inhibitors reviewed here (Figure 1) have the ability to enhance alkylating agents, platinating agents, topoI poisons and IR in a variety of cell lines and xenografts. Some of the PARP inhibitors were efficacious against BRCA1-deficient cell lines and BRCA2-deficient cell lines and xenografts as a single agent [24,25]. One study showed that PARP inhibitors were more effective in potentiating the activity of an alkylator, a topoI poison and IR in MMR-deficient cell lines and xenografts, as compared with those that are MMR-proficient [35]. The mechanism of potentiation by PARP inhibitors was demonstrated to be dependent, at varying levels, on the activity of the BER and the HR pathways, and was validated using several of the PARP inhibitors reviewed here [24,36,37], but no dependence upon p53 status was established [28]. We demonstrated that some of the PARP inhibitors were dependent on the BER pathway for the potentiation of the effect of various drugs and IR. In the following sections we explore what happens when we inhibit other components of the BER pathway.
Ape1 DNA repair & redox inhibitors
Ape1 is a critical component in the BER pathway that is able to process AP sites for repair that were created as a result of the action of DNA glycosylases on single base lesions [1,9]. Methoxyamine is an alkoxyamine derivative able to interact with, and thereby block, AP sites created by DNA glycosyases removing a damaged nucleotide [56]. The interaction between methoxyamine and the AP site is very strong. It prevents the lyase activity of Ape1 endonuclease cleavage and pol β – downstream members of the BER pathway (reviewed in [1,5,9,18]). Methoxyamine, or TRC102, which is produced by Tracon Pharmaceuticals (CA, USA), is currently being used in a clinical trial in combination with pemetrexed, a folate antimetabolite, in advanced solid cancers (Table 1). Methoxyamine has sensitized a wide variety of cancer cell lines to temozolomide and other alkylating chemotherapeutic agents [29,57–59]. It has recently been shown that the methoxyamine-bound AP sites created by the combination of temozolomide and methoxyamine treatment can act as topo II poisons, as it is often located on the preferential cleavage site of topo II. Topo II is an enzyme that cuts both strands of DNA, allowing it to unwind. Sabourin et al. suggested the possibility that the methoxyamine-bound AP site complexes with topo II, thereby prohibiting it from fully functioning and completing the religation step. This would cause a further induction of topo II, resulting in greater amounts of cleavage, and therefore cytotoxicity. An alternate explanation by the authors was that the methoxyamine-bound AP sites could be blocking replication, causing induction of more topo II [60]. Some cancer cells have elevated levels of topo II, while normal tissues tend to have lower levels of topo II [61]. This would be promising for the selectivity of this treatment to cancer cells.
Recently there were a few reports of the discovery of direct inhibitors of the endonuclease activity of Ape1, including lucanthone and 7-nitroindole-2-carboxylic acid (NCA) (Figure 1). Lucanthone was able to potentiate the effects of MMS and temozolomide in breast cancer cells [62] and IR in patients with brain metastasis [63], but is not considered to be highly useful clinically due to concern regarding its off-target effects [1]. NCA has been reported to be able to potentiate the cytotoxicity of MMS, temozolomide and other chemotherapeutics in cancer cells [64]. However, others have reported that this agent is less promising as a lead candidate, and levels required for Ape1 inhibition have been reported to be in the high μM range [1,65].
Discovery of new small-molecule inhibitors of the endonuclease (repair) function of Ape1 have been reported [1,66,67]. On of these small-molecule Ape1 inhibitors is the arylstibonic acid compound 13755, identified via a high-throughput screening methodology (Figure 1). 13755 was able to decrease the repair activity of Ape1, but could not potentiate the effect of a classic alkylating agent, MMS, in a human osterogenic sarcoma cell line [67]. A group from the University of Southern California (CA, USA) used a pharmacophore-guided method to discover potential candidates that would inhibit Ape1 activity. Although these compounds were found to be specific to Ape1, more soluble derivatives will need to be discovered for them to be used clinically [66]. Our laboratory is using the high-throughput screening methodology in order to screen a library of compounds. A total of 45 compounds that were shown to be able to inhibit the DNA repair activity of Ape1 with more activity than previously shown with NCA are currently being analyzed further [1].
In addition to the DNA repair activity of Ape1, it is active in redox signaling. Ape1 reduces, thereby activating, various transcription factors, leading to transcription of genes that are important in cancer advancement and cell survival [1,6]. (2E)-3-(5-[,3-dimethoxy-6-methyl-1,4-benzoquinoyl])-2-nonyl-2-propenoic acid (E3330) blocks the redox function of Ape1 (Figure 1). Our laboratory performed a series of studies with E3330 and demonstrated that E3330 inhibited the redox function of Ape1 without inhibiting the repair function. In addition, E3330 decreased cell survival in several cancer cell lines as a single-agent at doses that caused no cell killing in human CD34+ (normal) cells (Reed AM, Fishel MK, Kelley MR. Unpublished Data). E3330 was able to inhibit angiogenesis, measured using a Matrigel™-based tube-formation assay, of endothelial cells using sub-cytotoxic doses (<5 μM) [6]. In one study, E3330 was able to inhibit growth and migration of pancreatic cancer cell lines [68]. Although the details of the mechanism of how E3330 is affecting angiogenesis and migration are still under investigation, the redox function of Ape1 is a novel and interesting target to pursue in the treatment of cancer.
Pol β inhibitors
Although still in the preclinical setting, it is worth mentioning that inhibitors of pol β have been discovered and are being investigated. Oleanolic acid, edgeworin, betulinic acid, stigmasterol and kohamaic acid A (KA-A) all inhibit pol β (Figure 1). Pol β is the predominant polymerase in short-patch BER, and functions in long-patch BER as well. In addition to its polymerase function in BER, the 5′ dRPase activity is also important for completion of repair. KA-A, isolated from fertilized sea urchin eggs, and its derivatives were able to prevent growth of a promyelocytic leukemia cell line [69]. In one study, oleanolic acid, edge-worin, betulinic acid and stigmasterol were all able to potentiate bleomycin, which is thought to induce strand breaks by intercalating the DNA and not allowing thymidine incorporation, in carcinomic human alveolar basal epithelial cells. In the same study, stigmasterol was only able to inhibit the removal of the dRP by pol β which is left after processing by Ape1, while the remaining three inhibitors were able to inhibit both the lyase activity and ability of pol β to insert the correct base [70].
Conclusion
The DNA repair inhibitors reviewed in this article demonstrate the ability of these agents to work in a wide variety of cell lines and in combination with numerous existing chemotherapeutic agents and IR. This is important, as it is doubtful that chemotherapeutics or IR will be replaced as front-line therapies in the near future. It is becoming more evident that combination therapy with rational targets is showing promise in preclinical and clinical studies. Therefore, adding agents that enhance current front-line treatments to increase the therapeutic index and reduce acquired tumor cell drug resistance would dramatically enhance cancer therapeutic efficacy sooner rather than later. The most successful inhibitors reviewed had some commonalities:
Some inhibitors were able to highly inhibit the activity (>90% inhibition) of their target at doses that caused minimal toxicity to the cell lines or xenografted mice, except BRCA1- and BRCA2-deficient cells and xenografts, which showed significant cell growth delay with the treatment of some PARP inhibitors.
As low levels of the inhibitors could be used to obtain significant inhibition of activity, the inhibitors could often dramatically potentiate the growth delay effect of chemotherapeutic agents and IR in xenografts, with little increased toxicity to the mice. However, it should be reiterated that the agents potentiated by PARP are not all thought of as ‘BER agents’, indicating cross-talk between DNA repair pathways as well as PARP.
The preclinical in vitro and in vivo studies demonstrated some exciting potentiation of cancer cell treatments. The results of the ongoing clinical trials will be revealing for the fate of these inhibitors and inhibitors of the same genre that are currently in the preclinical pipeline.
Executive summary
PARP inhibitors
INO-1001 potentiated the effects of irradiation (IR) in CHO cells and doxorubicin in p53-deficient xenografts. INO-1001 also helped mismatch repair (MMR)-deficient xenografts partially overcome temozolomide resistance.
AG14361 potentiated the effects of temozolomide in MMR-deficient but not MMR-proficient cell lines, and in MMR-deficient and -proficient xenografts. AG14361 also potentiates the effect of IR in MMR-deficient and -proficient cell lines along with MMR-deficient xenografts.
Potentiation in combination with irinotecan was demonstrated in MMR-deficient and proficient xenografts.
AG14361 had no effect in IR-treated cells deficient in NF-κB.
Proficient base excision repair was needed to enhance camptothecin cytotoxicity in cell lines.
AG14361 was successfully used as a single agent to induce cytotoxicity and growth delay in BRCA1−/− cell lines and BRCA2−/− cell lines and xenografts. The same effect was not demonstrated in wild-type BRCA2 or BRCA2+/− cell lines or xenografts.
AG014699 was able to potentiate the effects of temozolomide and topotecan in neuroblastoma cell lines and xenografts. One patient in a clinical trial using temozolomide in combination with AG014699 showed a complete response. Other patient responses across a variety of cancer types ranged from partial response to some stabilization. The doses of AG014699 used demonstrated no symptomatic toxicities. Additionally, the patients were all able to tolerate full doses of temozolomide in combination with the AG014699.
ABT-888 was able to potentiate the cytotoxic effects of temozolomide, cisplatin, carboplatin and cyclophophamide in BRCA1-deficient breast carcinoma xenografts. ABT-888 was also able to potentiate the cytotoxic effects of IR in colon cancer cell lines and lung cancer cell lines and a lung cancer xenograft model. However, ABT-888 was not able to overcome temozolomide resistance in glioblastoma xenografts previously exposed to temozolomide.
AZD2281 was able to potentiate the effects of temozolomide in a colon cancer tumor model and methyl methanesulfonate (MMS) in a colon cancer cell line. AZD2281 was able to potentiate tumor growth delay in cisplatin- and carboplatin-treated mouse mammary tumors, but the effect was transient. AZD2281 was able to increase the sensitivity of some glioma cell lines to IR unless the cells were not replicating. AZD2281 caused cytotoxicity as a single agent in BRCA1-deficient cells and BRCA2-deficient cells. However, mammary tumors were able to develop resistance to AZD2281 over time.
Ape1 DNA repair & redox inhibitors
Methoxyamine potentiates the effects of temozolomide and other alkylating agents in several cancer cell lines. Topoisomerase II may be involved in the potentiation of the temozolomide effect by methoxyamine.
Lucanthone potentiates the effects of MMS, temozolomide and IR, but has too many off-target effects to be clinically relevant.
7-nitroindole-2-carboxylic acid (NCA) potentiates the effects of MMS and temozolomide, but other laboratories have not been able to demonstrate this effect. NCA is not a very viable, clinically relevant lead agent.
The arylstibonic acid compound 13755 can inhibit Ape1 repair activity, but was not able to potentiate MMS in HOS cells.
E3330 inhibits redox function of Ape1, but not repair function. E3330 causes single-agent cytotoxicity to several cancer cell lines, but not CD34+ (normal) cells at the same doses. The E3330 redox inhibitor inhibits growth and migration of pancreatic cancer cell lines and angiogenesis in a variety of endothelial cells at subtoxic doses.
Pol β inhibitors
KA-A and its derivatives, isolated from fertilized sea urchin eggs, specifically inhibit mammalian pol β, and KA-A prevented growth of a leukemia cell line.
Stigmasterol inhibits only the lyase activity of pol β.
Oleanolic acid, edgeworin and betulinic acid inhibit both the lyase and polymerase activity of pol β.
Future perspective
The long history and advances in the understanding of the basic science of DNA repair pathways has allowed us to better develop rationales for combinational treatments to potentiate tumor-cell killing. For example, with the knowledge that temozolomide generates lesions that are repaired by AGT and BER, we can thoughtfully pair AGT and BER inhibitors with this agent. This could widely expand the range of cancers that could be treated with temozolomide, where previous data would suggest temozolomide would not work. Hopefully, further elucidation of DNA repair pathways and their role in cancer versus normal cells will reveal many new potential targets for inhibition to potentiate tumor-cell response. Small-molecule inhibitor discovery is an intense and expensive process. New methods and strategies will certainly be developed to try to both streamline this discovery process and ensure that the end products will have the desired inhibitory effect, be soluble and deliverable. It may possibly even be important that future inhibitors contribute minimal toxicity to patients if they need to be used in combination with agents or IR that already cause toxicity to the patient. Several of the studies reviewed here demonstrated dramatic in vitro and in vivo potentiation of existing chemotherapeutic agents and IR when used in combination with BER-pathway inhibitors using an amount of chemotherapeutic agent or IR that as a single agent was minimally toxic to the cell lines or xenografts.
Another important area for the use of these and future inhibitors will be the ability to test for biomarkers. Some biomarkers, such as RAD51 and H2AX-γ foci formation levels in peripheral blood mononuclear cells before and after treatments to predict or understand tumor response and DNA repair phenotype linkage with genotypes, are already being employed in clinical trials [11,101]. The hope is that cancer therapeutics will become more individualized in the near future. Once diagnosed, biopsies or samples may be taken and sent to the laboratory to screen for hundreds of biomarkers that will better allow us to predict how the tumor would react to different treatments. It will be important to develop the tests that would quickly and accurately predict these biomarkers. As we learn more regarding cancer-cell signaling and the mechanisms they use to escape cell death and thrive, we will be better able to predict combinations and treatments that will be successful and allow us to elude the development of resistance in tumors. Alternatively, if resistance does arise due to cross-talk or compensation between not only the DNA repair pathways, but also between DNA repair, cell cycle, signaling and other important pathways, it will be important to have a molecular profile of the individual cancer to help us overcome resistance.
Acknowledgments
Financial & competing interests disclosure
Financial support for this work was provided by the National Institutes of Health, National Cancer Institute CA122298 to Melissa L Fishel, CA94025, CA106298, CA114571 and CA121168 to Mark R Kelley, IU Simon Cancer Center Translational initiative pilot funding (ITRAC) to Mark R Kelley and the Riley Children’s Foundation to Mark R Kelley. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Contributor Information
April M Reed, Department of Pediatrics (Section of Hematology/Oncology), Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, 1044 W. Walnut, Room 302C, Indianapolis, IN 46202, USA.
Melissa L Fishel, Department of Pediatrics (Section of Hematology/Oncology), Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, 1044 W. Walnut, Room 302C, Indianapolis, IN 46202, USA.
Mark R Kelley, Department of Pediatrics (Section of Hematology/Oncology), Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, 1044 W. Walnut, Room 302C, Indianapolis, IN 46202, USA Tel.: +1 317 274 2755 Fax: +1 317 278 9298 mkelley@iupui.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Bapat A, Fishel ML, Kelley MR. Going Ape as an approach to cancer therapeutics . Antioxid Redox Signal. 2009;11:651–668. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer . Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Damia G, D’Incalci M. Targeting DNA repair as a promising approach in cancer therapy. Eur J Cancer. 2007;43:1791–1801. doi: 10.1016/j.ejca.2007.05.003. ▪ Review of DNA repair pathway targeting with a helpful schematic of DNA repair pathways, with inhibitors listed alongside the proteins to which they are targeted. [DOI] [PubMed] [Google Scholar]
- 4.Belzile J, Choudhury S, Cournoyer D, Chow T. Targeting DNA repair proteins: a promising avenue for cancer gene therapy. Curr Gene Ther. 2006;6:111–123. doi: 10.2174/156652306775515538. [DOI] [PubMed] [Google Scholar]
- 5.Madhusudan S, Middleton MR. The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer . Cancer Treat Rev. 2005;31:603–617. doi: 10.1016/j.ctrv.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Luo M, Delaplane S, Jiang A, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1 . Antioxid Redox Signal. 2008;10:1853. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes . J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 8.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy . Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 9.Kelley MR, Fishel M. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med Chem. 2008;8:417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lord CJ, Garrett MD, Ashworth A. Targeting the double-strand DNA break repair pathway as a therapeutic strategy. Clin Cancer Res. 2006;12:4463–4468. doi: 10.1158/1078-0432.CCR-06-1269. [DOI] [PubMed] [Google Scholar]
- 11.Collins A, Gaivao I. DNA base excision repair as a biomarker in molecular epidemiology studies. Mol Aspects Med. 2007;28:307–322. doi: 10.1016/j.mam.2007.05.005. ▪ Overview of current assays for DNA repair biomarkers. [DOI] [PubMed] [Google Scholar]
- 12.Cho W. Contribution of oncoproteomics to cancer biomarker discovery. Mol Cancer. 2007;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadin SB, Vargas-Roig LM, Drago G, Ibarra J, Ciocca DR. DNA damage and repair in peripheral blood lymphocytes from healthy individuals and cancer patients: A pilot study on the implications in the clinical response to chemotherapy . Cancer Lett. 2006;239:84–97. doi: 10.1016/j.canlet.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Tnetori L, Graziani G. Recent approaches to improve the antitumor efficacy of temozolomide. Curr Med Chem. 2009;16:245–257. doi: 10.2174/092986709787002718. [DOI] [PubMed] [Google Scholar]
- 15.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways . Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 16.Klapman J, Malafa M. Early detection of pancreatic cancer: why, who, and how to screen. Cancer Control. 2008;15:280–287. doi: 10.1177/107327480801500402. [DOI] [PubMed] [Google Scholar]
- 17.Sheridan C. Genentech raises stakes on PARP inhibitors. Nat Biotech. 2006;24:1179–1180. doi: 10.1038/nbt1006-1179. [DOI] [PubMed] [Google Scholar]
- 18.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency . DNA Repair. 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells . Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 21.Aguilar-Quesada R, Munoz-Gamez J, Martin-Oliva D, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition . BMC Mol Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology . Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 23.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery . DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. ▪▪ Uses targeted inhibition to exploit the DNA repair deficiency that allows BRCA2-deficient cancers to be chemoresistant, in order to selectively kill the cancer cells. [DOI] [PubMed] [Google Scholar]
- 25.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. ▪▪ Using targeted inhibition to exploit the DNA repair deficiency that allows BRCA1- and BRCA2-deficient cancers to be chemoresistant in order to selectively kill the cancer cells. [DOI] [PubMed] [Google Scholar]
- 26.Brock WA, Milas L, Bergh S, et al. Radiosensitization of human and rodent cell lines by INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase . Cancer Lett. 2004;205:155–160. doi: 10.1016/j.canlet.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Cheng CL, Johnson SP, Keir ST, et al. Poly(ADP-ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft . Mol Cancer Ther. 2005;4:1364–1368. doi: 10.1158/1535-7163.MCT-05-0128. [DOI] [PubMed] [Google Scholar]
- 28.Mason K, Valdecanas D, Hunter N, Milas L. INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase, enhances tumor response to doxorubicin. Invest New Drugs. 2008;26:1–5. doi: 10.1007/s10637-007-9072-5. [DOI] [PubMed] [Google Scholar]
- 29.Fishel ML, He Y, Smith ML, Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide . Clin Cancer Res. 2007;13:260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 30.Denny B, Wheelhouse R, Stevens M, Tsang L, Slack J. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 31.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 32.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment . Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levesque AA, Eastman A. p53-based cancer therapies: is defective p53 the Achilles heel of the tumor? . Carcinogenesis. 2007;28:13–20. doi: 10.1093/carcin/bgl214. [DOI] [PubMed] [Google Scholar]
- 34.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin . Biochem Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 35.Calabrese CR, Almassy R, Barton S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361 . J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 36.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks . Clin Cancer Res. 2005;11:8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 37.Kyle S, Thomas HD, Mitchell J, Curtin NJ. Exploiting the Achilles heel of cancer: the therapeutic potential of poly(ADP-ribose) polymerase inhibitors in BRCA2-defective cancer . Br J Radiol. 2008;81:S6–S11. doi: 10.1259/bjr/99111297. [DOI] [PubMed] [Google Scholar]
- 38.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-κB transcriptional activation . Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 39.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-[κ]B activation requires PARP-1 function to confer radioresistance . Oncogene. 2008;28(6):832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2 . Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 41.Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plummer R, Jones C, Middleton M, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors . Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas HD, Calabrese CR, Batey MA, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial . Mol Cancer Ther. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 44.Daniel RA, Rozanska AL, Thomas HD, et al. Inhibition of poly(ADP-ribose) polymerase-1 enhances temozolomide and topotecan activity against childhood neuroblastoma . Clin Cancer Res. 2009;15(4):1241–1249. doi: 10.1158/1078-0432.CCR-08-1095. [DOI] [PubMed] [Google Scholar]
- 45.Donawho CK, Luo Y, Luo Y, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models . Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 46.Albert JM, Cao C, Kim KW, et al. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models . Clin Cancer Res. 2007;13:3033–3042. doi: 10.1158/1078-0432.CCR-06-2872. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Shi Y, Guan R, et al. Potentiation of temozolomide cytotoxicity by poly(ADP) ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks . Mol Cancer Res. 2008;6:1621–1629. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 48.Clarke MJ, Mulligan EA, Grogan PT, et al. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines . Mol Cancer Ther. 2009;8(2):407–414. doi: 10.1158/1535-7163.MCT-08-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menear KA, Adcock C, Boulter R, et al. 4-[3-(4-Cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 50.Evers B, Drost R, Schut E, et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin . Clin Cancer Res. 2008;14:3916–3925. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 51.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs . Proc Natl Acad Sci USA. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dungey FA, Löser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-ribose) polymerase: mechanisms and therapeutic potential . Int J Radiat Oncol Biol Phys. 2008;72:1188–1197. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 53.Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2 . Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 54.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 56.Liuzzi M, Talpeart-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem. 1985;260:5252–5258. [PubMed] [Google Scholar]
- 57.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer . Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 58.Rinne M, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol Cancer Ther. 2004;3:955–967. [PubMed] [Google Scholar]
- 59.Taverna P, Hwang H-s, Schupp JE, et al. Inhibition of base excision repair potentiates iododeoxyuridine-induced cytotoxicity and radiosensitization . Cancer Res. 2003;63:838–846. [PubMed] [Google Scholar]
- 60.Yan L, Bulgar A, Miao Y, et al. Combined treatment with temozolomide and methoxyamine: blocking apurininc/pyrimidinic site repair coupled with targeting topoisomerase IIα . Clin Cancer Res. 2007;13:1532–1539. doi: 10.1158/1078-0432.CCR-06-1595. [DOI] [PubMed] [Google Scholar]
- 61.Sabourin M, Osheroff N. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucl Acids Res. 2000;28:1947–1954. doi: 10.1093/nar/28.9.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone . Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 63.Del Rowe JD, Bello J, Mitnick R, et al. Accelerated regression of brain metastases in patients receiving whole brain radiation and the topoisomerase II inhibitor, lucanthone . Int J Radiat Oncol Biol Phys. 1999;43:89–93. doi: 10.1016/s0360-3016(98)00374-5. [DOI] [PubMed] [Google Scholar]
- 64.Madhusudan S, Smart F, Shrimpton P, et al. Isolation of a small molecule inhibitor of DNA base excision repair . Nucl Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target . Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 66.Zawahir Z, Dayam R, Deng J, Pereira C, Neamati N. Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem. 2009;52:20–32. doi: 10.1021/jm800739m. [DOI] [PubMed] [Google Scholar]
- 67.Seiple LA, Cardellina JH, II, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids . Mol Pharmacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou G-M, Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther. 2008;7:2012–2021. doi: 10.1158/1535-7163.MCT-08-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mizushina Y, Manita D, Takeuchi T, et al. The inhibitory action of kohamaic acid A derivatives on mammalian DNA polymerase β . Molecules. 2008;14:102–121. doi: 10.3390/molecules14010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Inhibitors of DNA polymerase β: activity and mechanism . Bioorg Med Chem. 2008;16:4331–4340. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
Website
- 101.Clinicaltrials.gov website www.clinicaltrials.gov